Abstract
The Pseudopimelodidae family comprises 35 species however, cytogenetic studies have been performed in only six species. This study uncovered karyotypic data on Pseudopimelodus pulcher and Microglanis cottoides. Both species possessed 2n = 54, with 20m + 16sm + 10st + 8a and FN = 100 for P. pulcher and 30m + 14sm + 6st + 4a and FN = 104 for M. cottoides. A female of M. cottoides with 45m + 21sm + 9st + 6a (2n = 81) plus two extra small chromosomes was found, indicating a natural triploidy with supernumerary chromosomes. The formation of the polyploid individual seems to have come from a diploid female gamete, due to the presence of a marker chromosome pair partially heterochromatic presents only in females and common to that exemplar. This triploid female showed three chromosomes with nitrate staining (AgNOR), 18S rDNA probe and chromomycin A3 (CMA3) staining. AgNORs were observed on pairs 12 and 23 in P. pulcher and pair 24 in M. cottoides, results that were confirmed with an 18S rDNA probe and CMA3 fluorochrome. These are the first chromosomal data for P. pulcher and provide the first description of natural triploidy with the presence of supernumerary chromosomes in this family and emphasizing well the chromosomal rearrangements diversification between this species.
Keywords: Extra chromosomes, Heterochromatin, Pisces, Polyploidy
Introduction
The family Pseudopimelodidae is widely distributed in South America and is considered the least known family among the naked Neotropical freshwater catfishes (Shibatta 2003). According to Ferraris (2007) the family consists of 30 species and recently new species have been described: Microglanis carlae (Alcaraz et al. 2008); Microglanis minutus (Ottoni et al. 2010); Microglanis robustus (Ruiz and Shibatta 2010); Microglanis oliveirai and Microglanis xylographicus (Ruiz and Shibatta 2011).
Karyotypic descriptions in the Pseudopimelodidae family have been performed on six species of four distinct genera: Cephalosilurus apurensis (Mees 1978); Microglanis aff. cottoides, M. cottoides (Boulenger 1891); cc (cited as M. cottoides) Shibatta and Benine (2005); Pseudopimelodus bufonius (Valenciennes 1840); Pseudopimelodus mangurus (Valenciennes 1835) and Lophiosilurus alexandri Steindachner 1876 (Table 1). These analyses revealed 2n = 54 in all of the specimens analyzed, indicating a conserved karyotypic evolution in relation to the diploid number.
Table 1.
Cytogenetics data on species of the family Pseudopimelodidae
| Species | Localty | 2n | Karyotypic formula | FN | NOR | 18S rDNA | Reference |
|---|---|---|---|---|---|---|---|
| Cephalosilurus apurensis | Orinoco river/Venezuela | 54 | 6m + 28sm + 14st + 6a | _ | Pair 19 st, p, terminal | _ | Martinez et al. (2008) |
| Lophiosilurus alexandri | São Francisco river | 54 | 16m + 18sm + 10st + 10a | _ | 1 pair sm, p, terminal | 1 pair sm, p, terminal | Marques et al. (2008) |
| Microglanis aff. cottoides | Cavalo stream, Jaraguá do Sul river/SC | 54 | 10m + 32sm + 10st + 2a | _ | Pair 23 st, p and pair 22 st, q, terminal | _ | Martinez et al. (2008) |
| Microglanis garavelloi (cited M. cottoides) | Araquá and Capivara river | 54 | 22m + 20sm + 12st | 96 | Pair 1 m, q, terminal | _ | Vissotto et al. (1999) |
| Microglanis cottoides | Forquetinha river/RS | 54 | 30m + 14sm + 6st + 4a | 104 | Pair 24 st, p, terminal | Pair 24 st, p, terminal | Present study |
| Pseudopimelodus bufonius | Trade aquarium/Amazonia | 54 | 12m + 30sm + 12st | _ | Pairs 9, 10 and 11 sm, p, terminal | _ | Martinez et al. (2008) |
| Pseudopimelodus mangurus | Mogi-Guaçu river/SP | 54 | 6m + 26sm + 12st + 10a | _ | Pair 19 st, p, terminal | _ | Martinez et al. (2004) |
| Pseudopimelodus pulcher | Laranjinha river/PR | 54 | 20m + 16sm + 10st + 8a | 100 | Pair 12 sm and pair 23 st, p, terminal | Pair 12 sm and pair 23 st, p, terminal | Present study |
2n diploid number, FN fundamental number, NOR nucleolus organizer regions type: m, metacentric, sm submetacentric, st subtelocentric, a, acrocentric, p short arm, q long arm
Heterochromatin in Pseudopimelodidae appears as heterochromatic blocks usually distributed in pericentromeric and terminal regions of some chromosomes (Vissotto et al. 1999; Martinez et al. 2008; Marques et al. 2008). The AgNORs in this group of fish can be single, as in L. alexandri (Marques et al. 2008), with only one pair bearing this site, or multiple, as in P. bufonius (Martinez et al. 2008) with two pairs AgNORs.
