Abstract
Sodium-coupled SLC12 cation chloride cotransporters play important roles in cell volume and chloride homeostasis, epithelial fluid secretion, and renal tubular salt reabsorption. These cotransporters are phosphorylated and activated indirectly by With-No-Lysine (WNK) kinases through their downstream effector kinases, Ste20- and SPS1-related proline alanine-rich kinase (SPAK) and oxidative stress-responsive kinase 1 (OSR1). Multiple WNK kinases can coexist within a single cell type, although their relative contributions to SPAK/OSR1 activation and salt transport remain incompletely understood. Deletion of specific WNKs from cells that natively express a functional WNK-SPAK/OSR1 network will help resolve these knowledge gaps. Here, we outline a simple method to selectively knock out full-length WNK1 expression from mammalian cells using RNA-guided clustered regularly interspaced short palindromic repeats/Cas9 endonucleases. Two clonal cell lines were generated by using a single-guide RNA (sgRNA) targeting exon 1 of the WNK1 gene, which produced indels that abolished WNK1 protein expression. Both cell lines exhibited reduced endogenous WNK4 protein abundance, indicating that WNK1 is required for WNK4 stability. Consistent with an on-target effect, the reduced WNK4 abundance was associated with increased expression of the KLHL3/cullin-3 E3 ubiquitin ligase complex and was rescued by exogenous WNK1 overexpression. Although the morphology of the knockout cells was indistinguishable from control, they exhibited low baseline SPAK/OSR1 activity and failed to trigger regulatory volume increase after hypertonic stress, confirming an essential role for WNK1 in cell volume regulation. Collectively, our data show how this new, powerful, and accessible gene-editing technology can be used to dissect and analyze WNK signaling networks.
Keywords: WNK1, SLC12 cotransporters, genome editing, CRISPR/Cas system
slc12 cotransporters mediate the electroneutral movement of cations with Cl− across membranes (10). To date, nine SLC12 family members have been identified. Among these is a subgroup of sodium-dependent cation chloride cotransporters, which include the bumetanide-sensitive Na+-K+-2Cl− cotransporters NKCC1 (SLC12A2) and NKCC2 (SLC12A1), and the thiazide-sensitive Na-Cl cotransporter NCC (SLC12A3). In the kidney, NKCC2 and NCC are expressed at the apical membrane of epithelial cells of the thick ascending limb of the loop of Henle (TAL) and the distal convoluted tubule (DCT), respectively. Both of these cotransporters reabsorb sodium with chloride from the tubular lumen and thus help control extracellular fluid volume and blood pressure (10). In contrast, NKCC1 is nearly ubiquitously expressed and plays an important role in the maintenance of cell volume and intracellular chloride concentration (24). NKCC1 is also enriched in the basolateral membranes of the renal collecting duct and polarized secretory epithelia, where it facilitates the vectorial movement of ions into the tubular lumen (11, 22, 24, 46).
For the sodium-dependent SLC12 cotransporters to be maximally active, they must be phosphorylated at specific serines and threonines harbored within their intracellular N termini (31). A network of serine threonine kinases mediates this process. Direct phosphorylation of the cotransporters is carried out by two homologous tonicity-responsive serine threonine kinases, Ste20- and SPS1-related proline alanine-rich kinase (SPAK) and oxidative stress-responsive kinase 1 (OSR1) (27, 44). Immediately upstream of SPAK and OSR1 are the With-No-Lysine (WNK) kinases, which activate SPAK and OSR1 via T-loop phosphorylation (48). Mammals harbor four WNK paralogs; three of these, WNK1, WNK3, and WNK4, are expressed in the kidney (25, 33). All of the WNKs share a similar domain architecture, consisting of an N-terminal serine-threonine kinase domain, an autoinhibitory domain (AID), and several coiled-coil and SPAK/OSR1 binding domains (31). The coiled-coil domains facilitate the formation of WNK homo- and heterooligomers (4, 21, 42). Once associated in complexes, the kinase domains can trans-autophosphorylate their partners, while the AIDs can cross-inhibit the activity of other associated WNKs (42, 51, 52). Thus WNK kinases assemble into higher order regulatory complexes to carry out their downstream effects on SPAK/OSR1 activity and electroneutral salt transport. The stability of these complexes is likely dependent on their oligomeric composition and stoichiometry, and through interactions with the Kelch-like 3/cullin 3 (KLHL3/CUL3) E3 ubiquitin ligase complex (29, 45).
A broad range of hormones and osmotic stressors that stimulate sodium-dependent cation chloride cotransport utilize the WNK-SPAK/OSR1 cascade to mediate their effects (31, 38). Some evidence suggests that these diverse physiological stimuli convergently trigger SPAK/OSR1 phosphorylation by signaling through specific WNKs (3, 41). However, it remains unclear whether a single WNK kinase, or whether the coordinated action of multiple WNKs is required to carry out a specific physiological response. Tackling this question in whole animal models is challenging due to a number of issues, including the heterogeneity of cell types in renal parenchyma, the discrepancies in WNK and SPAK/OSR1 expression in different segments of the nephron, and the embryonic lethality of deleting some components of the network, including WNK1 and OSR1 (49, 54). A commonly used alternative strategy is to knock down endogenous WNKs in cell lines that express an intact WNK-SPAK/OSR1 network by RNA interference (RNAi). A drawback of this approach, however, is that small interfering (si) RNA-mediated gene silencing is rarely 100% effective at eliminating the expression of a target gene product. Indeed, in our own studies, we have only been able to reduce the expression of a specific WNK kinase to about half of its native abundance (39); similar results have been reported by others (4). Thus RNAi experiments can only support, but not verify, whether a specific WNK kinase is essential for transducing a specific physiological response. To resolve this critical question, cell lines that completely lack the expression of one or more WNKs are needed.
