Abstract
Vertebrates possess two paralogs of cytochrome c oxidase (COX) subunit 4: a ubiquitous COX4-1 and a hypoxia-linked COX4-2. Mammalian COX4-2 is thought to have a role in relation to fine-tuning metabolism in low oxygen levels, conferred through both structural differences in the subunit protein structure and regulatory differences in the gene. We sought to elucidate the pervasiveness of this feature across vertebrates. The ratio of COX4-2/4-1 mRNA is generally low in mammals, but this ratio was higher in fish and reptiles, particularly turtles. The COX4-2 gene appeared unresponsive to low oxygen in nonmammalian models (zebrafish, goldfish, tilapia, anoles, and turtles) and fish cell lines. Reporter genes constructed from the amphibian and reptile homologues of the mammalian oxygen-responsive elements and hypoxia-responsive elements did not respond to low oxygen. Unlike the rodent ortholog, the promoter of goldfish COX4-2 did not respond to hypoxia or anoxia. The protein sequences of the COX4-2 peptide showed that the disulfide bridge seen in human and rodent orthologs would be precluded in other mammalian lineages and lower vertebrates, all of which lack the requisite pair of cysteines. The coordinating ligands of the ATP-binding site are largely conserved across mammals and reptiles, but in Xenopus and fish, sequence variations may disrupt the ability of the protein to bind ATP at this site. Collectively, these results suggest that many of the genetic and structural features of COX4-2 that impart responsiveness and benefits in hypoxia may be restricted to the Euarchontoglires lineage that includes primates, lagomorphs, and rodents.
Keywords: COX4-2, oxygen sensitivity, HRE, ORE
to survive in variable conditions, organisms must possess the ability to sense changes in the environment and modulate energy production accordingly. An important point of metabolic regulation is thought to occur at cytochrome c oxidase (COX) (2, 3, 13, 42), the final complex of the electron transport system. COX catalyzes the reduction of oxygen to water while translocating protons across the inner mitochondrial membrane, contributing to a proton gradient that drives ATP synthesis. A complete vertebrate COX complex is a dimer composed of 2 monomers, each of 13 subunits. The three largest subunits, which form the enzymatic core of the complex, are mitochondrially encoded and are highly conserved across eukaryotes (36). The remaining 10 nuclear-encoded subunits serve structural and regulatory functions. Many of these subunits exist as paralogs in vertebrates, some of which are lineage specific. For example, although COX4, 6A, 6B, 6C, 7A, and 7C have equal numbers of paralogs in fish and mammals, COX7B and 8 have more in mammals, and COX5A and 5B have more in fish (29). COX4 is of particular interest as the only subunit with a paralog pair that responds to environmental changes in mammals (22).
COX4 is the largest nuclear-encoded subunit and interacts directly with two of the three catalytic subunits within the complex (41). The subunit contains two sites that competitively bind ATP and ADP, allowing COX to sense cellular energy conditions and modulate turnover rates accordingly (3, 31, 32). In mammals, COX4 exists as an isoform pair and displays a major-minor pattern of isoform transcription, where COX4-1 mRNA is present in vast excess in most tissues and COX4-2 mRNA is present at detectable levels in brain and in highest amounts (about equal to COX4-1 transcripts) in respiratory tissues (23, 22). During exposure to hypoxia, the COX4 isoform pattern has been shown to reverse, with transcription switching from COX4-1 to COX4-2, while COX4-1 peptides are selectively degraded by the LON protease (14, 22, 25). The trigger for the oxygen-dependent switch in transcription has been attributed to either conventional hypoxia-responsive elements (HRE) (14) or oxygen-responsive elements (ORE) (25). Fukuda et al. (14) characterized two putative HRE in the human COX4-2 gene, one present in the proximal promoter region and one in the first intron. Through transfection experiments, the group was able to show that these regions conferred a hypoxic response in COX4-2 through transactivation by hypoxia-inducible factor-1 (HIF-1) (14). Conversely, Hüttemann et al. (25) noted that the putative HREs present in human COX4-2 were poorly conserved across mammals. They identified a 13-bp region of the COX4-2 proximal promoter that is more conserved. This ORE conferred hypoxia responsiveness independently of HIF-1 and the HREs (25). Three transcription factors have since been identified as ORE-binding proteins. Recombination signal sequence-binding protein Jκ (RBPJ) and coiled-coil-helix-coiled-coil-helix domain 2 increase transcription levels (CHCHD2), and CXXC finger protein 5 acts as an inhibitor (CXXC5) (1).
The COX4 paralog switch is thought to optimize electron transfer reactions in COX in response to changing energetic conditions. COX complexes containing COX4-2 have a turnover rate approximately twice that of complexes containing COX4-1 (24, 25) and are able to optimize the electron transfer reactions, resulting in a decrease of the production of reactive oxygen species (ROS) under hypoxic conditions (14). Although COX4-2 containing COX shows a normal response to ATP and ADP as was shown with purified enzyme from cow lung (24), there is some evidence to suggest that under hypoxic conditions adenylate regulation of COX is lost altogether (22). These differences may be caused by a pair of cysteine residues on COX4-2 that form a disulfide bond, preventing adenyate binding (23). The COX4 paralogs differ in efficiency of electron transfer and proton pumping and perhaps sensitivity to cytosolic adenylate concentrations. Thus subunit switching may enable a cell to fine tune COX to alter ATP production, respiration, and mitochondrial ROS production in response to tissue type, energetic status, and hypoxia (39).
While it seems clear that the COX4-2 protein and gene adaptations can play a role in the regulation of energy production in rodents and humans, as well as some tumors, the evolutionary origins and phylogenetic distribution have not been well characterized. Given that the COX4 paralog pair is present throughout vertebrates, it is likely that the isoforms arose from a whole genome duplication event early in vertebrate evolution, although one paralog has been secondarily lost in birds and the available evidence suggests that COX4-2 of fish may have a different origin than COX4-2 of tetrapods (29). If this pathway is central to oxygen sensing and/or hypoxic metabolism, natural selection may affect the gene both in regulatory and structural regions in lineages that have distinctive hypoxic adaptations. For example, it could be hypothesized that more hypoxia-tolerant species may express COX4-2 constitutively in more tissues or that these species may have a more responsive promoter. In this study we were interested in determining if the hypoxia-dependant COX4 paralog switch is present in lower vertebrate taxa and whether COX4-2 appears to be of importance in hypoxia-tolerant species. Interestingly, we find that the familiar features of the COX4-2 gene and protein may be restricted to the Euarchontoglires lineage that includes rodents and primates.
METHODS
COX4-2 Sequence Analysis
COX4-2 genes and their promoters were characterized from candidate vertebrate species. Sequences were obtained from the online databases Ensembl and NCBI, and the presence or absence of the putative elements responsible for the hypoxic response of COX4-2 was determined by alignments with the human element sequences found by Fukuda et al. (14) and Hüttemann et al. (25) using MultAlin (7).
Translated nucleotide sequences of COX4-2 were also examined from a wide array of vertebrate species, focusing on the conservation of amino acids suggested to be involved in the COX ATP/ADP-binding site (Arg-20, Arg-73, Tyr-75, Glu-77, and Trp-78) and the presence or absence of two cysteine residues thought to be important in the isoform-specific function of human COX4-2 (Cys-16 and Cys-30) (23). Polypeptide sequences were obtained from the online database Ensembl for candidate species from several vertebrate classes and aligned using ClustalW2 (16, 28). Accession numbers for all sequences used can be found in Table 1.
Table 1.
Species used for assessment of conservation of COX4-2 promoter elements (DNA sequences) and cysteine residues (AA)
| Common Name | Species Name | Ensembl Accession No. | Use (DNA, AA) |
|---|---|---|---|
| Anole | Anolis carolinensis | ENSACAG00000011459 | D, A |
| Cat | Felis catus | ENSFCAG00000014664 | D |
| Cavefish | Astyanax mexicanus | ENSAMXG00000016908 | A |
| Cod | Gadus morhua | ENSGMOG00000002985 | A |
| Coelacanth | Latimeria chalumnae | ENSLACG00000009141 | A |
| Cow | Bos taurus | ENSBTAG00000016171 | D, A |
| Dog | Canis lupus familiaris | ENSCAFG00000007049 | D, A |
| Dolphin | Tursiops truncatus | ENSTTRG00000007924 | D, A |
| Gar | Lepisosteus oculatus | ENSLOCG00000006091 | A |
| Gorilla | Gorilla gorilla | ENSGGOG00000009052 | D |
| Hedgehog | Erinaceus europaeus | ENSEEUG00000009547 | D, A |
| Human | Homo sapiens | ENSG00000131055 | D, A |
| Hyrax | Procavia capensis | ENSPCAG00000011518 | D, A |
| Macaque | Macaca mulatta | ENSMMUG00000007383 | D |
| Medaka | Oryzias latipes | ENSORLG00000001813 | A |
| Microbat | Myotis lucifugus | ENSMLUG00000004800 | D, A |
| Mouse | Mus musculus | ENSMUSG00000009876 | D |
| Pika | Ochotona princeps | ENSOPRG00000004657 | D, A |
| Platyfish | Xiphophorus maculatus | ENSXMAG00000016678 | A |
| Rat | Rattus norvegicus | ENSRNOG00000007827 | D, A |
| Stickleback | Gasterosteus aculeatus | ENSGACG00000004286 | A |
| Tilapia | Oreochromis niloticus | ENSONIG00000001633 | A |
| Xenopus | Xenopus tropicalis | ENSXETG00000027592 | D, A |
| Zebrafish | Danio rerio | ENSDARG00000022509 | A |
COX, cytochrome c oxidase; AA, amino acid sequences.
Animal Experiments
All animal experiments were carried out in accordance and approved by the Queen's University Animal Care Committee.
Zebrafish (Danio rerio, Hamilton) were donated from a wild-type population at Université de Montréal. They were held in a 140-liter aquarium with dechlorinated water maintained at 25°C for at least 6 wk. The fish were kept under a 12:12-h light-dark photoperiod and fed Omega One Freshwater Flakes (OmegaSea, Painesville, OH) daily ad libitum. Groups of eight fish were transferred to 8-liter jars at 25°C with aerated water flowing through overnight. After 12 h, the water flowing to the hypoxic fish was bubbled with 4% O2-96% N2 for 8 h. At the onset of treatment, gas mixtures were bubbled directly into the jar, with no flow through. After 2 h, flow through of gas-equilibrated water began at a rate of 4 ml/min. Control fish were treated in the same manner, but their water was bubbled with air. Immediately following the experiment, fish were lethally anesthetized in a solution of 0.4 g/l of tricaine methanesulphonate and 0.8 g/l sodium bicarbonate. Gills, liver, brain, and white muscle were dissected, flash frozen in liquid nitrogen, and stored at −80°C.
