Abstract
Genetically encoded voltage sensors expand the optogenetics toolkit into the important realm of electrical recording, enabling researchers to study the dynamic activity of complex neural circuits in real time. However, these probes have thus far performed poorly when tested in intact neural circuits. Hybrid voltage sensors (hVOS) enable the imaging of voltage by harnessing the resonant energy transfer that occurs between a genetically encoded component, a membrane-tethered fluorescent protein that serves as a donor, and a small charged molecule, dipicrylamine, which serves as an acceptor. hVOS generates optical signals as a result of voltage-induced changes in donor-acceptor distance. We expressed the hVOS probe in mouse brain by in utero electroporation and in transgenic mice with a neuronal promoter. Under conditions favoring sparse labeling we could visualize single-labeled neurons. hVOS imaging reported electrically evoked fluorescence changes from individual neurons in slices from entorhinal cortex, somatosensory cortex, and hippocampus. These fluorescence signals tracked action potentials in individual neurons in a single trial with excellent temporal fidelity, producing changes that exceeded background noise by as much as 16-fold. Subthreshold synaptic potentials were detected in single trials in multiple distinct cells simultaneously. We followed signal propagation between different cells within one field of view and between dendrites and somata of the same cell. hVOS imaging thus provides a tool for high-resolution recording of electrical activity from genetically targeted cells in intact neuronal circuits.
Keywords: voltage imaging, optogenetics, neural circuitry, synaptic integration
genetically encoded voltage sensors hold great promise in the investigation of circuit activity of targeted populations of neurons. With this approach one can isolate the activity of neurons within complex networks defined on the basis of gene expression (Huang and Zeng 2013; Taniguchi et al. 2011), synaptic connectivity (Miyamichi et al. 2011), or function (Barth 2007; Guenthner et al. 2013). By reporting voltage changes from many neurons simultaneously, such probes will enable neuroscientists to investigate fundamental questions about neural circuits. Genetically encoded voltage sensors initially employed a biological voltage-sensing motif fused to a fluorescent protein (Siegel and Isacoff 1997) and were extended to include fluorescence resonance energy transfer (FRET) pairs of fluorescent proteins (Akemann et al. 2012; Cao et al. 2013; Jin et al. 2012). The best of these gave fluorescence changes of ∼35%/100 mV, but their ∼10-ms response filtered rapid voltage changes such as action potentials. Further improvement of FRET sensors and sensors based on archaebacterial proton pumps (Kralj et al. 2012) resulted in probes with ∼2-ms response times (Gong et al. 2014; St-Pierre et al. 2014).
Hybrid voltage sensors (hVOS) harness a FRET interaction between a genetically encoded, membrane-tethered fluorescent protein and a lipophilic anion, dipicrylamine (DPA). Voltage sensitivity arises from changes in the distance between these two molecules (Fig. 1A) (Chanda et al. 2005). With the best probes, hVOS can give rise to a ΔF/F/100 mV of ∼25% (Wang et al. 2010). Because DPA crosses the membrane in ∼0.5 ms, hVOS can track action potentials with high temporal fidelity (Bradley et al. 2009; Chanda et al. 2005; Wang et al. 2010).
Fig. 1.
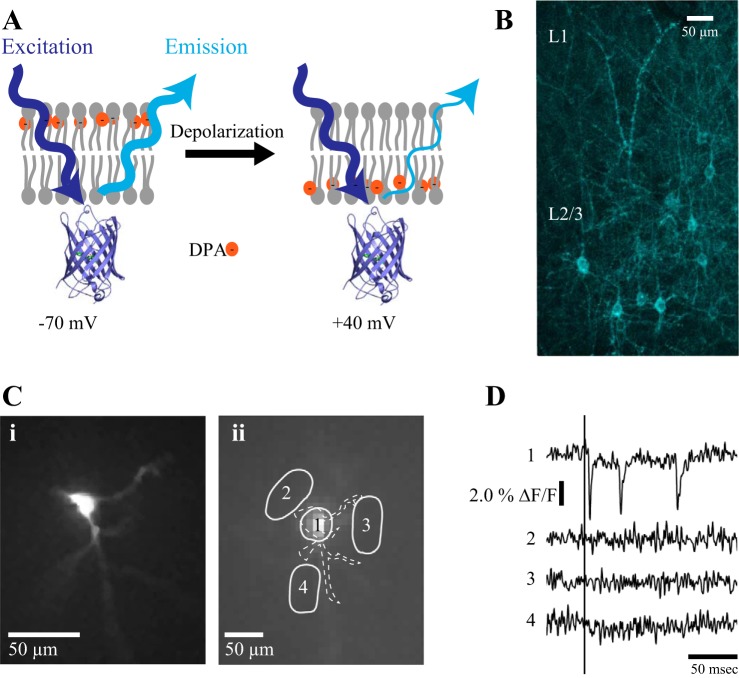
Hybrid voltage sensor (hVOS) expression and imaging. A: illustration of the hVOS mechanism. The probe (hVOS 1.5) cerulean fluorescent protein (CeFP) is tethered to the inner leaflet of the plasma membrane by a truncated farnesylation motif (Wang et al. 2010). Negatively charged lipophilic dipicrylamine (DPA) is bath applied and partitions into the lipid bilayer. Depolarization drives DPA towards the inner leaflet where it interacts with CeFP by fluorescence resonance energy transfer (FRET) and decreases fluorescence emission. B: cortical slice from an 11-day-old mouse expressing probe by in utero electroporation. Collapsed z-series of images from a 2-photon microscope reveals sparsely distributed hVOS probe-expressing neurons in layer 2/3 of the somatosensory cortex. Ci: resting light intensity (RLI) image of a single neuron in a slice from the somatosensory cortex taken with the CCD-SMQ camera used for voltage imaging. The resolution of this image was enhanced off-line with a fractal algorithm in Perfect Resize 8 (onOne Software). Cii: same neuron at a lower magnification, with soma and surrounding regions of interest (ROI) outlined in solid white. The dashed white contour outlines the cell and its main processes. D: single trial fluorescence responses from 4 ROI in Cii containing the soma (trace 1) and 3 surrounding ROI (traces 2–4). Stimulation with a single current pulse (200 μA) applied at the time indicated by the vertical line from an electrode positioned off column and out of view in L2/3 evoked a series of 3 voltage changes. The first event followed the stimulation within a few milliseconds, and the subsequent events reflect either circuit or repetitive activity. Traces from neighboring ROI showed no fluorescence changes.
