Abstract
Cryptochromes (Crys) and photolyases (Phrs) are flavoproteins that contain an identical cofactor (flavin adenine dinucleotide, FAD) within the same protein architecture, but whose physiological functions are entirely different. In this study, we investigated light-induced conformational changes of a cyanobacterium Cry/Phr-like protein (SCry-DASH) with UV–visible and Fourier transform infrared (FTIR) spectroscopy. We developed a system to measure light-induced difference spectra under the concentrated conditions. In the presence of reducing agent, SCry-DASH showed photoreduction to the reduced form, and we identified a signal unique for an anionic form in the process. Difference FTIR spectra enabled us to assign characteristic FTIR bands to the respective redox forms of FAD. An asparagine residue, which anchors the FAD embedded within the protein is conserved in not only the cyanobaterial protein, but also in Phrs and other Crys including the mammalian clock-related Crys. By characterizing an asparagine-to-cysteine (N392C) mutant of SCry-DASH, which mimics an insect specific Cry, we identified structural changes of the carbonyl group of this conserved asparagine upon light-irradiation. We also found that the N392C mutant is stabilized in the anionic form. We did not observe a signal from protonated carboxylic acid residues during the reduction process, suggesting that the carboxylic acid moiety would not be directly involved as a proton donor to FAD in the system. These results are in contrast to plant specific Crys represented by Arabidopsis thaliana Cry1 which carry Asp at the position. We discuss potential roles for this conserved asparagine position and functional diversity in the Cry/Phr frame.
The cryptochrome/photolyase family of proteins are found throughout all three kingdoms of life, from bacteria to archaea to animals including humans (1, 2). Cryptochromes (Crys)1 and photolyases (Phrs) share an identical chromophore (flavin adenine dinucleotide, FAD) within a homologous protein architecture termed a Phr-like domain (1) (Figure 1a-d). Despite structural homology, their physiological functions are quite distinct. Phrs repair ultraviolet (UV)-induced photoproducts in DNA using near UV and blue light with the cofactor. In higher organisms, Phr-like gene products, termed Cry to distinguish them from the DNA repair Phrs, also function in a light-dependent manner, but control growth or regulate flowering time in plants, and maintain circadian rhythms in animals (3). For Crys, redox of FAD has been debated (4, 5); however, contributions of FAD to the signaling still remain enigmatic. Even for the better-characterized Phrs, the means by which FAD redox changes result in downstream protein dynamics are still poorly understood.
Figure 1.
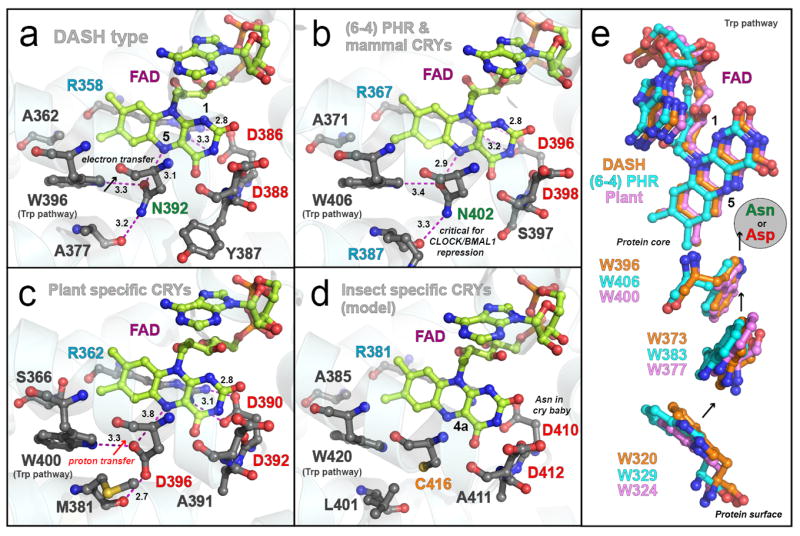
Conservation and unique tuning in FAD binding domains of the photolyase and cryptochrome family proteins. Crystal structures of SCry-DASH (a), Arabidopsis (6-4) Phr (b), Arabidopsis Cry1 (c) and a homology model of Drosophila Cry (d) show conservation at the FAD binding sites and indicate the unique tuning. The Cry/Phr proteins have two Asps at the FAD binding site (Asp386 and Asp388 in SCry-DASH). One of the Asps forms a salt bridge with Arg across the isoalloxazine ring system. The salt bridge is invariant in all Cry/Phr family members. Many Crys and Phrs have a conserved asparagine at the location of Asn392 in SCry-DASH, the region in which the polypeptide chains anchor the cofactor at N5 of isoalloxazine (a and b), whereas plant and insect specific Crys have Asp and Cys, respectively, at the position (c and d). FAD is drawn in yellow, and critical distances are shown in angstroms with magenta dotted lines. Representatives of each gene cluster of the family, a summary of the conservation at the region and coordinates used are listed in Table 1. The homology model of Drosophila Cry was built with SWISS-MODEL (swissmodel.expasy.org) based on Drosophila (6-4) Phr (PDB id: 3CVV). (e) The Trp triad pathway contributing to the FAD redox is also conserved in the Cry/Phr family; SCry-DASH is shown in orange, Arabidopsis (6-4) Phr in blue, and Arabidopsis Cry1 in pink. In Figure 1e, molecules were rotated 60 degrees from Figure 1a–c. Figures were drawn with PyMOL software (57).
The Cry/Phr family is categorized into two major classes. Cryptochrome-DASH (Cry-DASH) is a newly found cluster of proteins belonging to the class I family (6). While the gene clusters of animal and plant Crys homologous are literally found in the respective kingdoms, Cry-DASH type genes are found in all kingdoms, bacteria, plants (6) and also some vertebrates (7). Although specific physiological functions in individual organisms are still unclear, Cry-DASH proteins are likely photoreceptors, as the proteins contain two light-harvesting cofactors, FAD and 5,10-methenyltetrahydrofolate, and the light-dependent DNA repair activity is negligible in vivo (6, 8), retaining repair activity for single-stranded DNA in vitro (9, 10). On the other hand, Phr is a photoreceptor in a way (11). Originally, genes belonging to the cluster were discovered in Synechocystis sp. PCC6803 and Arabidopsis thaliana, however, the overall structural features of the encoded proteins are in fact more similar to Homo sapiens and Drosophila Crys, than to prototypical plant Crys containing characteristic long C-termini. These Cry-DASH proteins additionally provide relatively good yields in recombinant bacterial expression systems, and several X-ray crystal structures of Cry-DASH derived from Synechocystis sp. and Arabidopsis have been reported (12, 13) (Figure 1a). Cry-DASH proteins are therefore a good model for studying Crys and Phrs using spectroscopic methods.
The photoreduction and oxidation processes of Cry-DASH have previously been investigated by UV–vis spectroscopy for zebrafish (14), Xenopus laevis (15), and Synechocystis sp. (16). EPR analyses of mutant Cry-DASH from Xenopus laevis have also been successful (15, 17). Phrs appear to use the reduced FADH− as a redox-active cofactor in the DNA repairs, whereas the oxidized (FADox) and neutral semiquinoid (FADH•) forms are inactive (18). For at least a few Crys, on the other hand, different FAD chemistry has been reported. For example, in Arabidopsis Cry1, the FADox form has been proposed as an unphotolyzed state (4, 5). Light-dependent reduction from FADox to FADH• and/or FADH− is believed to trigger the signaling. Furthermore, the anionic form (FAD•−) has been observed in insect Cry (19, 20). Regardless of the cofactor redox chemistry though, all proteins belonging to the Cry/Phr family contain the conserved “Trp triad” and exhibit light-dependent function FAD reduction in the presence of reducing agent (Figure 1e) (15).
The amino acid adjacent to the N5 atom of the FAD isoalloxazine ring is considered a key signal transduction residue in flavoproteins. For example, in the LOV (light, oxygen and voltage) domain of the plant blue light photoreceptor phototropin, whose chromophore is FMN, a cysteine is located near the C4a position of the FMN isoalloxazine ring (21-24), and a neutral glutamine in the β-sheet anchors the N5 position (25). Upon light excitation, the cysteine residue forms a covalent bond with isoalloxazine ring at the C4a position. Simultaneously, the glutamine residue changes its hydrogen-bonding partner from the C4=O group to the N5–H group, resulting in local structural changes that transmit the light signal downstream of protein through a β-sheet (26) and/or so-called Jα helix (27-29). Though the Gln residue itself is not necessary for the normal photoreaction, it is important for the structural changes in the signal transduction. Similarly, the BLUF (sensor of blue-light using FAD) domain has a Gln nearby the FAD isoalloxazine N5 position, which has been shown to have a critical role in the light signal conversion (the formation of red-shifted intermediate) (30-32).
