Abstract
Genes in the protein secretion pathway have been targeted to increase productivity of monoclonal antibodies in Chinese hamster ovary cells. The results have been highly variable depending on the cell type and the relative amount of recombinant and target proteins. This paper presents a comprehensive study encompassing major components of the protein processing pathway in the endoplasmic reticulum (ER) to elucidate its role in recombinant cells. mRNA profiles of all major ER chaperones and unfolded protein response (UPR) pathway genes are measured at a series of time points in a high-producing cell line under the dynamic environment of a batch culture. An initial increase in IgG heavy chain mRNA levels correlates with an increase in productivity. We observe a parallel increase in the expression levels of majority of chaperones. The chaperone levels continue to increase until the end of the batch culture. In contrast, calreticulin and ERO1-l alpha, two of the lowest expressed genes exhibit transient time profiles, with peak induction on day 3. In response to increased ER stress, both the GCN2/PKR-like ER kinase and inositol-requiring enzyme-1alpha (Ire1α) signalling branch of the UPR are upregulated. Interestingly, spliced X-Box binding protein 1 (XBP1s) transcription factor from Ire1α pathway is detected from the beginning of the batch culture. Comparison with the expression levels in a low producer, show much lower induction at the end of the exponential growth phase. Thus, the unfolded protein response strongly correlates with the magnitude and timing of stress in the course of the batch culture.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-013-9678-8) contains supplementary material, which is available to authorized users.
Keywords: Productivity, Recombinant CHO cells, Unfolded protein response (UPR) pathway, ER stress, Gene expression
Introduction
Chinese hamster ovary (CHO) cell lines are extensively used as a host system for the production of recombinant therapeutic proteins (Jayapal et al. 2007). Engineering advancements in the field of bioreactor technology, coupled with high throughput cell line selection approaches have enabled high titers to be achieved in industry. However, understanding the molecular basis of productivity is still a challenge as productivity is a multifaceted phenomenon with many different pathways contributing to the creation of a high producer.
To increase productivity, many cellular pathways including energy metabolism, cell cycle, apoptosis and protein secretion have been extensively studied (Chen et al. 2001; Kim and Lee 2002; Watanabe et al. 2002; Ifandi and Al-Rubeai 2005; Seth et al. 2006; Jayapal et al. 2007; Kim et al. 2012). Increasing cell density by either reducing apoptosis (Meents et al. 2002; Figueroa et al. 2007) or altering cell metabolism (Chen et al. 2001) can lead to increase in product titers. Alternatively, the protein synthesis and secretion machinery can be targeted to increase the specific productivity of a cell. For example, overexpression of chaperones, calnexin and calreticulin, led to a two-fold increase in the productivity of thrombopoietin secreting CHO cells (Chung et al. 2004).
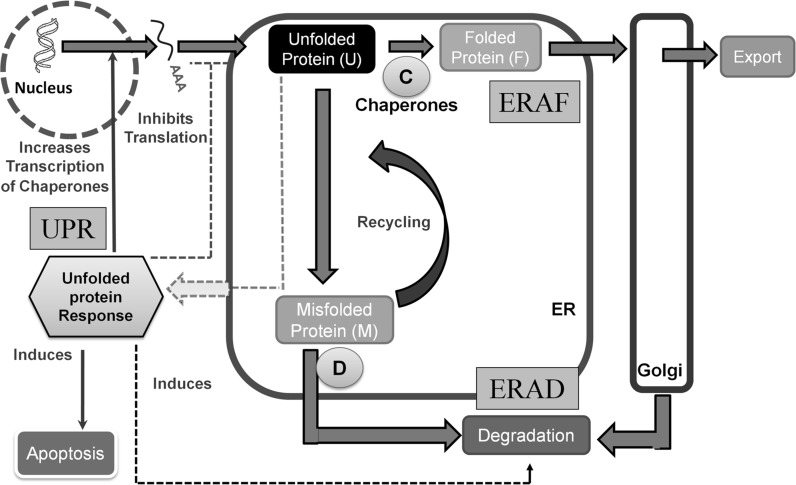
The protein secretion pathway in mammalian systems starts with translocation of the nascent protein into the endoplasmic reticulum (ER). Homeostasis in ER is maintained by coordinated regulation between three pathways (see Fig. 1): endoplasmic reticulum associated folding (ERAF), ER associated protein degradation (ERAD) and unfolded protein response pathway (UPR) (Schröder 2008). In the event of accumulation of unfolded protein in the ER, the UPR signaling cascade is activated in response to ER stress. The UPR pathway attempts to maintain homeostasis by shutting down protein translation so as to minimize the load on cells. Alternatively, the folding capacity is enhanced by increasing the transcriptional level of chaperones. At the same time, degradation enzymes are also up regulated to remove mis-folded proteins (Schröder and Kaufman 2005; Schröder 2006).
Fig. 1.
Engineering view of protein secretion pathway in endoplasmic reticulum. Under stress, accumulation of unfolded proteins inside ER leads to activation of the unfolded protein response (UPR) pathway. ER homeostasis is restored back by a co-ordinated activation of three different pathways; endoplasmic reticulum associated folding (ERAF), endoplasmic reticulum associated degradation (ERAD) and UPR. C denotes chaperones and D denotes degradation enzymes
The output of the UPR pathway varies between adaptation and apoptosis in response to intensity and duration of ER stress. While in the presence of low levels of stress, the cells can adapt and handle the increased ER load, high or prolonged levels of stress induce apoptosis (Hetz 2012). Another pathway that responds to protein accumulation in the ER membrane is the ER overload response (EOR), where NF-κB is activated on production of reactive oxygen intermediates and release of Ca2+ from the ER (Pahl and Baeuerle 1997; Teruya et al. 2005).
Different genes of the protein secretion pathway have been targeted in recombinant CHO cells, but the results have been highly variable. The outcome often depends on the cell line, protein product and the cell engineering target gene. For example, overexpression of protein disulfide isomerase protein (PDI) led to a significant increase in the secretion rates of human antibody in one study (Borth et al. 2005), whereas intracellular retention of TNFR:Fc protein was observed in other recombinant CHO cells (Davis et al. 2000). No effect of PDI overexpression was seen on productivity in thrombopoietin secreting CHO cells (Mohan et al. 2007).
The effectiveness of a cell-engineering approach often depends on the existing levels and relative amounts of chaperones and UPR proteins. A comprehensive study of these genes in recombinant CHO cells is essential for a better understanding of the pathway and its response. Majority of existing studies are limited to a few chosen genes from these pathways and to few time-points (Murphy et al. 2001; Underhill et al. 2005; Cudna and Dickson 2006; Doolan et al. 2008; Ku et al. 2008). In this work, we undertake dynamic profiling of all major ER chaperones and UPR pathway genes in a batch culture. Two recombinant CHO cell lines with varying productivity levels of a secretory IgG are compared to investigate the relation of UPR induction to growth and productivity related stress.
The response of UPR pathway to chronic stress is different from that to short term stress (DuRose et al. 2006; Merquiol et al. 2011). In a batch culture, cells are actively growing and also continuously produce recombinant proteins. This provides a unique environment of continuously increasing load on the ER. The current paper thus complements past studies, which usually focus on the short term response to either a pharmacological agent such as tunicamycin or overexpression of a secretory protein (Rutkowski et al. 2006).
Materials and methods
Cell culture conditions
The CHO cell lines used in this study secrete an anti-rhesus IgG. The cell lines obtained as a gift from Dr. Miranda Yap (BTI, Singapore) are derived from CHO DG44 and have been described earlier (Chusainow et al. 2009). The cells were cultured in a medium containing 50 % PF-CHO (Hyclone, Thermo Scientific, Rockford, IL, USA) and 50 % CD CHO (Gibco-Invitrogen, Carlsbad, CA, USA) supplemented with 2.0 g/l sodium bi-carbonate (Sigma-Aldrich, St. Louis, MO, USA), 4 mM l-Glutamine (Sigma-Aldrich), 0.10 % Pluronic (HiMedia Pvt. Ltd, Mumbai, India), 600 μg/ml G418 (Sigma-Aldrich) and 250 nM Methotrexate (Sigma-Aldrich). All cultures were run in replicate at 37 °C with 8 % CO2 in a total culture volume of 300 ml in 1 l Erlenmeyer flasks (Corning, Lowell, MA, USA) by shaking at 110 RPM. Cell concentration and viability were determined by the trypan blue exclusion method using a hemocytometer. Samples were taken regularly for RNA isolation and quantification of secreted antibody. The cumulative specific death rate was calculated as , where XD(t) and XV(t) refer to dead cell density and viable cell density at time t, respectively.
Primer design
Primers for quantitative PCR were designed in Primer 3 (http://frodo.wi.mit.edu/) using a consensus sequence obtained by performing multiple sequence alignment of mouse, human and rat cDNA. Wherever available, the designed primers were checked for their specificity by blasting with the CHO genome database (www.chogenome.org) and redesigned if necessary. The complete list of primers used in this study is shown in Table 1.
Table 1.