Genetically encoded voltage sensor performance depends on well-defined properties such as sensitivity, brightness, and kinetics, which do not depend on instrumentation or biological preparation and can readily be compared. However, performance also depends on properties that are more difficult to control, such as expression level, plasma membrane targeting, and background fluorescence. Variations in these parameters make it difficult to compare probes and extrapolate their performance from cultured cell models to intact neural circuits. Voltage sensors tested in intact preparations have thus far yielded weak signals with significant temporal distortion (Akemann et al. 2010; Cao et al. 2013; Gong et al. 2014; St-Pierre et al. 2014). hVOS generated robust signals at the population level in transgenic mice (Wang et al. 2012), but the labeling was too ubiquitous for testing single cells. Here we report hVOS probe targeting to sparsely distributed neurons where we could test performance in brain slices at the single-cell level. We expressed hVOS 1.5 (cerulean fluorescent protein tagged at the COOH terminus with a truncated farnesylation motif; Wang et al. 2010) in mouse brain either by in utero electroporation or by transgenic mouse generation and found sparse labeling of neurons in brain slices from young animals. Imaging with hVOS 1.5 reliably detected subthreshold voltage changes in single cells in single trials. Simultaneous patch clamping provided a calibration of probe performance and assessed the temporal fidelity of hVOS recordings. hVOS reported both subthreshold synaptic potentials and action potentials simultaneously from as many as 10 different neurons. In single neurons we monitored signal propagation between dendritic compartments and cell bodies. These results demonstrate that hVOS serves as a method for imaging electrical signals in genetically targeted populations of neurons in intact neural circuits.
MATERIALS AND METHODS
In utero electroporation.
All animal procedures were approved by the Animal Care and Use Committee of the University of Wisconsin School of Medicine and Public Health. Timed pregnant C57-black mice at 14–15 days of gestation were anesthetized by inhalation of a vaporized isoflurane/oxygen mixture while maintaining body temperature with a heat pad. The area over the abdomen was sterilized with 70% ethanol. Incisions spanning ∼2 cm were made in the skin, and subsequently the body wall, along the lower midline of the abdomen, to expose the abdominal cavity. Uterine horns were carefully pulled out of the abdomen and kept moist throughout the procedure by drops of 0.9% saline. The lateral ventricles on one side of each embryo were injected in utero with DNA encoding the hVOS 1.5 sequence in a syn-lox lentiviral vector (Wang et al. 2011) (1–3 μg/μl), using a glass micropipette and a picospritzer. Fast green dye was added to the DNA solution to allow for visualization of injections. Following injections, a homemade forceps-style electrode and square pulse electroporator (BTX; Harvard Apparatus) generated trains of five 50-ms square pulses at 37 volts to electroporate injected DNA into neural progenitor cells lining the subventricular zone. The uterine horns were carefully returned to the abdominal cavity, and the body wall and skin were sutured closed by absorbable synthetic sutures (Ethicon). Following surgery, animals were removed from anesthesia, monitored for an hour, and returned to the animal facility where they were monitored on a daily basis.
Slice preparation.
Animals were anesthetized by inhalation of isoflurane and decapitated using a guillotine. The brains were quickly removed and placed in ice-cold cutting solution (composition in mM: 124 NaCl, 3.2 KCl, 26 NaHCO3, 1.25 NaH2PO4, 1 CaCl2, 6 MgSO4, and 10 glucose) bubbled with 5% CO2-95% O2 (carbogen). Brains were mounted on a chilled cutting chamber using superglue, bathed in cold cutting solution, and sliced at 300–400 μm using a vibratome (Leica VT1200S). Coronal slices were made from brains expressing the probe by in utero electroporation, and horizontal slices were made from transgenic 1.5 mice. Slices were held in normal artificial cerebrospinal fluid (ACSF; same as cutting solution but with 2.5 mM CaCl2 and 1.2 mM MgCl2) for 30 min.