Goldfish (Carassius auratus, Linnaeus) were acquired from a local aquarium store. They were held in a 750-liter tank with dechlorinated water maintained at ∼15°C for at least 6 wk to allow for acclimation. The fish were kept under a 12:12-h light-dark photoperiod and fed Omega One Freshwater Flakes (OmegaSea, Painesville, OH) daily ad libitum. Anoxia experiments were carried out in air-tight 120-liter tanks with a flow-through water system. Eight fish were exposed to anoxia (100% nitrogen) for 21.5 h. The water was de-oxygenated by pumping nitrogen gas into the tank via an air stone while monitoring oxygen levels using a FOXY oxygen probe (Ocean Optics, Dunedin, FL). After 3 h, no oxygen was detected in the tank. Immediately following the experiment, tissues were collected from fish as described above for the zebrafish. Control fish were taken from the holding tank at the beginning of the anoxia treatment, and tissues were collected in the same manner described above.
Tilapia (Oreochromis niloticus, Linnaeus) were received from Red Fish Ranch of British Columbia. They were held in a 620-liter aquarium with dechlorinated water maintained at 28°C for at least 6 wk. The fish were kept under a 12:12-h light-dark photoperiod and fed crushed pellets once daily. Six fish were used in each of four treatment groups: 21% oxygen (control), 5% oxygen (hypoxia), 3% oxygen (hypoxia), and 1.5% oxygen (hypoxia). Each fish was transferred into individual 6-liter containers at 28°C with aerated heated water maintained for 18 h for each specific oxygen treatment. Water was bubbled with 5% O2-95% N2 for the 5% hypoxic treatment group, 3% O2-97% N2 for the 3% hypoxic treatment group, and 1.5% O2-98.5% N2 for the 1.5% hypoxic group. Water was gradually brought from 21% (control) oxygen to the correct percent oxygen level for each hypoxia treatment over half an hour to allow the fish to acclimatize. A time-course experiment was also completed at 5% oxygen where the fish were held for 4, 8, 12, 18, 24, 30, 36, 42, 48, 54, 60, 66, and 72 h in the hypoxic treatment. All control fish were taken from the holding tank at the beginning of the hypoxia treatment, and tissues from all fish were collected in the same manner described above.
Green anoles (Anolis carolinensis, Voigt) were acquired from Boreal Scientific. They were kept in an enclosure at ambient temperature (23°C) and supplied with an infrared heat lamp and fresh water supply for 2 wk. The anoles were held under a 12:12-h light-dark photoperiod and fed live crickets every other day ad libitum. Hypoxia experiments were carried out in plastic 500-ml containers maintained at 23°C in an incubation chamber. Anoles were exposed to either normoxia or 4% O2-96% N2. Immediately following the experiment, animals were euthanized using an injection of pentobarbital (150 mg/kg), and heart, brain, and lung tissues were collected and frozen as described above.
Western painted turtle (Chrysemys picta bellii, Schneider) liver and pectoralis muscle samples were generously donated by Dr. Leslie Buck (University of Toronto). Tissues were dissected from individuals that were from a control group or had been submerged in freshwater for 24 h and immediately frozen in liquid nitrogen, as described previously (35).
Tissue Culture Experiments
Two fish cell lines and two mammalian cell lines were employed in this study. Fish cell lines were grown in Leibovitz's L-15 media (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) under regular atmospheric conditions. ZEB2J cells (Danio rerio epithelium) were maintained at 26°C and RTG-2 cells (Oncorhynchus mykiss, Walbaum gonadal fibroblasts) were maintained at 19°C. Mammalian cell lines were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and held at 5% CO2-95% humidity. Sol8 mouse (Mus musculus, Linnaeus) myoblast cells and LNCaP human prostate cancer cells were all maintained at 37°C.
Hypoxia and anoxia treatments were done using gas mixtures of 2% oxygen-98% nitrogen or 100% nitrogen, respectively, and lasted for 8 or 24 h. Control treatments were done under normal atmospheric conditions for 24 h. Cells were grown in six-well plates until they were ∼90% confluent. The media were then switched to L-15 with 10% FBS that had been preequilibrated to the appropriate temperature and oxygen levels. Hypoxia treatment plates were kept in an air-tight chamber containing the appropriate gas mixture and control treatment plates were left at normal atmospheric conditions. All treatments were carried out at the temperatures at which the cells were previously grown. Anoxia caused cell death in LNCaP cells after 24 h and in Sol8 cells after 8 h.
Cells were harvested immediately following these treatments by removing the media and scraping in RLT buffer from Qiagen's RNeasy kit (Valencia, CA) containing 1% β-mercaptoethanol. Cell suspensions were lysed by vortexing for ∼10 s and then stored at −80°C.
RNA Extraction and cDNA Synthesis
RNA was extracted from whole animal tissues with TRIzol reagent (Invitrogen), using 1 ml of TRIzol per 50 mg of tissue. Samples were homogenized in TRIzol with a tissue homogenizer (Fisher, Ottawa, ON, Canada) and then centrifuged at 12 000 g for 10 min at 4°C. The supernatant of each sample was collected and mixed with 0.2 ml of chloroform per 1 ml of TRIzol. Samples were incubated at room temperature for ∼3 min and then centrifuged at 12,000 g for 15 min at 4°C. The upper aqueous layer was collected and mixed with 0.5 ml of isopropanol per 1 ml of TRI-zol. After 5 min of incubation at room temperature, the samples were centrifuged at 12 000 g for 10 min at 4°C. The supernatant was removed, and an equal volume of 75% ethanol was used to wash the pellet. Samples were resuspended in RNase-free water and quantified using a spectrophotometer reading at 260 nm. The quality of the RNA samples was also assessed during the quantification using the 260/280 ratio.
Extraction of RNA from cell lysates was done using an RNeasy Mini kit (Qiagen). Purified RNA samples were quantified by reading absorbance at 260 nm.
RNA was reverse-transcribed to cDNA using the QuantiTect Reverse Transcription kit (Qiagen) with 1 μg of RNA in a 20-μl reaction, as per the manufacturer's instructions.
Quantitative PCR
Primers for quantitative (q)PCR were designed to amplify 50–200 bp (Table 2) in a manner that was linear with respect to template concentration and produced a single dissociation peak. When possible, primers were designed using sequences available online. For those genes measured in this study that lack a published sequence, primers were designed from consensus sequences of published genes of related species. While consensus primers served for qPCR in most instances, some turtle genes required partial sequencing using consensus primers before qPCR primers could be designed from these resulting partial sequences. Thus for turtle fragments the transcripts of COX4-2 and vascular endothelial growth factor A were amplified using consensus primers (COX4-2: forward 5′-CCCTGAGCAGAAAGCCCTGAAA-3′, reverse 5′-CATTCGTTCTTCTATAATCCCACTTG-3′; VEGFA: forward 5′-GCAGCTTCTGCAGGACAATTGA-3′, reverse 5′-GCAAGTGCGCTCGTTTAACTCA-3′). These larger fragments were amplified, cloned, and sequenced. To confirm that the amplified fragment was of the desired gene, the obtained sequence was aligned with the consensus sequences used to design the primers. qPCR primers were then designed from the resulting sequence fragments.
Table 2.
Primers used for quantitative PCR quantification of target mRNAs and for pDrive plasmids
| Species/Gene | Forward (5′-3') | Reverse (5′-3') | Tann, °C | Source |
|---|---|---|---|---|
| Zebrafish (Danio rerio) | ||||
| COX4-1 | CAAGTTTGTGCAGCAGCTG | CAAAGAAGAAGATTCCTGCAA | 63 | 29 |
| COX4-2 | CGACAGACCTTACAAAGACATCC | GAAGAATAAGATCCCAGCCGT | 63 | 29 |
| IGFBP-1 | ACGCAAGAAACTGGTGGAAC | GGGCTGTCTGGAGTTCGATA | 59 | 30 |
| VegFA | AGTATCCCGATGAGATCGAGCACA | CACCTCCATAGTGACGTTTCGTGT | 59 | — |
| β-Actin† | TCCAGGCTGTGCTGTCCCTGTA | GTCAGGATCTTCATGAGGTAGTC | 59 | 12 |
| RPL13A | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG | 59 | 40 |
| Goldfish (Carassius auratus) | ||||
| COX4-1 | AGCTTCGCTGAAATGGAAA | CCTCATGTCCAGCATCCTCT | 59 | 14 |
| COX4-2 | AAACTTGCCTTGTACAGGCTTACG | AAGAAGAAGATGCCGCCCAC | 56 | — |
| IGFBP-1 | TGGTTGGGCAGGAACCAATC | TCTCGAAGCACCTCCTCACA | 59 | — |
| EF-1α | CAGGTCATCATCCTGAACCAC | AACGACGGTCGATCTTCTC | 59 | 5 |
| TBP | AGTGGTGCAGAAGTTGGGTTTTCC | ATGTGSSGCACCAGGCCCTCTAA | 59 | 12 |
| Rainbow trout (Oncorhynchus mykiss) | ||||
| COX4-1* | CACATGGTGTTGCCAAGGTAGAC | GCGCTCAGCTCTGTCACAAACT | 63 | — |
| COX4-2* | ACCACGAGGTGTCCGACTCA | AGGACGTCCTTCCATGGTCTGT | 63 | — |
| VegFA | AGACAGCCCACATACCCAAG | AAGACGTCCACCAGCATCTC | 59 | — |
| EF-1α | TCCTCTTGGTCGTTTCGCTG | ACCCGAGGGACATCCTGTG | 59 | — |
| Tilapia (Oreochromis niloticus) | ||||
| COX4-1 | GCAACCAGAGCCCTAAGCCTTATT | CTCTGCACTCAGGTTCTGCACA | 67 | — |
| COX4-2 | GCTGAGGTAGCACAGTCTGGA | TTCCCTAGAGAGGCTCCATCTTTC | 67 | — |
| IGFBP-1 | AGGAGAGCATGAAAGCCAAA | TCCGTCCAGAGAGGATTCAC | 59 | — |
| Human (Homo sapiens) | ||||
| COX4-1 | GAGCAATTTCCACCTCTGT | CAGGAGGCCTTCTCCTTCTC | 59 | 14 |
| COX4-2 | ATTTCCTCCAAAGCCGATCAC | GAGACAGCTGGGGATGCAAGTCA | 65 | — |
| VegFA | AGCTACTGCCATCCAATCGAGA | TACACGTCTGCATGGTGATGTTGG | 59 | — |
| EF-1α‡ | ATTGATGCTCCTGGACACAGA | CAGCAACAATCAGGACAGCACAGT | 59 | — |
| Mouse (Mus musculus) | ||||
| COX4-1 | GCAGCCTTTCCAGGGATG | GCTCTTCTCCCAAATCAGA | 59 | — |
| COX4-2 | AGAACTAGAGGGACAGGGACACA | TCGCTGAGCTCTGTGCAGAA | 59 | — |
| VegFA | CCAAGGCCAGCAAATCCCTGTGGGCC | CCGCCTCGGCTTGTCACA | 59 | 8 |
| β-Actin | AGCACCATGAAGATCAAGATCAT | ATCCACATCTGCTGGAAGGT | 59 | 27 |
| Anole (Anolis carolinensis) | ||||
| COX4-1 | ATGGAGTGCTCTTTCGCTGG | AAAGCCACTGAGGCCAATGA | 59 | — |
| COX4-2 | AGTTATCCGCTGCCAGACAC | ACGGTCTTCCATTCGTTGCT | 59 | — |
| VegFA§ | TTCACAGACTCACGTTGCAAGTCG | GCAAGTGCGCTCGTTTAACTCAAG | 59 | — |
| Turtle (Chrysemys picta bellii) | ||||
| COX4-1* | CCAGAGAATGCTGGATATGA | CCATTCCTTCTTTTCGTAGTCCCA | 59 | — |
| COX4-2 | AAGAGAAGGAGAAGGGCTCATGGAAG | TTCGTTGGATGGCCTGTTCATCTC | 59 | — |
| pDrive-specific | CCGGGTTGGACTCAAGACGATAGTTA | ACGCTGTAGGTATCTCAGTTCGGT | 59 | — |
Tann, annealing temperature; see text for additional definitions.