The amino acid adjacent to the FAD isoalloxazine N5 appears to be conserved in Cry/Phr proteins as well, and interestingly, most family members, including mammalian and DASH type Crys, have an Asn at this position (Figure 1a and b). However, plant Crys (e.g. Arabidopsis Cry1) have an Asp and insect specific Crys have Cys instead (Figure 1c and d, Table 1) (13, 19). A study comparing midpoint redox potentials among FAD redox species for CPD Phrs and Arabidopsis Cry1 suggested that this amino acid residue (Asn or Asp, respectively) plays an important role in the control of the FAD redox state (33). In addition, a mutant of Escherichia coli DNA Phr whose Asn was replaced to other amino acids displays defects in DNA repair activity (34). Our recent studies further showed that substitution of the Asn to Asp or Cys in mouse Crys, thus mimicking plant or insect specific Crys, disturbs the function as a clock protein (35). Therefore, the identity of this residue may contribute to cluster-, class- or species-specific funtion roles for Cry/Phr proteins.
Table 1.
Comparison of amino residues around FAD.
| Representative | Gene Cluster | Position386a) | 387 | 388 | 392 | PDB id |
|---|---|---|---|---|---|---|
| SCry-DASH | DASH type Cry | Asp | Tyr | Asp | Asn | 1NP7 |
| human Cry1 | (6-4) Phr/clock Cry | Asp | Ala | Asp | Asn | n.d. |
| Arabidopsis (6-4) Phr | (6-4) Phr/clock Cry | Asp | Ser | Asp | Asn | 3FY4 |
| Drosophila Cry | Insect specific Cry | Aspb) | Ala | Asp | Cys | n.d. |
| Arabidopsis Cry1 | Plant specific Cry | Asp | Ala | Asp | Asp | 1U3C |
| E. coli CPD Phr | CPD phr | Asp | Gly | Asp | Asn | 1DNP |
numbering in SCRY-DASH
Asn in the cry baby mutant identified in Drosophila Cry
Light-induced difference Fourier transform infrared (FTIR) spectroscopy is a useful and powerful method for characterizing structure-function relationships in photoreceptive proteins, as shown for rhodopsins (36-38). However, only a limited number of FTIR studies are reported for full-length photoreceptors containing flavin cofactors. FTIR was applied to E. coli DNA Phr and Arabidopsis Cry1, but the Cry/Phr family proteins have been not well assigned (39, 40), as compared to isolated LOV (25, 26, 41-47) and BLUF domains (32, 48-50) found in other flavoproteins with distinct folds and unique photoreactions. To apply to the spectroscopic system, a solid paradigm with detailed coordinates is essential. In this report, we characterized the photoreaction of Cry-DASH derived from Synechocystis sp. PCC6803 (SCry-DASH) by UV–vis and FTIR spectroscopy. In addition to the wild-type protein, we also examined the N392C mutant of SCry-DASH, which mimics insect specific Crys (Table 1, Figure 1a and d) (13, 19, 20) in our studies of the photoreaction.
Solution samples are generally not suitable for FTIR analysis; however, we were able to construct an FTIR system using a concentrated solution to measure light-induced difference spectra and applied it to SCry-DASH. Using this system, we identified a signal unique to an anionic form (FAD•−) of the protein during the photoreduction process. Substitution to cysteine at the position enriches the FAD•− form. By comparing difference spectra of FAD•− and FADox (FAD•−/FADox) with those of FADH− and FADox (FADH−/FADox), FTIR bands specific to FAD•− and FADH−, respectively, were assigned. We furthermore examined structural changes in the carbonyl group of Asn392 during the FAD•− formation process. In contrast to observations of Arabidopsis Cry1 (40), no significant signal corresponding to protonated carboxylic acid moiety was observed for SCry-DASH, suggesting that this contribution is negligible or minor. Thus, we also concluded that while Cry/Phr family proteins also conserve two Asps at the FAD binding site (Figure 1a-d, Table 1), amino acid difference at the Asn position of SCry-DASH is important for the functional diversity observed in plant or insect specific Crys. We discuss potential roles of the position in the Cry/Phr frame.
MATERIALS AND METHODS
Sample Preparation
Synechocysitis sp. Cry-DASH (SCry-DASH) was expressed in E. coli as a fusion protein with glutathione-S-transferase (GST) at the N-terminus (6). After purification of SCry-DASH by glutathione agarose resin (GE Healthcare), the GST tag was cleaved with thrombin. SCry-DASH (OD450nm~1.5) was dissolved in 50 mM Tris-HCl (pH 8), 300 mM NaCl, and 5% (w/v) glycerol and stored at −80°C until use. The N392C mutant of SCry-DASH was constructed by PCR using the QuickChange site-directed mutagenesis method (Stratagene). Nucleotide substitutions were confirmed by DNA sequencing.
UV–vis Spectroscopy in Dilute Solution
UV–vis spectra of the diluted SCry-DASH solution were measured with a V-550DS spectrophotometer (JASCO) at 277 K. SCry-DASH (OD450nm~0.15) in 50 mM Tris-HCl (pH 8), 300 mM NaCl, 100 mM dithiothreitol (DTT) was transferred into an optical cuvette (path length, 1 cm), deoxygenated by blowing N2 gas and sealed with parafilm (Pechiney Plastic Packaging). To initiate the photoreaction, the sample was illuminated with white light using a 1 kW halogen-tungsten lump, and an ND10 filter was used to prevent rapid photoreaction.
UV–vis and FTIR Spectroscopy in Concentrated Solution
SCry-DASH (OD450nm~1.5) was concentrated by about 20 times using a Microcon YM-30 unit (Millipore). After 1.8 μL of concentrated SCry-DASH and 0.2 μL of 1 M DTT were mixed, the solution was put on a BaF2 window (diameter, 18 mm), sandwiched with another BaF2 window directly and sealed with parafilm. H/D exchange was carried out by diluting the SCry-DASH sample with the buffer prepared in D2O and concentrating three times with an Amicon YM-30.
The concentrated solution in BaF2 windows was measured using UV-550DS and FTS-7000 (Bio-Rad) spectrophotometers equipped with a cryostat (Optistat DN, Oxford) and a temperature controller, respectively. Temperature was set at 277 K and illumination of white light was carried out with a 1 kW halogen-tungsten lump. For FTIR spectra, 128 interferograms at 2 cm−1 resolution were recorded before and after illumination, and 7 and 5 recordings for wild-type and N392C mutant were averaged, respectively.
RESULTS
In vitro Photoreduction of SCry-DASH
The FAD cofactor buried within the Cry/Phr protein fold (Figure 1) is subject to redox changes. In the presence of reducing agents, such as 2-mercaptoethanol or DTT in vitro, for example, the cofactor can be reduced in a light-dependent manner (14-17). In order to characterize the light-dependent redox changes of FAD in the SCry-DASH protein, we first recorded the light-induced UV–vis spectra of a dilute protein solution (Figure 2a). Under anaerobic conditions in the presence of DTT, white light illumination efficiently converted the redox state from FADox to FADH−, after only 4 min (Figure 2a, black solid line). After totally 9 min prolonged illumination, we observed an increase in the accumulation of the reduced form at 369 nm (Figure 2a, red line). Comparison of the two spectra allows us to estimate accumulation of the reduced form FADH−. The dotted line in Figure 2a shows a 1.45× magnified spectrum of the shorter exposed sample with negative peaks at 448 and 471 nm, indicative of FADox, overlaid onto the prolonged illumination spectrum. Peak intensity at 369 nm of the long exposure sample is significantly larger than that of the magnified spectrum, indicating an accumulation of the reduced form by the longer exposure. The difference in overall intensities may be due to a mixed sample population containing various redox stages of protein. Similar photoreaction could be also observed at the higher concentration used for the FTIR studies as well (Figure 2b). In the difference spectrum of the 2 min illumination, negative peaks at 349 and 383 nm were observed (black lines in Figure 2b).
Figure 2.
