Abstract
Chronic stimulation of β-adrenergic receptors (βAR) can promote survival signaling via transactivation of epidermal growth factor receptor (EGFR) but ultimately alters cardiac structure and contractility over time, in part via enhanced cytokine signaling. We hypothesized that chronic catecholamine signaling will have a temporal impact on cardiac transcript expression in vivo, in particular cytokines, and that EGFR transactivation plays a role in this process. C57BL/6 mice underwent infusion with vehicle or isoproterenol (Iso) ± gefitinib (Gef) for 1 or 2 wk. Cardiac contractility decreased following 2 wk of Iso treatment, while cardiac hypertrophy, fibrosis, and apoptosis were enhanced at both timepoints. Inclusion of Gef preserved contractility, blocked Iso-induced apoptosis, and prevented hypertrophy at the 2-wk timepoint, but caused fibrosis on its own. RNAseq analysis revealed hundreds of cardiac transcripts altered by Iso at each timepoint with subsequent RT-quantitative PCR validation confirming distinct temporal patterns of transcript regulation, including those involved in cardiac remodeling and survival signaling, as well as numerous cytokines. Although Gef infusion alone did not significantly alter cytokine expression, it abrogated the Iso-mediated changes in a majority of the βAR-sensitive cytokines, including CCL2 and TNF-α. Additionally, the impact of βAR-dependent EGFR transactivation on the acute regulation of cytokine transcript expression was assessed in isolated cardiomyocytes and in cardiac fibroblasts, where the majority of Iso-dependent, and EGFR-sensitive, changes in cytokines occurred. Overall, coincident with changes in cardiac structure and contractility, βAR stimulation dynamically alters cardiac transcript expression over time, including numerous cytokines that are regulated via EGFR-dependent signaling.
Keywords: βAR, heart, remodeling, cytokines, EGFR
during heart failure (HF) chronic increases in circulating catecholamines lead to sustained β-adrenergic receptor (βAR) activation, which has been shown to adversely impact cardiac function through changes in remodeling, including hypertrophy, fibrosis, and apoptosis (49). A major contributing factor toward adverse cardiac remodeling is the regulation of cardiac gene expression (19, 21), which has been reported in response to both acute and chronic infusion of catecholamines (3, 7, 11, 13, 29, 44). Although these studies have generally focused on expression changes via microarrays or RT-quantitative PCR (RT-qPCR) analysis of select genes at a single timepoint of infusion, few studies have assessed the dynamic regulation of cardiac transcript expression by chronic βAR stimulation over time. Because alterations in cardiac remodeling due to chronic βAR stimulation may have distinct temporal aspects and be reflected in alterations in transcript expression, we sought to determine how cardiac transcript expression becomes altered over time.
It is increasingly apparent that inflammation plays an important role in regulating cardiac remodeling in response to adverse events, and elevations in cardiac and systemic cytokines are associated with HF and contribute to cardiomyocyte dysfunction, hypertrophy, apoptosis, and extracellular matrix remodeling (17, 46). Of particular importance is the timing of inflammatory mediator production with acute and controlled inflammation being beneficial and excessive or prolonged inflammation being detrimental. Increases in the pro-inflammatory cytokines CCL2, TNF-α, IL-1β, and IL-6 have been shown to be influenced by chronic βAR stimulation (6, 12, 17, 30); however, a comprehensive assessment of how chronic βAR stimulation alters cytokine expression in the heart is lacking.
Although chronic βAR stimulation is ultimately detrimental to cardiac function through its impact on left ventricular (LV) remodeling (49), we and others have shown that βAR-mediated transactivation of epidermal growth factor receptor (EGFR) conveys cardioprotective signaling, in part via regulation of apoptotic gene expression (13, 32). Furthermore, using a more comprehensive RNAseq approach, we recently demonstrated that a substantial portion of cardiac transcripts regulated acutely by βAR stimulation is sensitive to the EGFR inhibitor gefitinib (42). However, the contribution of βAR-mediated EGFR transactivation to changes in cardiac transcript expression in response to chronic catecholamine stimulation has not been assessed. Here, we perform RNAseq analysis on murine left ventricular RNA samples at distinct timepoints of catecholamine infusion to determine the overall impact of chronic βAR stimulation on cardiac transcripts, with a focus on cytokine transcript expression in particular, and whether EGFR-dependent signaling influences this process.
MATERIALS AND METHODS
Mice.
Wild-type C57BL/6 mice (male, 12 wk old, attained from Jackson Laboratory) were administered vehicle (sterile PBS + 25% DMSO) or isoproterenol (Iso; I6504; Sigma, St. Louis, MO; 3 mg·kg−1·day−1) via mini osmotic pumps (Alzet; 1.0 μl/h for 7 days or 0.5 μl/h for 14 days), in conjunction with additional mini osmotic pumps containing either vehicle or Gefitinib (Gef; G-4408; LC Laboratories, Woburn, MA). Initial experiments were performed to ascertain the concentration of Gef required to inhibit Iso-mediated responses. Mice infused with increasing concentrations of Gef (1–25 mg·kg−1·day−1) for 1 day were treated with Iso (1 mg/kg) for 10 min and phosphorylated ERK1/2 (P-ERK1/2) levels assessed in their heart lysates, shown previously by us to be a reliable readout of βAR-mediated EGFR transactivation (13, 42). Complete inhibition of the Iso-mediated P-ERK1/2 response was attained at 25 mg·kg−1·day−1, which was used for the Gef ± Iso pumps administered for 1 or 2 wk. After 7 or 14 days mice were euthanized according to Animal Care and Use Protocol No. 4091, and hearts were excised and frozen in liquid N2 and stored at −80°C until biochemical analysis or fixed in paraformaldehyde for histological analysis. All animal procedures and experiments were carried out according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee, wherein all efforts were made to minimize suffering.
Primary cell isolation.
Cardiomyocyte and cardiac fibroblast cultures were prepared from 1- to 2-day-old Sprague Dawley rat pups (Harlan Laboratories, Indianapolis, IN) by enzymatic digestion. Hearts were excised and placed in sterile ADS solution containing 116 mM NaCl, 20 mM HEPES, 80 μM Na2HPO4, 56 mM glucose, 5.4 mM KCl, and 800 mM MgSO4-7H2O (pH 7.35). Blood and connective tissue were removed, and ventricles were minced and subjected to five 15-min enzymatic digestions using collagenase II (Worthington, Lakewood, NJ) and pancreatin. Fibroblasts and myocytes were separated by preplating for 2 h. After isolation, cardiomyocytes were cultured overnight in F-10 media containing 10% horse serum, 5% FBS, and 1% penicillin-streptomycin-fungizone (PSF) at 37°C in a humidified incubator with 5% CO2. The following day, media was replaced with F-10 media containing 5% FBS and 1% PSF. Fibroblasts were cultured in MEM containing 10% FBS. Cells were treated 3 h with 10 μM Iso with or without a 10-min pretreatment with 1 μM of the EGFR inhibitor AG1478.
Immunoblotting.
Left ventricular or HEK 293 cell samples were homogenized in lysis buffer containing 20 mM Tris (pH 7.4), 137 mM NaCl, 10% glycerol, 1 mM EDTA, 1% Nonidet P-40, 10 mM NaF (chemicals attained from Fisher Scientific, Pittsburgh, PA), 1× HALT protease inhibitor cocktail (78437; Thermo Scientific, Rockford, IL), and phosphatase inhibitor cocktail set IV (524628; Calbiochem). Equal amounts of lysates were resolved by SDS-PAGE (10% gels) and transferred to Immobilon-PSQ polyvinylidene fluoride 0.2 μm pore size membranes (Millipore, Billerica, MA). Odyssey Blocking Buffer (LI-COR Biosciences; Lincoln, NE) was used to prevent nonspecific binding. Immunoblotting was performed overnight at 4°C with diluted antibodies against P-ERK1/2 (1:1,000; Cell Signaling, Danvers, MA) or total-ERK1/2 (1:5,000; Cell Signaling). After being washed with TBS-T, membranes were incubated at room temperature for 60 min with the appropriate diluted secondary antibody [IRDye680 Donkey anti-rabbit IgG (H + L) at 1:20,000; IRDye800CW Goat anti-mouse IgG (H + L) at 1:15,000; LI-COR Biosciences]. Bound antibody was detected using the LI-COR Biosciences Odyssey System (LI-COR Biosciences). P-ERK1/2 intensities were normalized to corresponding T-ERK1/2 intensities.
Echocardiography.
Cardiac function was assessed via transthoracic two-dimensional echocardiography performed at 0, 7, or 14 days post-minipump implantation using a 12-mHz proble on mice anesthetized with isoflurane (1.5%). M-mode echocardiography was performed in the parasternal short-axis view to assess several cardiac parameters including left ventricular (LV) end-diastolic dimension, wall thickness, LV fractional shortening and ejection fraction. Percent fractional shortening was calculated using the this equation: [(LV internal dimension;diastole − LV internal dimension;systole)/LV internal dimension;systole] × 100%. Percent ejection fraction was calculated using this equation: [(LV volume;diastole − LV volume;systole)/LV volume;diastole] × 100%.
Histological analysis.
Excised hearts were fixed in 4% paraformaldehyde, paraffin embedded, and sectioned at 5 μm thickness. Deparaffinized sections were stained for hematoxylin-eosin (Sigma-Aldrich) or Masson trichrome (Sigma-Aldrich). NIS Elements software was used to measure fibrosis from 8 random fields per section. An in situ cell death detection kit, TMR Red (Roche Diagnostics, Mannheim, Germany), was used to measure apoptosis in deparaffinized heart sections via terminal deoxynucleotidyl-transferase-mediated dUTP nick-end labeling (TUNEL). Deparaffinized sections were incubated with proteinase K, and DNA strand breaks were labeled according to manufacturer's instructions using tetra-methyl-rhodamine-dUTP. Cells were visualized at 20× magnification using Nikon Eclipse microscope and the percentage of TUNEL-positive nuclei calculated in relation to the number of 4′,6-diamidino-2-phenylindole-stained nuclei from 10 random fields per section.