Voltage imaging.
Slices were incubated in ACSF containing 4 μM DPA for at least 40 min before experiments. Slices were transferred to a recording chamber mounted on the stage of an Olympus BX51 microscope equipped with epifluorescence and IR-DIC. The preparation was continuously perfused with carbogen-bubbled ACSF containing 4 μM DPA. An LED (435 nm; Prizmatix) provided fluorescent excitation light. A CCD camera (Hamamatsu) was used to obtain high-resolution (640 × 480) images of regions for recording and a CCD-SMQ camera (Redshirt Instruments) with 80 × 80 resolution was used to image hVOS signals at a frame rate of 2 kHz. A sliding mirror was used to direct the light from the preparation between the two cameras. Data were acquired in single trials or as averages of up to 10 trials, as noted in each case. The timing of illumination, stimulation, and acquisition was all controlled by the computer program Neuroplex provided with the CCD-SMQ camera.
Electrophysiology.
Patch pipettes were fabricated with a vertical pipette puller (Narishige) and had resistances of 3.5–5 MΩ when filled with a solution containing 130 mM K-gluconate, 10 mM HEPES, 7 mM KCl, 1 mM EGTA, 2 mM Na2ATP, and 2 mM MgATP (pH 7.2). Neurons expressing hVOS probe were selected for patch-clamp recording by fluorescence, and patch pipettes were position with a Sutter MP225 motorized micromanipulator under visual guidance with IR-DIC optics. Whole cell recordings were achieved after giga-seal formation by applying suction. Signals were recorded with an Axopatch 200B patch-clamp amplifier (Molecular Devices) coupled to a computer through a DigiData interface. Simultaneous fluorescence and electrical recordings were synchronized by triggering Neuroplex image acquisition with the patch-clamp software (Clampex 9.2; Molecular Devices).
For extracellular stimulation glass electrodes were filled with ACSF and positioned with a manual Narishige micromanipulator. Current pulses (0.18 ms, 50–320 μA) were delivered with a stimulus isolator (World Precision Instruments) triggered by the imaging software (Neuroplex).
Data analysis.
Fluorescence traces represent averages from a region of interest (ROI) selected manually using Neuroplex. All traces were divided by the resting light intensity to give ΔF/F and low-pass filtered at 400 Hz to reduce noise. Further analysis was performed with Neuroplex or Clampfit 10.2 (Molecular Devices). Subtraction of slow decays in fluorescence was performed by fitting to a fourth order polynomial using Origin 8.6 software (OriginLab). For measurements of signal-to-noise ratio (SNR), ΔF/F was divided by the root mean square noise, determined as the SD of a 200-ms segment of fluorescence before stimulation.
RESULTS
hVOS recording from single cells and multiple cells.
In utero electroporation with hVOS 1.5-encoding DNA in e14–e15 mouse embryos induced probe expression in layer 2/3 neurons in the somatosensory cortex (Fig. 1B), mirroring the pattern of excitatory neuron targeting reported previously with this protocol (Saito and Nakatsuji 2001). Cortical slices from 2- to 4-wk-old animals generally exhibited sparse hVOS probe expression, with distinct and well-separated labeled neurons. Electrical stimulation in the presence of 4 μM DPA evoked clear optical signals in labeled neurons. Figure 1C presents an enhanced fluorescence image of a labeled neuron with a bright soma and dendrites radiating outward in three directions. Figure 1Cii presents the resting light intensity (RLI) image from this neuron as it appeared in the CCD-SMQ camera used for hVOS imaging (with 80 × 80 resolution; the white dashed contour indicates the outline of the cell and major processes). Signal from an ROI containing the soma showed a robust change in response to stimulation that was clearly visible in a single trial (Fig. 1D, top trace). No responses appeared in the adjacent ROI, confirming the specificity of signal (Fig. 1D, bottom 3 traces). In this example electrical stimulation from an electrode outside the field of view elicited an initial event within a few milliseconds, and two more events ∼30 and 100 ms later that could reflect either repetitive spiking or reverberating circuit activity.