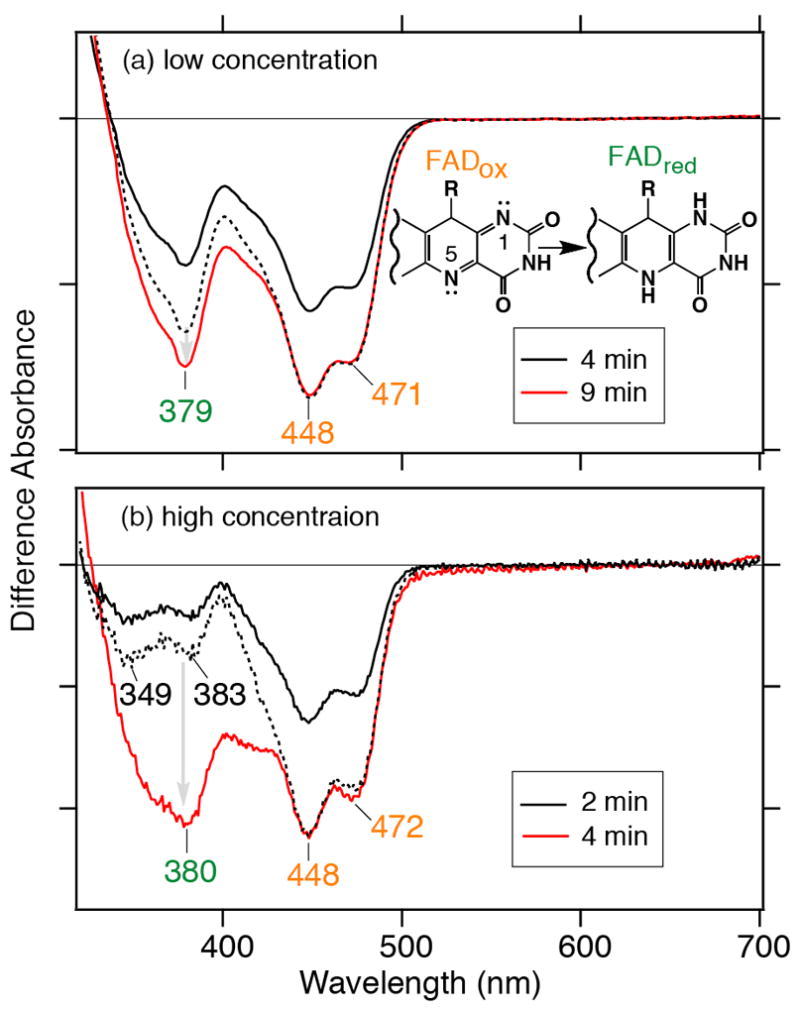
Light-induced UV–vis difference spectra upon photoreduction of SCry-DASH. White light illumination was carried out at (a) low and (b) high protein concentrations in the presence of 100 mM DTT. The low concentration sample was exposed for 4 or 9 minutes, while the higher concentration sample was exposed for 2 or 4 minutes. In the respective panels, the shorter exposure is shown in black, and the longer exposure is in red, respectively. The black dotted line indicates the magnified spectrum of the shorter exposure spectrum (black solid line), 1.45-times for the low exposure sample and 1.7-times for the high exposure sample (see details in the text). One division of the y-axis corresponds to 0.04 (a) or 0.005 (b) absorbance units.
Ideally, after a short period, various redox states of FAD should exist, while prolonged light exposure will result in homogenous accumulation of the FADH− form under anaerobic conditions. However, we did not clearly observe characteristic absorbance at 500–700 nm corresponding to the radical form FADH• or the anionic form FAD•− in the transition from the oxidized to reduced FADH− (Figure 2). In insect specific Crys, the anionic form, having characteristic absorbance at 400 and smaller peak at 500 nm, was detected in the photoreduction process (19, 20). These intermediate forms may not be stabilized under the conditions or may be masked by the dominant species. To extract the spectral component, we thus adjusted intensities with the following calculations;
where intensity ratio represents that ratio of the larger intensity to the smaller intensity at 448 nm for the oxidized form and 369 nm for the reduced form.
As shown in Figure 3a, the obtained spectra were distinct from those of the oxidized or reduced form, showing a characteristic peak for the anionic form at 400 nm, though the peak intensity around 500 nm is smaller than those reported (19, 20). Under the present conditions, based on the obtained spectrum, we concluded that the short illumination gave 7 % FAD•− over FADH− in diluted solution, and 62 % FAD•− and 38 % FADH− in concentrated solution (discussed further below). We could not extract FADH• spectra by changing parameters (not shown).
Figure 3.
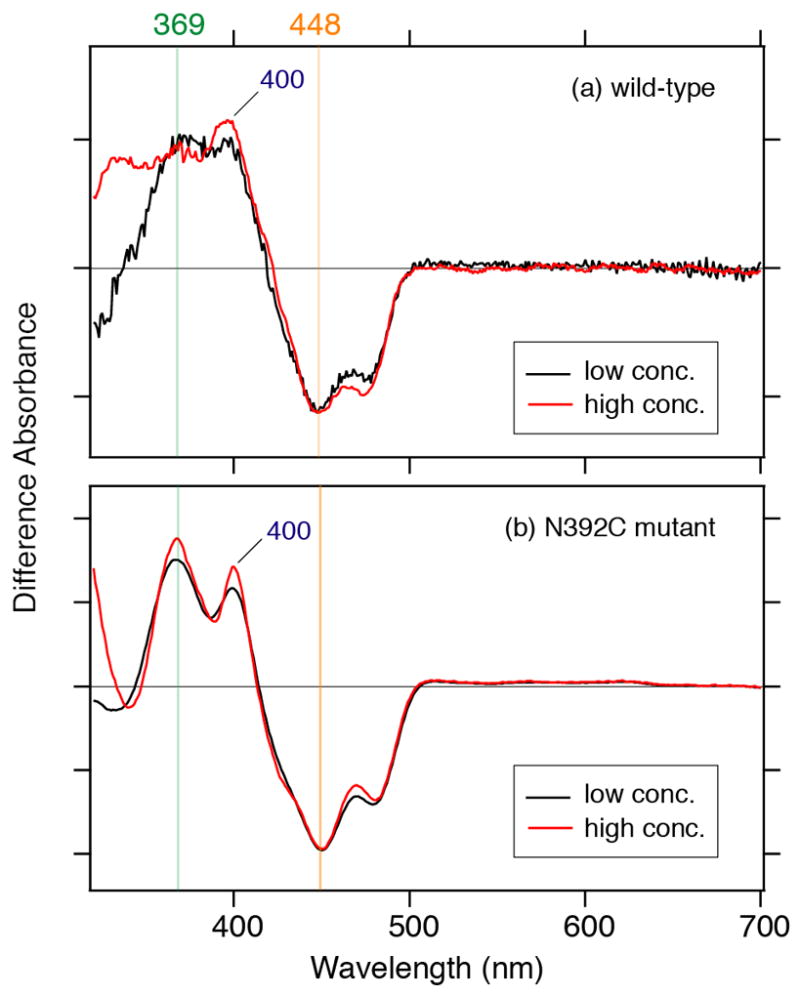
Extraction of the FAD•− form in SCry-DASH. (a) Subtraction of adjusted short illumination spectra, containing predominantly the oxidized form, from that of long exposure, enriched in the reduced form, revealed a peak characteristic for the anionic form at 400 nm. Diluted condition is in red, and concentrated sample in black. (b) The SCry-DASH N392C mutant stabilizes the anionic form. Illumination with white light was carried out in the presence of 100 mM DTT for 2 min for the low concentration sample (red line) and 4 min for the high concentration sample (black line).
Analysis of cysteine mutant SCry-DASH mimicking insect specific Crys
During the redox process in insect specific Crys, the anionic form of FAD is stabilized (19, 20). Notably, the N1 and N5 atoms of the FAD isoalloxazine ring are key for the redox chemistry. Interestingly, insect specific Crys have Cys residues in the vicinity of the N5 atom, such as in plant blue-light photoreceptor phototropins, while the N1 atom is exposed (Figure 1d). It is thereby suggested that the presence of cysteine could stabilize the anionic form FAD•− (19, 20). DASH type Cry proteins, as well as Phrs and mammalian Crys, have Asn at the position (Figure 1a and b, Table 1). So, to investigate the effect of this residue nearby the isoalloxazine N5 atom on the photoreaction, we measured the photoreaction of the N392C mutant, which mimicks insect specific Crys (Figure 1a and d). Figure 3b shows the light-induced UV–vis difference spectra of N392C SCry-DASH in diluted (red line) and concentrated (black line) solutions, respectively, using short illumination. Remarkably, we obtained spectra with FAD•− peak intensity well distinguished from FADox but matching that of insect specific Cry naturally containing Cys as the residue in question (19) (Figure 3b). Thus, we concluded that placement of cysteine at position 392 of SCry-DASH can stabilize the anionic form FAD•−. Because prolonged illumination resulted in sample precipitation (not shown), we could not confirm whether the N392C mutant is capable of forming the FADH−. Similar spectra between wild-type and N392C, however, suggest that both SCry-DASH proteins can undergo photoreduction. (Figure 3). These data implicate that the position neighboring the N5 atom of isoalloxazine is critical for the diversity of Cry/Phr family members (Figure 1a-d).