Fluorescence in situ hybridization (FISH) has been performed only on L. alexandri by Marques et al. (2008). The authors confirmed the occurrence of single NORs in this species using the 18S rDNA probe; the 5S rDNA site, which was found in another pair of chromosomes, is not syntenic to the NOR. Marques et al. (2008) also used chromomycin A3 (GC specific) and 4′-6-diamino-2-phenylindole (DAPI) (AT specific) fluorochromes in L. alexandri and Martinez et al. (2004) in Pseudopimelodus mangurus; and the two species showed CMA3+ signals corresponding to the AgNORs.
The current paper revealed karyotypic data for Pseudopimelodus pulcher (Boulenger 1887) and M. cottoides in comparison with other family species, and will aid in understanding the karyotypic diversity of the group. The data for P. pulcher are novel, and we present the first description of natural triploidy and supernumerary chromosomes in the family in M. cottoides.
Materials and methods
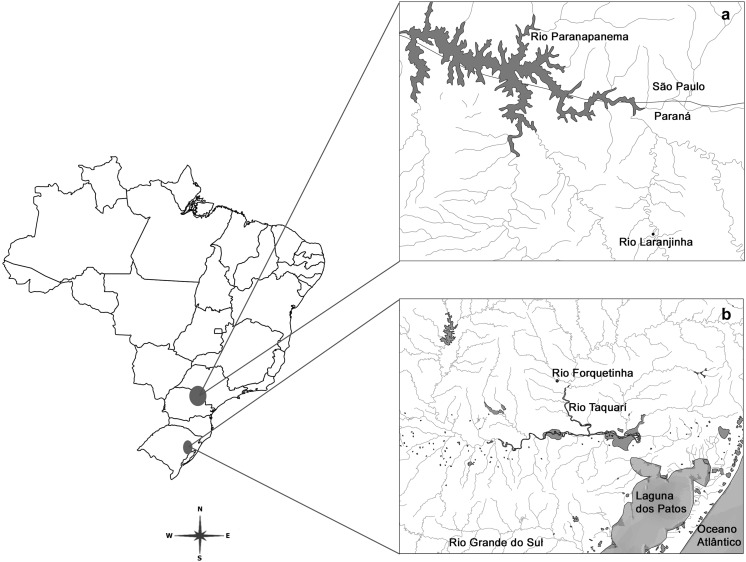
We analysed five specimens of P. pulcher (Boulenger 1887) (two males, two females and one sex unidentified) from the Laranjinha River of the Paranapanema Basin, located in the city of Ribeirão do Pinhal/PR/Brazil (23°24′9.11″S and 50°27′16.4″W) and twelve specimens of M. cottoides (Boulenger 1891) (five males and seven females) from the Forquetinha River, located in the city of Forquetinha/RS/Brazil (29°24′21.8″S and 52°03′18.3″W) in the Patos Lagoon Hydrographic System/RS (Fig. 1). Specimens were deposited in the Museum of Zoology of the Universidade Estadual de Londrina under vouchers 6033 (M. cottoides) and 5767 (P. pulcher). The samples were collected with the permission of the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis, protocol number 12,280/2.
Fig. 1.