Primers were designed from consensus sequences from closely related species.
Primer set was used for all other species listed except mouse.
Primer set was used for mouse, anole, and turtle as well.
Primer set was used for anole and turtle.
Cloning and sequencing were carried out for all primer sets used. They were first tested by PCR using an Eppendorf Mastercycler Gradient thermocycler (Eppendorf, Hamburg, Germany). All reactions were run in 25-μl volumes containing 1× PCR buffer, 1 mM MgCl2, 0.75 units Taq DNA polymerase (Qiagen), 0.4 μM deoxyribonucleotide triphosphates (dNTPs; Promega, Madison, WI), 0.3 μM of each primer, 50–100 ng of cDNA template, and the balance of nuclease-free water. PCR reactions involved a 3-min denaturation at 94°C followed by 35 cycles of 15 s at 94°C, 30 s at the appropriate annealing temperature, and 30 s at 72°C. There was a final extension period of 10 min at 72°C. PCR products were gel purified, ligated into the pDrive cloning vector (Qiagen), transformed into cloning-competent DH5α cells (Invitrogen), and grown on agar plates containing 0.05 mM IPTG, 0.2 mM bromo-chloro-indolyl-galactopyranoside (X-gal), and 50 μg/ml ampicillin. Colonies testing positive for inserts were grown overnight in Lysogeny Broth with 50 μg/ml ampicillin. Plasmids were purified using the QIAprep Spin Miniprep kit (Qiagen), quantified by spectrophotometric readings at 260 nm, and then sequenced (Robarts Research Institute, London, ON, Canada). Samples were sequenced with an Applied Biosystems 3730 Analyzer (Applied Biosystems, Carlsbad, CA) and aligned with published sequences to verify the amplification of the correct gene fragment.
Real-time quantitative PCR was performed on an ABI 7500 real-time PCR system (Applied Biosystems) using GoTaq qPCR Master Mix (Promega). Reactions were done in 25-μl volumes with 12.5 μl of the GoTaq Master Mix, 25–100 ng of cDNA template, 0.58 μM each of the forward and reverse primer, and the balance of water. qPCR reactions consisted of an initial 15-min denaturation at 95°C followed by 40 cycles of 15 s at 95°C, 15 s at the appropriate annealing temperature, and 36 s at 72°C. All samples were analyzed in duplicate, and each primer set was tested with a no-template control.
For every transcript level measured in qPCR, the threshold cycle (CT) of each cDNA sample was normalized to a housekeeping factor obtained from the geometric mean of the CT recorded for TATA-binding protein (TBP; goldfish only) or β-actin (all other species) and 60S ribosomal protein L13α (RPL13A; zebrafish only) or elongation factor-1α (EF-1α; all other species). Thus changes in CT levels for a particular transcript between samples from different treatments could be corrected for any changes due to the efficiency of cDNA synthesis. When comparing the relative amounts of COX4-1 and COX4-2 transcripts within single tissues, a further correction was made to ensure each primer pair was able to detect their target transcripts with a comparable accuracy. The correction factor was determined by comparing the CTs obtained for 1 pg of pDrive plasmid containing the appropriate insert using the target primers and pDrive specific primers (Table 2).
Transfection Experiments
To address the question of the hypoxia sensitivity of COX4-2 promoter elements across vertebrates, we used two approaches. First, we used plasmid constructs of tandem repeats of the human HRE1, HRE2, or ORE to assess which elements are hypoxia responsive in lower vertebrate taxa. Unfortunately, this approach limited our investigation to those species where the elements could be identified, notably excluding all teleost species. To broaden the investigative range of species, we then measured the ability of ∼2 kb of COX4-2 proximal promoters from candidate mammalian and teleost species to respond to hypoxia.
Element transfections.
Plasmid constructs were developed for human (Homo sapiens, Linnaeus), green anole, and Western clawed frog (Xenopus tropicalis, Wagler) and tested for hypoxic activity in a transfection experiment. The constructs consisted of four tandem repeats of each of the three putative elements responsible for the hypoxic response seen in mammalian COX4-2 (HRE1, HRE2, and ORE). Oligonucleotides of the tandem repeats were ordered, along with a universal reverse primer containing a MluI restriction site (Table 3). As a positive control, a tandem repeat of the human inducible nitric oxide synthase (iNOS) HRE (5′-GTGACTACGTGCTGCCTAG-3′) was also designed, as outlined in Table 3.
Table 3.
Element sequences used in transfection experiment
| Species | HRE1 (5′-3') | HRE2 (5′-3′) | ORE (5′-3′) | Accession No. |
|---|---|---|---|---|
| Human (Homo sapiens) | CTGTGCGCTCCCACGCC | CAGGCCTGTGTGCACGTA | GGACGTTCCCACGC | NG_012180.1 |
| Anole (Anolis carolinensis) | GTTTGCACTCTTTGGCC | CAGGCATGGGCCAACTTG | AGGCTATCACTCGC | NW_003339315.1 |
| Xenopus (Xenopus tropicalis) | ATTTTCGATACCACATA | CGGGTCTCTGGGCAGTAG | GGAGTTGCTCTAAT | NW_003171408.1 |
The plasmid construct for each element contained 4 tandem repeats of the element followed by a 12-bp sequence containing a MluI restriction site (5′-ACGCGTATCGAT-3′).
HRE, hypoxia-responsive elements; ORE, oxygen-responsive elements.
Double-stranded products were obtained through reactions using the Klenow fragment of DNA polymerase I and an Eppendorf Mastercycler Gradient thermocycler (Eppendorf, Hamburg, Germany). Reactions were run in 25-μl volumes containing 1× PCR NEBuffer 2 (50 mM NaCl, 10 mM Tris·HCl, 10 mM MgCl2, and 1 mM DTT), 12.5 units Klenow (New England Biolabs, Ipswich, MA), 0.4 μM dNTPs (iNtRON Biotechnology, Seongnam-Si, Gyeonggi-do, Korea), 20 mM each of an element oligonucleotide and the universal reverse primer, and the balance of nuclease-free water. The oligonucleotides were first allowed to anneal for 10 min before the addition of Klenow by decreasing the thermocycler temperature from 99 to 4°C at a rate of 0.3°C/s. The synthesis reaction was carried out at 25°C for 15 min, after which Klenow was inactivated by holding at 75°C for 20 min. To allow for the ligation of these elements into the target plasmid vector, the double-stranded products were phosphorylated by 2 units of T4 polynucleotide kinase per μg DNA (New England Biolabs).
The plasmid vector used in this experiment was the pGL4.23[luc2/minP] Vector (Promega), which contains a multiple cloning site upstream of a minimal promoter and the luciferase gene luc2. To prepare the vector for ligation with the element inserts, it was digested by 40 units of EcoRV per μg DNA (New England Biolabs). To prevent self-ligation of the vector, terminal phosphate groups were removed by 4 units of calf intestinal phosphatase per microgram of DNA (New England Biolabs). The resulting cut plasmid was electrophoresed in a 1% agarose gel, excised, and purified.
The ligation of the plasmid and inserts was performed with T4 DNA ligase (New England Biolabs). Plasmid constructs were then transformed into cloning-competent DH5α cells (Invitrogen) and grown on agar plates containing 0.13 mg ampicillin/l. Multiple colonies for each construct were grown overnight in LB broth with 50 μg ampicillin/ml. Plasmids were purified using the QIAprep Spin Miniprep kit (Qiagen), quantified by spectrophotometric readings at 260 nm, and tested for the presence of inserts by digestion reactions with MluI and XhoI followed by visualization on an agarose gel with ethidium bromide. Those plasmids showing inserts of the correct size were then sequenced as outlined in the qPCR section to verify the correct orientation of the insert.
Transfection experiments were carried out using the prostate cancer cell line PC3 in 24-well plates. Cells were grown and kept under the conditions outlined for the mammalian cell lines. When cells were confluent, the media were changed and a mixture of the experimental plasmid construct and a control pRL Renilla vector (Promega) was added to each well using the transfection agent FuGENE6.0 (Roche Applied Science, Laval, QC, Canada). Transfected cells were kept in an incubator under normal conditions for the control group or under 2% O2, 5% CO2, and the balance nitrogen for the hypoxia treatment. The anoxia treatment plates were incubated in an air-tight chamber containing 5% CO2 and the balance nitrogen. All plates were held at 37°C for the duration of the experiment.
Transfected cells were harvested after 24 h of treatment by incubating with 1× passive lysis buffer (Promega, Madison). The resulting cell suspensions were transferred to 1.5-ml tubes and frozen at −80°C for at least 12 h. Luciferase assays were carried out using a Dual-Luciferase Reporter Assay System (Promega) as per the manufacturer's instructions. Assays were measured using an Lmax Luminescence Microplate Reader (Molecular Devices, Sunnyvale, CA), and resulting values were determined relative to the control Renilla luminescence values for each sample.
Promoter fragment transfections.
Plasmid constructs containing ∼2 kb of sequence directly upstream of the first exon of COX4-2 were developed for rat (Rattus norvegicus, Fischer de Waldheim), goldfish, and zebrafish and tested for hypoxic activity in a transfection experiment. We used the human iNOS HRE construct developed for the element transfections as a positive control.
DNA was isolated from tissues collected from the hypoxia experiments for the teleost species; rat tissue was previously obtained from sentinel males from the animal care center at Queen's University. Samples were digested by proteinase K (0.2 mg/ml) in 200 mM NaCl, 20 mM Tris, 50 mM EDTA, and 0.10% SDS (pH 8.0), while incubating at 55°C. When tissues were completely dissolved, the samples were treated with RNase A (10 μg/ml). The DNA was extracted in a volume of phenol-chloroform-isoamyl alcohol (25:24:1) and centrifuged for 10 min at 1,700 g. The aqueous phase was reserved and DNA was precipitated by adding 0.1 ml of ammonium acetate (7.5 M) and 2 ml of 100% ethanol/ml of extraction buffer, followed by centrifugation for 3 min at 1,700 g. The pellet was washed with 70% ethanol, air-dried, and dissolved in 250 μl TE buffer (10 mM Tris·HCl and 1 mM EDTA, pH 8.0). The DNA was quantified, and the purity was assessed as described in RNA Extraction and cDNA Synthesis.