FTIR spectroscopic analyses of SCry-DASH
We confirmed that SCry-DASH is capable of responding to light at even relatively high concentrations using UV–vis (Figures 2 and 3). We next analyzed the photoreduction of SCry-DASH in a concentrated solution sandwiched between BaF2 windows using FTIR spectroscopy. Figure 4a and b show the difference FTIR spectra upon 2- and 4-min illumination, respectively, in the 1800–1350 cm−1 region. Significant peaks for SCry-DASH were detected at 1714 (−), 1695 (−), 1687 (−), 1675 (+), 1663 (−), 1647 (+), 1627 (+), 1607 (+), 1580 (−), 1545 (−), 1512 (+), 1486 (+), and 1396 (+) cm−1. Overall, the spectra look similar regardless of the exposure time. However, some noticeable differences were found upon longer illumination. When the spectra were normalized to the negative peak at 1714 cm−1, corresponding to the C4=O stretch of FAD (51-55), peaks in the 1700–1600 cm−1 region were sharper in the long exposure compared with the short time. The positive band at 1486 cm−1 disappeared, whereas the positive band at 1396 cm−1 increased in intensity with longer exposure. Based on the analyses of the UV–vis difference spectra (Figures 2 and 3), the FADH− state would be predominant with longer illumination, while the short exposure contains more information on the FAD•− state. Ideally, the results of the FTIR experiments would be reveal FAD•− and FADH− at a 62:38 ratio, as seen in UV–vis analyses (Figure 2b and 3a). So, to determine the FAD•−/FADox and FADH−/FADox ratios from the difference FTIR spectra, we used reference bands, as described below.
Figure 4.
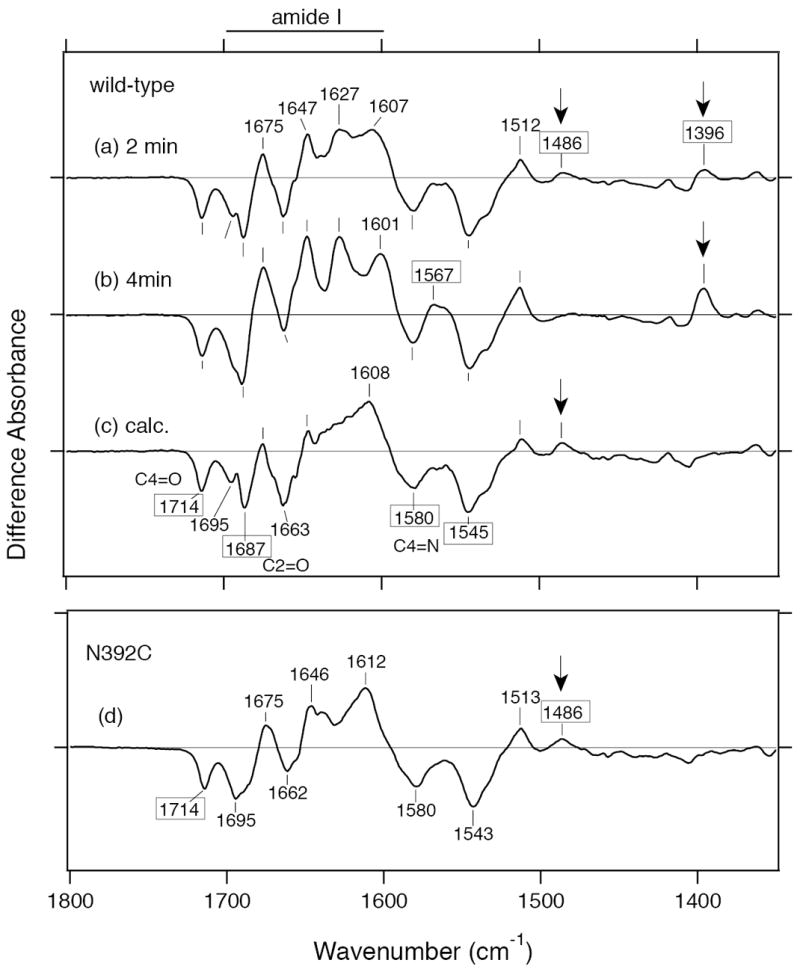
Light-induced FTIR difference spectra of wild-type SCry-DASH and the N392C mutant. Light-induced FTIR difference spectra (1800–1350 cm−1 region) illustrate additional details of photoreduction. Illumination was carried out at a concentration similar to that for the UV–vis studies for 2 min (a) and 4 min (b) in the presence of 100 mM DTT. Calculated spectrum of FAD•−/FADox (c) was obtained by subtraction of the magnified short exposure spectrum from the long exposure spectrum, while the exposed spectrum is considered sufficient to represent the difference spectrum of FADH−/FADox (see the text for more details). Based on the peak intensity at 1714 (−) cm−1, the short exposure spectrum was magnified by 1.3 times to normalize the height to that of the long exposure to calculate the difference spectrum of FADH−/FADox. Lane d shows the difference spectrum of N392C mutant. One division of the y-axis corresponds to 0.006 absorbance units.
The N392C mutant, which exhibits a stable anionic form, can be useful in comparing the FAD•− and FADox states of SCRY-DASH. As clearly seen in the UV–vis spectra (Figure 3b), an FTIR spectrum of this mutant could help to distinguish FAD•− from FADox under similar conditions. Figure 4d shows the light-induced difference FTIR spectrum of N392C. In the spectrum, there are peaks at 1714 (−), 1695 (−), 1675 (+), 1663 (−), 1646 (+), 1612 (+), 1607 (+), 1580 (−), 1545 (−), 1513 (+), and 1486 (+) cm−1. The FTIR spectrum of the N392C mutant is not entirely coincident with that of the wild-type, though some peaks are well overlapped. Remarkably, the spectra of short exposed wild-type and mutant proteins both exhibit a positive band at 1486 cm−1, which was not observed in the long illumination, whereas the mutant showed no positive band at 1396 cm−1 (Figure 4). These results imply that the positive band at 1486 cm−1 is originated from the FAD•−, while the positive band at 1396 cm−1 is characteristic of the FADH− form.
We then calculated the FAD•−/FADox difference FTIR spectrum of wild-type SCry-DASH so as to control for the peak at 1396 (+) cm−1 belonging to FADH− (Figure 4c). Calculation was carried out as follows:
Consequently, the spectrum of the wild-type protein contains 60 % FAD•−/FADox and 40 % FADH−/FADox upon short illumination. This estimation well matches to numbers obtained by UV–vis spectra (Figures 2b and 3a).
Detection of conformational changes of SCry-DASH in the vicinity of FAD
Figure 5 compares the two difference FTIR spectra of FAD•−/FADox (red line) and FADH−/FADox (black line) of wild-type SCry-DASH. Negative peaks at 1714, 1687, 1580, and 1545 cm−1 were common between the two spectra, which presumably originate from vibrations of the FADox form. The peaks are assigned as follows; C4=O stretch (1714 cm−1), C2=O stretch (1687 cm−1), and C=N stretches (1580 and 1545 cm−1) (51-55). As described above, positive bands at 1567 and 1396 cm−1 are specific to FADH−, while a positive band at 1486 cm−1 is specific to FAD•−. The origins of these positive bands have not yet been identified.
Figure 5.