Map of Brazil indicating the collection sites: a Laranjinha river—Paranapanema basin/Paraná, b Forquetinha river—Patos lagoon hydrographic system/Rio Grande do Sul
Mitotic chromosomes were obtained by direct preparation after removal of the kidney, as described by Bertollo et al. (1978). The chromosomes were organised according to Levan et al. (1964), with modifications, to determine the fundamental number (FN). Metacentric (m), submetacentric (sm) and subtelocentric (st) chromosomes were considered biarmed, and acrocentric (a) chromosomes were considered uniarmed. The distribution of heterochromatin was analysed by Giemsa C-banding (Sumner 1972). Silver nitrate staining of the active nucleolar organizer regions (AgNOR) was performed according to the method of Howel and Black (1980). The GC- and AT-rich bands were detected with CMA3 and DAPI, respectively, according to the technique described by Schweizer (1980). In addition, the FISH technique was carried out following the protocol reported by Pinkel et al. (1986), with modifications, along with an 18S rDNA probe designed for Prochilodus argenteus (Hatanaka and Galetti 2004).
Results
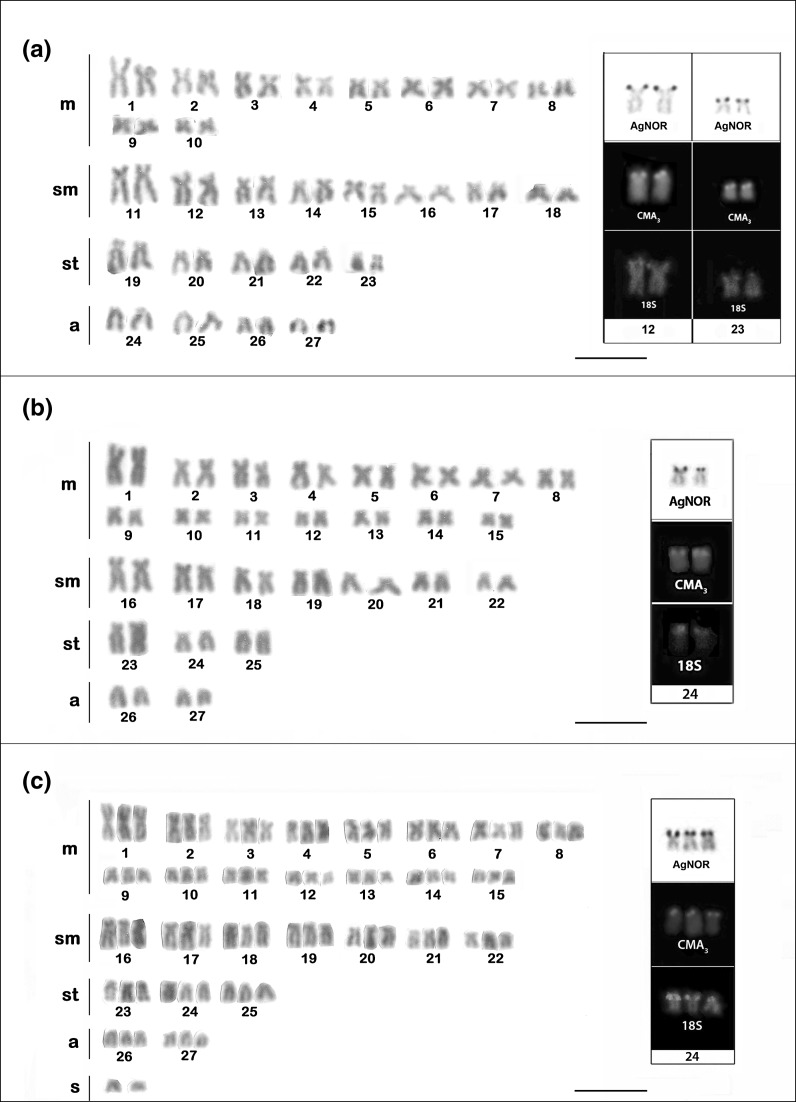
The diploid number of P. pulcher was 54 with a karyotypic formula of 20m + 16sm + 10st + 8a and FN = 100 (Fig. 2a).