Promoter fragments a∼2 kb long immediately upstream of the 5′-untranslated region (5′-UTR) were amplified using the isolated DNA. Primers were designed from sequences published online, except in the case of goldfish, where the promoter region was amplified using the “RAGE” protocol, as described in by Bremer et al. (5). Primers contained unique restriction site adapters on the outer ends to allow unidirectional cloning directional cloning into reporter plasmids (Table 4). Initial amplification, cloning into pDRIVE, and verification sequencing were accomplished as described in Quantitative PCR. The 2-kb fragments were then excised out of the pDRIVE plasmids with the appropriate restriction enzymes (New England Biolabs), ligated into a luciferase reporter plasmid (pGL2 basic; Promega), and cloned and sequenced as described in Element transfections. The vector used for this experiment is similar to the one used in our previous transfections but lacks any native promoters.
Table 4.
Primers used for long promoter transfection experiment
| Species | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Rat (Rattus norvegicus) | ACTGCAACGCGTGGCCTTGAACTCAGT | TGCAGTCTCGAGGTCTAGTTCAGCACC |
| Goldfish (Carassius auratus) | ACTGCAACGCGTGCATTAGCTTTTGG | TGCAGTCTCGAGGGAGAGACTGTGCA |
| Zebrafish (Danio rerio) | ACTGCAACGCGTGGTTGCCACAGCGGA | TGCAGTCTCGAGAAACTGTGAGAGACTGTG |
Accession numbers for rat and zebrafish are the same as those listed in Table 1. The promoter sequence for goldfish was generated using a “RAGE” protocol as described in methods. Restriction sites are shown in bold.
Transfections were carried out using the human liver hepatocellular carcinoma cell line HepG2 in 12-well plates. The procedures followed for the transfection and luciferase assays were similar to that described in Element transfections. The only variations were the use of Renilla-TK vector (Promega) as the internal control and an oxygen level of 4% for the hypoxia condition.
Statistical Analysis
All data are presented as means ± SE and are expressed relative to the control group for each treatment. To determine significance between control and treatment groups, Mann-Whitney U-tests were used. Differences were considered significant if P < 0.05.
RESULTS
Mammalian Cells in Culture
We investigated two mammalian cell lines from two different species (mouse and human) representing two different tissue types (myoblasts and prostate adenocarcinoma cells), yet neither displayed the expected shift from transcription of COX4-1 to COX4-2 during exposure to hypoxia. The results of these experiments are shown in Fig. 1. As neither cell type was able to survive exposure to anoxia, only results from exposure to 2% oxygen are displayed.
Fig. 1.
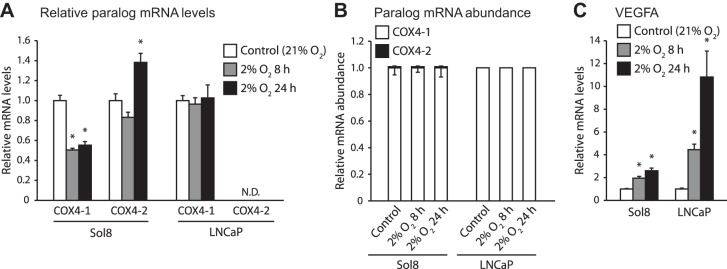
Transcriptional response to hypoxia in cytochrome c oxidase subunit 4 (COX4) paralogs in murine myoblast cells (Sol8) and human prostate cancer cells (LNCaP). Cells were exposed to 8 or 24 h of 2% O2; n = 6 per treatment. A: mRNA levels for COX4-1 and COX4-2, relative to control treatments. B: proportion of COX4 transcripts present as COX4-1 or COX4-2. C: relative mRNA levels for VEGFA, a known hypoxia-responsive gene. *Significant difference from the control treatment as determined by a Mann-Whitney U-test. Error bars depict SE.
The murine Sol8 cells exposed to hypoxia displayed significant increases of 2- to 2.6-fold in the mRNA for the hypoxia-responsive gene VEGFA (Fig. 1C). A significant decrease by half in COX4-1 mRNA levels was seen for both hypoxic treatments compared with the control group. There was a significant 1.4-fold increase in COX4-2 transcripts only after 24 h of exposure to hypoxia (Fig. 1A), but for all treatments, COX4-2 transcripts accounted for <0.1% of all COX4 transcripts (Fig. 1B).
As with Sol8 cells, LNCaP cells displayed significant increases of 4.4- to 11-fold in VEGFA mRNA levels when exposed to hypoxia (Fig. 1C). COX4-1 mRNA levels remained steady across treatment groups. Interestingly, although we were able to amplify fragments of the COX4-2 gene using LNCaP DNA (data not shown), we could not detect any COX4-2 transcripts under any condition, suggesting the COX4-2 paralog is not transcribed in this cell line (Fig. 1B).
Fish Tissues
Although we examined four species of teleost fish and employed approaches using both whole animals and tissue cultures, we found little indication that transcription switches from COX4-1 to COX4-2 during hypoxia treatments in teleosts (Fig. 2).
Fig. 2.
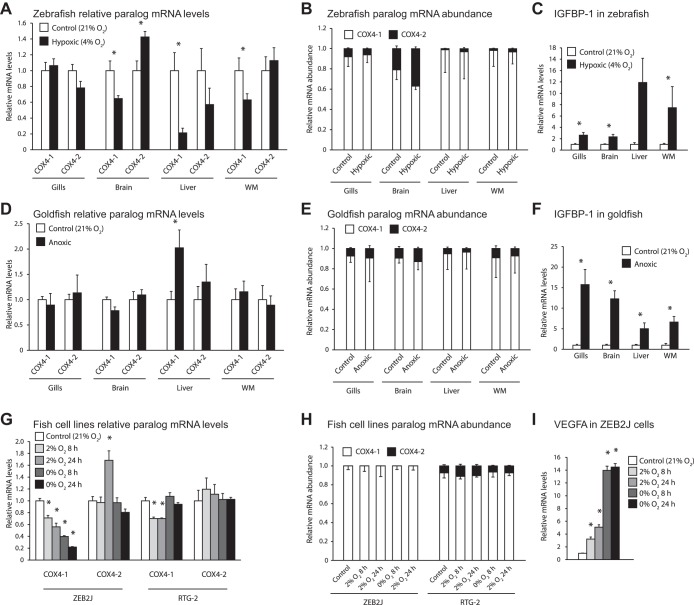
Transcriptional response of COX4 paralogs to hypoxic treatments in whole zebrafish and goldfish, as well as ZEB2J and RTG-2 cell lines. A–C: transcriptional response in gills, brain, liver, and white muscle (WM) of zebrafish exposed to 8 h of 4% O2; n = 8 per treatment. D–F: transcriptional response in the corresponding tissues of goldfish exposed to 21.5 h of anoxia (undetectable levels of O2). G–I: transcriptional response of zebrafish epithelial cells (ZEB2J) and rainbow trout gonadal fibroblasts (RTG-2) exposed to 8 or 24 h of 2% O2 or anoxia; n = 6 per treatment. A, D, and G: mRNA levels for COX4-1 and COX4-2 relative to control treatments. B, E, and H: proportion of COX4 transcripts present as COX4-1 or COX4-2. C, F, and I: relative mRNA levels for insulin-like growth factor binding protein-1 (IGFBP-1; whole zebrafish and goldfish) or VEGFA (ZEB2J), which are known hypoxia-responsive genes. No response was seen in any hypoxia-responsive gene we measured in RTG-2 cells (data not shown). *Significant difference from the control treatment as determined by a Mann-Whitney U-test. Error bars depict SE.
Adult zebrafish exposed to low oxygen levels displayed a hypoxia response at the transcript level, as indicated by the increases in the mRNA levels of the known hypoxia-responsive gene insulin-like growth factor binding protein-1 (IGFBP-1). With regards to the COX4 paralogs, brain, liver, and white muscle displayed significant decreases in COX4-1 mRNA levels, and there was a minor (40%) but significant increase in COX4-2 mRNA levels in zebrafish brain (Fig. 2A). When both paralogs are taken into account, however, COX4-1 remained well in excess of COX4-2 in all tissues during hypoxia, although in the brain COX4-2 accounted for almost 40% of the COX4 transcripts (Fig. 2B).
The highly hypoxia-tolerant goldfish required exposure to undetectable levels of oxygen to activate hypoxia response pathways, which we measured as significant increases in IGFBP-1 transcript levels in all examined tissues. No significant changes were seen for either COX4 paralog in any tissue, except for a twofold increase in COX4-1 transcripts in hypoxic liver tissues. The relative abundances of each COX4 paralog changed little between normoxia and hypoxia treatment groups. In agreement with transcript patterns seen in zebrafish, the highest abundance of COX4-2 was seen in brain tissues, although it remained between 9 and 13% of total COX4 transcripts (Fig. 2E).
Similar results were obtained for the experiments involving cultured fish tissues, also summarized in Fig. 2, G and H. ZEB2J cells showed dose- and time-dependent decreases in COX4-1 (Fig. 2G). COX4-2 mRNA levels remained unchanged except for a 70% increase in cells exposed to 2% O2 for 24 h. COX4-1 transcripts account for >99% of total COX4 transcripts across all treatment groups. In experiments with rainbow trout gonad (RTG-2) cells, COX4-1 mRNA levels decreased significantly by 40% with exposure to 2% O2 but did not change during anoxia. No significant changes were seen in COX4-2 mRNA levels for any treatment group. When taking both COX4 paralogs into account, COX4-1 accounted for between 88 and 94% of total COX4 transcripts (Fig. 2H).
While VEGFA transcripts increased significantly in ZEB2J cells with increasing severity of hypoxia treatment (Fig. 2I), no significant differences were seen VEGFA transcripts in RTG-2 cells. Given that the same incubation conditions resulted in a hypoxic response in ZEB2J cells, we believe the lack of response in RTG-2 cells reflects a different VEGF signaling pathway, rather than a failure to impose hypoxia.