Whole transcriptome analysis.
Total RNA was isolated from LV samples using a Qiagen RNeasy Fibrous Tissue Midi Kit with Proteinase K and DNAseI digestion according to manufacturer's protocol (Qiagen, Valencia, CA). Total RNA from four hearts per treatment condition were combined (as unreplicated pooled samples) and submitted for whole transcriptome analysis with the SOLiD 4 platform (Applied Biosystems, Foster City, CA) as a fee-for-service using the Cancer Genomics Shared Resource at the Kimmel Cancer Center (Philadelphia, PA). A detailed description of RNAseq fragment mapping to the mouse genome using Life Technologies/Applied Biosystems Bioscope software (version 1.3), where 1 copy number ≈ 3 reads per kilobase of exon per million mapped reads, and differential gene expression analysis of unreplicated samples using Cufflinks software by the Cancer Genomics Shared Resource has been described previously (22). P values attained in the differential expression analysis were adjusted for multiple testing using the Benjamini-Hockberg correction (36), yielding q values. Genes were considered significantly altered if q < 0.05 and the absolute fold-change in expression was ≥1.5. The raw unfiltered transcriptome data were available at Dryad (http://doi.org/10.5061/dryad.5s9r5). Hierarchical clustering was performed using Gene Cluster 3.0 (4). Functional annotation of data sets, network, and upstream regulator analyses were performed using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). Significance of association between sets of genes and related molecular and cellular functions was assessed using the Fisher's Exact Test with P values <0.05, indicating a statistically significant nonrandom association. Functional activation predictions were based on the directionality of transcripts altered in the various molecular categories of the data sets that yielded z-scores, with a z-score ≥2 indicating activation and a z-score ≤−2 indicating inhibition.
RT-qPCR.
cDNA was synthesized from the total RNA of individual LV samples or isolated cardiac myocytes and fibroblasts using the high capacity cDNA reverse transcription kit (Applied Biosystems), and RT-qPCR was performed with SYBR Select Master Mix (Applied Biosystems) in triplicate for each sample using primers listed in Table 1 at an annealing temperature of 60.1°C. All RT-qPCR data was analyzed using Applied Biosystems Comparative CT Method (ΔΔCT). Gene expression analysis was normalized to translationally controlled tumor protein -1 (TPT1). Validation of transcriptome results and IPA predictions with biological replicates for each condition were performed on 3–5 independent RNA samples isolated from mouse hearts.
Table 1.
Primer sequences used for RT-PCR
| Sequence |
|||
|---|---|---|---|
| Gene Name | Reference Sequence | Forward | Reverse |
| Ccl2 | |||
| Mouse | NM_011333 | 5′-CTC GGA CTG TGA TGC CTT AAT-3′ | 5′-TGG ATC CAC ACC TTG CAT TTA-3′ |
| Rat | NM_031531 | 5′-GTC TCA GCC AGA TGC AGT TAA T-3′ | 5′-CTG CTG GTG ATT CTC TTG TAG TT-3′ |
| Ccl5 | |||
| Mouse | NM_013653 | 5′-CCA GAG AAG AAG TGG GTT CAA G-3′ | 5′-AGC AAT GAC AGG GAA GCT ATA C-3′ |
| Ccl6 | |||
| Mouse | NM_009139 | 5′-GGA GGG AGA TGG GAC CAT ATA A-3′ | 5′-GCA CCA AGC TCA GAG GAA TAA-3′ |
| Rat | NM_001004202 | 5′-GGG CTC ATA CAA GAT ACG GTA AA-3′ | 5′-CAT GGG ATC TGT GAG GCA TAG-3′ |
| Ccl20 | |||
| Mouse | NM_001159739 | 5′-ACA GCC CAA GGA GGA AAT G-3′ | 5′-AGT CCA CTG GGA CAC AAA TC-3′ |
| Rat | NM_019233 | 5′-CTG CCT CAC GTA CAC AAA GA-3′ | 5′-TCG ACT TCA GGT GAA AGA TGA TAG-3′ |
| Col4a4 | |||
| Mouse | NM_007735 | 5′-AGC AAG CGG ATG ACA AAG A-3′ | 5′-AGC CAG AAG CCC AAT AGA TTA C-3′ |
| Cxcl12 | |||
| Mouse | NM_021704 | 5′-GCT AAG GTT TGC CAG CAT AAA G-3′ | 5′-CGA GGG CAG GGA TGA ATA TAA G-3′ |
| Cytl1 | |||
| Mouse | NM_001081106 | 5′-TTG GAG AGC AAG CAC CTT AG-3′ | 5′-GTG TAA GCA GAG ACC AGA AAG A-3′ |
| Dapk1 | |||
| Mouse | NM_029653 | 5′-CCG CTG TCA ACT ACG ACT TT-3′ | 5′-GTC CTG GAT TGT CAT CCT CTT C-3′ |
| Eln | |||
| Mouse | NM_007925 | 5′-CTC ATC CAT CCA TCC ATC CAT C-3′ | 5′-GAC AGG TGA ACC AGG TTG ATA G-3′ |
| Il1b | |||
| Mouse | NM_008361 | 5′-ATG GGC AAC CAC TTA CCT ATT T-3′ | 5′-TT CTA GAG AGT GCT GCC TAA TG-3′ |
| Rat | NM_031512 | 5′-CTA TGG CAA CTG TCC CTG AA-3′ | 5′-GGC TTG GAA GCA ATC CTT AAT C-3′ |
| Il2 | |||
| Mouse | NM_008366 | 5′-GCG GCA TGT TCT GGA TTT G-3′ | 5′-TGT GTT GTC AGA GCC CTT TAG-3′ |
| Rat | NM_053836 | Forward 5′-GCA GGC CAC AGA ATT GAA AC-3′ | 5′-CCA GCG TCT TCC AAG TGA A-3′ |
| Il6 | |||
| Mouse | NM_031168 | 5′-GAA GTT AGA GTC ACA GAA GGA GTG-3′ | 5′-GTT TGC CGA GTA GAC CTC ATA G-3′ |
| Rat | NM_012589 | 5′-CTA TGG CAA CTG TCC CTG AA-3′ | 5′-GGC TTG GAA GCA ATC CTT AAT C-3′ |
| Il17a | |||
| Mouse | NM_010552 | 5′-CGC AAT GAA GAC CCT GAT AGA T-3′ | 5′-CTC TTG CTG GAT GAG AAC AGA A-3′ |
| Rat | NM_001106897 | 5′-AAA CGC CGA GGC CAA TAA-3′ | 5′-GAA GTG GAA CGG TTG AGG TAG-3′ |
| Lif | |||
| Mouse | NM_001039537 | 5′-CTG CTC TCC CTC TTT CCT TTC-3′ | 5′-ACA TTC CCA CAG GGT ACA TTC-3′ |
| Nppb | |||
| Mouse | NM_008726 | 5′-ACC ACC TTT GAA GTG ATC CTA TT-3′ | 5′-GCA AGT TTG TGC TCC AAG ATA AG-3′ |
| Nr4a1 | |||
| Mouse | NM_010444 | 5′-CTC CAC GTC TTC TTC CTC ATC-3′ | 5′-CTG GAG GAT AGG GTC TCA TCT A-3′ |
| Osm | |||
| Mouse | NM_001013365 | 5′-GTG CTC TCT CTC ATG CCT TAT C-3′ | 5′-CCA GAT CAG TGT TCC TTG GTA G-3′ |
| Postn | |||
| Mouse | NM_015784 | 5′-TGT GTA TCG GAC GGC TAT CT-3′ | 5′-CTC TGC TGG TTG GAT GAT TTC T-3′ |
| Ppbp | |||
| Mouse | NM_023785 | 5′-TGT GCT GAT GTG GAA GTG ATA G-3′ | 5′-GAT GAA GCA GCT GGT CAG TAA-3′ |
| Thbs1 | |||
| Mouse | NM_011580 | 5′-CCT GGA CTT GCT GTA GGT TAT G-3′ | 5′-GTC ATC ATC TCT CTC GGT GTT G-3′ |
| Tnfa | |||
| Mouse | NM_013693 | 5′-CTA CCT TGT TGC CTC CTC TTT-3′ | 5′-GAG CAG AGG TTC AGT GAT GTA G-3′ |
| Rat | NM_012675 | 5′-ACC TTA TCT ACT CCC AGG TTC T-3′ | 5′-GGC TGA CTT TCT CCT GGT ATG-3′ |
| Tnfsf11 | |||
| Mouse | NM_01163 | 5′-GGA AGC GTA CCT ACA GAC TAT C-3′ | 5′-CTC CCT CCT TTC ATC AGG TTA T-3′ |
| Rat | NM_057149 | 5′-CAT CGC TCT GTT CCT GTA CTT-3′ | 5′-CGA GTC CTG CAA ACC TGT AT-3′ |
| Tnfsf12 | |||
| Mouse | NM_011614 | 5′-AGT CCT GTC CTC TCC TCA AA-3′ | 5′-TGG TGG GAT GGG ATG TTA AAG-3′ |
| Tslp | |||
| Mouse | NM_021367 | 5′-TCA TGA CCT GAC TGG AGA TTT G-3′ | 5′-AGC CAG GGA TAG GAT TGA GA-3′ |
| Tpt1 | |||
| Mouse | NM_009429 | 5′-ATC ATC TAC CGG GAC CTC ATC-3′ | 5′-CCC TCT GTT CTA CTG ACC ATC T-3′ |
| Rat | NM_053867 | 5′-TGG AGC TGC AGA GCA AAT TA-3′ | 5′-TCT TCA CGG TAG TCC AGT AGA G-3′ |
ELISA.