Fig. 2.
Karyotypes of a Pseudopimelodus pulcher, b, c Microglanis cottoides diploid and triploid females, respectively. The boxes contain the chromosome NOR-bearing pairs with silver nitrate staining, CMA3 and FISH with 18S rDNA probe. The scale bar represents 5 μm
AgNORs were observed on the short arm of a submetacentric chromosome pair (pair 12) and on one pair of subtelocentric chromosomes (pair 23). These regions were coincident with the 18S rDNA probe after the FISH (Fig. 2a-box). The fluorochrome staining also presented four chromosomes coincident the NORs, with fluorescent signals for CMA3 (Fig. 2a-box).
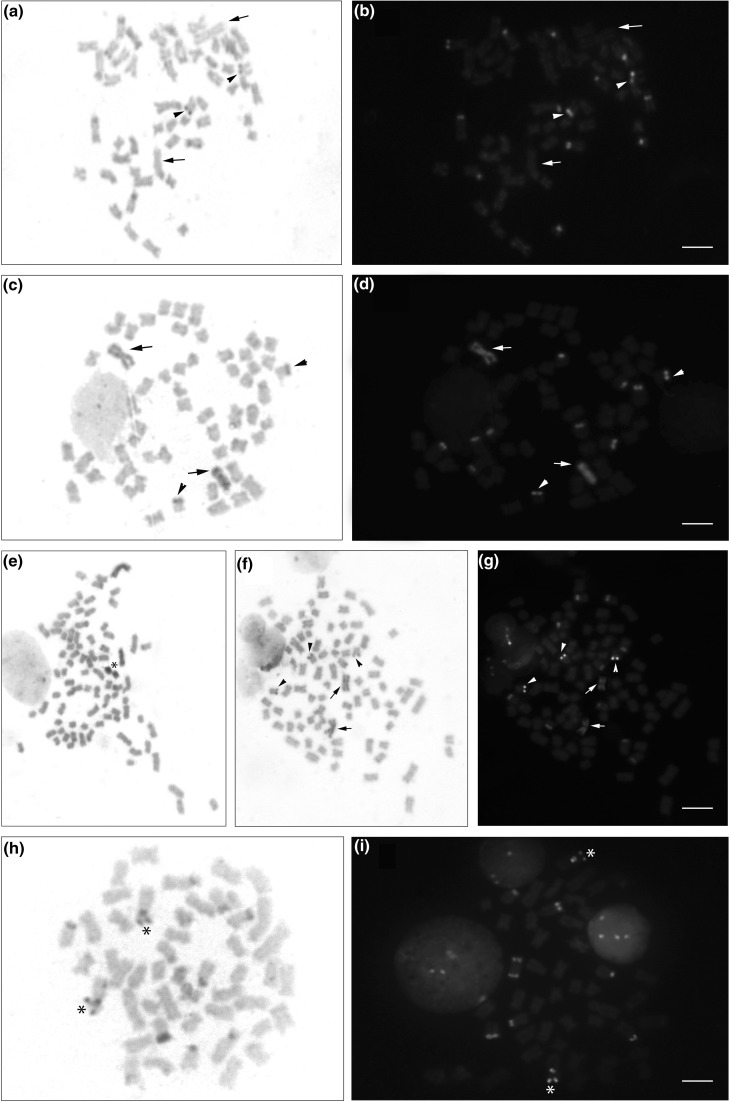
The heterochromatin was distributed in the pericentromeric and terminal regions of some chromosomes and was positive for both CMA3 and DAPI after fluorochrome staining (Fig. 3h and i); one large-sized submetacentric pair, probably pair 12 of the NOR, showed heterochromatic markings in the pericentromeric and terminal regions of the short arm (Fig. 3h); the pericentromeric region was DAPI+ and CMA3+ and the terminal region was CMA3+ (Fig. 3i).
Fig. 3.