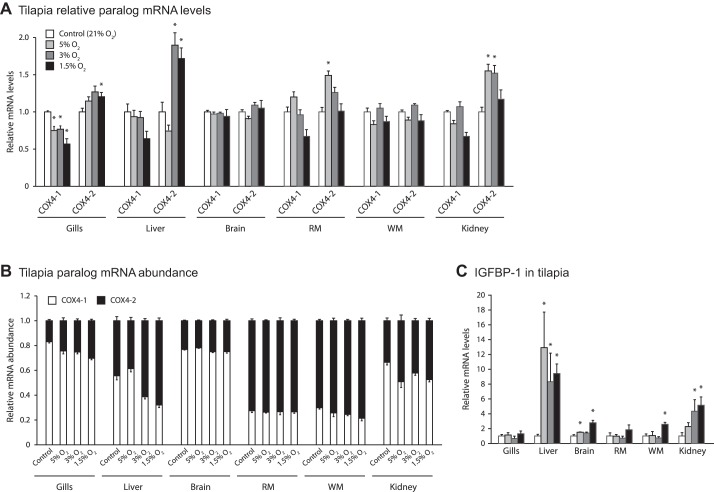
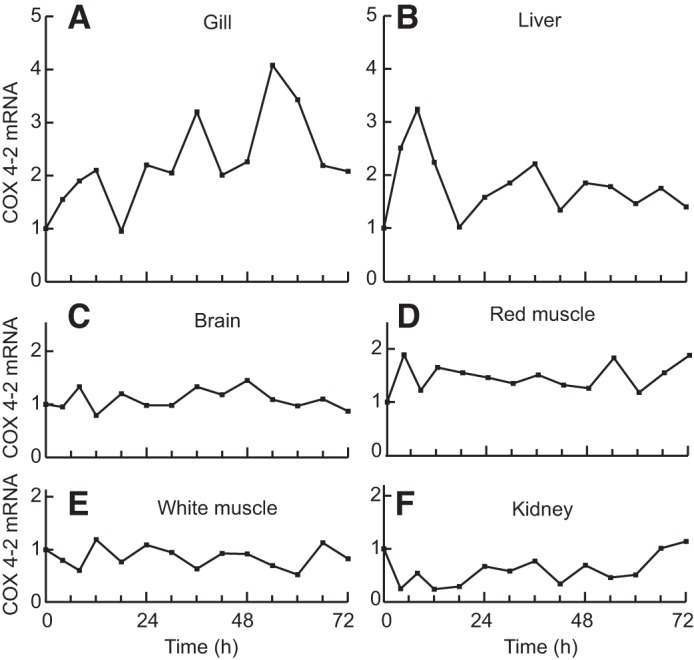
Since goldfish and zebrafish belong to the same family (Cyprinidae), we extended our studies to tilapia. We exposed tilapia to a range of hypoxic conditions and confirmed that hypoxia response pathways had been activated by measuring increases in IGFBP-1 mRNA levels in at least one tissue under each treatment (Fig. 3C). In hypoxia-treated tilapia, COX4 paralogs in liver and gill displayed slight decreases in COX4-1 mRNA levels, and there was a minor but significant increase in COX4-2 mRNA levels (Fig. 3A). COX4-2 increased 90% in the hypoxic liver tissue and 27% in the hypoxic gill tissue compared with the control treatment (Fig. 3A). White muscle, red muscle, kidney, and brain tissue were also examined and displayed no changes in mRNA transcript levels between COX4-1/COX4-2 during hypoxia. Minimal changes in COX4-1/2 were found in any of the tissues examined in the fish treated at 5% hypoxia over the time course of 0–72 h (Fig. 4).
Fig. 3.
Intertissue comparisons of the transcriptional response of COX4 paralogs to hypoxic treatments in tilapia. Transcriptional response in various tissues of tilapia exposed for 18 h to varying degrees of hypoxia; n = 6 per treatment, with exception of treatment group 1.5% where there was an n = 5 due to one mortality. A: mRNA levels for COX4-1 and COX4-2 relative to control treatments. B: proportion of COX4 transcripts present as COX4-1 or COX4-2. C: relative mRNA levels for IGFBP-1, a known hypoxia-responsive gene. *Significant difference from the control treatment as determined by a Mann-Whitney U-test. Error bars depict SE.
Fig. 4.
Time course of transcriptional response of COX4 paralogs to hypoxia in tilapia. Fish were exposed to hypoxia (5% O2) for 72 h, with single fish sampled throughout the time course. A: gill. B: liver. C: brain. D: red muscle. E: white muscle. F: kidney.
Reptile Tissues
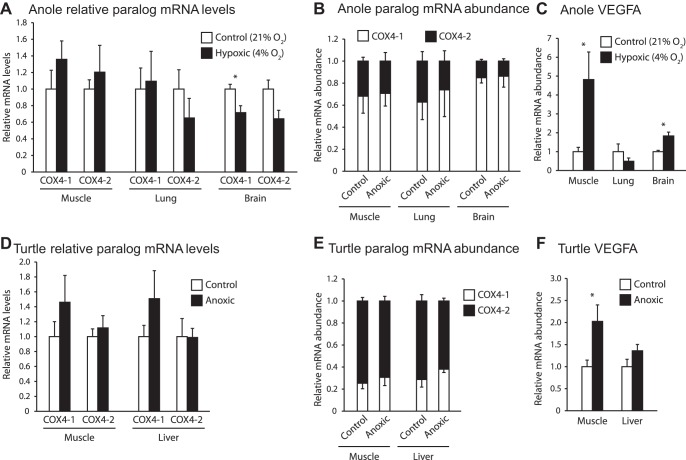
We also studied two reptiles, phylogenetically intermediate between mammals and fish. We failed to observe such a switch in either species we studied, although we found a uniquely high abundance of COX4-2 mRNA (Fig. 5).
Fig. 5.
Transcriptional response of COX4 paralogs to hypoxic treatments in reptilian species. A–C: response in muscle, lung and brain tissues of green anole exposed to 4% O2 for 24 h. D–F: response in muscle and liver tissues of Western painted turtles exposed to 24 h or anoxia; n = 6 per treatment. A and D: mRNA levels for COX4-1 and COX4-2 relative to control treatments. B and E: proportion of COX4 transcripts present as COX4-1 or COX4-2. C and F: relative mRNA levels for VEGFA, a known hypoxia-responsive gene. *Significant difference from the control treatment as determined by a Mann-Whitney U-test. Error bars depict SE.
Green anoles exposed to hypoxia for 24 h possessed VEGFA mRNA levels significantly higher than the control group in brain and muscle tissues, suggesting that a hypoxia response was occurring at the level of transcription (Fig. 5C). COX4 paralog mRNA levels remained largely unchanged, except for a significant 30% decrease in COX4-1 mRNA of brain tissues. COX4-2 transcript abundance ranged from 15 to 35% in all tissues examined (Fig. 5B).
Results of the turtle experiment were in agreement with our other findings. We found that although VEGFA increased significantly in hypoxic muscle tissue by twofold, no switch in either COX4 isoform was detected during exposure to hypoxia in either liver or muscle tissues (Fig. 5, D and E). As with anoles, turtles showed COX4-2 mRNA was higher than in fish, but in turtles COX4-2 was the dominant isoform under all treatments in both tissues, accounting for 60–75% of all COX4 transcripts (Fig. 5E).
Structure of the COX4-2 Promoter and Polypeptide
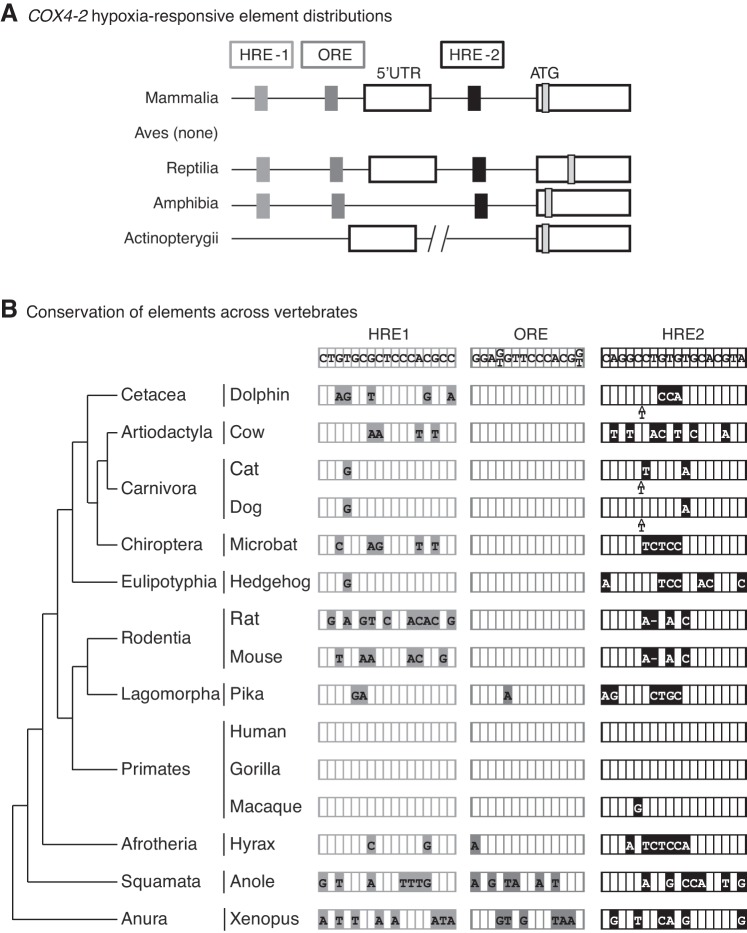
The promoter sequences of COX4-2 genes from a selection of vertebrate species were compiled, and the presence or absence of the putative oxygen-responsive elements was determined. The structures of vertebrate COX4-2 promoter regions as well as the results of our investigation are summarized in Fig. 6. Although homologous elements were identified in similar positions in the COX4-2 gene of tetrapods, none of the elements were recognized in the COX4-2 genes of any teleost species (Fig. 4A). In comparing elements across tetrapods, the ORE is better conserved across mammals than are either of the HREs (Fig. 6B). Single nucleotide differences are seen in the ORE for pika (Ochotona princeps, Link) of the order Lagomorpha and hyrax (Procavia capensis, Pallas) of the order Afrotheria. The HREs are conserved only within the primate family, although there is a single nucleotide substitution from C to G in macaques (Macaca sylvanus, Linnaeus). Outside of Mammalia, the elements are poorly conserved. Of the three elements, HRE2 is the most conserved in Xenopus and anole: 61 and 67% homology, respectively, for HRE2 compared with 53–59% for the other elements.
Fig. 6.
Comparison of putative hypoxia-responsive elements (HREs) in the COX4-2 gene across vertebrates. A and B: human sequences for the 2 HREs and the oxygen-responsive element (ORE) were used to search for orthologous elements in other vertebrate species representing several different lineages. A: location of the 3 elements in relation to the 5′-untranslated region (5′-UTR) and the ATG start site (ATG) of COX4-2 is shown for major classes of vertebrates. B determined elements are presented in relation to the vertebrate phylogeny. Differences from the consensus (human) sequence are indicated by filled boxes containing the alternative nucleotide or deletion (represented by a dash).
The polypeptide sequences of COX4-2 from vertebrate species were also examined to look for the conservation of two cysteine residues present in human COX4-2 (Cys-16, Cys-30). These are thought to form a disulfide bond, blocking an ATP-binding site seen in COX4-1 (23, 25). The two Cys residues are seen in the Euarchontoglires lineage that includes rodents, primates, lagomorphs, scandentians, and dermapterans (Fig. 7). Within this lineage, one of the Cys residues is absent in select species of rodents (sciuridae: ground squirrel, hystriocogathia: guinea pig) and primates (marmoset and galago). Outside Euarchontoglires, one of the Cys residues is replaced with either tyrosine or glycine. Likewise, nonmammalian vertebrates show either one or no Cys residue at this location. The amino acids that form the ATP-binding site were highly conserved even if the cysteine residues that disrupt it were not. However, in Xenopus, an insertion moves the conserved E-101, W-102 residues, and in fish, the first polar (R) residue shared in tetrapods is nonpolar (L or V).
Fig. 7.