Previous work in brain slices from transgenic thy-1-hVOS mice revealed robust fluorescence changes from populations of neurons (Wang et al. 2012). However, the ubiquitous nature of probe expression in these slices made it impossible to attribute signals to individual neurons. This previous study used mice >4 wk old. Because with in utero electroporation we found optimal single cell labeling in 1- to 3-wk-old animals, we turned to younger 1- to 2-wk-old transgenic thy-1-hVOS 1.5 mice and observed distinct labeled neurons in the pyramidal cell layers of the CA1 and CA3 regions of the hippocampus, as well as in the entorhinal cortex (Fig. 2A). [Figure 2A also shows denser labeling in the dentate gyrus in a pattern reported and discussed previously (Wang et al. 2012).] Figure 2B presents a high-resolution fluorescence image (480 × 640 pixels) of a labeled neuron in the entorhinal cortex, and Fig. 2C presents the RLI image from this neuron with the CCD-SMQ camera with the soma within an ROI. Patch-clamp recordings from this neuron showed that current injection evoked simultaneous voltage and fluorescence changes (n = 5 cells in entorhinal cortex; Fig. 2D). hVOS reliably reported both action potentials and subthreshold voltage changes in single trials. The fluorescence signal closely tracked the rapid rise and fall of voltage during action potentials, as can be seen more clearly when the traces were superimposed (Fig. 2E). The speed of the fluorescence changes reflects the rapid movement of DPA, which flips between the two surfaces of the membrane in ∼0.5 ms (Bradley et al. 2009; Chanda et al. 2005; DiFranco et al. 2007; Fernandez et al. 1983). The superimposed traces in Fig. 2E also revealed that the fluorescence change during the subthreshold phase of the response is disproportionately small compared with the fluorescence change during the spikes. This difference reflects the nonlinearity of hVOS responses to voltage (Chanda et al. 2005; DiFranco et al. 2007; Wang et al. 2010). hVOS provides a voltage-dependent signal that is well described by a Boltzmann function, reflecting the voltage-dependent distribution of DPA between the inner and outer surfaces (Fig. 1A). Below approximately −50 mV, DPA begins to accumulate at the outer surface and the effect of voltage on DPA distribution becomes smaller. Voltage changes above −50-mV drive the DPA to the inner membrane face so the hVOS probe will produce a larger signal in this range of voltage.
Fig. 2.
Simultaneous patch-clamp and hVOS recording. A: fluorescence image shows hVOS expression in the hippocampus and entorhinal cortex in a slice from a 2-wk-old thy1-hVOS 1.5 transgenic mouse. B: high-resolution fluorescence image of an hVOS labeled neuron in entorhinal cortex. C: CCD-SMQ RLI image of the neuron in B, with simultaneous whole cell patch-clamp recording indicated. D: depolarizing 200-ms current pulses of increasing amplitude (300, 500, 600, and 700 pA) elicited voltage changes visible in single trials. The smallest current pulse elicited a subthreshold voltage change and the larger pulses also elicited spikes. E: hVOS trace from the 2nd column of D was inverted, normalized, and superimposed on the simultaneously recorded voltage trace.
Brain slices often contained several probe-labeled neurons within a field of view, making simultaneous hVOS imaging from multiple cells possible. Figure 3 presents hVOS recordings from a somatosensory cortical slice (expression by in utero electroporation). Ten well-separated labeled neurons are clearly visible in layer 2/3 in a high-resolution fluorescence image (Fig. 3A). A superimposed fluorescence-DIC image shows the neurons and the stimulating electrode ∼300 μm away (Fig. 3B). Figure 3C shows these neurons circled and numbered in an RLI image from the CCD-SMQ camera. Electrical stimulation through the electrode in Fig. 3B, bottom left corner, evoked clear fluorescence changes in all 10 neurons (Fig. 3D). Responses were visible in all of these neurons in single trials. Increasing the stimulus produced highly variable increases in response amplitude, indicating that these neurons respond independently to stimulation.
Fig. 3.
hVOS recording from 10 labeled neurons. A: layer 2/3 of a slice of somatosensory cortex from an animal expressing hVOS probe by in utero electroporation, 10 labeled neurons were visible in a fluorescence image. B: superimposing fluorescence and DIC shows the labeled cells, the surrounding slice, and the stimulating electrode in layer 4. C: RLI image with the CCD-SMQ showed the 10 neurons with their somata circled and numbered. D: single-trial hVOS recordings from the neurons indicated in C show optical responses to 100- and 200-μA stimuli applied at the vertical lines.
Synaptic potentials and spike thresholds.
Increasing the stimulus current from 100 to 200 μA produced especially large increases in response amplitude in some neurons (Fig. 3D), with ΔF/F in cells 5, 6, and 8 increasing from ∼1 to >3%. In these cells the increases reflected the emergence of a rapid component that summated with the smaller, slower responses evoked by 100 μA. The sharp rise and fall of the fast components resembled fluorescence responses associated with action potentials seen under current clamp (Fig. 2, D and E) and had similar amplitudes of ∼3%. This suggested that the smaller slow components represented subthreshold synaptic potentials and the larger rapid components represented action potentials.
To distinguish action potentials from subthreshold synaptic depolarizations in hVOS responses, we performed thresholding experiments by stimulating with a series of pulses with small increments in current intensity. Figure 4A shows a high-resolution fluorescence image from a slice of somatosensory cortex with two neurons in view. A superimposed fluorescence-DIC image shows the stimulating electrode ∼250 μm from the nearest of the two neurons (Fig. 4B), and an RLI CCD-SMQ image shows the two somata of these neurons in ROI (Fig. 4C). Stimulus currents of 250 and 270 μA evoked small fluorescence responses in both neurons (Fig. 4D). Increasing the stimulus current to 290 μA evoked a sharp spike in cell 1 but not in cell 2. This stimulus current was very near threshold as a second stimulus of the same intensity produced a small response in cell 1 and a spike in cell 2. These small increments in stimulus current increased the fluorescence responses by approximately threefold. Both neurons in Fig. 4 exhibited threshold fluctuations with individual trials showing either a synaptic potential or a summation of a spike with a synaptic potential. Averages of five trials with 260 μA and five trials with 310 μA superimposed show this sharp difference between subthreshold and suprathreshold responses for neuron 1 and illustrate the summation of synaptic and action potentials (Fig. 4E).