Somatic metaphases with C-banding of Microglanis cottoides: (a–b) diploid male, (c–d) diploid female and (e–g) triploid female and Pseudopimelodus pulcher (h–i). C-banding stained with Giemsa (a, c, e, f, h) and overlapping DAPI/CMA3 (b, d, g). The arrowheads in cells of M. cottoides (a–g) indicate the NOR-bearing pairs, and the arrows indicate the two chromosomes with the heterochromatic block in the diploid (c–d) and triploid (f–g) females; in the diploid male (a–b), the arrows indicate chromosome pair one without the heterochromatin block. In (e), the asterisk indicates the two supernumerary chromosomes that are totally heterochromatic; in (h), the asterisk indicates the heterochromatic chromosome pair with AT and GC sites in the pericentromeric region and GC in the terminal region. The scale bar represents 5 μm
The diploid number of M. cottoides also was of 2n = 54 with 30m + 14sm + 6st + 4a and a FN equal to 104 (Fig. 2b). A female of M. cottoides showed a karyotypic formula of 45m + 21sm + 9st + 6a totalizing 81 chromosomes corresponding to a triploid genome, plus two extra chromosomes of the small metacentric type (Fig. 2c).
The AgNOR of the M. cottoides diploid individuals was localized on the short arm of subtelocentric pair 24, and of the triploid individual was located in three subtelocentric chromosomes (24). In both karyotypes (diploid and triploid), the NOR was coincident with 18S rDNA probe, and was also positive for CMA3 fluorochrome staining (Fig. 2 b and c-box).
The heterochromatin in diploid individuals of M. cottoides was distributed in the pericentromeric and terminal regions of some chromosomes in both sexes (Fig. 3 a and c), however, all females presented a large block of interstitial heterochromatin in the 1st chromosome pair (Fig. 3c), not observed in any of the male individuals of M. cottoides (Fig. 3a). All heterochromatic regions of diploid individuals were DAPI+ and CMA3−, including interstitial heterochromatin in the 1st chromosome pair of the female diploid (Fig. 3 b and d).
The heterochromatin in triploid individual also was distributed in the pericentromeric and terminal regions of some chromosomes and in two out of three chromosomes number one of the complement was observed a large block of heterochromatin (Fig. 3 e, f, g), as the 1st chromosome pair of the diploid female; these chromosomes after staining with DAPI proved to be much more evident and positive for this fluorochrome and negative for CMA3 (Fig. 3g). The supernumerary chromosomes of the triploid individual appeared totally heterochromatic (Fig. 3e).
The NOR of pair 24 was C-band positive in all individuals (diploid and triploid), revealing to be heterochromatic in the terminal region of the short arm (Fig. 3 a–g).
Discussion
Both P. pulcher and M. cottoides showed diploid numbers of 54 chromosomes, corroborating the 2n described for the Pseudopimelodidae species that have been cytogenetically studied to date (Table 1). Although only a few species have already been karyotypically analyzed (7 of the 35 described species in the family), the diploid number observed in the analyzed species indicates conservative chromosome evolution in this group of fish.
Previously, Pseudopimelodidae belonged to the family Pimelodidae, divided into three monophyletic subfamilies: Pimelodinae, Heptapterinae and Pseudopimelodinae according to Shibatta (2003). As they have different morphological characteristics, according to the classification of Reis et al. (2003), most Pimelodidae were divided into two other families: Heptapteridae described by Bockmann and Guazzelli (2003) and Pseudopimelodidae described by Shibatta (2003). Swarça et al. (2007) in a cytogenetic revision showed that cytogenetic data corroborate this separation into three distinct families, separating them according to the diploid number, being most Pimelodidae species with 2n = 56, most Heptapteridae species with 2n = 58 and Pseudopimelodidae with predominance of 2n = 54, as confirmed in the present study (Table 1).
The exemplar of M. cottoides triploidy (3n = 81 + 2), is the first report of such an event in the Pseudopimelodidae family. Natural triploidy has been previously described in several species of neotropical fish (Malacrida et al. 2003; Garcia et al. 2003; Kantek et al. 2007; Tsuda et al. 2010). This event might be related to changes in environmental temperatures, facilitating the retention of the second polar body during the meiotic division and leading to the formation of a diploid gamete, which, after being fertilised, would give rise to a triploid individual (Cuellar and Uyeno 1972). Triploidy is induced in fish farms, usually by heat shock, with the aim of increasing growth in juveniles, extending the survival of individuals and improving growth in adult fish, and it is also useful for controlling overpopulation (Tiwary et al. 2004).