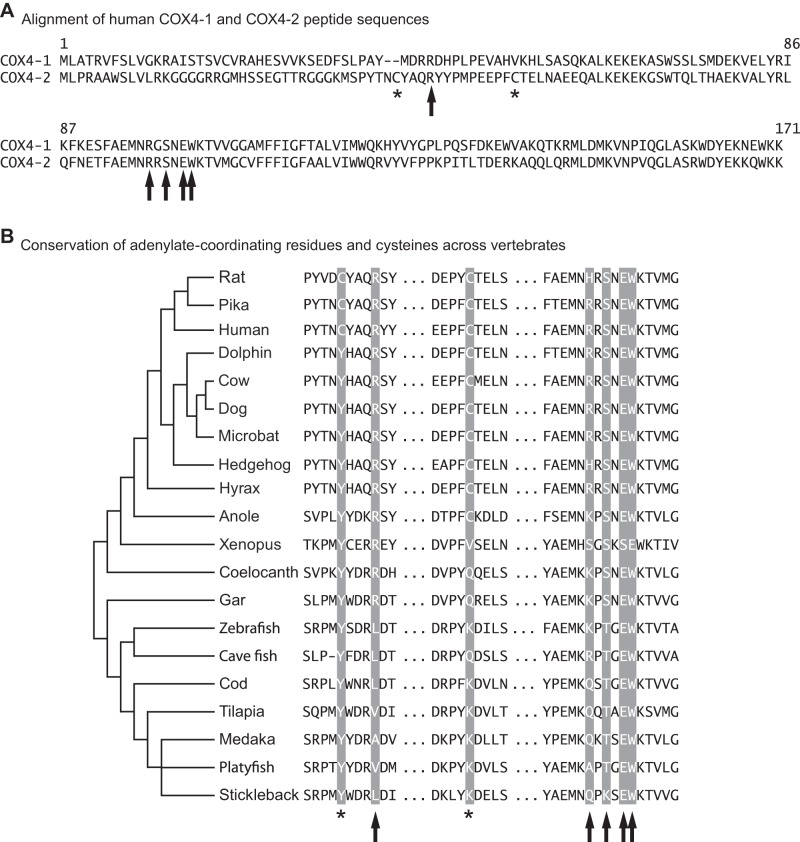
Conservation of features in vertebrate COX4-2 peptides. A: alignment of human COX4-1 and COX4-2 amino acid sequences. The COX4 amino acids suggested to be involved in the ATP/ADP binding site are denoted by arrows (Arg-20, Arg-73, Tyr-75, Glu-77, and Trp-78), and the COX4-2 cysteines that may disrupt the site are denoted by asterisks (Cys-16 and Cys-30). B: conservation of COX4-2 adenylate-coordinating amino acids and cysteines across vertebrates. Species names and accession numbers for each polypeptide sequence used can be found in Table 1.
Reporter Analyses of the COX4-2 Promoter
Element transfection.
The three putative elements in the COX4-2 promoter were developed into reporter constructs in four tandem repeats and analyzed for their response to hypoxia in a reporter experiment. The elements from human, anole, and Xenopus were used in the experiment. Relative to the normoxic control, the well-characterized HRE from the human iNOS gene responded to 24 h of hypoxia and anoxia by increasing 11- and 15-fold, respectively. Of the COX4-2 elements, only the human HRE2 was seen to respond to the oxygen treatments: 8-fold in hypoxia, 12-fold in anoxia. The other element constructs were unaffected by oxygen (Fig. 8A).
Fig. 8.
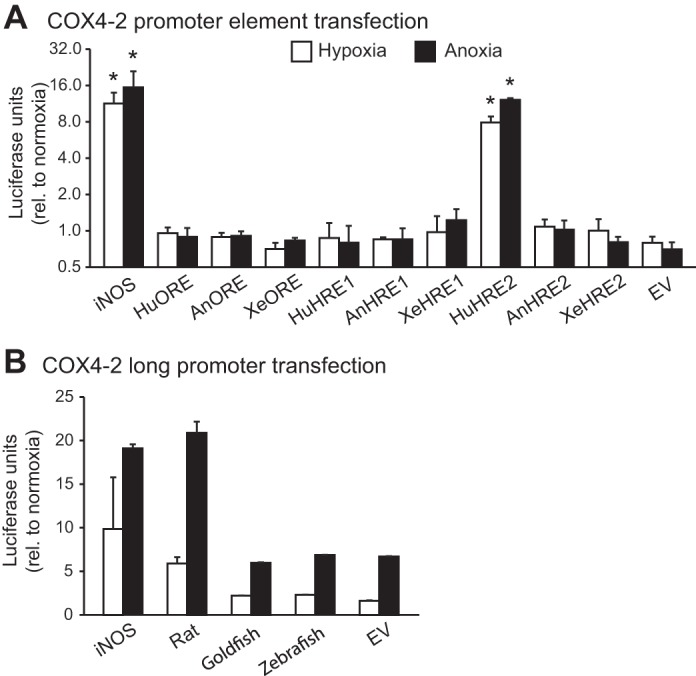
Reporter gene activities for luciferase (pGL4.23) under control of the COX4-2 promoter and hypoxia responsive elements from candidate vertebrate species. A: PC3 cells were transfected with constructs containing 4 tandem repeats of HRE1 and 2 and the ORE from human (Hu), anole (An), and Xenopus (Xe), and were exposed to normoxia, 2% O2 (hypoxia), or anoxia for 24 h. B: HepG2 cells were transfected with constructs containing ∼2 kb of promoter sequence directly upstream of the COX4-2 5′-UTR from rat, goldfish, and zebrafish and were treated in the same way as the PC3 cells for A. Also included in both experiments were constructs containing four tandem repeats of a known hypoxia-responsive element from the human iNOS gene and the baseline activities for the empty pGL4.23 vector (EV). *Significant difference from the control treatment as determined by a Mann-Whitney U-test. Results were normalized to cotransfected Renilla reporter activity levels and are presented relative to normoxic activities (+SE).
Long promoter transfection.
We used 2-kb segments of COX4-2 promoters from rat, goldfish, and zebrafish to produce plasmid constructs which we tested in normoxic, hypoxic, and anoxic conditions. The human iNOS HRE construct was used as a positive control and increased by 10-fold under hypoxic treatment and 19-fold under anoxic treatment. Of the long promoters, only the rat construct was activated more than the empty vector, by 6-fold and 21-fold under hypoxia and anoxia, respectively. The effects of hypoxia on the goldfish and zebrafish promoter constructs under hypoxic and anoxic treatments were indistinguishable from the response of the empty vector and thus may be related to effects on luciferase degradation rather than synthesis (Fig. 8B).
DISCUSSION
COX regulation plays a central role in the oxidative phosphorylation regulation, and it is well accepted that the nuclear encoded genes play a major role in the regulation of COX function in mammals. COX4 has been of particular interest because the genetic paralogs of this subunit appear to be oxygen sensitive, and their peptides cause the ATP turnover rates of the COX complex to differ by almost twofold (14, 22, 25).
COX4-1 is the ubiquitously transcribed paralog under normoxic conditions, while transcription switches to COX4-2 during bouts of hypoxia. The presence of COX4-2 in the COX complex appears to benefit mammalian tissues by allowing the COX complex to function at high turnover rates regardless of the current energy demands of the cell, thus decreasing the production of mitochondrial ROS under low oxygen conditions. These unique functions are thought to be conferred by the two cysteine residues that preclude ATP-mediated inhibition of COX activity at this subunit. The transcriptional regulation of the COX4-2 paralog is believed to be controlled by some combination of three putative oxygen-sensitive elements present in the gene (termed HRE1 and 2, and ORE) (14, 25). Uncertainties remain about the COX4 paralog function and mechanism even in mammals, but we were interested the evolution of the protein and gene regulation more broadly in vertebrates.
Sensitivity of COX4 Paralogs to Hypoxia
The hypoxia-responsive switch in COX4 paralogs was first reported in 2007, and several shortfalls in the collective knowledge of this mechanism remain. To date, the transcriptional response of COX4 paralogs to hypoxia has only been reported for one whole animal model (mouse), a select few number of human cancer cell lines, and most recently in human leg muscles (14, 22, 38, 9). Within the mouse hypoxia experiments, a transcriptional response in COX4-2 was seen in only lung and liver out of the five tissues tested, and only one of two types of brain cells when mouse primary cell cultures were examined (14, 22). The use of human cancer cell lines, while common in functional studies, allows for the possibility of generating results which are not characteristic of whole organisms, as these cells have been immortalized through some combination of genetic and epigenetic alterations. The small number of models that have been used to measure this mechanism raises questions about the pervasiveness of the COX4 hypoxia response across mammalian taxa and tissues, as well as other vertebrate lineages. To address this, we used both cellular and whole animal approaches to assess the transcript levels of COX4-1 and COX4-2 in candidate species of fish, reptiles, and mammals. Overall, we found little evidence of the hypoxia-induced switch from COX4-1 to COX4-2 in any of the nonmammalian models, suggesting that hypoxia responsiveness seen in rodents and humans is a derived trait.
COX4 transcriptional response in mammalian cells.
Previous studies by other groups have involved many cell lines that respond to hypoxia with an increase in COX4-2 mRNA, including rat pulmonary smooth muscle cells (1), mouse astrocytes (22), and cell lines including human lung H460 cells (25), human embryonic kidney 293 cells (1), human cervical cancer HeLa (14), human liver cancer Hep3B (14), human colon cancer Hct116 cells (14), and human lung cancer A594 cells (14). The impression from these studies is that the COX4-2 response is common within cultured cells, although variability in the response between cells lines is reported (1, 22). Of the human cell types previously used in studies on COX4 hypoxia responsiveness, all have displayed a pronounced increase in COX4-2 transcription during hypoxia, and all but one, a primary culture of pancreatic tissue, have been derived from tumors (14, 38). However, tumor cells differ in their dependence on hypoxia, and even lineages of tumor cells can evolve in culture (19). For many tumors, evolution in response to hypoxia in vivo can alter their metabolism, and it has been suggested that COX4 paralog switching may play a role in hypoxic metabolism and tumor survival (15, 39).
In the course of exploring the utility of cell culture models for transfections, we noted that several common models showed no response in COX4-2. Mouse muscle cells (Sol8) expressed COX4-2 and showed a 40% increase in COX4-2 (Fig. 1A), but the fraction of COX4-2 transcripts present in the COX4 pool amounted to <0.1%. With human prostate cancer cells (LNCaP), there was no evidence that the gene was expressed at all (Fig. 1A), although the same primers detected the presence of the gene.
Another tissue of interest is striated muscle, given the importance of oxygen in metabolism and adaptive remodeling. Fukuda et al. (14) similarly found no increase in COX4-2 mRNA levels in the cardiac muscle of adult mice exposed to hypoxia, although whether this achieved tissue hypoxia is unclear. Thoroughbred horses (Equus ferus caballus, Linnaeus) were found to decrease the transcription of COX4-2 in response to bouts of sprint exercise (20). Nonetheless, a recent study by Desplanches et al. (9) reported that COX4-2 transcription increases in the vastus lateralis of adult human males after endurance training under hypoxic conditions. Whether the differences in response are linked to species, striated muscle type (cardiac vs. skeletal), or skeletal fiber type remains unclear but collectively, these findings highlight the variability in the responsiveness of COX4-2. When responses are seen, it is not clear if the driving force is hypoxia directly, or whether the changes are driven by other signals. Furthermore, none of the studies have assessed whether the changes in muscle lead to changes in COX subunit composition or function.