List of genes and their primer sequences used in this study
| Gene name | Gene ID | Sense primer | Anti-sense primer | Tm (°C) | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| Beta actin | β-actin | AGCTGAGAGGGAAATTGTGCG | GCAACGGAACCGCTCATT | 62 | 163 | Ku et al. 2008 |
| Ero1p-like alpha | ERO1α | GCTGCTTCTGCCAGGTTAGT | ACGGCACAGTCTCTTCTTCC | 60 | 202 | GenBank Acc. No.: NC_000080.6 |
| Glucose regulated protein 78 | GRP78 | CTATTGCGTATGGCCTGGAT | AAAGTCTTCCCCACCGAGAT | 60 | 158 | CHO-K1 GenBank Acc. No.: EGV94255 |
| X-Box binding protein 1 | XBP1 | CTCCAGAGACGGAGTCCAAG | AAAGGGAGGCTGGTAAGGAA | 60 | 299 | CHO-K1 GenBank Acc. No.: EGW12286 |
| Unspliced X-Box binding protein 1 | XBP1u | CTCCAGAGACGGAGTCCAAG | CAGAGGTGCACGTAGTCTGAGTGCTG | 60 | 86 | Replaced reverse primer of XBP1 with the splice site |
| Spliced X-Box binding protein 1 | XBP1s | CTCCAGAGACGGAGTCCAAG | GAGTCCGCAGCAGGTG | 62 | 69 | Replaced reverse primer of XBP1 with regions adjoining the splice site |
| Endoplasmic reticulum-localized DnaJ homologue 4 | ERDJ4 | CACTTCCAGACACACCAGGA | TTCCTCTTCGCTGAGTGACA | 60 | 214 | Ku et al. 2008 |
| Calnexin | CNX | CCTCAGATTGCCAACCCTAA | AGCACTGAAAGGCGTCATCT | 60 | 214 | Ku et al. 2010 |
| Glucose regulated protein 94 | GRP94 | TGGGAAGAGGTTCCAGAATG | CGTGAGACGCTGAGATACCA | 60 | 172 | CHO-K1 GenBank Acc. No.: EGW09701 |
| Immunoglobulin heavy chain | IgGHC | GCTTCTATCCCAGCGACATC | CATCACGGAGCATGAGAAGA | 60 | 173 | Kantardjieff et al. 2010 |
| Immunoglobulin light chain | IgGLC | CTGTTGTGTGCCTGCTGAAT | CCTAGCACTCTCCCCTGTTG | 60 | 251 | Kantardjieff et al. 2010 |
| Growth arrest and DNA damage gene 34 | GADD34 | CCTGGTCTGCAAAGTGCTGAT | CCAGCTCAGTCACTCCCTCTTC | 62 | 70 | Ohya et al. 2008 |
| Activating Transcription Factor 4 | ATF4 | TTCTCCAGCGACAAGGCTAAG | GCACTGACCAACCCATCCA | 62 | 65 | Ohya et al. 2008 |
| CCAAT/enhancer-binding protein (C/EBP) homologous protein | CHOP | GGGAGCTGGAAGCCTGGTAT | GGGACCCCCATTTTCATCTG | 60 | 65 | Ohya et al. 2008 |
| Calreticulin | CRT | ACGTGAAGCTGTTTCCGAG | TGTTGATCAGCACATTCTTGC | 60 | 150 | CHO-K1 GenBank Acc. No.: EGW00614 |
| Nuclear factor erythroid 2-related factor 2 | NRF2 | GACATCCTTTGGAGGCAAGA | AAAAGGCCTTCTCCTGTTCC | 60 | 152 | CHO-K1 GenBank Acc. No.: EGW02666 |
| UDP-glucose:glycoprotein glucosyltransferase-variant 1 | UGGT1-V1 | GCTGGAAGCCAGTGAGTTCT | GCTGGAAGGCTTGAATTGAG | 60 | 224 | CHO-K1 GenBank Acc. No.: EGW06464 |
Quantitative real time PCR
Total RNA was isolated from 107 cells using Trizol reagent (Sigma-Aldrich) as per the manufacturer’s instructions. RNA was quantified by measuring absorbance at 260 and 280 nm using a nanospectrophotometer (Implen, Munich, Germany). The quality of RNA was checked on a 1 % denaturing agarose (Sigma-Aldrich) gel at 70 volts. 4 μg of total RNA was converted to cDNA (in replicate for selected RNA samples) using a reverse transcriptase kit (Thermo Fischer Scientific, formerly Fermentas, Burlington, ON, Canada) in a total reaction volume of 20 μl as per the manufacturer’s protocol. 100 ng of cDNA was used for quantitative PCR using either Maxima Sybr Green/ROX Master Mix (Fermentas) or QuantiFast SYBR Green PCR kit (Qiagen, Germantown, MD, USA) in a total reaction volume of 25 μl supplemented with 0.5 μM gene specific primers in duplicates. The reaction conditions were as follows: Initial denaturation of 95º C for 3 min followed by 40 cycles of 95º C for 1 min, annealing temperature for 1 min and 72 °C for 1 min. gDNA contamination and formation of primer-dimers was checked by running a no RT control and no template control, respectively. Primer specificity was checked by melting curve analysis. β-Actin was used as the housekeeping gene for normalization. The ΔΔCT method was used for computing expression levels (Livak and Schmittgen 2001). The expression levels is defined as where ΔΔCT = [(Cgenet in test cell line – Cgenet in reference cell line on day 1) – (Cβ-Actint in test cell line – Cβ-Actint in reference cell line on day 1). The reference cell line has the lowest productivity (3 pg/cell/day) and expression levels of all the genes. Note that this reference cell line is also derived from the same parental cell line.
X-Box binding protein 1 (XBP1) splicing assay
Primers were designed for the total form of X-Box binding Protein 1 (XBP1T). Unspliced X-Box binding Protein 1 (XBP1U) form was specifically amplified by just replacing the reverse primer of XBP1T with the splice site sequence. The threshold cycle (CT) value of spliced X-Box binding Protein 1 (XBP1S) were calculated from that of XBP1T and XBP1U using the following relation: , where , and refers to the CT value for XBP1S, XBP1U and XBP1T respectively. Note that levels. Assuming 100 % efficiency and adjusting to same threshold values, we arrive at the relation . Rearranging this equation will give which leads to the above expression for calculating CT value for XBP1s. Specific primers were also designed for XBP1s and few samples repeated.
Antibody quantification using ELISA
Antibody titers in the culture supernatant were quantified by ELISA using the protocol as described by Chusainow et al. (2009). Goat Anti-human IgGA + IgG + IgM (H + L) (KPL, Gaithersburg, MD, USA) was used as the primary coating antibody. Alkaline phosphatase conjugated anti-human IgG (Fc specific) was used as the secondary antibody (Sigma-Aldrich) while p-Nitrophenyl Phosphate (pNNP) (Sigma-Aldrich) was used as the substrate. The absorbance was read at 405 nm using a multi plate reader (Spectramax M5e, Molecular Devices, Downingtown, PA, USA). Human IgG (Sigma-Aldrich) was used as a standard. The specific productivity was calculated as where X(t) is the viable cell density and P(t) is the concentration of IgG at time t as determined by ELISA.
Data analysis
All data are expressed as mean ± SD from three replicate cultures for the high producer and two replicate cultures for low producer cell line. Outliers were removed by neglecting data points that did not lie within mean ± 1.5 SD. Student’s t test was employed to estimate statistical significance of expression levels at different days with respect to day 1 levels.
Results
Growth and productivity profile
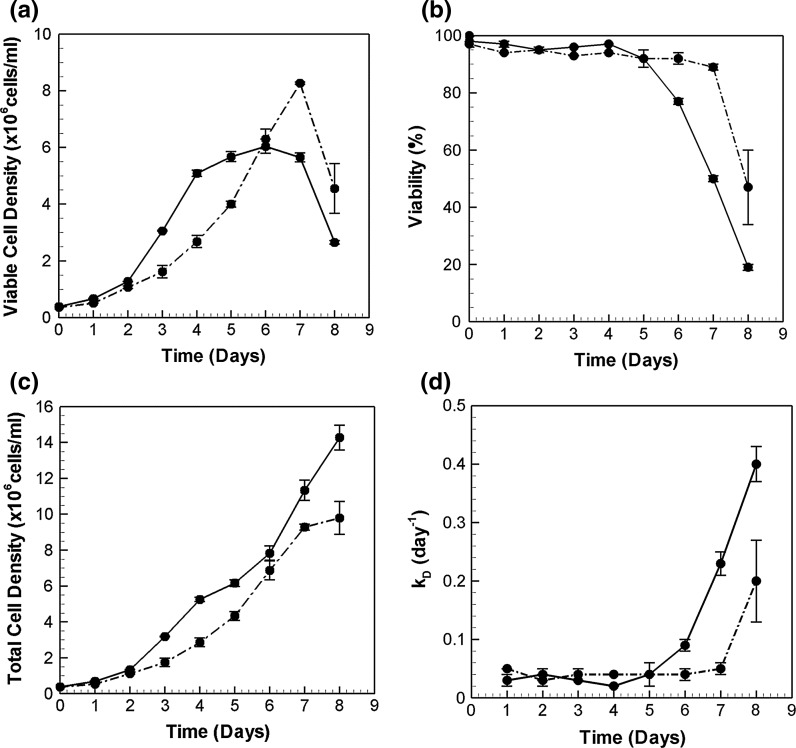
The two cell lines chosen for this study were primarily distinguished based on their IgG productivities, and are referred to as low (LP) and high producer (HP). Figure 2a, b present their growth and viability profiles. The low producer attains a maximum viable cell density of 8 million cells/ml. The exponential phase lasts until day 7 with viability over 90 % and a specific growth rate of 0.3 day−1. The high producer, on the other hand, exhibits a higher specific growth rate (0.6 day−1) until day 4, beyond which the viability of HP cells declines gradually. The viable cell density reaches a maximum of 6 million cells/ml. The viable cell density data suggests cessation of growth on day 4 for the HP. However, the total cell density plot (Fig. 2c) shows continual growth past day 4. This growth is however accompanied by an increased death rate, kD (Fig. 2d), leading to an apparent stationary phase from day 4 to day 7.