Despite its conserved diploid number, the family Pseudopimelodidae shows great variability, with the occurrence of multiple and single NORs (both in species of different genera and different species within the same genus) as well as variation in their location (Table 1). The differences observed among species of the same genus, such as the single NORs of P. mangurus (Martinez et al. 2004) and the multiple NORs of P. pulcher, confirm that these regions are suitable chromosome markers that may be species-specific, and the significant variability indicates that these regions do not follow a pattern within the family. According to Galetti (1998), a single chromosome pair bearing 45S rDNA is considered a primitive character shared among fish. Despite the small number of species studied, most Pseudopimelodidae present single NORs, which may be considered a plesiomorphic character shared among the groups from which the multiple NORs are derived. This possibility could be confirmed by the analysis of additional species from this family.
The NORs of M. cottoides and P. pulcher were confirmed by FISH using an 18S rDNA probe. These regions were also positive for CMA3 staining and are therefore GC-rich. Fluorochrome staining had previously been performed only on P. mangurus (Martinez et al. 2004) and L. alexandri (Marques et al. 2008), in which the NORs were also coincident, a finding that was confirmed in the latter by FISH with an 18S rDNA probe.
The triploid M. cottoides specimen presented three NOR-bearing chromosomes, similar to previously reported cases of triploidy in fish (Maistro et al. 1994; Tsuda et al. 2010; Silva et al. 2011) confirmed either by impregnation with silver nitrate and CMA3 or by hybridization with an 18S rDNA probe without the inactivation of ribosomal genes.
The triploid M. cottoides female exhibited an interstitial heterochromatic block on two of the three copies of chromosome one, similar to that observed on the first chromosome pair of the diploid female, and both individuals presented DAPI+ heterochromatin. This feature may be a chromosome marker, confirming the female as a bearer of a diploid gamete because there was no evidence of interstitial heterochromatin on the first chromosome pair in any of the males. This finding suggests that the two heterochromatic chromosomes of the triploid female originated from a diploid female gamete and the euchromatic chromosome one from the haploid male gamete.
Supernumerary chromosomes were observed only in triploid individuals of M. cottoides, which were totally heterochromatic. Cases of triploidy associated with B chromosomes have already been reported in Astyanax scabripinnis Jenyns 1842 (Maistro et al. 1994) and Curimata modesta Walbaum 1792 (Venere and Galetti 1985). The fact that these B chromosomes are shown only in the triploid M. cottoides individual, indicate that the occurrence of extra chromosomes constitute a unique condition present in the triploid specimen and that they are a consequence of the same event that led to the formation of the triploid fish, as suggested by Pansonato-Alves et al. (2011) in a triploid Characidium cf. zebra (Crenuchidae) specimen.
The presence of a small amount of heterochromatin in M. cottoides and P. pulcher, where only a few chromosomes showed positive bands, has also been described for Microglanis garavelloi (cited as M. cottoides in Vissotto et al. 1999), P. mangurus (Martinez et al. 2004), C. apurensis, Microglanis aff. cottoides and P. bufonius (Martinez et al. 2008), suggesting that this may be a characteristic of the family. All of the M. cottoides individuals (diploid and triploid) exhibited C-band-positive NORs, which were previously observed by Martinez et al. (2008) in C. apurensis and P. mangurus as well as in Microglanis aff. cottoides (Martinez et al. 2004).
After C-band treatment with CMA3 and DAPI fluorochromes, both GC and AT-rich heterochromatin were observed in P. pulcher; however, a large submetacentric pair, most likely pair 12 of the NOR, revealed pericentromeric GC and AT-rich and terminal, only GC-rich, showing that these regions differ from one another in relation to the heterochromatin base composition in this chromosome, which might suggest a chromosome marker in this species.