Levels of secreted CCL2 or TNF-α were detected using a mouse CCL2 DuoSet ELISA kit (R&D Systems, Minneapolis, MN) or mouse TNF-α DuoSet ELISA kit (R&D Systems) according to manufacturer's instructions using plasma collected from the mice at time of euthanization. In brief, 96 well plates were coated overnight with Capture Antibody. Plates were blocked with Reagent Diluent and incubated with plasma. After incubations with the Detection Antibody and Streptavidin-HRP, substrate solution was added and absorbance was measured at 450 nm with a 540 nm wavelength correction.
Statistical analysis.
Data presented are expressed as mean or relative quantity (RQ) ± standard error (SE). Statistical analysis was performed using unpaired two-tailed Student t-tests and one- and two-way ANOVA with a Newman-Keuls multiple comparison test where appropriate using Prism 5.0 software (GraphPad Software; San Diego, CA) with P values indicated in figure legends.
RESULTS
Effects of chronic βAR stimulation on cardiac function, structure, and survival.
To assess changes in cardiac transcriptome expression in response to chronic catecholamine stimulation at distinct timepoints, C57BL/6 mice underwent osmotic minipump implantation to deliver a constant infusion of Iso, Gef, or Gef plus Iso for 1 or 2 wk. Cardiac function of the mice was monitored via weekly echocardiography (Fig. 1A and Table 2) and cardiac structure assessed postinfusion to assess the impact of chronic Iso ± Gef stimulation. Although contractility was maintained following 1 wk of Iso infusion, contractile dysfunction was apparent by 2 wk, with significant decreases in both percent fractional shortening (Fig. 1B) and percent ejection fraction (Fig. 1C) in comparison with those of vehicle controls. Combined Gef plus Iso infusion prevented Iso-mediated decreases in percent fractional shortening and percent ejection fraction, whereas Gef did not induce changes in contractile function on its own. Although decreased contractility occurred only after 2 wk of Iso infusion, LV hypertrophy was evident after 1 wk of Iso infusion as indicated via both echocardiography (increased LV wall thickness and LV dilation) (Table 2), as well as cardiac morphometric analysis (Fig. 2A) and gravimetric analysis of heart weight to tibia length (Fig. 2B) or body weight (Table 2). Although Gef did not impact these parameters after 1 wk of Iso infusion, cardiac function was preserved and LV hypertrophy reduced at the 2-wk timepoint compared with that of Iso alone. In addition to hypertrophy, LV fibrosis, as estimated by Masson trichrome staining (Fig. 3, A and B), and apoptosis, as estimated via TUNEL-positive nuclei (Fig. 4, A and B), were significantly increased at both 1 and 2 wk following Iso treatment. Interestingly, Gef treatment prevented Iso-induced increases in apoptosis but had no effect on Iso-induced fibrosis while inducing fibrosis on its own at both 1 and 2 wk of infusion.
Fig. 1.
Effects of chronic isoproterenol (Iso) ± gefitinib (Gef) stimulation on cardiac contractility. A: representative M-mode echocardiography from C57BL/6 mice before treatment and following 1 or 2 wk. Vehicle (Veh) ± Gef or Iso ± Gef. Left ventricular fractional shortening (FS; B) and ejection fraction (EF; C) were measured at the short axis from M mode using VisualSonic Analysis Software. *P ≤ 0.05 vs. Veh; n = 8–16 each, 2-way ANOVA.
Table 2.
Echocardiography dimensions
| Vehicle |
Iso |
Gef |
Gef + Iso |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 1 Week | 2 Weeks | Baseline | 1 Week | 2 Weeks | Baseline | 1 Week | 2 Weeks | Baseline | 1 Week | 2 Weeks | |
| n | 14 | 14 | 7 | 14 | 14 | 7 | 16 | 16 | 8 | 16 | 16 | 8 |
| Diastolic | ||||||||||||
| LV volume, μl | 69.4 ± 3.3 | 71.3 ± 2.9 | 69.5 ± 3.6 | 67.4 ± 4.9 | 75.8 ± 3.9 | 91.8 ± 3.1 | 72.6 ± 3.2 | 65.2 ± 3.4 | 74.2 ± 2.8 | 74.3 ± 2.2 | 64.6 ± 3.6 | 71.4 ± 2.4 |
| LVAW, mm | 0.74 ± 0.02 | 0.68 ± 0.03 | 0.64 ± 0.05 | 0.73 ± 0.02 | 0.81 ± 0.05* | 0.70 ± 0.06 | 0.65 ± 0.03 | 0.69 ± 0.04 | 0.75 ± 0.03 | 0.71 ± 0.02 | 0.75 ± 0.03 | 0.80 ± 0.04 |
| LVEDD, mm | 3.72 ± 0.10 | 3.81 ± 0.11 | 3.60 ± 0.16 | 3.74 ± 0.10 | 4.02 ± 0.11 | 4.28 ± 0.16* | 4.03 ± 0.07 | 3.81 ± 0.08 | 4.13 ± 0.10 | 4.09 ± 0.05 | 4.00 ± 0.13 | 4.04 ± 0.07 |
| LVPW, mm | 0.68 ± 0.04 | 0.66 ± 0.03 | 0.71 ± 0.04 | 0.67 ± 0.02 | 0.75 ± 0.03 | 0.85 ± 0.08 | 0.63 ± 0.03 | 0.85 ± 0.07 | 0.76 ± 0.06 | 0.70 ± 0.02 | 0.93 ± 0.07 | 0.80 ± 0.02 |
| LVID, mm | 3.84 ± 0.10 | 3.88 ± 0.11 | 3.69 ± 0.16 | 3.80 ± 0.11 | 3.94 ± 0.12 | 3.96 ± 0.31 | 4.05 ± 0.08 | 3.86 ± 0.09 | 4.09 ± 0.07 | 4.09 ± 0.05 | 3.88 ± 0.10 | 4.02 ± 0.06 |
| Systolic | ||||||||||||
| LV volume, μl | 29.3 ± 2.0 | 31.07 ± 2.2 | 28.17 ± 2.6 | 31.4 ± 3.8 | 31.5 ± 2.3 | 43.3 ± 2.9 | 29.7 ± 1.9 | 26.4 ± 2.0 | 33.5 ± 2.1 | 30.0 ± 1.5 | 27.9 ± 2.6 | 29.6 ± 2.2 |
| LVAW, mm | 1.14 ± 0.03 | 1.09 ± 0.04 | 1.02 ± 0.06 | 1.13 ± 0.04 | 1.26 ± 0.07* | 1.12 ± 0.03 | 1.06 ± 0.04 | 1.10 ± 0.04 | 1.16 ± 0.04 | 1.16 ± 0.04 | 1.23 ± 0.04 | 1.29 ± 0.07 |
| LVESD, mm | 2.76 ± 0.08 | 2.83 ± 0.07 | 2.73 ± 0.10 | 2.79 ± 0.14 | 2.86 ± 0.08 | 3.26 ± 0.10 | 2.80 ± 0.07 | 2.63 ± 0.09 | 2.52 ± 0.61 | 2.82 ± 0.05 | 2.55 ± 0.10 | 2.82 ± 0.12 |
| LVPW, mm | 1.05 ± 0.04 | 1.02 ± 0.03 | 1.07 ± 0.05 | 1.10 ± 0.04 | 1.05 ± 0.04 | 1.15 ± 0.03 | 0.94 ± 0.03 | 1.24 ± 0.09 | 1.12 ± 0.10 | 1.08 ± 0.04 | 1.31 ± 0.12 | 2.78 ± 0.08 |
| LVID, mm | 2.65 ± 0.09 | 2.70 ± 0.09 | 2.62 ± 0.14 | 2.78 ± 0.12 | 2.62 ± 0.16 | 2.75 ± 0.30 | 2.78 ± 0.07 | 2.63 ± 0.09 | 2.93 ± 0.07 | 2.80 ± 0.06 | 2.61 ± 0.09 | 1.17 ± 0.03 |
| Heart weight/body weight, mg/g | — | 4.73 ± 0.06 | 4.92 ± 0.12 | — | 5.85 ± 0.19* | 5.88 ± 0.20* | — | 4.76 ± 0.11 | 4.76 ± 0.10 | — | 5.40 ± 0.16* | 4.84 ± 0.07‡ |
Values are means ± SE.
Gef, gefitinib; Iso, isoproterenol; LV, left ventricular; LVAW, LV anterolateral wall; LVEDD, LV end-diastolic diameter; LVESD, LV end-systolic diameter; LVPW, LV posterior wall; LVID, LV internal dimension.
P < 0.05 vs. vehicle;
P < 0.05 vs. Iso.
Fig. 2.
Effect of chronic Iso ± Gef stimulation on cardiac hypertrophy. Representative hematoxylin-eosin (H&E) staining (A), and gravimetric analysis of heart weight to tibia length (HW/TL; B), of hearts from mice treated with Veh ± Gef or Iso ± Gef for 1 or 2 wk is shown. *P ≤ 0.05, ‡P ≤ 0.001; n = 8–16 each; 1-way ANOVA. ns, Not significant.
Fig. 3.
Effect of chronic Iso ± Gef stimulation on cardiac fibrosis. Representative Masson trichrome staining (A), and quantification of Masson trichrome staining (B), of hearts from mice treated with Veh ± Gef or Iso ± Gef for 1 or 2 wk is shown. *P ≤ 0.05, †P ≤ 0.01, ‡P ≤ 0.001; n = 8–16 each; 1-way ANOVA.