Fig. 2.
Growth characteristics of the two CHO cell lines. a Viable cell densities (×106cells/ml), b Viability (%), c Total cell densities (×106 cells/ml) and d Specific death rate (day−1) of the high and low producer in batch culture. Solid lines denote high producers (HP) while dash dotted lines denote low producers (LP)
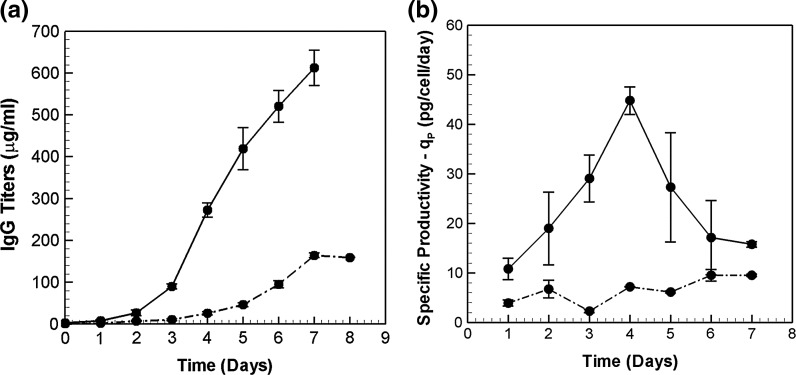
Secreted IgG concentrations measured for the two cell lines are plotted in Fig. 3a. The antibody titers steadily increase for both high and low producers, with a maximum IgG concentration of 600 μg/ml on day 7 for the high-producer cell line. The low producer could produce only 150 μg/ml by the end of the culture. Figure 3b presents the specific productivities. The specific productivity of the low producer remains at relatively low values between 3 and 9 pg/cell/day over the duration of the batch culture. By comparison, the high-producer has a much higher productivity with a peak value of 45 pg/cell/day on day 4.
Fig. 3.
Comparison of a Antibody titers (μg/ml) and b Specific productivities—qP (pg/cell/day) between HP (solid lines) and LP (dash dotted lines) in a batch culture
Transcriptomic profiling of ER resident chaperones and UPR genes
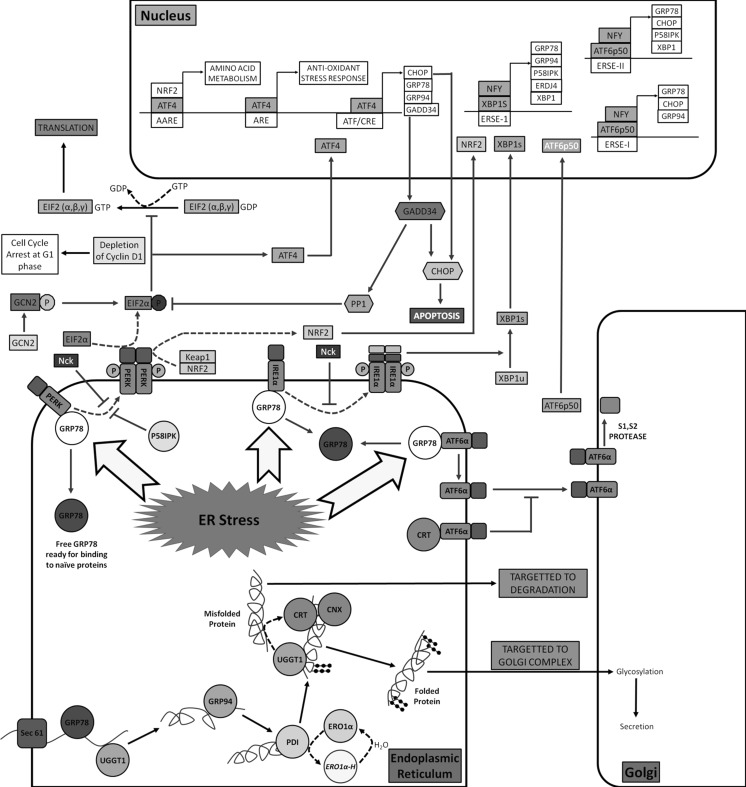
Multiple ER resident and UPR pathway genes were chosen for quantitative real time analysis. Figure 4 shows a detailed view of the pathway. Six major chaperones, namely, Glucose Regulated Protein 78 (GRP78), Glucose Regulated Protein 94 (GRP94), ERO1-l alpha (ERO1α), ER-localized DnaJ homologue 4 (ERDJ4), Calreticulin (CRT) and Calnexin (CNX) of the ERAF pathway were measured. In addition, UDP-glucose glycoprotein glucosyltransferase 1 (UGGT1), an enzyme responsible for recognizing misfolded proteins was profiled.
Fig. 4.
Detailed view of protein secretion pathway. On translocation of the unfolded protein into the ER, multiple chaperones help in folding, glycosylation and formation of disulfide bridges. In the event of an ER overload, accumulation of unfolded proteins leads to ER stress. Three membrane transducers, PERK, Ire1α and ATF6, are bound to GRP78 under normal conditions. ER stress leads to dissociation of GRP78 and subsequent activation of PERK and Ire1α by dimerization and phosphorylation. Activation of ATF6 is by translocation to golgi complex after dissociation from GRP78 where it is cleaved by serine proteases to an active transcription factor ATF6p50. Downstream targets of each of these pathways are shown. AARE amino acid response element; ARE anti-oxidant response element; ERSE endoplasmic reticulum stress element; ATF/CRE activating transcription factor/cAMP responsive element. Abbreviations for the genes are explained in the text
The figure also shows the three different arms of the UPR signaling cascade, initiated by PKR-like ER kinase (PERK), inositol-requiring enzyme-1alpha (Ire1α) and activating transcription factor 6 (ATF6) transducers. Transcription factors from the PERK and Ire1α branch of the UPR pathway were selected for the study. In addition, IgG light chain (IgG LC) and heavy chain (IgG HC) mRNA were also measured. The complete list of genes is presented in Table 1. Overall, 15 genes were analyzed at 6 time points along the growth curves of the low and high producer cell lines.
IgG HC mRNA limits productivity in early batch culture
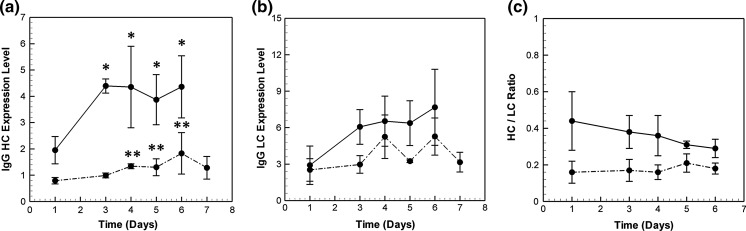
Figure 5a, b present the variation in the mRNA levels of heavy and light chain during the course of the batch culture. In the high producer, the mRNA levels of both HC and LC increase by about two fold from their respective day 1 values. The increase occurs as early as day 3, after which the levels are fairly constant. IgG HC mRNA levels are significantly higher in the high producer than those in the low producer at all time points. On the other hand, IgG LC levels are comparable in the two cell lines over the entire culture, except on days 3 and 5.
Fig. 5.
mRNA profile of a IgG heavy chain (HC) and b light chain (LC) in HP (solid lines) and LP (dash dotted lines).*Denotes p value < 0.05 with respect to day 1 expression levels in high producer, whereas **refers to low producer data. The ratio of IgG HC mRNA to LC mRNA is shown in c
We next compare the time profile of IgG heavy chain mRNA (Fig. 5a) with that of specific productivity (Fig. 3b) in the two cell lines. On day 1, both the productivity and IgG HC levels in the high producer are significantly higher than those in the low producer. Also, an increase in IgG HC levels on day 3 parallels with an increase in productivity in the HP cell line. Similarly, a two-fold increase in IgG HC levels on day 6 is accompanied by an increase in the corresponding productivity values in the low producer. Both the IgG HC levels and the specific productivity values in LP are at a level much lower than that of the high producer.