This study provides novel information on this fish family, including the first cytogenetic data on P. pulcher and supernumerary chromosomes and natural triploidy in M. cottoides population from the Patos Lagoon Hydrographic System/RS. The study of the heterochromatin allowed to infer about possible formation of triploidy, with fertilization of the diploid female gamete, due to the identification of a cytogenetic marker in this specie. Furthermore, this demonstrates the importance of cytogenetic analysis of other species of this group, suggesting different evolutionary paths in which chromosomal rearrangements may have played an important role in the karyotypic evolution of the family.
Acknowledgments
The authors are grateful to Dra. Lucia Giuliano Caetano and Dr. Alberto Sergio Fenocchio for the review of this manuscript and to MSc Bruno Ambrozio Galindo for the collection of Pseudopimelodus pulcher specimens. This research was supported by a grant from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The researcher received permission from Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) to collect fish specimens.
References
- Alcaraz HSV, Da Graça WJ, Shibatta OA. Microglanis carlae, a new species of bumblebee catfish (Siluriformes: pseudopimelodidae) from the rio Paraguay basin in Paraguay. Neotrop Ichthyol. 2008;6:425–432. doi: 10.1590/S1679-62252008000300016. [DOI] [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae) Bras J Genet. 1978;1:103–120. [Google Scholar]
- Bockmann FA, Guazzelli GM (2003) Family Heptapteridae. In: ReisRE, KullandeR SO, Ferraris jr CJ. Check list of the Freshwater Fishes of South and Central America.Edipurcs, Porto Alegre, pp 406–431
- Cuellar O, Uyeno T. Triploidy in rainbow trout. Cytogenetics. 1972;11:508–515. doi: 10.1159/000130217. [DOI] [PubMed] [Google Scholar]
- Ferraris Jr CJ (2007) Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of Siluriformes primary types. In: Zootaxa, pp 352–356
- Galetti PM., Jr Chromosome diversity in neotropical fishes: NOR studies. Ital J Zool. 1998;65:53–56. doi: 10.1080/11250009809386795. [DOI] [Google Scholar]
- Garcia C, Moreira-Filho O, Bertollo LAC, Centofante L. B chromosomes and natural triploidy in Rhamdia sp. (Pisces, Siluriformes, Heptapteridae) Cytologia. 2003;68:403–411. doi: 10.1508/cytologia.68.403. [DOI] [Google Scholar]
- Hatanaka T, Galetti PM., Jr Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus, Agassiz 1829 (Characiformes, Prochilodontidae) Genetica. 2004;122:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. Controled silver staining of nucleous organizer regions with a protective colloidal developer: a 1-step method. Experientia. 1980;36:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Kantek DLZ, Noleto RB, Fenocchio AS, Cestari MM. Cytotaxonomy, heterochromatic polymorphism and natural triploidy of a species of Astyanax (Pisces, Characidae) endemic to the Iguaçu river basin. Braz Arch Biol Technol. 2007;50:67–74. doi: 10.1590/S1516-89132007000100008. [DOI] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x. [DOI] [Google Scholar]
- Maistro EL, Dias AL, Foresti F, Oliveira C, Moreira-Filho O. Natural triploidy in Astyanax scabripinis (Pisces, Characidae) and simultaneous occurrence of macro B-chromosomes. Caryologia. 1994;47:233–239. doi: 10.1080/00087114.1994.10797301. [DOI] [Google Scholar]
- Malacrida ACCP, Dias AL, Giuliano-Caetano L. Natural triploidy in Astyanax aff. scabripinnis (Pisces, Characidae) of the Tibagi river bay-PR. Cytologia. 2003;68:267–270. doi: 10.1508/cytologia.68.267. [DOI] [Google Scholar]