Fig. 4.
Effect of chronic Iso ± Gef stimulation on cardiac survival. Representative transferase-mediated dUTP nick-end labeling (TUNEL) staining (A), and percentage of TUNEL-positive (TUNEL +ve) cells (B), in hearts from mice treated with Veh ± Gef or Iso ± Gef for 1 or 2 wk is shown. *P ≤ 0.05, †P ≤ 0.01, ‡P ≤ 0.001; n = 8–16 each; 1-way ANOVA.
Differential temporal regulation of the cardiac transcriptome by chronic βAR stimulation.
Having confirmed the deleterious effects of chronic βAR stimulation on cardiac structure and contractility over time, we next subjected total RNA isolated from LV samples to whole transcriptome analysis. Initially, 1,006 and 811 cardiac transcripts were identified as being regulated in response to chronic Iso infusion for 1 or 2 wk, respectively (Fig. 5A). Interestingly, the vast majority of transcripts altered at either 1 or 2 wk were unique to that timepoint, whereas relatively few transcripts were similarly altered at both timepoints, only 113 out of a total of 1,817 transcripts (Fig. 5B). Further analysis was performed to filter out false discovery rate-adjusted transcripts with q values >0.05 (36), leaving 780 and 689 transcripts significantly altered at 1 and 2 wk in response to chronic Iso infusion, respectively (Supplemental Tables S. 1 and S2). The relative proportion of filtered transcripts up- or downregulated at each timepoint differed slightly, with 64% up/36% down at 1 wk and 55% up/45% down at 2 wk.
Fig. 5.
Differential temporal regulation of the cardiac transcriptome by chronic β-adrenergic receptor (βAR) stimulation. A: heatmap depicting significant (P ≤ 0.05) changes in cardiac transcript expression in mouse hearts treated with Iso compared with Veh for 1 vs. 2 wk as detected by transcriptome analysis. Each RNAseq sample (Veh 1 wk, Iso 1 wk, Veh 2 wk, and Iso 2 wk) contained total RNA from 4 individual hearts per condition. B: Venn diagram depicting the number of cardiac transcripts significantly (P ≤ 0.05) increased or decreased following 1 or 2 wk Iso treatment. C: ingenuity analysis revealed the 10 most significantly associated cellular function categories of the cardiac transcripts regulated by 1 or 2 wk of Iso infusion.
Ingenuity analysis was used to analyze the disease and cellular functional categories associated with the altered cardiac transcripts following chronic Iso infusion. For the 1-wk transcripts, the top associated disease category was cardiovascular disease (p = 2.08e-11), and, in line with the data above, cardiac hypertrophy was the top associated cardiotoxicity-related category for both the 1- and 2-wk timepoints (p = 1.46e-4 and 1.61e-5, respectively). The relative quantity of up- and downregulated transcripts following 1 or 2 wk of Iso was determined in the most significantly associated cellular function categories (Fig. 5C). Although some categories, including cell proliferation, function, adhesion, and development, were represented by transcripts altered in both Iso infusion cohorts, several functional categories, such as cell morphology, movement, organization, death, gene expression, and lipid metabolism, were only represented by transcripts from either the 1-wk or the 2-wk group. Interestingly, although the cell death category only accounted for ∼5% of the cardiac transcripts regulated at the 1-wk timepoint, this category contained the highest activation prediction score of any other category, specifically for organismal death (z-score of 7.757, p = 2.52e-6), which corresponds well with the increased TUNEL staining we observed in the LV sections. To independently confirm the whole transcriptome findings, changes in expression of transcripts from the datasets that are involved in cardiac remodeling and survival (Col4a4, Dapk1, Eln, Nppb, Nr4a1, Postn, Thbs1) were assessed via RT-qPCR (Fig. 6, A and B), attaining similar results. The significantly enhanced expression of transcripts encoding collagen, elastin, periostin, BNP, and Nur77 fits well with the increased fibrosis and hypertrophy observed following chronic Iso infusion.
Fig. 6.
Temporal regulation of transcripts involved in cardiac remodeling and survival signaling by chronic βAR stimulation. RT-quantitative PCR (RT-qPCR) validation of changes in expression of transcripts predicted by transcriptome analysis to be involved in cardiac remodeling and survival following Iso infusion for either 1 wk (A) or 2 wk (B) is shown. *P ≤ 0.05, †P ≤ 0.01 vs. Veh of same transcript; n = 4–6 each; 2-tailed t-test. RQ, relative quantity.
Impact of chronic βAR stimulation on cardiac cytokine expression.
Because inflammation has been shown to negatively regulate cardiac structure and function in response to adverse conditions, such as chronic catecholamine signaling (6, 17, 35, 46), and levels of select inflammatory cytokines have been shown to become altered in response to βAR stimulation (30), we sought to more broadly assess changes in cardiac cytokine expression following 1 or 2 wk of Iso infusion. Thus we performed ingenuity upstream analysis to identify cytokines with predicted involvement in the regulation of the cardiac transcripts in our datasets. This analysis resulted in the identification of 48 cytokines (prediction p values <0.05; Table 3), 37 of which were predicted to be involved in the regulation of cardiac transcripts following 1 wk of Iso infusion, two of which following 2 wk and 9 of which at both timepoints. Network analysis based on experimentally observed relationships in various models indicated that βAR have been shown to regulate the expression of 35 of these 48 cytokines (Fig. 7A), including Il-6 and Il-1β, the cytokines most highly activated in response to chronic Iso infusion (p = 5.52e-12, z-score of 3.923; and p = 2.96e-5, z-score of 4.144, respectively). Changes in expression of cytokine transcripts selected from each category (predicted regulation in Table 3) or that were present in the βAR-regulation network or transcriptome datasets were confirmed via RT-qPCR at 1 wk (Fig. 7B) and 2 wk (Fig. 7C), reinforcing the potential importance of dynamic temporal-dependent cytokine responses to catecholamine overstimulation in the heart.
Table 3.
P values of cytokines predicted to be involved in Iso-mediated changes in cardiac transcript expression
| Cytokine | 1 Week | 2 Weeks |
|---|---|---|
| IL6 | 5.52E-12 | 1.40E-02 |
| OSM | 7.32E-09 | 4.18E-03 |
| CSF2 | 8.71E-09 | P > 0.05 |
| TNF | 1.70E-06 | 1.36E-02 |
| EDN1 | 3.22E-06 | 2.51E-03 |
| IL1A | 9.69E-06 | P > 0.05 |
| LIF | 1.63E-05 | P > 0.05 |
| IFNG | 1.95E-05 | P > 0.05 |
| WNT3A | 2.16E-05 | P > 0.05 |
| IL1B | 2.96E-05 | 6.03E-03 |
| IL13 | 3.07E-05 | P > 0.05 |
| CCL5 | 3.69E-05 | 2.89E-02 |
| IL11 | 4.73E-05 | P > 0.05 |
| CSF1 | 1.04E-04 | P > 0.05 |
| IL1 | 2.56E-04 | P > 0.05 |
| TNFSF11 | 2.94E-04 | P > 0.05 |
| IL4 | 6.90E-04 | P > 0.05 |
| WNT1 | 6.97E-04 | P > 0.05 |
| IL3 | 6.97E-04 | 1.78E-04 |
| IL15 | 7.20E-04 | 2.67E-02 |
| CD40LG | 9.74E-04 | P > 0.05 |
| PRL | 1.33E-03 | P > 0.05 |
| CTF1 | 1.35E-03 | P > 0.05 |
| FAM3B | 4.00E-03 | P > 0.05 |
| SPP1 | 5.27E-03 | P > 0.05 |
| IL17A | 6.72E-03 | P > 0.05 |
| CCL2 | 8.37E-03 | P > 0.05 |
| IL5 | 9.57E-03 | P > 0.05 |
| IFNB1 | 1.06E-02 | P > 0.05 |
| TNFSF14 | 1.22E-02 | P > 0.05 |
| Il3 | 1.49E-02 | P > 0.05 |
| TSLP | 1.63E-02 | P > 0.05 |
| CX3CL1 | 1.92E-02 | P > 0.05 |
| IL9 | 1.98E-02 | P > 0.05 |
| Ifn | 2.17E-02 | P > 0.05 |
| TNFSF12 | 2.57E-02 | P > 0.05 |
| CRH | 2.62E-02 | P > 0.05 |
| CYTL1 | 2.90E-02 | P > 0.05 |
| PPBP | 3.24E-02 | P > 0.05 |
| CXCL12 | 3.41E-02 | P > 0.05 |
| THPO | 3.55E-02 | P > 0.05 |
| IL7 | 3.59E-02 | P > 0.05 |
| EPO | 3.65E-02 | 4.31E-02 |
| IL16 | 4.57E-02 | P > 0.05 |
| CSF3 | 4.85E-02 | P > 0.05 |
| MIF | 4.98E-02 | P > 0.05 |
| IL2 | P > 0.05 | 2.28E-03 |
| CCL20 | P > 0.05 | 4.79E-02 |
Fig. 7.
Impact of chronic βAR (ADRB) stimulation on cardiac cytokine transcript expression. A: network analysis between ADRB and the 48 cytokines predicted to be regulated by Iso. RT-qPCR analysis of 17 cytokine transcripts were predicted to become altered following Iso infusion for 1 wk (B) or 2 wk (C). *P ≤ 0.05, †P ≤ 0.01, ‡P ≤ 0.001 vs. Veh of same transcript; n = 3–6 each; 2-tailed t-test.
Chronic βAR-mediated regulation of cardiac cytokine expression is sensitive to EGFR inhibition.