By comparison, the IgG LC mRNA time profile (Fig. 5b) does not show any distinct similarity with the specific productivities of the two cell lines (Fig. 3b). This is probably because IgG HC levels are stoichiometrically limiting, as the light chain is present in higher amounts (see Fig. 5c). The high producer has an HC/LC ratio of around 0.5, whereas the LP maintains a ratio of 0.2. An excess of LC over HC seems to be beneficial for productivity. Similar findings have been reported in a study by Schlatteret al. (2005) where a HC:LC ratio between 1:2 and 1:5 is found to be optimal for maximizing IgG productivity in stable CHO cells. Therefore, it can be argued that productivity is better correlated with IgG HC than to IgG LC levels.
Early upregulation of chaperones in high producer
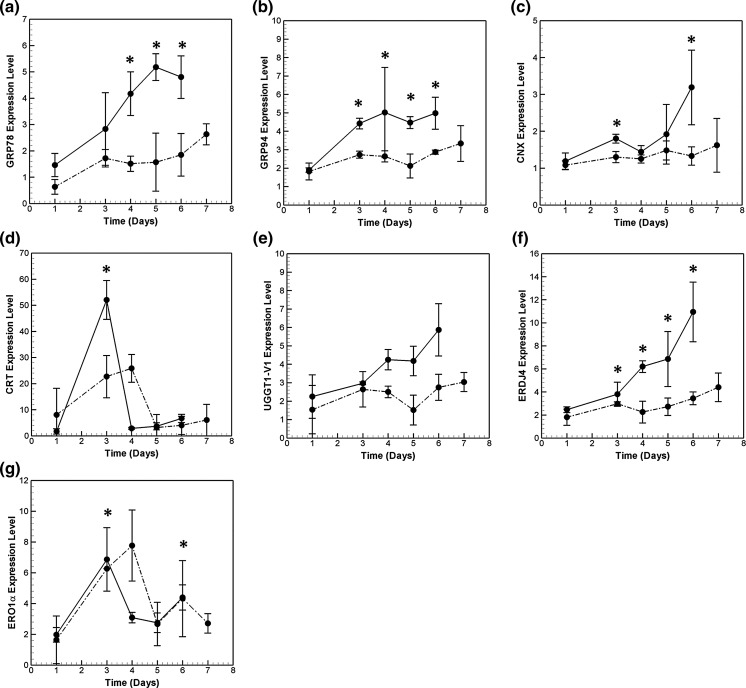
Time profile data for the HP cell line presented in Fig. 6a–g show chaperone upregulation compared to day 1 levels. The primary chaperone, GRP78, is induced to a modest two-fold on day 3 (Fig. 6a) and 3.3-folds on day 6 with respect to day 1. In comparison, mRNA of GRP94, which binds to partial folded proteins and acts sequentially after GRP78 (Schröder 2008), shows an increase of 2.3-fold on day 3 (Fig. 6b). As expected, GRP94 and GRP78 are the most abundant chaperones (see Fig. 1s).
Fig. 6.
Transcriptional profile of chaperones a GRP78 b GRP94 c CNX d CRT e UGGT1-V1 f ERDJ4 and g ERO1α in the high and low producer cell lines during the course of batch culture. Symbols used are same in Fig. 5
Calreticulin (CRT) and calnexin (CNX) are calcium dependent glycosylation chaperones involved in imparting glycosylation residues to the partially folded proteins, thus making them stable (Schröder and Kaufman 2005). CNX mRNA is present in high numbers at all the time points measured for the high-producer cell line. It is induced 2.7-fold on day 6 (Fig. 6c) compared to its day 1 value. CRT is present in very low concentrations except very high induction on day 3 (Fig. 6d).
The UGGT1 variant 1 (UGGTI-V1) chaperone recognizes unfolded proteins which are partially degraded and are not recognized by GRP78. This enhances the protein quality control mechanism inside the ER. The corresponding mRNA levels are found to increase steadily with time and reach a maximum of 3.5-fold increase on day 6 compared to its day 1 level (Fig. 6e). This may indicate an increase of misfolded proteins. Similarly, the co-chaperone ERDJ4 mRNA level increases steadily with an induction of 4.5-fold (Fig. 6f) on day 6. ERDJ4 helps in the folding of proteins by GRP78 by stimulating the ATP hydrolysis and ATP exchange.
Protein disulfide isomerases catalyze the formation of disulfide bonds in proteins. ERO1α is involved in the reoxidation of PDI for its continuous function. Among all the chaperones profiled in this study, ERO1α mRNA is present in the lowest amounts (Fig. 1s). It shows transient induction on day 3 (Fig 6g), similar to the CRT profile. It is interesting to note that although both CRT and ERO1α mRΝΑ return to basal levels, their concentration is still higher as compared to day 1.
The time profiles of the chaperones for the low producer are also plotted in Fig. 6a–g for comparison. Similar to the high producer, ERO1α and CRT show transient induction in the low producer. While ERO1α mRNA levels are similar in the two cell lines, CRT induction is comparatively low in LP cell line. The other chaperones, in general, show higher induction in the high productivity cell line. See, for example, UGGT-V1 and ERDJ4.
Dynamics of PERK signaling branch of UPR pathway
The PERK branch responds to the increase in ER stress by shutting down protein translation, so as to minimize the load on the cell. This is achieved by phosphorylation of EIF2α, a translation initiation factor involved in protein synthesis.
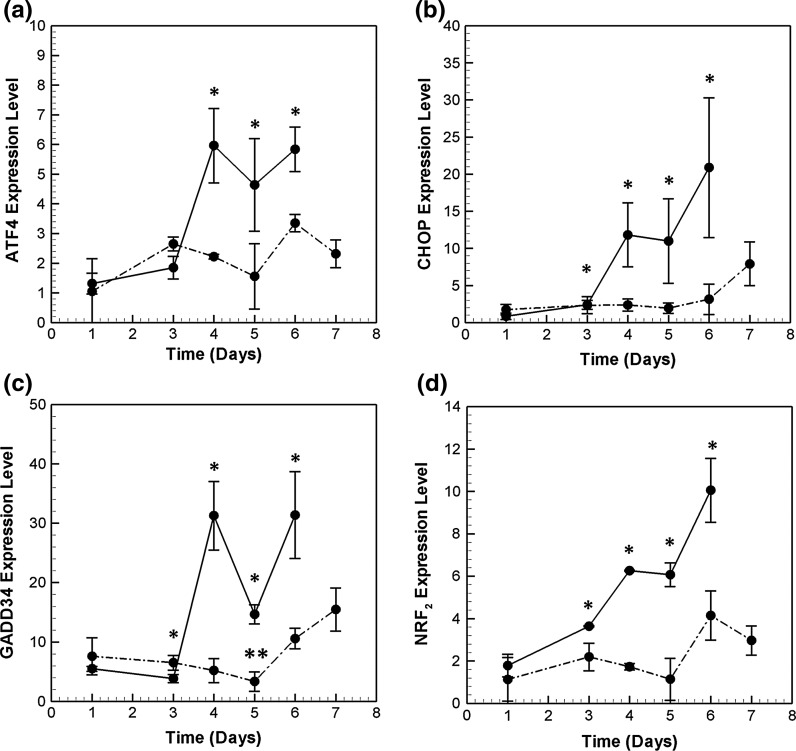
While the translation of most proteins is attenuated, few transcription factors, such as ATF4, are selectively translated. ATF4 has also been shown to be transcriptionally regulated under many stress conditions such as exposure to thapsigargin (Tg), hypoxia and oxidative stress (Harding et al. 2000a, b, 2003). Among the PERK pathway genes profiled in this study, ATF4 was the most abundant in both the high and low producers. As seen in Fig. 7a, ATF4 mRNA is induced 4.7-fold in the high producer on day 4 compared to its level on day 1, and continues to stay at high levels until the end of the culture.
Fig. 7.
Transcriptional profile of key PERK/GCN2 pathway genes, a ATF4, b CHOP, c GADD34 and d NRF2. Refer to Fig. 5 for symbols
ATF4 targets CHOP, a bZIP transcription factor that induces the expression of other pro-apoptotic proteins. On day 4 and 5 of the culture, CHOP mRNA is induced almost 13-fold in the HP cell line with respect to its day 1 levels. At day 6, CHOP mRNA levels reach a new state, with 24-fold induction compared to day 1 levels (see Fig. 7b).
CHOP and ATF4 together regulate the expression of GADD34 gene, which forms a negative feedback loop by de-phosphorylating EIF2α so as to restore translation (Novoa et al. 2001). GADD34 is induced by 5.7-fold on day 4 (Fig. 7c). Interestingly, GADD34 is repressed on day 5, only to be induced back again on day 6. GADD34 mRNA has been shown to have a very short half life, and needs continuous stress signals for its sustained activation (Rutkowski et al. 2006; Brush and Shenolikar 2008).