Fig. 4.
Spike thresholds in somatosensory cortex neurons. A: fluorescence image from a slice of somatosensory cortex from an animal expressing hVOS probe by in utero electroporation with 2 labeled neurons in layer 2/3. B: fluorescence-DIC image shows the labeled neurons, surrounding tissue, and stimulating electrode in layer 4. C: CCD-SMQ RLI image of the 2 labeled neurons in A and B with ROI containing their somata outlined and numbered. D: graded increases in stimulus current revealed the thresholds for action potentials in the 2 neurons. The stimulus currents are on the left, and time of stimulation is indicated by the vertical lines. E: 5 trial averages of responses to 260 an 310 μA from the soma of cell 1 were superimposed to show the subthreshold and suprathreshold responses.
We also observed sharp thresholds in the CA3 region of hippocampal slices from hVOS 1.5 transgenic mice. In a slice with four labeled pyramidal cells in view (Fig. 5A), stimulating the mossy fibers outside the field of view evoked responses that varied dramatically depending on the stimulus current (Fig. 5B). A current of 30 μA evoked a response in neuron 2 but not in the other three neurons. Increasing the stimulus current to 35 μA evoked spikes in all four neurons. It is noteworthy that we rarely saw subthreshold synaptic responses in the CA3 region. Figure 5B shows a small subthreshold response in neuron 4 with 33 μA. By contrast, subthreshold responses were common in somatosensory cortex (Figs. 3 and 4). This difference may reflect the unusual strength of mossy fiber synapses on CA3 pyramidal cells, where a single input has been reported to “detonate” a postsynaptic cell to fire (Henze et al. 2002). Such a detonator mechanism would indicate that the thresholds evident in Fig. 5B reflect the threshold for firing of single presynaptic mossy fibers.
Fig. 5.
Spike thresholds in hippocampal CA3 pyramidal cells. A: CCD-SMQ RLI image (i) revealed labeled pyramidal cells in the pyramidal cell layer of a hippocampal slice from a 13 day old thy1-hVOS 1.5 transgenic mouse. ROI containing cell somata were outlined (ii). B: single-trial hVOS responses to 30-, 33-, and 35-μA pulses applied to the mossy fibers from an electrode out of the field of view at times indicated by vertical lines. Individual cells had distinct thresholds.
We compared hVOS signals with action potentials in patch-clamped neurons evoked by direct current injection as in Fig. 2 with action potentials evoked by activation of synaptic inputs (Figs. 4–5). Table 1 presents the mean ΔF/F and SNR for single-trial action potential recordings in single neurons in somatosensory cortex, entorhinal cortex, and hippocampus. Mean ΔF/F for action potentials ranged from 2.54 to 2.94% in patch-clamped neurons and 3.00 to 3.47% in neurons activated by extracellular stimulation. Although the nonlinearity of hVOS discussed above makes it difficult to calibrate fluorescence in terms of membrane potential, these results indicate that action potentials generally produce a rapid ΔF/F of ∼3%. The SNR in these recordings ranged from 9.16 to 16.00. The slightly larger signals in extracellularly stimulated neurons may reflect either summation with signals from neighboring cells, differences in the Na+ driving force with our pipette solution, or a reduction in spike amplitude due to current loading by the patch-clamp amplifier in current-clamp mode (Magistretti et al. 1996).
Table 1.
hVOS ΔF/F and SNR ratio in single neurons during action potentials
| Entorhinal |
Somatosensory |
Hippocampus |
|||
|---|---|---|---|---|---|
| Cortex (Patched) (n = 5) | Cortex (Ex) (n = 34) | Cortex (Patched) (n = 3) | Cortex (Ex) (n = 16) | CA1-CA3 (Ex) (n = 36) | |
| ΔF/F % | 2.54 ± 0.49 | 3.00 ± 0.15 | 2.94 ± 0.72 | 3.47 ± 0.37 | 3.18 ± 0.18 |
| SNR | 15.26 ± 2.85 | 13.57 ± 0.87 | 10.62 ± 3.00 | 16.00 ± 1.52 | 9.16 ± 0.45 |
Mean ΔF/F and signal-to-noise ratio (SNR) of single-trial action potential recordings from individual neurons in deep entorhinal cortex, L2/3 somatosensory cortex, and the CA1-CA3 regions of the hippocampus. hVOS, hybrid voltage sensors. Entorhinal cortex and hippocampus recordings were from thy 1.5 transgenic mice (Wang et al. 2012) and somatosensory cortex recordings were from in utero electroporation. Signals were evoked by current injection in simultaneous fluorescence and whole cell recordings (Patched) or by extracellular stimulation (Ex) of neurons without patch clamping. All signals were low-pass filtered at 400 Hz (see materials and methods).