Network analysis between EGFR and the 48 cytokines predicted to be involved in Iso-mediated cardiac transcript regulation indicates that EGFR has been experimentally shown to modulate the expression of 37 of the cytokines (Fig. 8A), several of which were validated to be regulated by chronic Iso infusion via RT-qPCR (Fig. 7, B and C). To determine whether EGFR transactivation plays a role in the regulation of cardiac cytokine transcripts in response to chronic βAR stimulation, we compared changes in the transcript levels in the hearts of Iso-treated versus Gef + Iso-treated mice. We previously demonstrated the importance of EGFR transactivation with regard to acute βAR-mediated increases in Thbs1 transcript expression (42). Because Thbs1 was upregulated in response to chronic Iso at both the 1 and 2 wk timepoints (Fig. 6, A and B), we tested whether concurrent Gef infusion resulted in predicted blockade of Iso-induced Thbs1 expression. Indeed, Iso-mediated increases in Thbs1 expression at both 1 and 2 wk were blocked by co-infusion with Gef (Fig. 8B). Furthermore, several of the 12 cytokine transcripts shown to be significantly altered by chronic βAR signaling were differentially sensitive to Gef at the 1 and 2 wk timepoints (Table 4). Iso-mediated changes in Ccl2 (Fig. 8C), Ccl6, Ccl20, Il2, Il17a, and Tnfa (Fig. 8D) levels were blocked by concurrent Gef infusion at both 1 and 2 wk, whereas Iso-regulated Ccl5, Il1b, and Osm expression were completely insensitive to Gef. Interestingly, a handful of cytokine transcripts had distinct temporal sensitivity to Gef, where Iso-induced changes in Il6 and Tnsfs11 expression were sensitive to Gef at 1 wk, but not at 2 wk. Conversely, the Iso-mediated increase in Tslp expression was sensitive to Gef at 2 wk, but not at 1 wk. Importantly, Gef infusion did not induce significant alterations in transcript expression alone, although there was a trend toward Iso-independent Gef-sensitivity for a few transcripts, especially at the 2-wk timepoint.
Fig. 8.
Chronic βAR-mediated regulation of cardiac cytokine expression is sensitive to epidermal growth factor receptor (EGFR) inhibition. A: network analysis between EGFR and the 48 cytokines predicted to be regulated by Iso. ●, Cytokines validated to be regulated by chronic Iso infusion via RT-qPCR (in Fig. 7, B and C). The effects of Gef on changes in Iso-mediated transcript expression were assessed by RT-qPCR at 1 and 2 wk timepoints for Thbs1 (B), Ccl2 (C), and Tnfa (D). *P ≤ 0.05, †P ≤ 0.01, ‡P ≤ 0.001; n = 4–6 each; 1-way ANOVA. ELISAs for CCL2 (E) or TNF-α (F) were performed on serum from mice treated for 1 or 2 wk with Veh ± Gef or Iso ± Gef. *P ≤ 0.05; n = 4 each; 1-way ANOVA.
Table 4.
RQ values from qRT-PCR analysis of left ventricular cytokine expression
| 1 Week |
2 Weeks |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene Name | Vehicle | Iso | Gef | Gef + Iso | Vehicle | Iso | Gef | Gef + Iso |
| n | 4 | 4 | 3 | 4 | 5 | 5 | 4 | 4 |
| Ccl2 | 1.00 ± 0.05 | 1.91 ± 0.09* | 1.33 ± 0.07 | 1.27 ± 0.16‡ | 1.00 ± 0.08 | 0.70 ± 0.10 | 0.86 ± 0.04 | 1.18 ± 0.11‡ |
| Ccl5 | 1.00 ± 0.08 | 0.67 ± 0.08* | 0.91 ± 0.09 | 0.59 ± 0.06* | 1.00 ± 0.07 | 0.99 ± 0.18 | 1.14 ± 0.05 | 0.99 ± 0.20 |
| Ccl6 | 1.00 ± 0.12 | 1.89 ± 0.40* | 0.95 ± 0.12 | 1.45 ± 0.10‡ | 1.00 ± 0.11 | 2.26 ± 0.45* | 0.68 ± 0.08 | 0.69 ± 0.15‡ |
| Ccl20 | 1.00 ± 0.06 | 0.33 ± 0.15* | 0.85 ± 0.05 | 1.19 ± 0.25‡ | 1.00 ± 0.09 | 0.25 ± 0.07* | 0.99 ± 0.06 | 1.14 ± 0.26‡ |
| Il1b | 1.00 ± 0.07 | 2.52 ± 0.60* | 0.97 ± 0.16 | 2.81 ± 0.49* | 1.00 ± 0.03 | 1.64 ± 0.07* | 1.10 ± 0.19 | 1.98 ± 0.14* |
| Il2 | 1.00 ± 0.17 | 1.70 ± 0.28* | 0.72 ± 0.08 | 0.83 ± 0.04‡ | 1.00 ± 0.18 | 1.78 ± 0.07* | 0.78 ± 0.12 | 0.41 ± 0.04*‡ |
| Il6 | 1.00 ± 0.05 | 1.82 ± 0.22 | 0.82 ± 0.17 | 2.77 ± 0.42*‡ | 1.00 ± 0.12 | 0.80 ± 0.04 | 1.28 ± 0.17 | 1.15 ± 0.20 |
| Il17a | 1.00 ± 0.13 | 0.43 ± 0.14* | 1.07 ± 0.11 | 3.10 ± 0.28*‡ | 1.00 ± 0.08 | 0.20 ± 0.07* | 1.63 ± 0.40 | 1.96 ± 0.40*‡ |
| Osm | 1.00 ± 0.06 | 1.40 ± 0.14 | 1.03 ± 0.07 | 1.30 ± 0.17 | 1.00 ± 0.08 | 0.81 ± 0.02 | 1.20 ± 0.11 | 0.99 ± 0.18 |
| Tnfa | 1.00 ± 0.07 | 1.94 ± 0.09* | 0.84 ± 0.05 | 0.83 ± 0.17‡ | 1.00 ± 0.04 | 1.51 ± 0.15* | 0.55 ± 0.18 | 0.52 ± 0.07‡ |
| Tnfsf11 | 1.00 ± 0.14 | 0.68 ± 0.02* | 1.15 ± 0.09 | 1.30 ± 0.21‡ | 1.00 ± 0.13 | 0.50 ± 0.08* | 1.46 ± 0.14 | 0.33 ± 0.06* |
| Tslp | 1.00 ± 0.13 | 3.84 ± 1.16* | 0.34 ± 0.03 | 3.59 ± 0.56* | 1.00 ± 0.30 | 2.15 ± 0.41* | 0.74 ± 0.21 | 1.06 ± 0.27‡ |
Values are relative quantity (RQ) ± SE.
qRT-PCR, quantitative reverse transcription PCR.
P < 0.05 vs. vehicle;
P < 0.05 vs. Iso, one-way ANOVA with Newman-Keuls multiple comparison test.
To determine whether changes in cytokine transcript levels correlate with protein levels, we measured the expression of CCL2 and TNF-α, which were the two most significantly Iso-regulated transcripts that were also sensitive to Gef infusion. Secreted CCL2 and TNF-α were measured in serum obtained from mice under each treatment condition at both 1 and 2 wk. As observed at the transcript level, Gef alone did not alter expression of either cytokine but completely abrogated the Iso-mediated changes in the expression of both CCl2 (Fig. 8E) and TNF-α (Fig. 8F).
βAR-dependent and EGFR-sensitive cytokine transcript regulation predominates in cardiac fibroblasts.