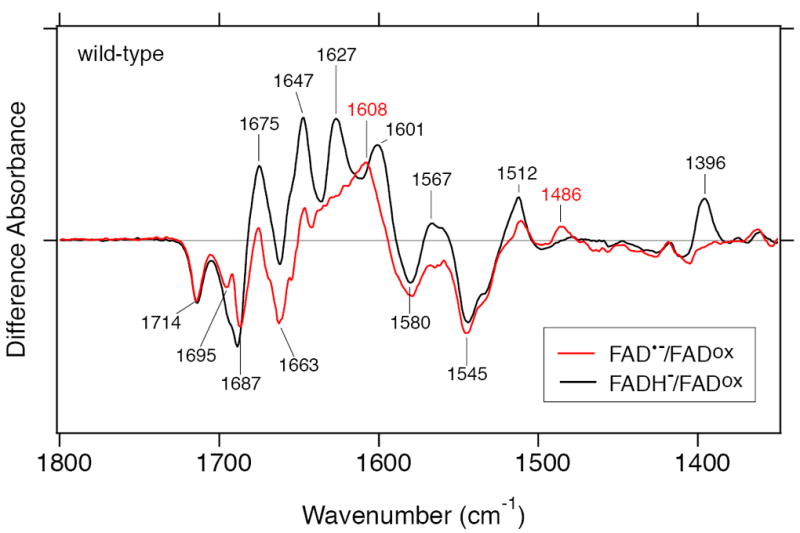
Comparison of two difference FTIR spectra of wild-type SCry-DASH. To identify significant peaks, the two different spectra representing FAD•−/FADox (red line) and FADH−/FADox (black line), taken from Figure 4c and b, respectively, were overlaid in the 1800–1350 cm−1 region.
Notably, very different spectral features were observed in the 1700–1600 cm−1 region (amide I region), where characteristic frequencies of the C=O stretches of peptide backbone are commonly observed. The FAD•−/FADox spectrum shows peaks at 1695 (−), 1687 (−), 1663 (−), 1647 (+), and 1608 (+) cm−1, while the FADH−/FADox spectrum has peaks at 1688 (−), 1675 (+), 1647 (+), 1627 (+), and 1601 (+) cm−1. These differences may reflect a secondary structural alteration between the FAD•− and FADH− states, while some of positive bands may simply indicate C=O stretches of FAD independent of the redox status. Interestingly, although FAD•− and FADH− states are both negatively charged, different FTIR signals strongly suggest that hydrogen-bonding network near the chromophore is significantly affected to induce conformational changes in the vicinity. The key difference between the two states would be due to protonation status of N5 atom of isoalloxazine in FAD (Figure 1).
Regulation of N5 Atom of isoalloxazine ring of FAD in the Cry/Phr architecture
To characterize the contribution of the isoalloxazine N5 atom to the conformational change, we compared difference FTIR spectra of the SCry-DASH proteins. Figure 6 compares the FAD•−/FADox difference FTIR of the wild-type (red line) and N392C mutant (blue line) SCry-DASH. Overall, spectra of the mutant are similar to wild-type, possessing peaks at 1714 (−), 1695 (−), 1675 (+), 1663 (−), 1647 (+), 1580 (−), 1545 (−), 1512 (+), and 1486 (+) cm−1. We observed significant differences in the amide I region (1700–1600 cm−1 region), indicating that the substitution at Asn392 also influenced the light-induced structural changes in the peptide backbone upon the formation of FAD•−.
Figure 6.
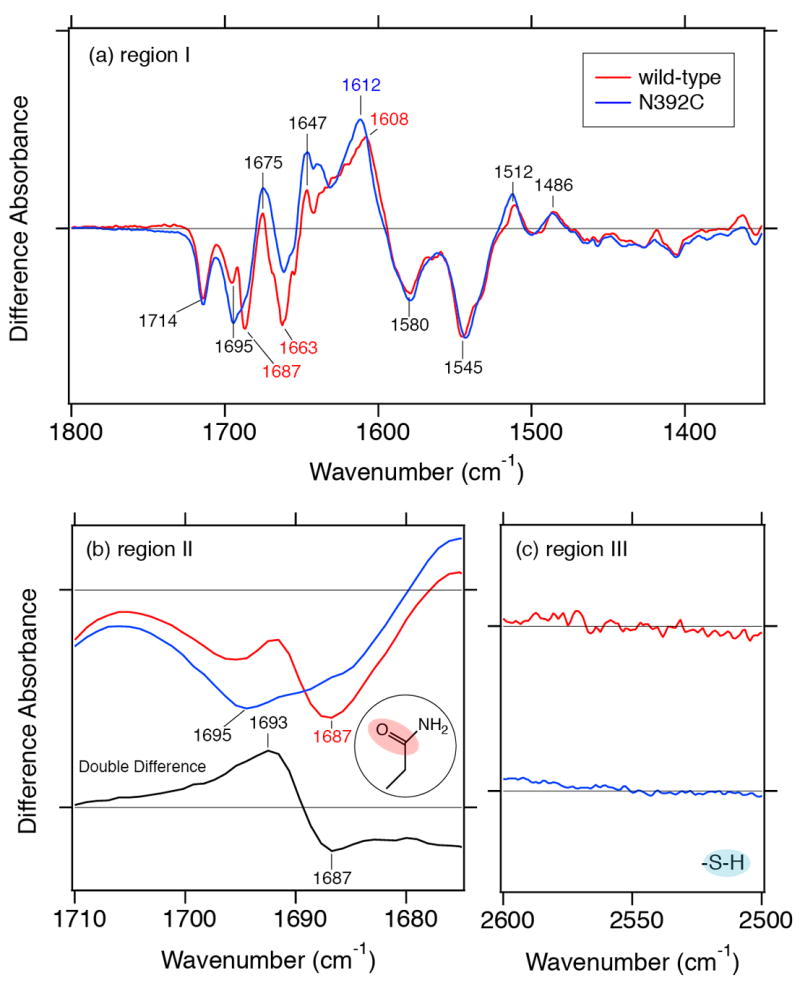
Impact of mutations on difference FTIR spectra. Difference FTIR spectra of FAD•−/FADox of wild-type SCry-DASH (red line) and N392C mutant (blue line) were compared in three ranges: (a) 1800–1350 cm−1, (b) 1720–1670 cm−1, and (c) 2600–2500 cm−1. Respective data for wild-type and the N392C mutant are identical to data shown in Figure 4c and d, respectively. Bottom black trace in panel b shows the double-difference spectrum between wild-type and the N392C mutant, where the N392C mutant spectrum (blue line) is subtracted from the wild-type spectrum (red line).
The wild-type spectrum shows negative peaks at 1695 and 1687 cm−1, while the N392C mutant lacks the corresponding negative peak at 1687 cm−1 (Figure 6b, upper traces). The double-difference spectrum between wild-type and N392C revealed peaks at 1693 (+) and 1687 (−) cm−1 (Figure 6b, bottom trace). Generally, C=O stretches of side chains of asparagine and glutamine appear in this region as well as peptide backbone (amide-I vibration). We interpret that the bands at 1693 (+)/1687 (−) cm−1 originate from the C=O stretch of side chain of Asn392. A shift of the stretching vibrational mode to higher frequency implies that hydrogen bonding strength of the C=O group is weakened upon the FAD•− formation. Figure 6c shows 2600–2500 cm−1 region of the spectra of wild-type and N392C. Normally, the S–H stretch appears in the 2575–2525 cm−1; however, no such band is observed for the N392C mutant (Figure 6c, blue line). In contrast to the wild-type, whose Asn392 weakens the hydrogen bond of its C=O side chain, the data indicates that the hydrogen bonding network of the mutant protein Cys392 does not change upon FAD•− formation. It should be noted that the C4=O stretching frequencies are identical between the N392C mutant and the wild-type (1714 cm−1, Figure 6a). This suggests that Asn392 does not interact with the N5 atom of FAD in the FADox form, because the interaction affects the C4=O band of FAD.