Profile of NRF2 mRNA (see Fig. 7d), another transcription factor involved in the PERK pathway, is very similar to that of CHOP. It is induced on day 3 in HP and reaches a maximum of 8.2-folds on day 6. NRF2 is is phosphorylated by PERK, and it later auto-regulates its own mRNA expression (Kaspar and Jaiswal 2010; Stępkowski and Kruszewski 2011). NRF2 also induces the expression of anti-oxidant responsive genes (Chen and Kunsch 2004). This is possibly the response to high oxidative stress in these recombinant CHO cells as a result of continuous and repeated folding processes inside the stressed ER (Cullinan et al. 2003).
The time profiles of PERK pathway genes in the low producer cell line are also plotted in Fig. 7. Initially, the mRNA levels of all genes are comparable between the two cell lines. Post day 3, while induction is observed in the HP cell line, the mRNA levels in the LP cell line continue to be maintained at basal levels. ATF4, CHOP, NRF2 and GADD34 mRNA are significantly induced in the low producer cell line only on day 6, but the induction levels are much lower than HP.
Dynamics of Ire1α signaling branch of UPR pathway
The Ire1α branch of UPR responds to high levels of unfolded protein in ER by increasing both survival and pro-apoptotic signals. The survival response is mediated by transcriptional induction of chaperones and ERAD pathway proteins (Travers et al. 2000; Lee et al. 2003), whereas apoptotic response is mediated by either transcriptional induction of CHOP or JNK pathway (Urano et al. 2000; Rutkowski et al. 2006).
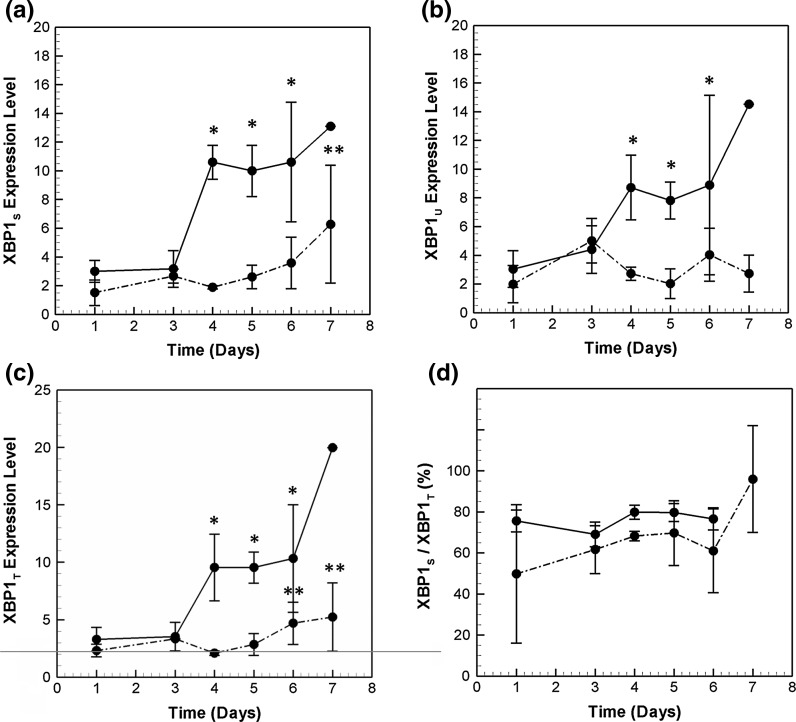
Induction of the Ire1α branch begins with the dissociation of membrane bound transducer Ire1α from GRP78. Post dissociation, the endoribonuclease activity of Ire1α acts on the mRNA of the transcription factor XBP1. A spliced form, XBP1S, is thus formed, which induces the expression of both chaperones and pro-apoptotic genes such as CHOP. The active form, XBP1S, also auto-regulates its own transcription. Figure 8a, b present the time profile of XBP1S and XBP1U, the unspliced form, in the high producer. A 3.8-fold induction of XBP1S is observed on day 4, with increasing trends until day 6. A similar transcriptional profile is also seen for XBP1U mRNA, although to slightly lower levels. The copy numbers of XBP1S are comparable to NRF2 and ATF4 transcription factors studied earlier. Also, XBP1S mRNA is always present in higher copy numbers as compared to the unspliced form.
Fig. 8.
Transcriptional profile of a XBP1s. b XBP1u and c XBP1T in the two cell lines. d Shows the ratio of XBP1s to XBP1T during the course of the batch culture. Refer to Fig. 5 for symbols
Figure 8c shows the transcriptional profile of XBP1T mRNA. It is induced both by XBP1s and ATF6p50, the active cleaved form of ATF6, which forms the third branch of UPR signaling. XBP1T follows a very similar time profile as that of XBP1S. An induction of 3.25-fold is observed for XBP1 mRNA on day 4, followed by a steady increase until day 6. The kinetic profiles in the low producer plotted in Fig. 8a–c show that the mRNA levels of XBP1T, XBP1U and XBP1S are always less as compared to those in the high producer.
Transcriptional profiling of XBP1S indicates the presence of spliced form mRNA at the early stages of the culture. Figure 8d plots the percentage of XBP1S wrt XBP1T mRNA in the two cell lines. On day 1, 76 % of XBP1T mRNA is present as XBP1S mRNA in the high producer cell line. This indicates that Ire1α pathway is active at this time point. The fraction of XBP1S to XBP1T mRNA increases slightly from day 3 to day 4, indicating further activation of the pathway. Presence of XBP1s has been observed in other recombinant CHO cell lines as well (Tigges and Fussenegger 2006; Ohya et al. 2008). The presence of high levels of XBP1S in an unstressed cell is a possible reason for the ineffectiveness of XBP1 overexpression on productivity (Ku et al. 2008). The fraction of XBP1S/XBP1T mRNA is similar in the low producer, with slightly lower levels on day 4 as compared to the HP data.
Note that the profile of XBP1S mRNA is highly correlated (correlation >0.95) with the mRNA of a number of downstream target chaperones, such as ERDJ4, UGGT1 and GRP78. While many chaperones are known to be regulated by coordinated action of XBP1S and ATF6, ERDJ4 has been shown to be dependent on XBP1s alone for its induction (Lee et al. 2003). In contrast, the mRNA profiles of chaperones, ERO1α and CRT are distinct from XBP1s, indicating a different regulatory mechanism.
Discussion
Many studies have targeted the protein secretion pathway in an attempt to increase productivity of recombinant proteins in mammalian cells. The results have been highly variable, especially pertaining to the folding and UPR components. The protein folding pathway and the response to ER stress are complex processes with multiple steps. In addition, UPR pathway has been shown to be highly dynamic with distinct kinetic outcomes depending on the input (DuRose et al. 2006; Gass et al. 2008; Merquiol et al. 2011; Pena and Harris, 2011). A mechanistic understanding of this pathway in recombinant cell lines will therefore aid in designing rational metabolic engineering strategies. Understanding its kinetics in a batch culture is a step towards this process. We have chosen two recombinant CHO cell lines from the same parental CHO DG44 cells for comparison. The two cell lines have over four-fold difference in productivity, where the peak productivity of the high producer is comparable to that seen in many industrial and fed-batch processes.
Different productivity profiles have been observed in recombinant mammalian cells in batch culture. For example, Jiang et al. report a specific productivity profile that is high in the initial stages of batch culture and later decreases (Jiang et al. 2006). In contrast, we observe that the specific productivity of the high producer cell line is initially low. It increases until day 4 and decreases subsequently. A similar profile for productivity has been reported earlier in a study with GS-NSO cells (Stansfield et al. 2007), where productivity of MaB increased up to day 8, and declined thereafter in a fed-batch culture. Other studies have also shown a similar productivity profile (Kaufmann et al. 1999; Hendrick et al. 2001; Yoon et al. 2006a, b).
The observed specific productivity profiles can be determined by many different factors such as how the cells are maintained and the expression system used in these cells. In the early stages of batch culture, the increase in qP is correlated to a corresponding increase in the IgG HC mRNA levels on day 3. This is similar to that reported by Barnes et al. and others (Dorai et al. 2006; Barnes et al. 2007; Li et al. 2007). The observed dynamics of IgG HC and LC genes is potentially due to some specific elements present in the hCMV-MIE promoter, which drives their expression. Note that this promoter has been reported to possess sites for many different transcription factors (Sambucetti et al. 1989; Yurochko et al. 1997). It is interesting to note that IgG HC and LC were also under the control of the same promoter in the GS-NSO cells used in the study mentioned above (Stansfield et al. 2007). The expression from this promoter seems to be inversely related to growth. When cells from day 4 are re-suspended in fresh medium (data not shown) we observe a drop in their productivity. This is accompanied by a decline in IgG HC mRNA levels.