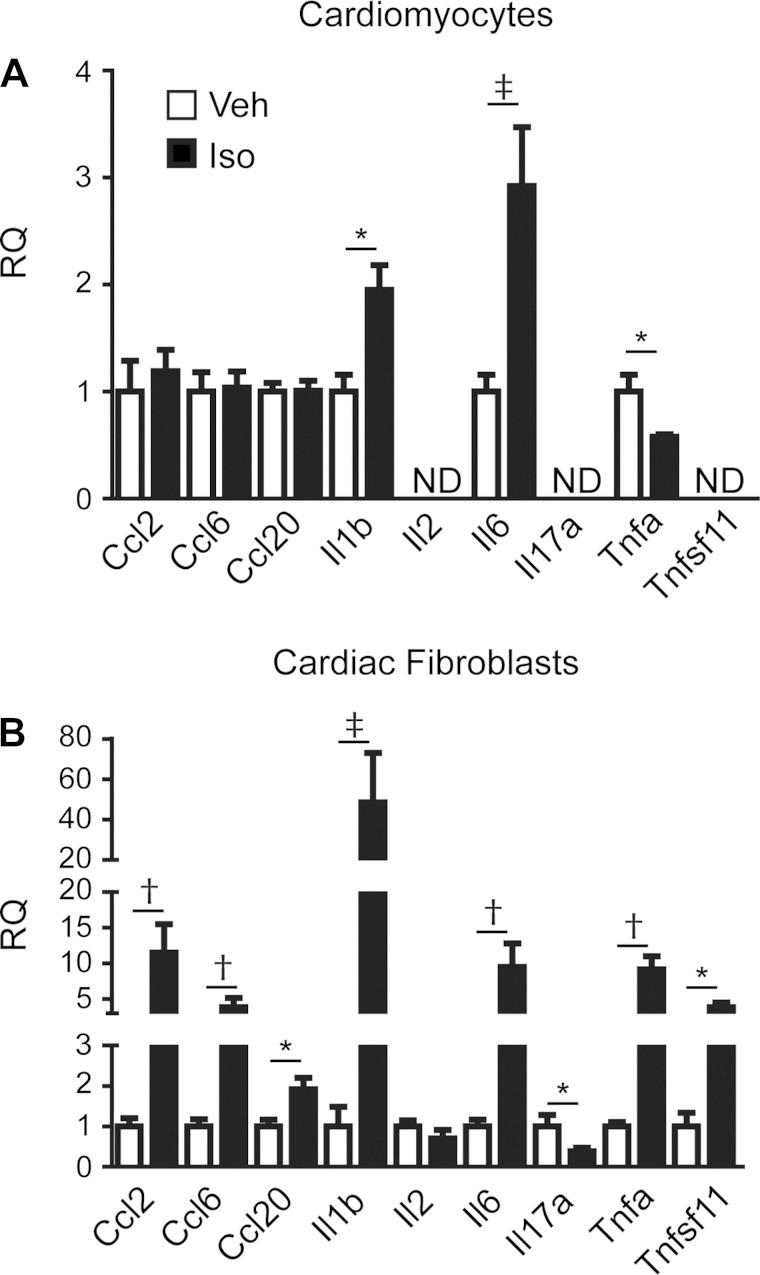
Finally, to assess whether EGFR-sensitive βAR-mediated changes in cardiac cytokine expression are cell-type dependent, and whether these changes occur in the absence of chronic alterations in cardiac structure and function, RT-qPCR was performed on RNA from primary rat neonatal cardiomyocytes or cardiac fibroblasts treated acutely with Iso (3 h). The cardiomyocytes did not express all of the transcripts examined, and Iso-induced changes in expression were only observed for Il1b and Il6, which were increased as observed in the heart, and Tnfa, which was decreased in opposition of the in vivo results (Fig. 9A). Each of the transcripts examined were detected in the cardiac fibroblasts, and aside from Il2, expression levels for all were significantly altered following Iso stimulation (Fig. 9B). EGFR-sensitivity of the Iso-induced changes in transcript expression was evaluated via pretreatment of the cells with the EGFR antagonist AG1478 (Table 5). As observed in the whole heart, pretreatment of the cardiomyocytes with the EGFR antagonist had no impact on Iso-enhanced Il1b and Il6 expression but did block Iso-mediated changes in Tnfa expression. In the cardiac fibroblasts, Iso-induced changes in Ccl6, Il1b, and Il6 expression were unaltered by EGFR inhibition, whereas Iso-mediated changes in Ccl2, Ccl20, Il17a, Tnfa, and Tnfsf11 expression were significantly inhibited by AG1478. These findings suggest that a majority of the EGFR-sensitive cytokine responses to Iso stimulation in the heart may be primarily mediated in cardiac fibroblasts.
Fig. 9.
Acute βAR-mediated and EGFR-sensitive regulation of cytokine expression occurs primarily in cardiac fibroblasts. RT-qPCR analysis of changes in cytokine expression following acute Iso stimulation (3 h) in primary isolated rat neonatal cardiomyocytes (A) or rat neonatal cardiac fibroblasts (B) is shown. *P ≤ 0.05, †P ≤ 0.01, ‡P ≤ 0.001 vs. Veh of same transcript; n = 4–8 each; 2-tailed t-test. ND, not detected.
Table 5.
RQ values from qRT-PCR analysis of isolated primary cardiac cell cytokine expression
| Gene Name | Vehicle | Iso | AG1478 | AG1478 + Iso |
|---|---|---|---|---|
| Cardiomyocyte | ||||
| n | 4 | 8 | 4 | 7 |
| Il1b | 1.00 ± 0.03 | 1.95 ± 0.23* | 0.81 ± 0.12 | 1.92 ± 0.24* |
| Il6 | 1.00 ± 0.16 | 2.92 ± 0.55* | 1.28 ± 0.23 | 3.44 ± 0.89* |
| Tnfa | 1.00 ± 0.16 | 0.58 ± 0.22* | 1.12 ± 0.07 | 1.21 ± 0.07‡ |
| Cardiac Fibroblast | ||||
| n | 5 | 6 | 4 | 5 |
| Ccl2 | 1.00 ± 0.20 | 11.51 ± 4.02* | 0.77 ± 0.26 | 1.68 ± 0.21‡ |
| Ccl6 | 1.00 ± 0.18 | 3.88 ± 1.31* | 1.51 ± 0.28 | 4.03 ± 0.82* |
| Ccl20 | 1.00 ± 0.17 | 1.92 ± 0.28* | 0.61 ± 0.07 | 0.97 ± 0.14‡ |
| Il1b | 1.00 ± 0.49 | 48.49 ± 24.55* | 1.34 ± 0.32 | 44.66 ± 18.03* |
| Il6 | 1.00 ± 0.17 | 9.50 ± 3.30* | 0.73 ± 0.11 | 8.78 ± 2.25* |
| Il17a | 1.00 ± 0.28 | 0.38 ± 0.09* | 0.84 ± 0.19 | 3.06 ± 1.14*‡ |
| Tnfa | 1.00 ± 0.11 | 9.17 ± 1.86* | 1.16 ± 0.20 | 2.63 ± 0.35‡ |
| Tnfsf11 | 1.00 ± 0.34 | 3.86 ± 0.64* | 0.99 ± 0.34 | 0.87 ± 0.41‡ |
Values are relative quantity (RQ) ± SE.
P < 0.05 vs. vehicle;
P < 0.05 vs. Iso, one-way ANOVA with Newman-Keuls multiple comparison test.
DISCUSSION
Chronic Iso infusion in mice is a common model system that mimics the elevated catecholamines and sustained βAR stimulation observed during HF that lead to structural and functional cardiac deficits (3, 11, 12, 29). Indeed, we found in our current study that Iso infusion in C57BL/6 mice increased cardiac hypertrophy, cardiomyocyte death, interstitial fibrosis, and cardiac dysfunction over time. Although the mechanisms responsible for relaying long-term structural and functional changes in response to chronic Iso are multivariate, alterations in cardiac gene expression, as detected via RT-PCR or microarray analysis, have been associated with cardiac dysfunction (3, 11, 29, 44). To more comprehensively assess the differential impact of chronic Iso infusion over time on cardiac transcript expression, we used RNAseq analysis, which identified 780 and 689 cardiac transcripts significantly altered in response to chronic Iso at 1 and 2 wk, respectively. The majority of the transcripts regulated at the two timepoints were distinct from one another and associated with cell growth and proliferation, cellular maintenance and cell death and survival pathways, each of which have been demonstrated to be regulated by cytokine signaling. Indeed, elevations in proinflammatory cytokines are associated with chronic heart failure and are correlated with disease severity and progression (6, 17, 35, 46), and although inflammation is necessary for healing, it can become maladaptive if prolonged and excessive. Thus orchestration of the inflammatory response is crucial for determining the outcome of cardiac injury (14). In particular, TNFα, IL-6, IL-1β, and IL-18 are known to contribute to cardiomyocyte hypertrophy, contractile dysfunction, cardiomyocyte apoptosis, and extracellular matrix remodeling in human HF and rodent models of HF (6, 17). Furthermore, chronic Iso infusion has been demonstrated to elevate expression of CCL2, TNF-α, IL-1β, and IL-6 in rodent hearts (12, 30). Transcriptome and network analysis of hearts from the Iso-infused mice revealed 48 cytokine transcripts altered in response to 1 or 2 wk of continuous βAR stimulation. Several of these genes, including Ccl2, Tnfa, Il1b, and Il6, have been shown to change with chronic Iso treatment while others have not been previously reported to be regulated by βAR, such as Ccl6, Ccl20, Osm, and Tslp.
EGFR activation is known to influence cytokine expression in several cell types including keratinocytes, epidermal, and inflammatory cells; however, the role of EGFR in the production of inflammatory mediators in the heart has not been investigated (20, 25, 34). Inhibition of EGFR both pharmacologically and using epidermis-selective EGFR knockout mice have shown that EGFR regulates production of a number of inflammatory mediators including CCL2, CCL5, TNF-α, and IL-1β (20, 27, 33, 34, 37). Furthermore, cytokines can increase MMP activity in inflamed tissue, leading to the cleavage of HB-EGF, further perpetuating EGFR involvement in inflammation (5). Previously we have shown that βAR-dependent EGFR transactivation acutely regulates cardiomyocyte apoptotic gene expression in vitro as well as cardiac transcript expression in vivo (13, 42). EGFR-sensitive cardiac transcripts acutely regulated by Iso in vivo were primarily involved in cell signaling, cell cycle, cell death, and cell development, including Thbs1 (42), which we show in this study to be similarly EGFR-sensitive even under conditions of chronic catecholamine stimulation. Thus we sought to determine the contribution of EGFR signaling in altering cardiac cytokine transcript expression following chronic βAR activation. Although the influence of βAR-mediated EGFR transactivation on cardiac cytokine expression has not been previously reported, here we have demonstrated that several Iso-regulated cytokine transcripts are differentially sensitive to EGFR inhibition, with some transcripts up- or downregulated in a temporally distinct manner (Tables 4 and 5 and Fig. 10). In some instances, as observed with IL-17a, EGFR transactivation actually acts to repress βAR-mediated transcript regulation, but under conditions of EGFR inhibition the remaining βAR signaling branches act overwhelmingly to increase transcript expression.
Fig. 10.
Temporal- and Gef-sensitive changes in cardiac transcript expression in response to chronic βAR stimulation. βAR stimulation induces differential changes in cardiac transcript expression following Iso infusion for 1 wk (left) vs. 2 wk (right). Transcripts are regulated via EGFR-dependent and -independent mechanisms. *Transcript (Il17a), whose expression is dominantly repressed by βAR-mediated EGFR transactivation but is significantly enhanced in the presence of Gef via EGFR-independent βAR signaling.