Protonated carboxylic acid residues are unnecessary in the photoreduction of SCry-DASH
For plant specific Cry, carboxylic acid deprotonation was observed in the formation process of FADH• (40). However, it is unclear which Asp in plant specific Cry contributes to the photoexcitation. Plant specific Crys have Asp, insect specific Crys have Cys, and the rest of the family contain Asn at the position corresponding to Asn392 of SCry-DASH, while the entire family additionally contains two conserved Asps (Asp386 and Asp388 in SCry-DASH) near the FAD (Figure 1a-d, Table 1). In Drosophila, a mutation at Asp410 to Asn (“cry baby”) abolishes the function as a circadian photoreceptor (Figure 1d) (56). To investigate the contribution of aspartic acids to the photoreduction, we performed deuterium exchange experiments (Figure 7). Difference FTIR spectra of FADH−/FADox of SCry-DASH in H2O (black line) and D2O (green line) buffer look similar in the 1800–1350 cm−1 region, but with a slight shift in the amide I region, reduced intensity at 1675 (+) cm−1 and newly appearing band at 1669 (−) cm−1 (Figure 7a). In general, C=O stretches of peptide backbone are slightly affected (downshift by 2–3 cm−1) by the H/D exchange in the N–H groups. However, the C2=O stretch of FADox shows a downshift by >10 cm−1 from the deuteration of N3–H group (53-55), and the negative band at 1669 cm−1 might originate from a C2=O stretch. The band intensity at 1396 (+) cm−1 is slightly reduced in the D2O buffer. H/D exchangeable groups in the FAD are N3–H (in the FADox and FADH−forms) and N5–H (in the FADH− form) groups. The positive band at 1396 cm−1 may include a N–H bending vibration of FAD. The negative band at 1714 cm−1 probably originates from a C4=O stretch of FADox form (Figure 7b). The 4 cm−1 spectral shift in the D2O buffer may be due to an indirect effect of the deuteration of N3–H group (53-55). For SCry-DASH, no significant signal was observed in the 1800–1720 cm−1 region. This implies that the FAD reduction process of SCry-DASH does not involve structural changes of protonated carboxylic acid residues by protonaton/deprotonation or hydrogen-bonding alteration. The difference would likely arise from the distinct residues in the vicinity of N5 atom of FAD (Figure 1a and c).
Figure 7.
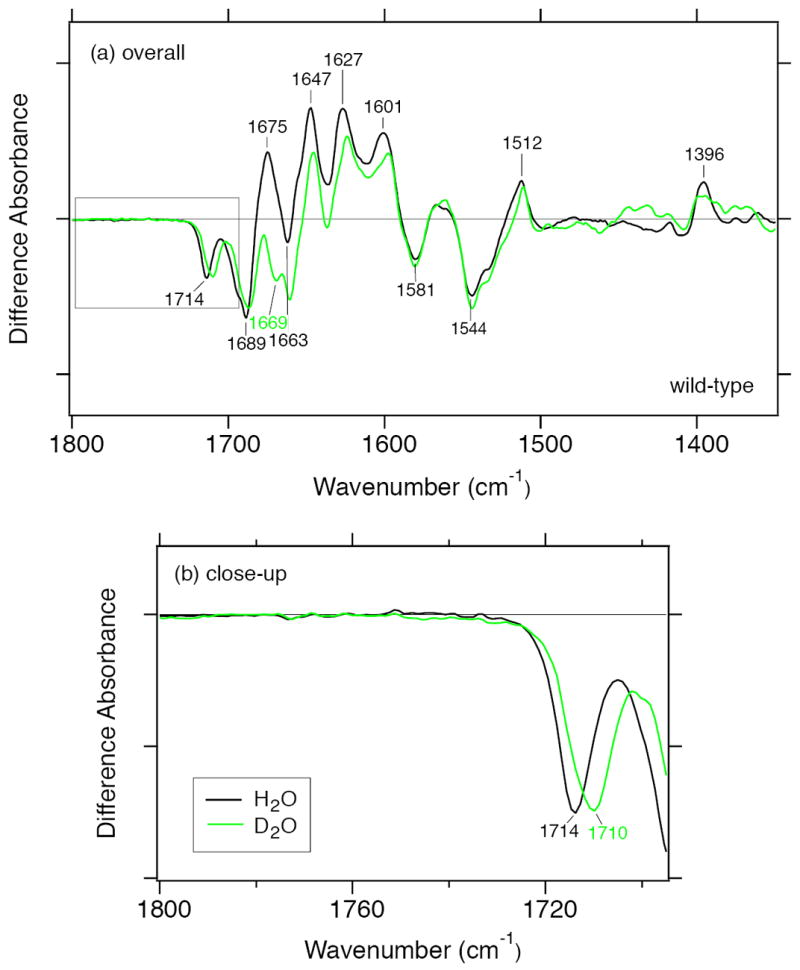
Deuterium effects on photoreduction of SCry-DASH. Difference FTIR spectra of FADH−/FADox of wild-type SCry-DASH in H2O (black line) and D2O (green line) buffer were compared in the 1800–1350 cm−1 (a) and 1800–1710 cm−1 (b) regions.
DISCUSSION
Adaptations to light are diverse in different organisms, and chemical reactions triggered by light are subtle but critical. Unlike other photoreceptors, such as rhodopsin (retinal), phytochrome (bilin) or photoactive yellow protein (coumaric acid), drastic cis-trans chemical conformational change is not expected in flavin-bound blue-light photoreceptors containing planer isoalloxazine rings. Light-dependent conformational assembly changes and dissociations have been proposed for the flavin-based photoreceptors such as Crys and LOV and BLUF domain proteins. To fully understand how light is captured, resulting in such structural rearrangement for signalling, however, detailed characterizations of the chromophore and its vicinity in both the ground and excited states are required. Three-dimensional structural information on Cry/Phr family proteins has revealed a similar overall fold and conserved mode of chromophore binding, and light activation of FAD has been discussed, but signal conversion signal (i.e. FAD-driven protein dynamics) remain unclear. For this aim, it is essential to couple information on the FAD redox state to contributions of identified interaction sites. In a light- and time-dependent process (Figure 2), we extracted a signal for the anionic form FAD•− as one of the intermediates (Figure 3a).
Several recent studies of FAD redox states in Cry-DASHs have been reported (14-16). Damiani et al. showed that SCry-DASH can form FADH• (16). Biskup et al. and Damiani et al. also both showed that FADH• accumulation occurs not by photoreaction but by oxidation of the light-induced reduced form (14, 15). In contrast, here we detected FAD•−, not FADH•, upon light illumination. We infer that the stable redox states that arise from the processes of photoreduction and oxygen-induced oxidation are distinct. Our data is also in contrast to that of Zikihara et al. (14), in which they observed FADH• upon light illumination. Indeed, stable intermediates may different between Cry-DASHs of Synechocystis and zebrafish. The molecular mechanisms of these unique reaction pathways are intriguing and await further characterization.
Mutation of Asn392 to Cys, which mimics Drosophila Cry, stabilized the FAD•− form (Figure 3b). As represented by the structure of SCry-DASH (Figure 1a), in many Cry and Phr proteins, the neutral Asn (Asn392 of SCry-DASH) interacts with the N5 position of the FAD isoalloxazine, whereas insect and plant specific Crys have Cys and Asp, respectively (Figure 1b-d). Phrs and most Crys are believed to form FADH• and FADH− (2, 14-17, 33, 40), while insect Crys form stable FAD•− (19, 20). As is suggested that Asn or Asp at the position can control the redox state of FAD (33), the Cys is also positioned aptly to be involved in the redox state of FAD.
The Asn is highly conserved in DASH type Crys, Phrs and also mammalian clock-related Crys (Table 1). This key conserved Asn is crucial both for (i) DNA repair Phrs in restoring normal bases from UV-induced photoproducts in DNA strands and also for (ii) mammalian clock-related Crys in repressing transcriptional activation by the BMAL1/CLOCK complex through the E-box contents in DNA as part of a negative feedback (Figure 1, Table 1). Substitution of the Asn in mammalian Crys to Cys or Asp, mimicking insect or plant specific Crys, respectively, disturbs the suppression activity in the clock system, indicating that the residue is one of keys for the functional diversity of Cry/Phr family. Genetic approaches have revealed that Drosophila Cry and mammalian Crys are both involved in maintenance of robust daily rhythms but their essential roles in the circadian rhythms are quite different. The insect specific Cry has been considered as a primary photoreceptor. This was supported through an identified mutant named cry baby, which has a substitution at one of the Asps conserved among the Cry/Phr family (Figure 1d) (56).