- Marques MBA, Moreira-Filho O, Garcia C, Margarido VP. Cytogenetic analyses of two endemic fish species from the São Francisco river basin: Conorhynchus conirostris and Lophiosilurus alexandri (Siluriformes) Genet Mol Biol. 2008;31:215–221. doi: 10.1590/S1415-47572008000200008. [DOI] [Google Scholar]
- Martinez ERM, Oliveira C, Foresti F. Cytogenetic analyses of Pseudopimelodus mangurus (Teleostei: Siluriformes: Pseudopimelodidae) Cytologia. 2004;69:419–424. doi: 10.1508/cytologia.69.419. [DOI] [Google Scholar]
- Martinez ERM, Nirchio M, Granado A, Foresti F, Oliveira C. Cytogenetic analysis of three catfish species of the family Pseudopimelodidae (Teleostei, Siluriformes) Genet Mol Biol. 2008;31:692–696. doi: 10.1590/S1415-47572008000400015. [DOI] [Google Scholar]
- Ottoni FP, Mattos JLO, Barbosa MA. Description of a new species of Microglanis from the rio Barra Seca basin, southeastern Brazil (Teleostei: Siluriformes: Pseudopimelodidae) Vertebrate Zoology. 2010;60:187–192. [Google Scholar]
- Pansonato-Alves JC, Oliveira C, Foresti F. Karyotypic conservatism in samples of Characidium cf. zebra (Teleostei, Characiformes, Crenuchidae): physical mapping of ribossomal genes and natural triploidy. Genet Mol Biol. 2011;34:208–213. doi: 10.1590/S1415-47572011005000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RE, Kullander SO, Ferraris jr CJ. Check list of the freshwater fishes of South and Central America. Porto Alegre: Edipurcs; 2003. p. 742. [Google Scholar]
- Ruiz WBG, Shibatta OA. A new species of Microglanis (Siluriformes, Pseudopimelodidae) from lower Rio Tocantins basin, Pará, Brazil, with description of superficial neuromasts and pores of lateral line system. Zootaxa. 2010;2632:53–66. [Google Scholar]
- Ruiz WBG, Shibatta OA. Two new species of Microglanis (Siluriformes: Pseudopimelodidae) from the upper-middle rio Araguaia basin, Central Brazil. Neotrop Ichthyol. 2011;9:697–707. doi: 10.1590/S1679-62252011000400002. [DOI] [Google Scholar]
- Schweizer D. Simultaneous fluorescent staining of R bands and specific heterochromatic regions (DA-DAPI bands) in human chromosomes. Cytogenet Cell Genet. 1980;27:190–193. doi: 10.1159/000131482. [DOI] [PubMed] [Google Scholar]
- Shibatta OA. Family Pseudopimelodidae. In: Reis RE, Kullander SO, Ferraris Jr CJ, editors. Check list of the freshwater fishes of south and Central America. Porto Alegre: Edipurcs; 2003. pp. 406–431. [Google Scholar]
- Silva M, Matoso DA, Ludwig LAM, Gomes E, Almeida MC, Vicari MR, Artoni RF. Natural triploidy in Rhamdia quelen identified by cytogenetic monitoring in Iguaçu basin, southern Brazil. Environ Biol Fish. 2011;91:361–366. doi: 10.1007/s10641-011-9794-2. [DOI] [Google Scholar]
- Sumner ATA. Simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Swarça AC, Fenocchio AS, Dias AL (2007) An update cytogenetic review for species of the families pseudopimelodidae, pimelodidae and heptapteridae (pisces, siluriformes) suggestion of a cytotaxonomical classification. Caryologia 60:338–348
- Tiwary BK, Kirubagaran R, Ray AK. The biology of triploid fish. Rev Fish Biol Fish. 2004;14:391–402. doi: 10.1007/s11160-004-8361-8. [DOI] [Google Scholar]
- Tsuda JR, Moraes VPO, Giuliano-Caetano L, Dias AL. Occurrence of natural triploidy in Rhamdia quelen (Siluriformes, Heptapteridae) Genet Mol Res. 2010;9:1929–1935. doi: 10.4238/vol9-3gmr949. [DOI] [PubMed] [Google Scholar]
- Venere PC, JrPM Galetti. Natural triploidy and chromosome B in the fish Curimata modesta (Curimatidae, Characiformes) Rev Brasil Genet. 1985;7:681–687. [Google Scholar]
- Vissotto PC, Foresti F, Oliveira C. Karyotype description of five species of Pimelodidae (Teleostei, Siluriformes) Chromosom Sci. 1999;3:1–7. [Google Scholar]