Although we have observed significant inhibition of βAR-mediated alterations in cardiac cytokine expression by concurrent EGFR inhibition, there exists the possibility that these changes occurred secondary to alterations in cardiac structure and function. Although we cannot rule out this possibility in the context of our in vivo experiments, our in vitro data demonstrates that acute βAR-mediated EGFR transactivation does directly influence cytokine transcript expression in the absence of chronic changes in cardiac structure and function. In particular, and consistent with their known role in inflammation, cardiac fibroblasts predominantly mediate βAR-dependent changes in cytokine expression in an EGFR-sensitive manner. However, some cytokines, such as IL2, were not altered by Iso stimulation in isolated cardiomyocytes or cardiac fibroblasts, suggesting a different cell population, likely immune cells, may be responsible for these changes.
Of the cytokines identified within the transcriptome analysis, Tnfa and Ccl2 are known to be regulated by both βAR stimulation and EGFR activation (3, 5, 29, 33); however, we show for the first time that βAR-mediated EGFR transactivation can regulate the expression of these transcripts, both acutely in isolated cardiac cells and chronically in vivo, where a sustained increase in Tnfa and Ccl2 expression in the heart and TNF-α and CCL2 levels in the plasma with continued Iso infusion were prevented with EGFR inhibition. The role of TNF-α in HF has been extensively studied and although it has been shown to be beneficial at earlier timepoints, excessive TNF-α becomes detrimental with chronic inflammation and promotes hypertrophy (39, 43, 47). TNF-α also contributes to cardiomyocyte apoptosis due to its ability to induce apoptosis through both intrinsic and extrinsic apoptotic pathways (15). Excessive TNF-α influences collagen synthesis in cardiac fibroblasts by causing an imbalance between extracellular matrix synthesis and degradation by dysregulation of degradative enzymes like MMPs, leading to LV dilation (24, 40, 41). CCL2 (monocyte chemoattractant protein-1, MCP-1) is also known to play an important role in the heart, primarily through its ability to recruit monocytes/macrophages to the site of inflammation. Overexpression of CCL2 leads to cardiac hypertrophy, dilation, and increased LV dysfunction (31). In addition to its role in chemotaxis, CCL2 has a direct effect on cardiomyocytes through the induction of ER-stress-response proteins, which protects cardiomyocytes against further stress (1). CCL2 has been found to promote fibrosis through several different mechanisms in different tissues including production of MMPs in immune cells and myocytes and stimulation of TGF-β secretion to increase extracellular matrix production (9, 16, 45). Thus our findings identify βAR-mediated EGFR transactivation as a mechanism of cytokine regulation in the heart and may help to explain why in this study we observe an ameliorative effect of pharmacologic EGFR inhibition on cardiac hypertrophy and apoptosis in response to chronic Iso infusion. The earliest in vivo timepoint examined in this study followed 1 wk of Iso infusion, and although cardiac function was maintained, structural changes were already evident. Due to the importance of temporal expression changes in directing a balance between beneficial versus detrimental effects of cytokines, and with consideration of the rapid changes in cytokine expression in response to βAR-mediated EGFR transactivation we observed in isolated cardiac fibroblasts, it may be useful to examine in vivo cytokine changes at earlier timepoints before substantial cardiomyocyte death and fibrosis has occurred.
Concurrent EGFR inhibition prevented chronic Iso-mediated cell death, hypertrophy, and contractile dysfunction; however, Gef alone induced interstitial fibrosis that was independent of Iso-mediated fibrosis. Thus we have shown that EGFR-transactivation influences some, but not all, cardiac remodeling associated with chronic Iso-induced HF. Other studies have shown that transactivation of EGFR by distinct GPCRs including EP4, α1AR, angiotensin II, and urotensin II promotes hypertrophy (10, 18, 23, 26, 28, 48). Although global EGFR genetic ablation is embryonic lethal, EGFR inhibition alone in adult animals has been demonstrated to impact cardiac structure and function. Female C57BL/6 mice treated orally for 3 mo with the EGFR inhibitors EKB-569 or AG-1478 displayed decreased cardiac function with increases in LV wall thickness, cell death, and as we observe in our study, fibrosis (2). Furthermore, SM22 promoter-mediated deletion of EGFR in vascular smooth muscle cells and partial EGFR deletion in cardiomyocytes was recently shown to increase cardiac fibrosis, LV wall thickness and LV diameter, supporting the observation that EGFR inhibition can increase fibrosis (38). Our findings contrast with previously reported enhancement of chronic catecholamine-induced cardiac dysfunction and apoptosis with the EGFR antagonist erlotinib, administered via daily intraperitoneal injection (32). In our study, we have used the EGFR antagonist gefitinib, with a different selectivity profile than erlotinib (8), administered via constant subcutaneous infusion, which may account for the differences in pathophysiologic outcomes of the two EGFR inhibitors under conditions of chronic catecholamine stimulation. A more refined genetic approach, such as conditional cardiac-specific deletion of EGFR, would be useful in future studies to definitively attribute changes in cardiac pathophysiology specifically to βAR-mediated EGFR transactivation.
In summary, using an RNASeq approach, we have demonstrated that chronic βAR stimulation induces temporally dynamic alterations in cardiac transcript expression associated with increased cardiac remodeling and decreased cardiac contractility. In particular, changes in cytokine transcript expression in response to βAR activation over time may reflect a worsening phenotype and are differentially sensitive to inhibition of EGFR signaling. Although βAR-mediated EGFR transactivation has been shown to promote survival signaling, especially at acute timepoints (13, 42), EGFR inhibition in this study led to long-term decreases in cardiac hypertrophy and apoptosis, as well as altered cytokine expression, in response to chronic Iso stimulation. Acutely, these effects appear to predominate in cardiac fibroblasts. Overall, the potential benefit of targeting EGFR-dependent βAR signaling in the treatment of HF may best be considered temporally, such that specific windows of opportunity for selective activation or inhibition of this pathway and resulting cytokine expression can adequately relay cardioprotective outcomes.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grants HL-105414 (to D. G. Tilley) and HL-091799 (to W. J. Koch) and the American Heart Association postdoctoral fellowship (to L. A. Grisanti).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: L.A.G. and D.G.T. conception and design of research; L.A.G., A.A.R., J.A.T., J.I.G., and R.L.C. performed experiments; L.A.G. and D.G.T. analyzed data; L.A.G. and D.G.T. interpreted results of experiments; L.A.G. and D.G.T. prepared figures; L.A.G. and D.G.T. drafted manuscript; L.A.G. and D.G.T. edited and revised manuscript; W.J.K. and D.G.T. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
Present address of J. A. Talarico: Clinical Trials Office, Children's Hospital of Philadelphia.
Present address of J. I. Gold: University of Chicago.
REFERENCES
- 1.Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol 291: H1411–H1420, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrick CJ, Yu M, Chao HH, Threadgill DW. Chronic pharmacologic inhibition of EGFR leads to cardiac dysfunction in C57BL/6J mice. Toxicol Appl Pharmacol 228: 315–325, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boluyt MO, Long X, Eschenhagen T, Mende U, Schmitz W, Crow MT, Lakatta EG. Isoproterenol infusion induces alterations in expression of hypertrophy-associated genes in rat heart. Am J Physiol Heart Circ Physiol 269: H638–H647, 1995. [DOI] [PubMed] [Google Scholar]
- 4.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 20: 1453–1454, 2004. [DOI] [PubMed] [Google Scholar]
- 5.El-Abaseri TB, Hammiller B, Repertinger SK, Hansen LA. The epidermal growth factor receptor increases cytokine production and cutaneous inflammation in response to ultraviolet irradiation. ISRN dermatol 2013: 848705, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Menyar AA. Cytokines and myocardial dysfunction: state of the art. J Card Fail 14: 61–74, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Errami M, Galindo CL, Tassa AT, Dimaio JM, Hill JA, Garner HR. Doxycycline attenuates isoproterenol- and transverse aortic banding-induced cardiac hypertrophy in mice. J Pharmacol Exp Ther 324: 1196–1203, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Fabian MA, Biggs WH 3rd Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat biotechnol 23: 329–336, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Frangogiannis NG, Dewald O, Xia Y, Ren G, Haudek S, Leucker T, Kraemer D, Taffet G, Rollins BJ, Entman ML. Critical role of monocyte chemoattractant protein-1/CC chemokine ligand 2 in the pathogenesis of ischemic cardiomyopathy. Circulation 115: 584–592, 2007. [DOI] [PubMed] [Google Scholar]