Dendritic integration.
Imaging with synthetic voltage sensitive dyes in single cells can reveal voltage changes in dendrites, and this approach provides a powerful method for exploring synaptic integration (Antic and Zecevic 1995; Popovic et al. 2012). In probe-labeled neurons we were often able to see extensive dendritic arbors and follow the spread of voltage changes through different compartments. Figure 6A presents an RLI image of a neuron in the somatosensory cortex with one ROI containing the cell body and one RLI containing a segment of dendrite. One pulse from a stimulating electrode elicited a train of eight events that continued for several hundred milliseconds (Fig. 6B). Thus this single trial revealed a succession of responses to a single stimulus, similar to that in Fig. 1, but also revealing the timing of the voltage changes in distinct cellular compartments. This example showed many more events than Fig. 1. Again, we cannot distinguish between intrinsic repetitive firing or circuit activity. For each event, the fluorescence change initiated in the cell body and propagated to the dendritic ROI 70 μm away with a delay of 1.5 ms (Fig. 6C). The dendritic depolarization was somewhat slower than the somatic depolarization.
Fig. 6.
Propagation between compartments. A: CCD-SMQ RLI image of a labeled neuron in the somatosensory cortex (expressing probe by in utero electroporation). B: hVOS imaging revealed a burst of activity evoked by stimulation (140 μA, at the arrow). Each event propagated from the soma (black trace) to the dendritic branch (red trace) (ROI labeled in A). C: later events in the soma and dendritic branch were averaged and superimposed to reveal initiation in the soma and propagation to the dendritic compartment with a delay of 1.5 ms between times to half-maximum. D: CCD-SMQ RLI image of a neuron in the somatosensory cortex with several clear dendritic branches. E: dendritic branches were demarcated in ROI and labeled. White arrows indicate direction of propagation used for the x-axis in G. F: stimulus evoked fluorescence responses from ROI indicated in E. Ten trial averages were superimposed to show variable delays in dendrites following onset in the soma. Traces normalized to RLI are plotted above and traces normalized to their maxima are plotted below. G: spatiotemporal map of responses from dendritic branches numbered in E, with the horizontal axis representing increasing distance from the soma (along corresponding white arrows in E) and the vertical axis representing time. Color scale encodes response amplitude from dark blue (zero) to yellow (maximum), normalized to the maximal signal from the soma. The horizontal red dashed line represents t = 0, and the diagonal red dashed lines highlight the propagation front to indicate velocity.
Figure 6D shows an RLI image of another somatosensory cortical neuron with an extensive dendritic arbor; an enhanced color image highlights the morphology of this neuron more clearly (Fig. 6E). Extracellular stimulation evoked hVOS responses in the soma and in several distinct dendritic branches. Averages of 10 trials from 8 distinct dendrites illustrated that responses initiated in the soma and propagated with delays to different dendritic branches (Fig. 6F). Spatiotemporal maps were constructed with distance along the branch as the x-axis and time as the y-axis (Fig. 6G). The downward movement in these plots toward the right reflects the spread of the voltage change along each dendrite, and the slopes indicate the speed of this spread. The greater downward slopes in maps 4 and 7 indicate slower spread in these processes compared with maps 1, 2, 3, 5, and 6. Voltage changes in the secondary dendritic branches (map 7) were weaker and delayed.
The neuron shown in Fig. 6, D and E, depolarized first in the soma and then in the dendrites, suggesting that our stimulation activated this cell antidromically. We saw the opposite sequence in another example shown in Fig. 7. The action potential was triggered in the dendritic compartment labeled d3 (light blue outline in Fig. 7B and light blue trace in Fig. 7C). The hVOS signal from that compartment indicated that voltage rose sharply to a much higher level, and this peak preceded the peaks in the other dendrites and soma. Other compartments exhibited two component responses, an early synaptic potential and a later surge of variable amplitude that reflected the spread of the spike originating in compartment d3. The response in the soma peaked over a millisecond after the peak in compartment d3, and the peak in d2 followed the peak in d3 by 2–3 ms, indicating back propagation of the spike initiated in d3. Thus hVOS tracks the onset of a synaptically triggered spike in d3 and sequential propagation to the soma and other dendritic branches.
Fig. 7.

Spike initiation in dendrites. A: CCD-SMQ RLI image of a labeled neuron in somatosensory cortex (in utero electroporation). B: ROI containing soma and 5 dendritic branches. C: hVOS traces (5 trial averages) in colors corresponding to the ROI in B. Traces normalized to RLI are presented on the left, and traces normalized to their maxima are presented on the right. The spike initiated in dendritic compartment d3 (light blue).
Circuit responses to extracellular stimulation.