The initial increase in specific productivity until day 3 is related to the expression system that controls the IgG HC and LC mRNA. By comparison, specific productivity increases further from day 3 to day 4 without any corresponding increase in IgG HC or LC mRNA levels. Interestingly, mRNA levels of chaperones such as GRP94, ERDJ4 and Calnexin (CNX) are upregulated in this period, which possibly enhances the capacity of ER to process unfolded proteins. Note that most UPR pathway genes show only a minimal increase in the initial phase of batch culture. However, we observe a steep induction of ATF4, CHOP and GADD34 (Fig. 7a–c) mRNA levels on day 4. Similar upregulation is also seen for the XBP1T and XBP1S mRNA levels. This probably is a response to the increased stress due to higher productivity. It may also reflect the adaptation of the cells to handle increased productivity levels. Factors such as nutritional status may also play a role and are discussed later. Note that, other studies have also observed a positive correlation of mRNA levels of chaperones GRP78, GRP94 and CRT and XBP1S with that of secreted antibody concentration in production clones (Smales et al. 2004; Kober et al. 2012).
The maximum specific productivity observed in the high producer is 45 pg/cell/day. In comparison the low producer has only a maximum specific productivity of 9 pg/cell/day. This compares well with other studies on cell engineering of various chaperones and UPR pathway genes. For example, over-expression of GADD34, specific productivity of AT-III increased from 15 to 30 pg/cell/day (Ohya et al. 2008). Similarly, over-expression of PDI in CHO cells increased productivity to about 22 pg/cell/day (Borth et al. 2005). In another study with CHO cells expressing anti-apoptotic genes, a productivity of 100 pg/cell/day is reported on treatment with sodium butyrate and a shift in temperature (Kantardjieff et al. 2010). Microarray analysis suggested that the increased productivity could be because of an increase in secretory capacity. An upregulation of chaperones and other protein secretion related genes such as golgi apparatus is seen in these cells.
What is the stress signal: growth or IgG?
The UPR pathway is activated under many different stress conditions. It can be activated directly by high amounts of unfolded protein in the ER. The UPR also senses the nutritional state of the cell. For example, the PERK pathway has been shown to be activated by glucose deprivation and under hypoxic conditions (Koumenis et al. 2002; Elanchezhian et al. 2012). Similarly, amino-acid limitation also leads to attenuation of protein synthesis by phosphorylation of EIF2α phosphorylation via GCN2 kinase (Zhang et al. 2002). Studies have also shown upregulation of CHOP in response to glucose and glutamine deprivation in both CHO and NSO cell lines (Murphy et al. 2001; Lengwehasatit and Dickson 2002). In response to simultaneous deprivation of both glucose and glutamine, CHOP mRNA expression was further enhanced and was comparable to that in response to tunicamycin (Lengwehasatit and Dickson 2002). In contrast, addition of substrate failed to suppress the expression of chaperones in another case (Downham et al. 1996). Similarly, in a proteomics based comparison of four NS0 cell lines of varying productivity and similar growth rates, many chaperones were found to be upregulated in higher productivity cultures (Smales et al. 2004).
Limitation in glucose, oxygen or amino-acids can be caused by either high growth or high productivity of cells. While limitation of nutrients and oxygen due to high-growth rate is easily understandable, production of IgG also leads to similar stress. For example, secreted proteins cause amino-acid limitation since they deplete the intracellular amino-acid levels. Similarly, enhanced productivity will place a higher load on the energy metabolism of the cell due to the increased demands for ATP for protein folding.
The data reported in this paper clearly shows that HP is under higher stress as compared to the other cell line. The chaperones and transcriptional regulators from PERK/GCN2 and Ire1α branch of UPR are induced to a much higher level. The upregulation of XBP1s and chaperones in the high producer strongly suggests the activation of the Ire1α and possibly ATF6 pathways in the HP cell line. While the PERK/GCN2 branch could be activated by either nutrient limitation or ER stress, the Ire1α and ATF6 branches are activated through ER stress. Higher % spliced XBP1 in the high producer cell line is also indicative of enhanced Ire1α-P activity. Thus, the overall stress in the high producer is a combination of the factors discussed above, and their relative contribution needs further study.
Comparison with previous studies
The upregulation of protein secretion related genes in recombinant CHO cells has been reported earlier, but the studies have usually been limited to only a few chaperones primarily GRP78, GRP94 and occasionally PDI and ERP72 (Downham et al. 1996; Lambert and Merten 1997; Nissom et al. 2006). Further, in these cell lines there is limited information on the transcription control of these proteins through the UPR signalling pathway (Ku et al. 2010; Du et al. 2013). An integrated study of the role of UPR and its dynamics in mammalian bioprocesses is lacking.
A constitutively high expression of the chaperones through the course of the batch culture has been reported in literature. For example, in a study with CHO cells expressing TIMP-1 protein, constant levels of Bip and PDI were detected, whereas CRT was upregulated after initial lag phase (Underhill et al. 2005). In contrast, we find that the expression of chaperones is highly dynamic through the course of batch culture. The dynamics further correlates with increase in productivity. Many factors can contribute to the observed trend in specific productivity. These include how the cells are maintained, the expression system used and its relation to growth, as discussed above.
Chaperones including CNX and PDI and XBP1 were found to be upregulated in the producer culture of HEK293 cells when compared with a non-producer that grew at the same rate (Smales et al. 2004, Dietmair et al. 2012). Abundance of protein folding genes and IgG HC mRNA were also observed in recombinant NS0 cell line with a higher qP indicating that the stressed cells require the highly active folding machinery to maintain a higher productivity status (Smales et al. 2004).
The expression of the transcription factor CHOP has also been studied in NS0 myeloma cell lines and in CHO DG44 recombinant cell lines (Murphy et al. 2001; Lengwehasatit and Dickson 2002). The CHOP mRNA profiles were transient in the NS0 cell line. In another study, the CHOP expression levels were found to be induced at the end of the batch culture and correlated well with apoptosis. In contrast, CHOP is induced earlier in our culture, a reflection of higher nutritional and ER stress. Also, the levels of CHOP continue to increase until the end of the batch culture.
Distinct kinetic profiles of chaperones
Folding of IgG molecules in the ER consists of a series of steps. GRP78 and GRP94 act sequentially to initiate the folding process. Then the disulphide bonds are formed through the PDI protein. PDI is continuously reoxidized through ERO1α and ERO1β proteins. The removal of oligosaccharides occurs via the calnexin and calreticulin cycle, along with the recognition of mis-folded proteins by UGGT1.
On exposure to ER stress, the chaperones are upregulated in the HP cell line. However, the data suggest that not all chaperones are regulated similarly. Similar results have been observed earlier. For example, in a recent study, silencing of GRP94 was found to have a distinct UPR response when compared to that in response to BiP silencing or tunicamycin treatment (Eletto et al. 2012). The calreticulin gene was not upregulated in response to GRP94 silencing. In response to dengue virus, only GRP78 is upregulated but a similar increase was not detected in the case of GRP94 or PDI protein levels.
We show that the dynamics of chaperones in recombinant CHO cells producing IgG are distinct. While most chaperones’ levels continue to increase with time in the batch culture, expression of CRT and ERO1α is found to be transient. There is high fold increase in the expression levels of these genes, presumably because of the sudden increase in demand and very low basal expression levels. Interestingly, the functionally related protein CNX is present at higher concentrations and is induced by only two-fold. However, the kinetic trend is similar to that of CRT. The expression profiles of many chaperones including ERDJ4, GRP78, GRP94 and UGGT1 are found to be strongly correlated (R2 > 0.86) with that of XBP1S. However, the cause of transient regulation of CRT and ERO1α needs further investigation. CRT protein has been shown to be differentially expressed under various conditions. In response to amino acid starvation it is found to be induced (Heal and McGivan 1998), whereas it is selectively repressed in response to many differentiating agents such as Me2SO and retinoic acid (Clark et al. 2002).
The observed continuous upregulation of chaperones appears to be a response to chronic stress, which is different from that of short-term stress. For example, in cells infected with hepatitis C virus, all branches of UPR were induced post-infection. Peak induction was observed on day 3 (Merquiol et al. 2011), and the genes were significantly induced even at day 14. In contrast, on treatment of CHO cells with pharmacological agents such as dithiothreitol (DTT), thapsigargin (Tg) and tunicamycin (Tm), the activation of UPR occurs as early as 15 min (DuRose et al. 2006), and levels of many genes such as CHOP and GADD34 return to basal levels by 24 h (Rutkowski et al. 2006).
Dynamics of UPR
It has been reported that the UPR pathway exhibits a multi-phase response of alarm, adaptation and apoptosis in the presence of stress signals (Hetz 2012). The alarm phase is usually characterized by translation attenuation implemented by an increase in EIF2α-P levels. The adaptive phase is initiated by activation of the Ire1α/ATF6 pathways. An increase in chaperone levels occurs in this phase. Activation of GADD34 from the PERK signaling branch is also a marker for adaptation. In response to chronic stress, when homeostasis cannot be maintained, an increase in CHOP levels beyond a threshold marks the onset of the apoptosis phase.