Our FTIR spectral data illustrate that SCry-DASH can exist in several distinct FAD states including FADox, and FADH− (Figures 4-6). The detection of the anion form indicates that proton transfer to the chromophore, but not primary electron transfer, would likely take place in SCry-DASH. Successfully, we obtained FAD•−/FADox and FADH−/FADox FTIR difference spectra of SCry-DASH in the 1800–1350 cm−1 and 2600–2500 cm−1 regions, enabling to assign characteristic peaks. The C=O stretch of Asn392 side chain weakened the hydrogen-bonding strength in the FAD•− state of wild-type (Figure 6a and b). Because the C=O side chain of Asn392 forms hydrogen bond with Trp396 (Figure 1), the simplest interpretation is that the distance between Asn392 and Trp396 is further in the FAD•− state. Near the chromophore, the Cry/Phr family proteins contain a highly conserved Trp triad chain (Trp396-Trp373-Trp320 of SCry-DASH), originally identified in E. coli DNA Phr (Figure 1e) (15), that is considered to be an electron transfer pathway. Trp396 of the SCry-DASH triad is closest to the FAD. The negative charge of FAD•− could affect the surrounding environment. On the other hand, S–H group of Cys392 in the N392C mutant did not change its hydrogen-bonding environment (Figure 6c). In the N392C mutant, S–H group may face Ala377 and Trp393 (see Figure 1). Asn392 plays an important role in the electron and proton transfer into FAD•− for the 2nd electron transfer process, conversion from FAD•− to FADH−. The hydrogen-bonding environment of Asn392 should be crucial for the step. The substituted Cys residue may not directly interact with FAD chromophore. This does not necessarily mean that there would be no interaction between the cysteine and FAD in insect specific Crys (Figure 1d) as seen in the N392C mutant of SCry-DASH, while the anion form is stabilized rather than the radical form in the insect Crys (19). Figure 8 compares the FAD•−/FADox spectrum of N392C mutant (blue line) with FAD•−/FADox (black line) and FADH−/FADox (red line) spectra of wild-type SCry-DASH. The FAD•−/FADox spectrum of N392C is located between the FAD•−/FADox and FADH−/FADox spectra of wild-type in the 1680–1640 cm−1 region while quite different features appear in the 1640–1600 cm−1 region, especially between FAD•−/FADox of N392C and FADH−/FADox of wild-type. It would be reasonable that the spectral shapes are different between them if the C=O stretches of FAD•− or FADH− appear in the 1640–1600 cm−1 region. Nevertheless, judging from the spectra in the 1640–1600 cm−1 region, the N392C mutant structure in the FAD•− state may have hybrid features between the FAD•− and FADH− states of wild-type SCry-DASH, though the difference must originate from the interaction of the amino acid residue at the position 392.
Figure 8.
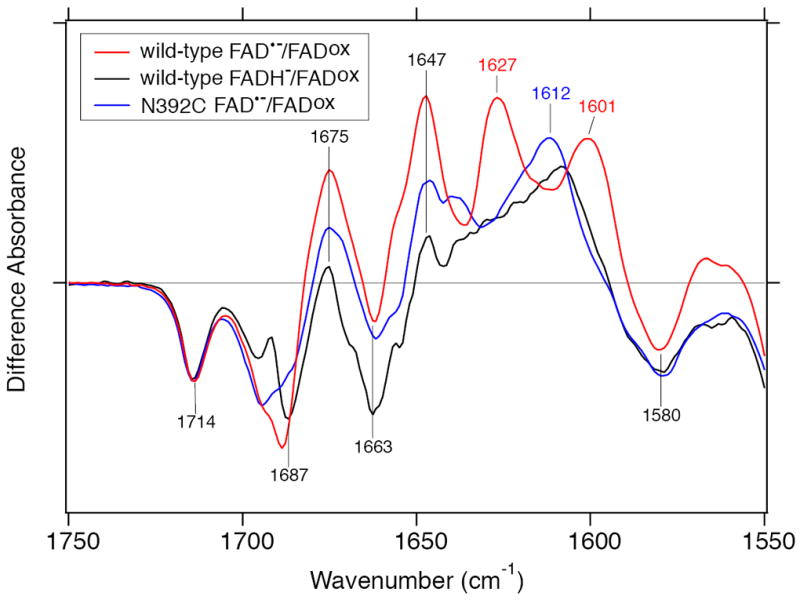
Overlaid difference FTIR spectra of wild-type SCry-DASH and the N392C mutant. Three different spectra in Figure 4 (lane b-d) were overlaid in the 1750–1550 cm−1 region. FAD•−/FADox and FADH−/FADox of wild-type SCry-DASH are shown by red and black lines, respectively, while FAD•−/FADox of N392C mutant is shown by a blue line.
We assume that the N392C mutant cannot proceed further to the FADH− state, because the N392C structure in the FAD•− state may be closer to that of FADH− state of wild-type. Unlike the wild-type, the S–H group of the substituted Cys would not interact with the neighbor Trp residue in the N392C mutant. Therefore, the mutant would not change its hydrogen-bonding environment much upon the formation of FAD•− state (Figure 6c). Notably, this structure may partially mimic that of FADH− state of wild-type. If it is the case, the hydrogen-bonding interaction between Asn392 and Trp396 may be weaker in the FADH− state than in the FAD•− state. That is, protonation at N5 atom of FAD in the FADH− state would strengthen the interaction between the polypeptide and FAD by forming a hydrogen bond between Asn392 and N5 position of isoalloxazine.
Our studies of SCry-DASH indicate that FAD is not protonated from a nearby carboxylic acid residue upon the formation of FADH− (Figure 7b). This may seem contrast to the FTIR study on Arabidopsis Cry1, whose aspartic acid residue donates proton for the FADH• formation (40). Kottke et al. suggested that the origin of the proton is Asp396 whose position corresponds to Asn392 in SCry-DASH, but other possibilities cannot be excluded, as the Cry/Phr family proteins (including DASH type and plant specific Crys) contain two highly conserved Asps at the FAD binding site (Figure 1a and c; Asp386 and Asp388 in SCry-DASH; Asp390 and Asp392 in Arabidopsis Cry1). However, our results best support such a role for Asp396 of Arabidopsis Cry1, corresponding to Asn392 of SCry-DASH, whereas the other two Asps would function uniquely for the light response.
CONCLUSION
We examined different FAD redox states of SCry-DASH by UV–vis and FTIR spectroscopy. The results not only identified a key role of Asn highly conserved among Cry/Phr proteins including mammalian clock-related Crys, but also ruled out two conserved Asps at the FAD binding site as hydrogen donors, demonstrating functional diversity and a critical role at the site in plant- and insect specific Crys.
Acknowledgments
We thank Dr. Haruki Nakamura for modeling advice of Drosophila Cryptochrome; Chiharu Hitomi for technical assistance; Ashley Pratt for critical reading of the manuscript.
Footnotes
This work was supported by grants from Japanese Ministry of Education, Culture, Sports, Science, and Technology to H. K. (20050015, 20108014) by NIH grant GM37684 (E. D. G.). K. H. was supported by the Skaggs Institute for Chemical Biology.
Abbreviations: Cry, crptochrome; Phr, photolyase; FAD, flavin adenine dinucleotide; FADH−, reduced form of FAD; FADox, oxidized form of FAD; FADH•, neutral semiquinoid form; FAD•−, anionic form of FAD; FTIR, Fourier transform infrared; BLUF, sensor of blue light using FAD; LOV, light, oxygen and voltage; FMN, flavin mononucleotide; UV–vis, UV–visible.