In fields of view with multiple neurons, the activation sequence can provide insight into circuit relations. Slices from the entorhinal cortex from a 2-wk-old thy-1 hVOS 1.5 transgenic mouse contained many neurons expressing probe. In one field of view, we noted four labeled neurons by arrowheads in an RLI image (Fig. 8A, left), and a high-resolution fluorescence image (Fig. 8A, right). Five trial averages from each ROI revealed a sequence of activation of these neurons (Fig. 8B). Figure 8C summarizes this sequence of activation. Stimulation was applied in the bottom left corner (Fig. 8Ci) and depolarized the red-and orange-labeled neurons almost simultaneously (Fig. 8Cii). The response of the blue- and green-labeled neurons followed with a delay of ∼1.5 ms. The blue-labeled neuron spiked rapidly (Fig. 8Ciii). A simultaneous depolarization of the green-labeled neuron was followed by a spike after a ∼2.5-ms delay (Fig. 8Civ). Given the relatively brief ∼0.5-ms transit time of DPA within the membrane (Bradley et al. 2009; Chanda et al. 2005; DiFranco et al. 2007; Fernandez et al. 1983), and the excellent temporal fidelity of fluorescence and voltage changes (Fig. 1E), these delays primarily represent the sum of the conduction time from the site of stimulation and the synaptic delay. To the extent that this sequence reflects synaptic connectivity, Fig. 8C also represents a hypothetical circuit that can account for this sequence. However, unlabeled neurons also contribute to polysynaptic activity and can serve as relay neurons in later responses. This experiment illustrates that we can see activation sequences in complex neural networks and that detailed experiments with an hVOS probe targeted to specific cell types have the potential to elucidate functional relations within complex networks in brain slices.
Fig. 8.
Propagation through a network. A, left: RLI image with the CCD-SMQ camera showing four labeled neurons indicated by colored arrowheads. A, right: high-resolution fluorescence image of the same field shows the neurons more clearly. B: hVOS traces from 4 ROI containing the somata show the temporal sequence of voltage changes in the 4 neurons highlighted in A; colors correspond to the arrowheads in A. Traces normalized to RLI are at left, and traces normalized to their maxima are at right. The red-labeled and orange-labeled neurons responded almost simultaneously, followed by the blue-labeled and the green-labeled neurons. The green-labeled neuron spiked ∼2 ms after the synaptic response. C: schematic representation of the sequence of responses in the 4 labeled neurons. Ci: stimulation from beyond the bottom left corner activated the red- and orange-labeled neurons at almost the same time. Cii: red-labeled and orange-labeled neurons responded with a delay of ∼1.5 ms. Ciii: blue-labeled and green-labeled neurons responded ∼2 ms later. Civ: green-labeled neuron spiked ∼2.5 ms after its initial synaptic response. The arrows indicate the sequence of activation and a hypothetical circuit that could include the labeled neurons along with unlabeled members of the populations represented by the labeled neurons.
DISCUSSION
The present study has demonstrated that hVOS 1.5 performs effectively in voltage imaging in intact brain slices, generating robust signals from individual neurons and tracking action potentials with high temporal fidelity. Action potentials produced mean ΔF/F signals ranging from 2.54% in patch-clamped neurons of the entorhinal cortex to 3.47% in synaptically activated neurons of the somatosensory cortex (Table 1). These responses could be clearly resolved in single trials. The SNR of an action potential is an important benchmark in voltage probe performance. In slices from cortex and hippocampus, hVOS 1.5 reported action potentials with mean SNRs ranging from 9.16 to 16.0 (Table 1). These values are significantly higher than recent estimates for two new genetically encoded voltage sensors. Using probe performance parameters determined from ASAP1, St-Pierre et al. (2014) estimated a spike SNR of 2–4. Gong et al. (2014) performed a similar calculation for FRET-opsin and obtained a spike SNR of ∼5. Our direct measurements of the spike SNR in cortical and hippocampal neurons in intact brain slices exceed these estimates, despite the added handicap of tissue background fluorescence. The poorest spike SNR with hVOS was obtained in the pyramidal cell layer of hippocampal slices from transgenic mice where the use of a pan-neuronal promoter (thy-1) produced general neuronal targeting. The higher background, as illustrated in Fig. 5, Ai and Aii, is the most likely explanation for the lower SNR of spikes in CA1 and CA3 region recordings in Table 1. Variations in background from site to site will create some spatial heterogeneity in the SNR. It is likely that more specific probe targeting will increase SNR values. The ratio of plasma membrane to cytoplasmic membrane expression will also influence background and SNR. Thus engineering probes with more efficient plasma membrane targeting should improve performance.
In addition to providing clear recordings of action potentials, hVOS 1.5 performed well enough to report much smaller subthreshold synaptic potentials in single trials. This represents an important new milestone in genetically encoded voltage sensor performance that adds a new dimension to the investigation of neural circuit mechanisms. Subthreshold synaptic potentials can fail to drive Ca2+ entry through voltage-gated Ca2+ channels and N-methyl-d-aspartate receptor channels, making them invisible to Ca2+ sensors. Thus hVOS can reveal nonspiking activity and thus elucidate synaptic connectivity based on monosynaptic outputs of single neurons. The rapid submillisecond response time of hVOS will enable the study of synchrony and small timing differences between many individual neurons within a circuit. In such studies of timing relations, it will be important to recognize the 32% increase in membrane capacitance and anticipated 15% reduction in action potential propagation velocity in the presence of 4 μM DPA (Wang et al. 2010). To investigate timing at this level of resolution, it will be important to test the limit of how far one can reduce the DPA concentration and still obtain useful signals.