- 10.Frias MA, Rebsamen MC, Gerber-Wicht C, Lang U. Prostaglandin E2 activates Stat3 in neonatal rat ventricular cardiomyocytes: a role in cardiac hypertrophy. Cardiovasc Res 73: 57–65, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Galindo CL, Skinner MA, Errami M, Olson LD, Watson DA, Li J, McCormick JF, McIver LJ, Kumar NM, Pham TQ, Garner HR. Transcriptional profile of isoproterenol-induced cardiomyopathy and comparison to exercise-induced cardiac hypertrophy and human cardiac failure. BMC physiol 9: 23, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garlie JB, Hamid T, Gu Y, Ismahil MA, Chandrasekar B, Prabhu SD. Tumor necrosis factor receptor 2 signaling limits beta-adrenergic receptor-mediated cardiac hypertrophy in vivo. Basic Res Cardiol 106: 1193–1205, 2011. [DOI] [PubMed] [Google Scholar]
- 13.Grisanti LA, Talarico JA, Carter RL, Yu JE, Repas AA, Radcliffe SW, Tang HA, Makarewich CA, Houser SR, Tilley DG. β-Adrenergic receptor-mediated transactivation of epidermal growth factor receptor decreases cardiomyocyte apoptosis through differential subcellular activation of ERK1/2 and Akt. J Mol Cell Cardiol 72: 39–51, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gullestad L, Ueland T, Vinge LE, Finsen A, Yndestad A, Aukrust P. Inflammatory cytokines in heart failure: mediators and markers. Cardiology 122: 23–35, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Haudek SB, Taffet GE, Schneider MD, Mann DL. TNF provokes cardiomyocyte apoptosis and cardiac remodeling through activation of multiple cell death pathways. J Clin Invest 117: 2692–2701, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haudek SB, Xia Y, Huebener P, Lee JM, Carlson S, Crawford JR, Pilling D, Gomer RH, Trial J, Frangogiannis NG, Entman ML. Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proc Natl Acad Sci USA 103: 18284–18289, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedayat M, Mahmoudi MJ, Rose NR, Rezaei N. Proinflammatory cytokines in heart failure: double-edged swords. Heart Fail Rev 15: 543–562, 2010. [DOI] [PubMed] [Google Scholar]
- 18.Kagiyama S, Eguchi S, Frank GD, Inagami T, Zhang YC, Phillips MI. Angiotensin II-induced cardiac hypertrophy and hypertension are attenuated by epidermal growth factor receptor antisense. Circulation 106: 909–912, 2002. [DOI] [PubMed] [Google Scholar]
- 19.Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation 122: 2727–2735, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb DJ, Modjtahedi H, Plant NJ, Ferns GA. EGF mediates monocyte chemotaxis and macrophage proliferation and EGF receptor is expressed in atherosclerotic plaques. Atherosclerosis 176: 21–26, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Lewin G, Matus M, Basu A, Frebel K, Rohsbach SP, Safronenko A, Seidl MD, Stumpel F, Buchwalow I, Konig S, Engelhardt S, Lohse MJ, Schmitz W, Muller FU. Critical role of transcription factor cyclic AMP response element modulator in beta1-adrenoceptor-mediated cardiac dysfunction. Circulation 119: 79–88, 2009. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Ertel A, Portocarrero C, Barkhouse DA, Dietzschold B, Hooper DC, Faber M. Postexposure treatment with the live-attenuated rabies virus (RV) vaccine TriGAS triggers the clearance of wild-type RV from the Central Nervous System (CNS) through the rapid induction of genes relevant to adaptive immunity in CNS tissues. J Virol 86: 3200–3210, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Zhang H, Liao W, Song Y, Ma X, Chen C, Lu Z, Li Z, Zhang Y. Transactivated EGFR mediates α1-AR-induced STAT3 activation and cardiac hypertrophy. Am J Physiol Heart Circ Physiol 301: H1941–H1951, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Li YY, Feng YQ, Kadokami T, McTiernan CF, Draviam R, Watkins SC, Feldman AM. Myocardial extracellular matrix remodeling in transgenic mice overexpressing tumor necrosis factor alpha can be modulated by anti-tumor necrosis factor alpha therapy. Proc Natl Acad Sci USA 97: 12746–12751, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichtenberger BM, Gerber PA, Holcmann M, Buhren BA, Amberg N, Smolle V, Schrumpf H, Boelke E, Ansari P, Mackenzie C, Wollenberg A, Kislat A, Fischer JW, Rock K, Harder J, Schroder JM, Homey B, Sibilia M. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci Transl Med 5: 199ra111, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Liu JC, Chen CH, Chen JJ, Cheng TH. Urotensin II induces rat cardiomyocyte hypertrophy via the transient oxidization of Src homology 2-containing tyrosine phosphatase and transactivation of epidermal growth factor receptor. Mol Pharmacol 76: 1186–1195, 2009. [DOI] [PubMed] [Google Scholar]
- 27.Mascia F, Mariani V, Girolomoni G, Pastore S. Blockade of the EGF receptor induces a deranged chemokine expression in keratinocytes leading to enhanced skin inflammation. Am J Pathol 163: 303–312, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez M, LaPointe MC. PGE2-induced hypertrophy of cardiac myocytes involves EP4 receptor-dependent activation of p42/44 MAPK and EGFR transactivation. Am J Physiol Heart Circ Physiol 288: H2111–H2117, 2005. [DOI] [PubMed] [Google Scholar]
- 29.Molojavyi A, Lindecke A, Raupach A, Moellendorf S, Kohrer K, Godecke A. Myoglobin-deficient mice activate a distinct cardiac gene expression program in response to isoproterenol-induced hypertrophy. Physiol Genomics 41: 137–145, 2010. [DOI] [PubMed] [Google Scholar]
- 30.Murray DR, Prabhu SD, Chandrasekar B. Chronic beta-adrenergic stimulation induces myocardial proinflammatory cytokine expression. Circulation 101: 2338–2341, 2000. [DOI] [PubMed] [Google Scholar]
- 31.Niu J, Kolattukudy PE. Role of MCP-1 in cardiovascular disease: molecular mechanisms and clinical implications. Clin Sci (Lond) 117: 95–109, 2009. [DOI] [PubMed] [Google Scholar]
- 32.Noma T, Lemaire A, Naga Prasad SV, Barki-Harrington L, Tilley DG, Chen J, Le Corvoisier P, Violin JD, Wei H, Lefkowitz RJ, Rockman HA. β-Arrestin-mediated beta1-adrenergic receptor transactivation of the EGFR confers cardioprotection. J Clin Invest 117: 2445–2458, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastore S, Mascia F, Mariani V, Girolomoni G. The epidermal growth factor receptor system in skin repair and inflammation. J Invest Dermatol 128: 1365–1374, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Paul T, Schumann C, Rudiger S, Boeck S, Heinemann V, Kachele V, Steffens M, Scholl C, Hichert V, Seufferlein T, Stingl JC. Cytokine regulation by epidermal growth factor receptor inhibitors and epidermal growth factor receptor inhibitor associated skin toxicity in cancer patients. Eur J Cancer 50: 1855–1863, 2014. [DOI] [PubMed] [Google Scholar]
- 35.Petersen JW, Felker GM. Inflammatory biomarkers in heart failure. Congest heart fail 12: 324–328, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375, 2003. [DOI] [PubMed] [Google Scholar]
- 37.Scholes AG, Hagan S, Hiscott P, Damato BE, Grierson I. Overexpression of epidermal growth factor receptor restricted to macrophages in uveal melanoma. Arch Ophthalmol 119: 373–377, 2001. [DOI] [PubMed] [Google Scholar]
- 38.Schreier B, Rabe S, Schneider B, Bretschneider M, Rupp S, Ruhs S, Neumann J, Rueckschloss U, Sibilia M, Gotthardt M, Grossmann C, Gekle M. Loss of epidermal growth factor receptor in vascular smooth muscle cells and cardiomyocytes causes arterial hypotension and cardiac hypertrophy. Hypertension 61: 333–340, 2013. [DOI] [PubMed] [Google Scholar]
- 39.Sharma R, Anker SD. Cytokines, apoptosis and cachexia: the potential for TNF antagonism. Int J Cardiol 85: 161–171, 2002. [DOI] [PubMed] [Google Scholar]
- 40.Sivasubramanian N, Coker ML, Kurrelmeyer KM, MacLellan WR, DeMayo FJ, Spinale FG, Mann DL. Left ventricular remodeling in transgenic mice with cardiac restricted overexpression of tumor necrosis factor. Circulation 104: 826–831, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Siwik DA, Colucci WS. Regulation of matrix metalloproteinases by cytokines and reactive oxygen/nitrogen species in the myocardium. Heart Fail Rev 9: 43–51, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Talarico JA, Carter RL, Grisanti LA, Yu JE, Repas AA, Tilley DG. β-Adrenergic receptor-dependent alterations in murine cardiac transcript expression are differentially regulated by gefitinib in vivo. PloS one 9: e99195, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tovey MG. Expression of the genes of interferons and other cytokines in normal and diseased tissues of man. Experientia 45: 526–535, 1989. [DOI] [PubMed] [Google Scholar]
- 44.Wong J, Chang C, Agrawal R, Walton GB, Chen C, Murthy A, Patterson AJ. Gene expression profiling: classification of mice with left ventricle systolic dysfunction using microarray analysis. Crit Care Med 38: 25–31, 2010. [DOI] [PubMed] [Google Scholar]
- 45.Xia Y, Frangogiannis NG. MCP-1/CCL2 as a therapeutic target in myocardial infarction and ischemic cardiomyopathy. Inflamm Allergy Drug Targets 6: 101–107, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Yndestad A, Damas JK, Oie E, Ueland T, Gullestad L, Aukrust P. Systemic inflammation in heart failure—the whys and wherefores. Heart Fail Rev 11: 83–92, 2006. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama T, Nakano M, Bednarczyk JL, McIntyre BW, Entman M, Mann DL. Tumor necrosis factor-alpha provokes a hypertrophic growth response in adult cardiac myocytes. Circulation 95: 1247–1252, 1997. [DOI] [PubMed] [Google Scholar]
- 48.Zeng SY, Chen X, Chen SR, Li Q, Wang YH, Zou J, Cao WW, Luo JN, Gao H, Liu PQ. Upregulation of Nox4 promotes angiotensin II-induced epidermal growth factor receptor activation and subsequent cardiac hypertrophy by increasing ADAM17 expression. The Can J Cardiol 29: 1310–1319, 2013. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Szeto C, Gao E, Tang M, Jin J, Fu Q, Makarewich C, Ai X, Li Y, Tang A, Wang J, Gao H, Wang F, Ge XJ, Kunapuli SP, Zhou L, Zeng C, Xiang KY, Chen X. Cardiotoxic and cardioprotective features of chronic beta-adrenergic signaling. Circ Res 112: 498–509, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.