In the current study of CHO cells in batch culture, we observe the adaptation and apoptosis phases of UPR pathway. The upregulation of CHOP and GADD34 is a hallmark of activation of the PERK pathway. The induction of GADD34 probably helps the cell in adapting to ER stress by restoring translation. Induction of GADD34 in HP can potentially reverse translational attenuation leading to increase in product titres and productivity. Corroborating this hypothesis is a recent study (Ohya et al. 2008), where over-expression of ATF4 and GADD34 led to increase in productivity of thrombopoietin. Similarly, increase in XBP1S levels indicates activation of Ire1α pathway. The occurrence of these phenomenons around day 3–4 marks the adaptation phase in the high producer cell line. On day 6, the apoptotic phase of UPR is activated in the HP, as illustrated by a further increase in CHOP levels, and a corresponding decrease in cell viability. On the other hand, in the LP cell line, the UPR pathway is activated at the end (day 7) of the batch culture. In the absence of high productivity, the corresponding response is also much subdued as compared to the HP.
Conclusion
This paper presents a comprehensive study of UPR pathway in recombinant CHO cells. Time profile of all major chaperones and signalling pathway genes are measured in a batch culture for two cell lines with varying productivity and growth profiles. A four-fold difference in productivity and a two-fold higher specific growth rate, place a higher load on the high producer. Increased stress results in much higher induction of the UPR pathway. The presence of chronic stress is further reinforced by the UPR response profiles that exhibit an adaptation phase followed by apoptosis at the end of the batch culture. Further, the data suggests a strong correlation between enhanced ER capacity and IgG productivity, although additional studies are required to establish the cause and effect relationship. Understanding this relationship between growth, ER stress and productivity will enhance our ability to target this pathway for increased production of recombinant proteins.
Electronic supplementary material
Estimate of relative abundance of various genes profiled in this study in HP cell line. Time profiles of ΔCT are plotted for a) IgG HC and LC b) Chaperones c) PERK pathway genes and d) Ire1α pathway genes. ΔCT is defined as , Positive values imply abundance greater than β-actin whereas negative values imply lower abundance. Note that β-actin is a moderately abundant gene (TIFF 1084 kb)
Acknowledgments
This work was partially supported by a grant from Department of Biotechnology, Government of India. We would like to thank Dr. Miranda Yap and Dr. Niki Wong, Bioprocessing Technology Institute, Singapore for providing the CHO cell lines. We would also like to thank Prof. Wei Shou Hu of University of Minnesota for useful discussions and comments on the manuscript.
References
- Barnes LM, Bentley CM, Moy N, Dickson AJ. Molecular analysis of successful cell line selection in transfected GS-NS0 myeloma cells. Biotechnol Bioeng. 2007;96:337–348. doi: 10.1002/bit.21119. [DOI] [PubMed] [Google Scholar]
- Borth N, Mattanovich D, Kunert R, Katinger H. Effect of increased expression of protein disulfide isomerase and heavy chain binding protein on antibody secretion in a recombinant CHO cell line. Biotechnol Prog. 2005;21:106–111. doi: 10.1021/bp0498241. [DOI] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S. Control of cellular GADD34 Levels by the 26S proteasome. Mol Cell Biol. 2008;28:6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XL, Kunsch C. Induction of cytoprotective genes through Nrf2/antioxidant response element pathway: a new therapeutic approach for the treatment of inflammatory diseases. Curr Pharm Des. 2004;10:879–891. doi: 10.2174/1381612043452901. [DOI] [PubMed] [Google Scholar]
- Chen K, Liu Q, Xie L, Sharp PA, Wang DIC. Engineering of a mammalian cell line for reduction of lactate formation and high monoclonal antibody production. Biotechnol Bioeng. 2001;72:55–61. doi: 10.1002/1097-0290(20010105)72:1<55::AID-BIT8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Chung JY, Lim SW, Hong YJ, Hwang SO, Lee GM. Effect of doxycycline-regulated calnexin and calreticulin expression on specific thrombopoietin productivity of recombinant chinese hamster ovary cells. Biotechnol Bioeng. 2004;85:539–546. doi: 10.1002/bit.10919. [DOI] [PubMed] [Google Scholar]
- Chusainow J, Yang YS, Yeo JHM, Toh PC, Asvadi P, Wong NSC, Yap MG. A study of monoclonal antibody-producing CHO cell lines: what makes a stable high producer? Biotechnol Bioeng. 2009;102:1182–1196. doi: 10.1002/bit.22158. [DOI] [PubMed] [Google Scholar]
- Clark RA, Li S-L, Pearson DW, Leidal KG, Clark JR, Denning GM, Reddick R, Krause KH, Valente AJ (2002) Regulation of calreticulin expression during induction of differentiation in human myeloid cells: evidence for remodeling of the endoplasmic reticulum. J Biol Chem 277:32369–32378 [DOI] [PubMed]
- Cudna RE, Dickson AJ. Engineering responsiveness to cell culture stresses: growth arrest and DNA damage gene 153 (GADD153) and the unfolded protein response (UPR) in NS0 myeloma cells. Biotechnol Bioeng. 2006;94:514–521. doi: 10.1002/bit.20861. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Schooley K, Rasmussen B, Thomas J, Reddy P. Effect of PDI overexpression on recombinant protein secretion in CHO cells. Biotechnol Prog. 2000;16:736–743. doi: 10.1021/bp000107q. [DOI] [PubMed] [Google Scholar]
- Dietmair S, Hodson MP, Quek L-E, Timmins NE, Gray P, Nielsen LK. A multi-omics analysis of recombinant protein production in Hek293 cells. PLoS one. 2012;7:e43394. doi: 10.1371/journal.pone.0043394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan P, Melville M, Gammell P, Sinacore M, Meleady P, McCarthy K, Francullo L, Leonard M, Charlebois T, Clynes M (2008) Transcriptional profiling of gene expression changes in a PACE-transfected CHO DUKX cell line secreting high levels of rhBMP-2. Mol Biotechnol 39:187–199 [DOI] [PubMed]
- Dorai H, Csirke B, Scallon B, Ganguly S. Correlation of heavy and light chain mRNA copy numbers to antibody productivity in mouse myeloma production cell lines. Hybridoma. 2006;25:1–9. doi: 10.1089/hyb.2006.25.1. [DOI] [PubMed] [Google Scholar]
- Downham MR, Farrell WE, Jenkins HA. Endoplasmic reticulum protein expression in recombinant NS0 myelomas grown in batch culture. Biotechnol Bioeng. 1996;51:691–696. doi: 10.1002/(SICI)1097-0290(19960920)51:6<691::AID-BIT7>3.3.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Du Z, Treiber D, McCoy RE, Miller AK, Han M, He F, Domnitz S, Heath C, Reddy P (2013) Non-invasive UPR monitoring system and its applications in CHO production cultures. Biotechnol Bioeng 110:2184–2194 [DOI] [PubMed]
- DuRose JB, Tam AB, Niwa M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell. 2006;17:3095–3107. doi: 10.1091/mbc.E06-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanchezhian R, Palsamy P, Madson CJ, Mulhern ML, Lynch DW, Troia AM, Usukura J, Shinohara T (2012) Low glucose under hypoxic conditions induces unfolded protein response and produces reactive oxygen species in lens epithelial cells. Cell Death Dis 3:e301 [DOI] [PMC free article] [PubMed]
- Eletto D, Maganty A, Dersh D, Makarewich C, Biswas C, Paton JC, Paton AW, Doroudgar S, Glembotski CC, Argon Y (2012) Limitation of individual folding resources in the ER leads to outcomes distinct from the unfolded protein response. J Cell Sci 12:4865–4875 [DOI] [PMC free article] [PubMed]
- Figueroa B, Ailor E, Osborne D, Hardwick JM, Reff M, Betenbaugh MJ. Enhanced cell culture performance using inducible anti-apoptotic genes E1B-19 K and aven in the production of a monoclonal antibody with Chinese hamster ovary cells. Biotechnol Bioeng. 2007;97:877–892. doi: 10.1002/bit.21222. [DOI] [PubMed] [Google Scholar]
- Gass JN, Jiang H-Y, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45:1035–1043. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633 [DOI] [PubMed]
- Heal R, McGivan J. Induction of calreticulin expression in response to amino acid deprivation in Chinese hamster ovary cells. Biochem J. 1998;329:389–394. doi: 10.1042/bj3290389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick V, Winnepenninckx P, Abdelkafi C, Vandeputte O, Cherlet M, Marique T, Renemann G, Loa A, Kretzmer G, Werenne J (2001) Increased productivity of recombinant tissular plasminogen activator (t-PA) by butyrate and shift of temperature: a cell cycle phases analysis. Cytotechnology 36:71–83 [DOI] [PMC free article] [PubMed]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Ifandi V, Al-Rubeai M. Regulation of cell proliferation and apoptosis in CHO-K1 cells by the coexpression of c-Myc and Bcl-2. Biotechnol Prog. 2005;21:671–677. doi: 10.1021/bp049594q. [DOI] [PubMed] [Google Scholar]