COX4 transcriptional response in fish.
We were interested in the COX4 paralog hypoxia response in fish for two reasons. First, fish possess a COX4 isoform pair that is orthologous to those seen in mammals; the COX4 paralogs appear to have arisen from an ancient whole genome duplication event which occurred before the divergence of Actinopterygians and tetrapods. We have previously found these genes to possess similar tissue-specific transcription patterns to the mammalian COX4 paralogs, although COX4-2 transcripts generally contributed a greater fraction to the COX4 pool in fish than mammals (29). Second, many fish reside in environments with highly variable oxygen content, making them necessarily more tolerant to hypoxia than most tetrapods. To investigate the pervasiveness of the hypoxic switch in COX4 paralogs across teleost fish, we employed species that differ markedly in hypoxia tolerance (the highly intolerant rainbow trout, moderately tolerant zebrafish, and highly tolerant goldfish and tilapia) and used both cellular and whole animal-based approaches. The results suggest that the COX4-1/COX4-2 switch seen during hypoxia in some mammalian tissues is not present in the teleost lineage.
We studied three species of fish, including both hypoxia-tolerant and -sensitive species, and subjected them to a range of hypoxia levels (befitting their tolerance) and various times of sufficient duration to capture changes in mRNA levels. For the most part, there were very few instances of a hypoxic response, and where statistically significant responses were seen, the response was rarely >50%, and there was no tissue that typically responded. For example, brain showed a 40% increase with hypoxia in zebrafish (Fig. 2A), but no response in tilapia (Fig. 3A) or goldfish (Fig. 2D). In liver, tilapia (Fig. 3A) responded with a 90% increase in COX4-2 mRNA, but neither goldfish (Fig. 2D) nor zebrafish (Fig. 2A) responded.
Another feature that differed among fish was the abundance of COX4-2 mRNA, expressed relative to COX4-1 mRNA. Mammals generally show relatively low levels of COX4-2/COX4-1, with lung as the only tissue where COX4-2 levels approach COX4-1 levels (23). We had expected gill to have the highest relative expression of COX4-2, given its functional homology with mammalian lung; however, this was not found. Zebrafish and goldfish had generally low levels of COX4-2 mRNA in most tissues, but the brain stood out with relatively high COX4-1 mRNA. Furthermore, tilapia was unlike the two minnow species in showing very high levels of COX4-2 mRNA in all tissues (Fig. 3B). The physiological significance of these differences in tissue abundance between mammals and fish and among fish species remains to be established. What is clear though is that even in tissues that showed a modest but statistically significant increase in COX4-2 mRNA in hypoxia, the changes are even less impressive when the relative abundance of COX4-2 to COX4-1 mRNA is considered.
Although our positive controls showed that there was sufficient oxygen stress to initiate a genetic response, the depth of hypoxia in a given tissue is uncertain because of the capacity for physiological changes in oxygen delivery. Thus we sought to augment these whole animal studies using cultured cells. In zebrafish ZEB2J cells, hypoxia led to an increase (70%) in COX4-2 mRNA levels in only one of four combinations of oxygen and duration (24 h at 2% O2; Fig. 2G). However, this effect is reduced when it is considered that the relative abundance of COX4-2 transcripts is extremely low, <0.1% of COX4-1 mRNA levels (Fig. 2H). In rainbow trout gonad cells, the relative abundance of COX4-2 mRNA was higher than in zebrafish cells but it showed no response to hypoxia or anoxia (Fig. 2G).
To better understand the control of the COX4-2 gene in fish, we created promoter constructs under the control of the proximal promoters from the COX4-2 genes from both goldfish and zebrafish and compared the response to a rat COX4-2 promoter. Although we observed a significant increase in luciferase activity driven by the rate proximal promoter, the response seen for the COX4-2 promoters from fish was similar to the empty vector with minimal promoter (Fig. 8B). Combined with our transcriptional data, there bulk of the evidence suggests that COX4-2 is not hypoxia responsive in teleost fish.
COX4 transcriptional response in reptile tissues.
To bridge the gap in the hypoxia responsiveness of COX4 paralogs between teleost fish and mammals, we looked at tissues of hypoxic and normoxic Western painted turtles and green anoles. Reptiles are ectotherms and are able to survive more extreme bouts of hypoxia than other tetrapods through behavioral lowering of body temperatures, resulting in lowered metabolic rates (18, 21). This class also possesses COX4 paralogs that are orthologs of those found in mammals (29).
Green anoles have become a popular model for laboratory-based research since becoming the first reptile species to have its genome published. In our study, we found no transcriptional response in COX4-2 to 24 h of exposure to hypoxia (Fig. 5A). Interestingly, COX4-2 accounted for a higher proportion of COX4 transcripts in anole tissues compared with the mammalian and fish tissues we looked at (15% in brain to 35% in lung; Fig. 5B).
The freshwater turtle group to which the Western painted turtle belongs is remarkably tolerant to anoxia, being obligate air breathers that are able to survive anoxia for up to 5 mo, and is often used to study the cellular mechanisms of hypoxia response in higher vertebrates (26). While we failed to see any significant changes in the mRNA levels of the COX4 paralogs when exposed to anoxia, we found that COX4-2 was the predominant transcript in both tissues (Fig. 5E). COX4-2 transcripts were two- to fourfold more abundant than COX4-1 under both normoxic and hypoxic conditions. Although anoles showed a higher COX4-2 abundance than other species, the trend is more extreme in turtles. With only two reptiles studied, the evolutionary origin of the greater expression in turtles is unclear, but it is consistent with the superior ability to survive hypoxia in this species. Increased constitutive expression of COX4-2 in turtles could create a COX enzyme that is better suited to hypoxic metabolism, a condition that could arise at any point in the daily life of the animal. This argument is predicated on the assumption that COX4-2 confers kinetic advantages to COX in hypoxia, which has not been shown experimentally in nonmammalian vertebrates. The isoform profile would have little consequence to anoxic metabolism given that any kinetic properties would only manifest if there was flux through the enzyme.
When energetically challenged through hypoxic exposure, reptiles conserve energy via a coordinated downregulation of both ATP producing and consuming processes within their tissues. In Western painted turtles, this results in a decrease in energy metabolism by at least 49% (6, 10). During the hypoxic stress and contrary to what occurs in mammalian tissues, ROS production in the brains of Western painted turtles decreases during bouts of anoxia compared with normoxia (33). While this may be due simply to the global decrease in oxygen availability, it also could be caused in part by the greater reliance on COX4-2, particularly during the transitioning between normoxia and anoxia, when tissues would be experiencing hypoxic conditions. In mammalian tissues, the presence of COX4-2 lowers ROS production during hypoxia, but increases ROS production during normoxia, relative to complexes containing COX4-1 (14). Anoxia-tolerant turtles, and by extension other reptile species, may have made a trade-off between increased ROS production under normoxic conditions and decreased production during hypoxic conditions as a result of their frequent exposure to low levels of oxygen.
This is the first study to our knowledge that has measured the normoxic levels of COX4 paralogs or the response of COX4 paralogs to hypoxia in reptiles, and we have only examined four tissue types from two species. Furthermore, we were only able to sequence a portion of each COX4 gene in Western painted turtle, so it is yet unknown if they possess similar gene and polypeptide structures to their mammalian counterparts, which may give some indication of the functional consequences each isoform has on the COX complex. It cannot be ruled out that the switch in the dominant COX4 paralog to COX4-2 found in turtles may have arisen from an alternative evolutionary subfunctionalization of the COX4 paralogs. That is, the COX4 paralogs in turtles and other reptiles may have evolved to both serve the same function in the COX complex, or they may have evolved to preform complimentary functions that differ from other vertebrates.
The finding that COX4-2 replaces COX4-1 transcription during hypoxic stress in mammalian tissues has led to the hypothesis that this occurs as a cellular survival mechanism to combat increased ROS production. We were interested in determining if this mechanism is widespread across vertebrates. Our results indicate that this may be a unique mammalian feature and, furthermore, that it may be limited to a select number of species or tissue types within the mammalian lineage. COX4-1 transcript levels remained dominant during hypoxia in all fish tissues we examined, from both whole animals and cultured cells. Similar results were obtained for cultured cells from mouse muscle and human prostate cancer. Additionally, no functional COX4-2 gene is found in the Avian class (29).
Evolution of COX4-2 in Vertebrates
Because of the presence of orthologous COX4 paralogs across most vertebrate lineages, we hypothesized that the hypoxia responsiveness of COX4-2 would be pervasive in vertebrates. However, our transcriptional data suggest that this is not the case. To gain a better understanding of the function and regulation of COX4-2 in mammals and lower vertebrates, we examined the evolution of the COX4-2 gene and polypeptide across vertebrates.
Regulatory elements in COX4-2.
The regulation of the COX4 paralogs has proven difficult to ascertain. One study concluded the oxygen sensitivity of COX4-1 and COX4-2 was due to the well-defined HIF-1 pathway. The authors showed that COX4-2 transcription was activated through the binding of HIF-1 to a pair of HREs while COX4-1 proteins were selectively degraded through the action of the protease LON, which is also activated through the HIF-1 pathway (14). A second study argued that COX4-2 is regulated by an unknown pathway independently of HIF-1. This group was able to show that the COX4-2 promoter was activated not by its putative HREs but by an element they termed the ORE (25). Although our studies do not address which of these contradictory conclusions is valid, we were able to explore the evolution of the elements in vertebrates.
We compared the promoters of the COX4-2 gene from a wide range of taxa to determine the conservation of the two HREs and the ORE across vertebrates. The first HRE is present in the proximal promoter, the second is present within the first intron of the gene, and the ORE is found slightly downstream of HRE1 within the proximal promoter (Fig. 6A). None of the putative elements were completely conserved across mammals, although the ORE was far more conserved than either HRE (Fig. 6B). This suggests that the ORE is of more regulatory importance than the HREs. Outside of the mammalian class, there was little conservation of the three elements. No recognizable sequence homology for any element could be found in any teleost species, and the elements found in amphibian and reptilian species were as low as 41% conserved compared with the human element sequences. In doing the transfection work in mammalian cells, we do assume that the mammalian DNA-binding proteins have the same sequence preference as their nonmammalian homologs (11).
To test the ability of the three HREs to activate COX4-2 during exposure to hypoxia, we conducted reporter gene experiments. This allowed for the elimination of the cell type- and species-specific differences discussed above, placing the reporter constructs containing the putative elements in a common context. We first compared the efficacy of the hypoxia response of the two HREs and the ORE from the human, anole, and Xenopus COX4-2 genes to a well-known HRE from the human iNOS gene. Only the human HRE2 reporter construct displayed a response to the hypoxic and anoxic treatments comparable to the iNOS HRE response (Fig. 8A). Furthermore, the ORE is the most conserved across mammals while the HRE2 tends to be the least conserved, although HRE2 was more conserved relative to the human sequence in anole and Xenopus than the other elements (61 and 67%, respectively, for HRE2 compared with 53–59% for the other elements; Fig. 6B). Our results support the hypothesis (14) that these HRE elements are responsible for the hypoxia responsiveness of COX4-2. Although the machinery for a HIF-1 response was present in these cells, as indicated by the iNOS and HRE2 responses, we saw no response to similar constructs based on the first HRE site.