An additional milestone achieved in the present study is the resolution of voltage changes in subcellular compartments. Single trial recordings revealed these changes but averaging five trials provided especially clear views of the spread of signals between the cell body and dendrites. We could distinguish between action potential initiation in the soma vs. dendrites and in other cases identify the dendritic compartment where a spike initiated (Fig. 7). The resolution of hVOS signals from subcellular compartments should motivate the development of probes that target expression to the soma, dendritic branches, dendritic spines, and other specialized regions. In this regard, it is promising that the probe hVOS 2.0 preferentially targets axons (Wang et al. 2010). This finding provides encouragement for efforts to target other cellular compartments such as somata and dendrites.
Imaging with genetically encoded voltage sensors represents the sensing counterpart to optical activation of targeted neurons with channelrhodopsins and related proteins. The nervous system is thought to encode information in patterns of active neurons and to recall information by enabling an appropriate retrieval signal to reactivate these patterns. At the population level, long-term potentiation can serve as an encoding mechanism in the storage and recall of patterns of activity (Jackson 2013). The imaging of voltage signals from individual neurons within intact circuits may ultimately permit the investigation of the storage and recall of patterns of activity at the single-cell level.
GRANTS
This research was supported by National Institute of Neurological Disorders and Stroke Grant NS-078301.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: N.G., P.O.B., and M.B.J. conception and design of research; N.G., P.O.B., and Y.M. performed experiments; N.G., P.O.B., and Y.M. analyzed data; N.G., P.O.B., Y.M., and M.B.J. interpreted results of experiments; N.G., P.O.B., and Y.M. prepared figures; N.G. drafted manuscript; N.G., P.O.B., and M.B.J. edited and revised manuscript; N.G., P.O.B., Y.M., and M.B.J. approved final version of manuscript.
REFERENCES
- Akemann W, Mutoh H, Perron A, Park YK, Iwamoto Y, Knopfel T. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J Neurophysiol 108: 2323–2337, 2012. [DOI] [PubMed] [Google Scholar]
- Akemann W, Mutoh H, Perron A, Rossier J, Knopfel T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat Methods 7: 643–649, 2010. [DOI] [PubMed] [Google Scholar]
- Antic S, Zecevic D. Optical signals from neurons with internally applied voltage-sensitive dyes. J Neurosci 15: 1392–1405, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr Opin Neurobiol 17: 567–571, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Luo R, Otis TS, DiGregorio DA. Submillisecond optical reporting of membrane potential in situ using a neuronal tracer dye. J Neurosci 29: 9197–9209, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell 154: 904–913, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci 8: 1619–1626, 2005. [DOI] [PubMed] [Google Scholar]
- DiFranco M, Capote J, Quinonez M, Vergara JL. Voltage-dependent dynamic FRET signals from the transverse tubules in mammalian skeletal muscle fibers. J Gen Physiol 130: 581–600, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JM, Taylor RE, Bezanilla F. Induced capacitance in the squid giant axon. Lipophilic ion displacement currents. J Gen Physiol 82: 331–346, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat Commun 5: 3674, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78: 773–784, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze DA, Wittner L, Buzsaki G. Single granule cells reliably discharge targets in the hippocampal CA3 network in vivo. Nat Neurosci 5: 790–795, 2002. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci 36: 183–215, 2013. [DOI] [PubMed] [Google Scholar]
- Jackson MB. Recall of spatial patterns stored in a hippocampal slice by long-term potentiation. J Neurophysiol 110: 2511–2519, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75: 779–785, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 9: 90–95, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti J, Mantegazza M, Guatteo E, Wanke E. Action potentials recorded with patch-clamp amplifiers: are they genuine? Trends Neurosci 19: 530–534, 1996. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, Callaway EM, Horowitz MA, Luo L. Cortical representations of olfactory input by trans-synaptic tracing. Nature 472: 191–196, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Gao X, Zecevic D. Voltage-sensitive dye recording from axons, dendrites and dendritic spines of individual neurons in brain slices. J Vis Exp 69: e4261, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol 240: 237–246, 2001. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron 19: 735–741, 1997. [DOI] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci 17: 884–889, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71: 995–1013, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, McMahon S, Zhang Z, Jackson MB. Hybrid voltage sensor imaging of electrical activity from neurons in hippocampal slices from transgenic mice. J Neurophysiol 108: 3147–3160, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, Chanda B, Jackson MB. Improved probes for hybrid voltage sensor imaging. Biophys J 99: 2355–2365, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, Dong M, Sun S, Chapman ER, Jackson MB. Syntaxin requirement for Ca2+-triggered exocytosis in neurons and endocrine cells demonstrated with an engineered neurotoxin. Biochemistry 50: 2711–2713, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]