- Jayapal K, Wlaschin K, Hu W-S, and Yap M (2007) Recombinant protein therapeutics from Cho cells—20 years and counting. CHO Consortium, SBE Special Edition, pp 40–47
- Jiang Z, Huang Y, Sharfstein ST. Regulation of recombinant monoclonal antibody production in chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnol Prog. 2006;22:313–318. doi: 10.1021/bp0501524. [DOI] [PubMed] [Google Scholar]
- Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok Y-J, Philp R, et al. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J Biotechnol. 2010;145:143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Kaspar JW, Jaiswal AK. An autoregulatory loop between Nrf2 and Cul3-Rbx1 controls their cellular abundance. J Biol Chem. 2010;285:21349–21358. doi: 10.1074/jbc.M110.121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann H, Mazur X, Fussenegger M, Bailey JE. Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol Bioeng. 1999;63:573–582. doi: 10.1002/(SICI)1097-0290(19990605)63:5<573::AID-BIT7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Kim NS, Lee GM. Inhibition of sodium butyrate-induced apoptosis in recombinant Chinese hamster ovary cells by constitutively expressing antisense RNA of caspase-3. Biotechnol Bioeng. 2002;78:217–228. doi: 10.1002/bit.10191. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim Y-G, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- Kober L, Zehe C, Bode J. Development of a novel ER stress based selection system for the isolation of highly productive clones. Biotechnol Bioeng. 2012;109:2599–2611. doi: 10.1002/bit.24527. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol Cell Biol 22:7405–7416 [DOI] [PMC free article] [PubMed]
- Ku SCY, Ng DTW, Yap MGS, Chao S-H. Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol Bioeng. 2008;99:155–164. doi: 10.1002/bit.21562. [DOI] [PubMed] [Google Scholar]
- Ku SCY, Toh PC, Lee YY, Chusainow J, Yap MGS, Chao S-H. Regulation of XBP-1 signaling during transient and stable recombinant protein production in CHO cells. Biotechnol Prog. 2010;26:517–526. doi: 10.1002/btpr.322. [DOI] [PubMed] [Google Scholar]
- Lambert N, Merten O-W. Effect of serum-free and serum-containing medium on cellular levels of ER-based proteins in various mouse hybridoma cell lines. Biotechnol Bioeng. 1997;54:165–180. doi: 10.1002/(SICI)1097-0290(19970420)54:2<165::AID-BIT8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Lee A-H, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengwehasatit I, Dickson AJ. Analysis of the role of GADD153 in the control of apoptosis in NS0 myeloma cells. Biotechnol Bioeng. 2002;80:719–730. doi: 10.1002/bit.10422. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang C, Jostock T, Dübel S. Analysis of IgG heavy chain to light chain ratio with mutant Encephalomyocarditis virus internal ribosome entry site. Protein Eng Des Sel. 2007;20:491–496. doi: 10.1093/protein/gzm038. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meents H, Enenkel B, Eppenberger HM, Werner RG, Fussenegger M. Impact of coexpression and coamplification of sICAM and antiapoptosis determinants bcl-2/bcl-xL on productivity, cell survival, and mitochondria number in CHO-DG44 grown in suspension and serum-free media. Biotechnol Bioeng. 2002;80:706–716. doi: 10.1002/bit.10449. [DOI] [PubMed] [Google Scholar]
- Merquiol E, Uzi D, Mueller T, Goldenberg D, Nahmias Y, Xavier RJ, Tirosh B, Shibolet O (2011) HCV causes chronic endoplasmic reticulum stress leading to adaptation and interference with the unfolded protein response. PLoS one 6:e24660 [DOI] [PMC free article] [PubMed]
- Mohan C, Park SH, Chung JY, Lee GM. Effect of doxycycline-regulated protein disulfide isomerase expression on the specific productivity of recombinant CHO cells: thrombopoietin and antibody. Biotechnol Bioeng. 2007;98:611–615. doi: 10.1002/bit.21453. [DOI] [PubMed] [Google Scholar]
- Murphy TC, Woods NR, Dickson AJ. Expression of the transcription factor GADD153 is an indicator of apoptosis for recombinant chinese hamster ovary (CHO) cells. Biotechnol Bioeng. 2001;75:621–629. doi: 10.1002/bit.1190. [DOI] [PubMed] [Google Scholar]
- Nissom PM, Sanny A, Kok Y, Hiang Y, Chuah S, Shing T, Lee YY, Wong KT, Hu WS, Sim MY, Philp R (2006) Transcriptome and proteome profiling to understanding the biology of high productivity CHO cells. Mol Biotechnol 34:125–140 [DOI] [PubMed]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T, Hayashi T, Kiyama E, Nishii H, Miki H, Kobayashi K, Honda K, Omasa T, Ohtake H (2008) Improved production of recombinant human antithrombin III in Chinese hamster ovary cells by ATF4 overexpression. Biotechnol Bioeng 100:317–324 [DOI] [PubMed]
- Pahl HL, Baeuerle PA. The ER-overload response: activation of NF-κB. Trends Biochem Sci. 1997;22:63–67. doi: 10.1016/S0968-0004(96)10073-6. [DOI] [PubMed] [Google Scholar]
- Pena J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem. 2011;286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi, Akha AA, Raden D, Kaufman RJ (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 4:e374 [DOI] [PMC free article] [PubMed]
- Sambucetti LC, Cherrington JM, Wilkinson GW, Mocarski ES. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8(13):4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter S, Stansfield SH, Dinnis DM, Racher AJ, Birch JR, James DC. On the optimal ratio of heavy to light chain genes for efficient recombinant antibody production by CHO cells. Biotechnol Prog. 2005;21:122–133. doi: 10.1021/bp049780w. [DOI] [PubMed] [Google Scholar]
- Schröder M. The unfolded protein response. Mol Biotechnol. 2006;34:279–290. doi: 10.1385/MB:34:2:279. [DOI] [PubMed] [Google Scholar]
- Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Seth G, Hossler P, Yee J, Hu W-S. Engineering Cells for cell culture bioprocessing—physiological fundamentals. Adv Biochem Eng Biotechnol. 2006;101:119–614. doi: 10.1007/10_017. [DOI] [PubMed] [Google Scholar]
- Smales CM, Dinnis DM, Stansfield SH, Alete D, Sage EA, Birch JR, Racher AJ, Marshall CT, James DC (2004) Comparative proteomic analysis of GS-NS0 murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng 88:474–488 [DOI] [PubMed]
- Stansfield SH, Allen EE, Dinnis DM, Racher AJ, Birch JR, James DC. Dynamic analysis of GS-NS0 cells producing a recombinant monoclonal antibody during fed-batch culture. Biotechnol Bioeng. 2007;97:410–424. doi: 10.1002/bit.21263. [DOI] [PubMed] [Google Scholar]
- Stępkowski TM, Kruszewski MK. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radical Biol Med. 2011;50:1186–1195. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- Teruya K, Daimon Y, Dong X-Y, Katakura Y, Miura T, Ichikawa A, Fujiki T, Yamashita M, Mori T, Ohashi H, Shirahata S (2005) An approach to further enhance the cellular productivity of exogenous protein hyper-producing Chinese hamster ovary (CHO) cells. Cytotechnology 47:29–36 [DOI] [PMC free article] [PubMed]
- Tigges M, Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab Eng. 2006;8:264–272. doi: 10.1016/j.ymben.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Underhill MF, Birch JR, Smales CM, Naylor LH. eIF2α phosphorylation, stress perception, and the shutdown of global protein synthesis in cultured CHO cells. Biotechnol Bioeng. 2005;89:805–814. doi: 10.1002/bit.20403. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664–666 [DOI] [PubMed]
- Watanabe S, Shuttleworth J, Al-Rubeai M. Regulation of cell cycle and productivity in NS0 cells by the over-expression of p21CIP1. Biotechnol Bioeng. 2002;77:1–7. doi: 10.1002/bit.10112. [DOI] [PubMed] [Google Scholar]
- Yoon SK, Hong JK, Choo SH, Song JY, Park HW, Lee GM. Adaptation of Chinese hamster ovary cells to low culture temperature: cell growth and recombinant protein production. J Biotechnol. 2006;122:463–472. doi: 10.1016/j.jbiotec.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Yoon SK, Kim SH, Song JY, Lee GM. Biphasic culture strategy for enhancing volumetric erythropoietin productivity of Chinese hamster ovary cells. Enzyme Microbial Technol. 2006;39:362–365. doi: 10.1016/j.enzmictec.2005.11.029. [DOI] [Google Scholar]
- Yurochko AD, Mayo MW, Poma EE, Baldwin AS, Huang ES. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-kappaB promoters. J Virol. 1997;71:4638–4648. doi: 10.1128/jvi.71.6.4638-4648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, Cavener DR (2002) The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 22:6681–6688 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estimate of relative abundance of various genes profiled in this study in HP cell line. Time profiles of ΔCT are plotted for a) IgG HC and LC b) Chaperones c) PERK pathway genes and d) Ire1α pathway genes. ΔCT is defined as , Positive values imply abundance greater than β-actin whereas negative values imply lower abundance. Note that β-actin is a moderately abundant gene (TIFF 1084 kb)