The possibility of false negative results in our transfection experiments cannot be excluded. The cell lines used may have lacked the necessary factors for the induction of ORE response.
Polypeptide structure of COX4-2 in vertebrates.
The functional significance of COX4-2 is believed to be an escape from inhibition by ATP at this subunit and an increase in turnover rate of the enzyme (22). This is thought to be due to the presence of two cysteine residues in the peptide sequence of COX4-2. These cysteines may form a disulfide bond in the matrix ATP-binding site of the subunit, which prevents the binding of adenylates (25). We examined the COX4-2 polypeptide sequences from a selection of lineages to determine the conservation of the cysteine pair and the conservation of the residues that coordinate ATP binding across vertebrates.
We found that the two cysteine residues were only found in mammals, and even within mammals they are not universally conserved (Fig. 7). Although mammals outside the rodent/primate lineage appear to lack the capacity to form the disulfide bond that disrupts an otherwise functional ATP-binding site, other taxa appear to have mutations in the coordinating ligands, effectively disrupting ATP binding by another mechanism. The Xenopus COX4-2 protein appears to possess an insertion that shifts the conserved EW residues of the ATP-binding site. In fish, the first polar (R) residue shared in tetrapods is nonpolar (L or V). While these changes are likely to influence the ability of the protein to bind ATP at this site, the functional consequences of these structural variants remain to be established experimentally. It is worth noting that it remains possible that the changes in regulation of this subunit in rodents may depend more on reversible phosphorylation and less on the disulphide bridge (17).
In summary, although the importance of the COX4-2 gene paralogs is widely accepted in human and rodent models, and likely plays an important role in hypoxic metabolism, the pervasiveness of the genetic and protein adaptations do not seem ubiquitous and therefore the evolutionary origins are uncertain. There is some evidence of coevolution in that rodents and mammals may impair ATP binding via a disulfide bridge, whereas in amphibians and teleosts, mutations may disrupt ATP binding directly. The divergence in the COX4-2 promoter is also striking, with no evidence of hypoxia responsiveness in fish, but in both fish and reptiles' greater constitutive expression of COX4-2, with turtle showing COX4-2 expression to dominate over COX4-1. The transcriptional activation of COX4-2 in hypoxic conditions may be limited to select tissues within select mammalian species.
GRANTS
This study was funded by a Discovery Grant (to C. D. Moyes) from the National Sciences and Engineering Research Council Canada.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. conception and design of research; K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. performed experiments; K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. analyzed data; K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. interpreted results of experiments; K.M.K., K.R., D.S.P., J.M., and C.D.M. prepared figures; K.M.K. and C.D.M. drafted manuscript; K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. edited and revised manuscript; K.M.K., K.R., D.S.P., J.M., T.S., and C.D.M. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Niels Bols and Lucy Lui for providing the ZEB2J and RTG-2 cell lines, Les Buck for providing turtle tissues, and Gary Armstrong and Pierre Drapeau for providing wild-type zebrafish. We also thank Emily Boniferro, Katharina Bremer, and Rhiannon Campden for assistance with the cloning and sequencing of goldfish genes.
REFERENCES
- 1.Aras S, Pak O, Sommer N, Finley R, Hüttemann M, Weissmann N, Grossman LI. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res 41: 2255–2266, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold S. The power of life-cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 12: 46–56, 2012. [DOI] [PubMed] [Google Scholar]
- 3.Arnold S, Kadenbach B. The intramitochondrial ATP/ADP-ratio controls cytochrome c oxidase activity allosterically. FEBS Lett 443: 105–108, 1999. [DOI] [PubMed] [Google Scholar]
- 4.Bickler PE, Buck LT. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu Rev Physiol 69: 145–170, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Bremer K, Monk CT, Gurd BJ, Moyes CD. Transcriptional regulation of temperature-induced remodeling of muscle bioenergetics in goldfish. Am J Physiol Regul Integr Comp Physiol 303: R150–R158, 2012. [DOI] [PubMed] [Google Scholar]
- 6.Buck LT, Land SC, Hochachka PW. Anoxia-tolerant hepatocytes: model system for study of reversible metabolic suppression. Am J Physiol Regul Integr Comp Physiol 265: R49–R56, 1993. [DOI] [PubMed] [Google Scholar]
- 7.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehne N, Kerkweg U, Otto T, Fandrey J. The HIF-1 response to simulated ischemia in mouse skeletal muscle cells neither enhances glycolysis nor prevents myotube cell death. Am J Physiol Regul Integr Comp Physiol 293: R1693–R1701, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Desplanches D, Amami M, Dupré-Aucouturier S, Valdivieso P, Schmutz S, Mueller M, Hoppeler H, Kreis R, Flück M. Hypoxia refines plasticity of mitochondrial respiration to repeated muscle work. Eur J Appl Physiol 114: 405–417, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doll CJ, Hochachka PW, Hand SC. A microcalorimetric study of turtle cortical slices: insights into brain metabolic depression. J Exp Biol 191: 141–153, 1994. [DOI] [PubMed] [Google Scholar]
- 11.Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet 26: 468–475, 2010. [DOI] [PubMed] [Google Scholar]
- 12.Duggan AT, Kocha KM, Monk CT, Bremer K, Moyes CD. Coordination of cytochrome c oxidase gene expression in the remodelling of skeletal muscle. J Exp Biol 214: 1880–1887, 2011. [DOI] [PubMed] [Google Scholar]
- 13.Erecinska M, Wilson DF. Regulation of cellular energy metabolism. J Membr Biol 70: 1–14, 1982. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda R, Zhang H, Kim J, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129: 111–122, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 18: 165–173, 2008. [DOI] [PubMed] [Google Scholar]
- 16.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–699, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helling S, Hüttemann M, Ramzan R, Kim SH, Lee I, Müller T, Langenfeld E, Meyer HE, Kadenbach B, Vogt S, Marcus K. Multiple phosphorylations of cytochrome c oxidase and their functions. Proteomics 12: 950–959, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks JW, Wood SC. Temperature regulation in lizards: effects of hypoxia. Am J Physiol Regul Integr Comp Physiol 248: R595–R600, 1985. [DOI] [PubMed] [Google Scholar]
- 19.Higgins LH, Withers HG, Garbens A, Love HD, Magnoni L, Hayward SW, Moyes CD. Hypoxia and the metabolic phenotype of prostate cancer cells. Biochim Biophys Acta 1787: 1433–1443, 2009. [DOI] [PubMed] [Google Scholar]
- 20.Hill EW, Eivers SS, McGivney B, Fonseca RG, Gu J, Smith N, Browne J, MacHugh DE, Katz LM. Moderate and high intensity sprint exercise induce differential responses in COX4I2 and PDK4 gene expression in Thoroughbred horse skeletal muscle. Equine Vet J 42: 576–581, 2010. [DOI] [PubMed] [Google Scholar]
- 21.Hochachka PW, Lutz PL. Mechanism, origin, and evolution of anoxia tolerance in animals. Comp Biochem Physiol B Biochem Mol Biol 130: 435–459, 2001. [DOI] [PubMed] [Google Scholar]
- 22.Horvat S, Beyer C, Arnold S. Effect of hypoxia on the transcription pattern of subunit isoforms and the kinetics of cytochrome c oxidase in cortical astrocytes and cerebellar neurons. J Neurochem 99: 937–951, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Hüttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 267: 111–123, 2001. [DOI] [PubMed] [Google Scholar]
- 24.Hüttemann M, Lee I, Gao X, Pecina P, Pecinova A, Liu J, Aras S, Sommer N, Sanderson TH, Tost M, Neff F, Aguilar-Pimentel JA, Becker L, Naton B, Rathkolb B, Rozman J, Favor J, Hans W, Prehn C, Puk O, Schrewe A, Sun M, Höfler H, Adamski J, Bekeredjian R, Graw J, Adler T, Busch DH, Klingenspor M, Klopstock T, Ollert M, Wolf E, Fuchs H, Gailus-Durner V, Hrabě de Angelis M, Weissmann N, Doan JW, Bassett DJ, Grossman LI. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J 26: 3916–3930, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hüttemann M, Lee I, Liu J, Grossman LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J 274: 5737–5748, 2007. [DOI] [PubMed] [Google Scholar]
- 26.Jackson DC. Hibernating without oxygen: physiological adaptations of the painted turtle. J Physiol 543: 731–737, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocha KM, Genge CE, Moyes CD. Origins of interspecies variation in mammalian muscle metabolic enzymes. Physiol Genomics 43: 873–883, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Little AG, Kocha KM, Lougheed SC, Moyes CD. Evolution of the nuclear-encoded cytochrome oxidase subunits in vertebrates. Physiol Genomics 42: 76–84, 2010. [DOI] [PubMed] [Google Scholar]
- 30.Marques IJ, Leito JT, Spaink HP, Testerink J, Jaspers RT, Witte F, van den Berg S, Bagowski CP. Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. J Comp Physiol B 178: 77–92, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Napiwotzki J, Kadenbach B. Extramitochondrial ATP/ADP-ratios regulate cytochrome c oxidase activity via binding to the cytosolic domain of subunit IV. Biol Chem Hoppe Seyler 379: 335–339 1998. [DOI] [PubMed] [Google Scholar]
- 32.Napiwotzki J, Shinzawa-Itoh K, Yoshikawa S, Kadenbach B. ATP and ADP bind to cytochrome c oxidase and regulate its activity. Biol Chem Hoppe Seyler 378: 1013–1021, 1997. [DOI] [PubMed] [Google Scholar]
- 33.Pamenter ME, Richards MD, Buck LT. Anoxia-induced changes in reactive oxygen species and cyclic nucleotides in the painted turtle. J Comp Physiol B 177: 473–481, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Poyton RO, Trueblood CE, Wright RM, Farrell LE. Expression and function of cytochrome c oxidase subunit isologues. Modulators of cellular energy production? Ann NY Acad Sci 550: 289–307, 1988. [DOI] [PubMed] [Google Scholar]
- 35.Ramaglia V, Buck LT. Time-dependent expression of heat shock proteins 70 and 90 in tissues of the anoxic western painted turtle. J Exp Biol 207: 3775–3784, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys 23: 331–366, 1990. [DOI] [PubMed] [Google Scholar]
- 37.Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 24: 97–106, 2009. [DOI] [PubMed] [Google Scholar]
- 38.Shteyer E, Saada A, Shaag A, Al-Hijawi FA, Kidess R, Revel-Vilk S, Elpeleg O. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am J Hum Genet 84: 412–417, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh KK, Russell J, Sigala B, Zhang T, Williams J, Keshav KF. Mitochondrial DNA determines the cellular response to cancer therapeutic agents. Oncogene 18: 6641–6646, 1999. [DOI] [PubMed] [Google Scholar]
- 40.Tang R, Dodd A, Lai D, McNabb WC, Love DR. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin (Shanghai) 39: 384–390, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272: 1136–1144, 1995. [DOI] [PubMed] [Google Scholar]
- 42.Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in human cells. Free Radic Biol Med 29: 202–210, 2000. [DOI] [PubMed] [Google Scholar]