References
- 1.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-5-220. Art. No. 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 3.Cashmore AR. Cryptochromes: enabling plants and animals to determin circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 4.Ahmad M, Grancher N, Heil M, Black RC, Giovani B, Galland P, Lardemer D. Action spectrum for cryptochrome-dependent hypocotyl growth inhibition in Arabidopsis. Plant Physiol. 2002;129:774–785. doi: 10.1104/pp.010969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouly J-P, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, Ahmad M. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 6.Hitomi K, Okamoto K, Daiyasu H, Miyashita H, Iwai S, Toh H, Ishiura M, Todo T. Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res. 2000;28:2353–2362. doi: 10.1093/nar/28.12.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daiyasu H, Ishikawa T, Kuma K, Iwai S, Todo T, Toh H. Identification of cryptochrome DASH from vertebrates. Genes Cells. 2004;9:479–495. doi: 10.1111/j.1356-9597.2004.00738.x. [DOI] [PubMed] [Google Scholar]
- 8.Froehlich AC, Chen C-H, Belden WJ, Madeti C, Roenneberg T, Merrow M, Loros JJ, Dunlap JC. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryotic Cell. 2010;9:738–750. doi: 10.1128/EC.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pokorny R, Klar Tobias, Hennecke U, Carell T, Batschauer A, Essen L-O. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA. 2008;105:21023–21027. doi: 10.1073/pnas.0805830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coesel1 S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Todo T, Bowler C, Falciatore A. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10:655–661. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED. Identification of a new cryptochrome class: Structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implication for photolyase activity. Proc Natl Acad Sci USA. 2006;103:17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zikihara K, Ishikawa T, Todo T, Tokutomi S. Involvement of electron transfer in the photoreaction of zebrafish cryptochrome-DASH. Photochem Photobiol. 2008;84:1016–1023. doi: 10.1111/j.1751-1097.2007.00364.x. [DOI] [PubMed] [Google Scholar]
- 15.Biskup T, Schleicher E, Okafuji A, Link G, Hitomi K, Getzoff ED, Weber S. Direct observation of a photoinduced radical pair in a cryptochrome blue-light photoreceptor. Angew Chem Int Ed. 2009;48:404–407. doi: 10.1002/anie.200803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damiani MJ, Yalloway GN, Lu J, McLeod NR, O’Neill MA. Kinetic stability of the flavin semiquinone in photolyase and cryptochrome-DASH. Biochemistry. 2009;48:11399–11411. doi: 10.1021/bi901371s. [DOI] [PubMed] [Google Scholar]
- 17.Weber S, Biskup T, Okafuji A, Marino AR, Berthold T, Link G, Hitomi K, Getzoff ED, Schleicher E, Norris JR., Jr Origin of light-induced spin-correlated radical pairs in cryptochrome. J Phys Chem B. 2010 doi: 10.1021/jp103401u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne G, Heelis PF, Rohrs BR, Sancar A. The active form of Escherichia coli DNA photolyase contains a fully reduced flavin and not a flavin radical, both in vivo and in vitro. Biochemistry. 1987;26:7121–7127. doi: 10.1021/bi00396a038. [DOI] [PubMed] [Google Scholar]
- 19.Berndt A, Kottke T, Breitkreuz H, Dvorsky R, Hennig S, Alexander M, Wolf E. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J Biol Chem. 2007;282:13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 20.Öztürk N, Song S-H, Selby CP, Sancer A. Animal type 1 cryptochromes. J Biol Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 21.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 22.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 23.Salomon M, Eisenreich W, Durr H, Schleicher E, Knieb E, Massey V, Rudiger W, Muller F, Bacher A, Richter G. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc Natl Acad Sci USA. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 26.Iwata T, Nozaki D, Tokutomi S, Kagawa T, Wada M, Kandori H. Light-induced structural changes in the LOV2 domain of Adiantum phytochrome3 studied by low-temperature FTIR and UV-visible spectroscopy. Biochemistry. 2003;42:8183–8191. doi: 10.1021/bi0345135. [DOI] [PubMed] [Google Scholar]
- 27.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 28.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto A, Iwata T, Sato Y, Matsuoka D, Tokutomi S, Kandori H. Light signal transduction pathway from flavin chromophore to Jα helix of Arabidopsis phototropin1. Biophys J. 2009;96:2771–2778. doi: 10.1016/j.bpj.2008.12.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349:1–5. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 31.Ito S, Murakami A, Sato K, Nishina Y, Shiga K, Takahashi T, Higashi S, Iseki M, Watanabe M. Photocycle features of heterologously expressed and assembled eukaryotic flavin-binding BLUF domains of photoactivated adenylyl cyclase (PAC), a blue-light receptor in Euglena gracilis. Photochem Photobiol Sci. 2005;4:762–769. doi: 10.1039/b505792b. [DOI] [PubMed] [Google Scholar]
- 32.Okajima K, Fukushima Y, Suzuki H, Kita A, Ochiai Y, Katayama M, Shibata Y, Miki K, Noguchi T, Itoh S, Ikeuchi M. Fate determination of the flavin photoreceptions in the cyanobacterial blue light receptor TePixD (Tll0078) J Mol Biol. 2006;363:10–18. doi: 10.1016/j.jmb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Balland V, Byrdin M, Eker APM, Ahmad M, Brettel K. What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. J Am Chem Soc. 2009;131:426–427. doi: 10.1021/ja806540j. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Mu W, Ding Yanwei, Luo Z, Han Q, Bi F, Wang Y, Song Y. Active site of Escherichia coli DNA photolyase: Asn378 is crucial both for stabilizing the neutral flavin radical cofactor and for DNA repair. Biochemistry. 2008;47:8736–8743. doi: 10.1021/bi800391j. [DOI] [PubMed] [Google Scholar]
- 35.Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci USA. 2009;106:6962–6967. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siebert F. Infrared spectroscopy applied to biochemical and biological problems. Methods Enzymol. 1995;246:501–526. doi: 10.1016/0076-6879(95)46022-5. [DOI] [PubMed] [Google Scholar]
- 37.Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 38.Kandori H. Hydrogen switch model for the proton transfer in the Schiff base region of bacteriorhodopsin. Biochim Biophys Acta. 2004;1658:72–79. doi: 10.1016/j.bbabio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Schleicher E, Hessling B, Illarionova V, Bacher A, Weber S, Richter G, Gerwert K. Light-induced reactions of Escherichia coli DNA photolyase monitored by Fourier transform infrared spectroscopy. FEBS J. 2005;272:1855–1866. doi: 10.1111/j.1742-4658.2005.04617.x. [DOI] [PubMed] [Google Scholar]
- 40.Kottke T, Batschauer A, Ahmad M, Heberle J. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry. 2006;45:2472–2479. doi: 10.1021/bi051964b. [DOI] [PubMed] [Google Scholar]
- 41.Swartz TE, Wenzel PJ, Corchnoy SB, Briggs WR, Bogomolni RA. Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry. 2002;41:7183–7189. doi: 10.1021/bi025861u. [DOI] [PubMed] [Google Scholar]
- 42.Iwata T, Tokutomi S, Kandori H. Photoreaction of the cysteine S–H group in the LOV2 domain of Adiantum phytochrome3. J Am Chem Soc. 2002;124:11840–11841. doi: 10.1021/ja020834c. [DOI] [PubMed] [Google Scholar]
- 43.Ataka K, Hegemann P, Heberle J. Vibrational spectroscopy of an algal phot-LOV1 domain probes the molecular changes associated with blue-light reception. Biophys J. 2003;84:466–474. doi: 10.1016/S0006-3495(03)74866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwata T, Nozaki D, Tokutomi S, Kandori H. Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry. 2005;44:7427–7434. doi: 10.1021/bi047281y. [DOI] [PubMed] [Google Scholar]
- 45.Iwata T, Nozaki D, Sato Y, Sato K, Nishina Y, Shiga K, Tokutomi S, Kandori H. Identification of the C=O stretching vibrations of FMN and peptide backbone by 13C-Labeling of the LOV2 Domain of Adiantum phytochrome3. Biochemistry. 2006;45:15384–15391. doi: 10.1021/bi061837v. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto A, Iwata T, Tokutomi S, Kandori H. Role of Phe1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- 47.Koyama T, Iwata T, Yamamoto A, Sato Y, Matsuoka D, Tokutomi S, Kandori H. Different Role of the Jα Helix in the Light-Induced Activation of the LOV2 Domains in Various Phototropins. Biochemistry. 2009;48:7621–7628. doi: 10.1021/bi9009192. [DOI] [PubMed] [Google Scholar]
- 48.Masuda S, Hasegawa K, Ishii A, Ono T. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sense of blue-light using FAD (BLUF); slr1694 of Synechocystis sp. PCC6803. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 49.Masuda S, Hasegawa K, Ono T. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 50.Hasegawa K, Masuda S, Ono T. Light induced structural changes of a full-length protein and its BLUF domain in YcgF (Blrp), a blue-light sensing protein that use FAD (BLUF) Biochemisty. 2006;45:3785–3793. doi: 10.1021/bi051820x. [DOI] [PubMed] [Google Scholar]
- 51.Bowman WD, Spiro TG. Normal mode analysis of lumiflavin and interpretation of resonance Raman spectra of flavoproteins. Biochemistry. 1981;20:3313–3318. doi: 10.1021/bi00514a051. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt J, Coudron P, Thompson AW, Watters KL, McFarland JT. Assignment and the effect of hydrogen bonding on the vibrational normal modes of flavins and flavoproteins. Biochemistry. 1983;22:76–84. doi: 10.1021/bi00270a011. [DOI] [PubMed] [Google Scholar]
- 53.Abe M, Kyogoku Y. Vibrational analysis of flavin derivatives: normal coordinate treatments of lumiflavin. Spectrochim Acta. 1987;43A:1027–1038. [Google Scholar]
- 54.Livery CR, McFarland JT. Assignment and the effect of hydrogen bonding on the vibrational normal modes of flavins and flavoproteins. J Phys Chem. 1990;94:3980–3994. [Google Scholar]
- 55.Zhang W, Vivoni A, Lombard JR, Birke RL. Time-resolved SERS study of direct photochemical charge transfer between FMN and a Ag electrode. J Phys Chem. 1995;99:12846–12857. [Google Scholar]
- 56.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]