To determine the role of the individual WNKs in signal transduction and cation chloride cotransporter activation, we are using clustered regularly interspaced short palindromic repeats (CRISPR) RNA-guided Cas9 nucleases to knock out components of the WNK-SPAK/OSR1 signaling pathway in mammalian cells. In the present study, we describe how we used this novel method to generate a mammalian WNK1 knockout cell line. In our experience, the technique is straightforward, cost effective, and efficient, permitting the generation of multiple genetically and biochemically validated knockout clones within weeks. We report that WNK1 knockout cells exhibit a substantial reduction in WNK4 expression and SPAK/OSR1 activity. As expected, they also display impaired volume regulation in response to hypertonic stress, consistent with a critical role for WNK1 in mediating NKCC1-dependent regulatory volume increase (RVI). We also report that total WNK1 deletion increases the abundance of the KLHL3/CUL3 complex, suggesting a new layer of feedback regulation between WNK1 and its cognate E3 ubiquitin ligase.
MATERIALS AND METHODS
Antibodies, plasmids, and reagents.
We used the following antibodies in this study: rabbit polyclonal anti-N-terminal WNK1 (exon 1; Sigma), rabbit polyclonal C-terminal WNK1 (exon 28; Sigma), rabbit polyclonal anti-WNK4 (Novus Biologicals) (39), rabbit polyclonal anti-WNK2 (ab28852; Abcam), rabbit polyclonal anti-SPAK (C-terminal; Cell Signaling Technology) (34), rabbit polyclonal anti-OSR1 (no. 3729; Cell Signaling Technology); rabbit polyclonal anti-cullin 3 (ab1871; Abcam) (1), rabbit polyclonal anti-Kelch-like 3 (ab66655; Abcam) (2, 26), mouse monoclonal anti-actin (Sigma); and rabbit polyclonal anti-tubulin (Bethyl). The rabbit polyclonal anti-WNK3 antibody was manufactured by Millipore (no. 07-2262) in collaboration with the University of Dundee's Division of Signal Transduction Therapy. The immunogen was a glutathione-S-transferase (GST) fusion protein to residues 1142–1461 of the human protein. The immunogenicity of this recombinant protein was confirmed in previous studies (42). The rabbit polyclonal anti-phospho S373/S325 SPAK/OSR1 (S motif) antibody, validated by immunoblotting in lysates of hypotonic low Cl−-stimulated HEK-293 cells treated with and without λ phosphatase, was from Millipore (no. 07-2273). Secondary horseradish peroxidase-conjugated antibodies raised in goat were from Jackson ImmunoResearch. The bicistronic pX330 vector containing cDNAs encoding human codon-optimized Streptococcus pyogenes Cas9 (hSpCas9) and an adaptable CRISPR RNA (crRNA)/trans-activating crRNA chimera containing adjacent Bbs I cloning sites for protospacer “guide sequence” insertion was purchased from Addgene (plasmid no. 42230). To generate the N-terminal hemagglutinin (HA)-tagged L-WNK1-pcDNA3.1 construct, a 5′ Eco RI-Pac I L-WNK1 fragment encoding the HA tag was swapped with the corresponding 5′-end of the original N-terminal myc-tagged L-WNK1 cDNA (50) in pcDNA3.1, using standard subcloning methods. All reagents were purchased from Sigma unless otherwise noted.
WNK1 single-guide RNA expression vector construction.
A 20-bp guide sequence (5′-GCACTCTGCGGGACAGCCGC-3′) targeting DNA within the first exon of WNK1 was selected from a published database of predicted high-specificity protospacer adjacent motif (PAM) target sites in the human exome (23). Two complementary oligos (5′-CACCGCACTCTGCGGGACAGCCGC-3′ and 5′-AAACGCGGCTGTCCCGCAGAGTGC-3′) containing the WNK1 guide sequence and BbsI ligation adapters were synthesized by IDT. One hundred micromolar of each oligo was annealed using T4 polynucleotide kinase (New England Biolabs) and 1 μl 10× T4 Ligation Buffer in a total volume of 10 μl in a Bio-Rad thermal cycler. The cycling conditions were 37°C for 30 min, then 95°C for 5 min, followed by a ramp to 25°C at 5°C/min. The annealed oligo was ligated into the BbsI-digested pX330 vector using 5 μl of 2× QuickLigation Buffer and 1 μl of QuickLigase (New England Biolabs). The ligation mixture was treated with PlasmidSafe exonuclease (Epicentre) and transformed in OneShot chemically competent Stbl3 cells (Life Technologies). After plasmid DNA extraction (Qiagen), the sequence of the construct was verified by automated DNA sequence analysis performed at the University of Pittsburgh Genomics and Proteomics Core Laboratories (GPCL).
DNA mismatch-specific (T7E1) endonuclease assay.
HEK293T cells in six- well dishes were cultured as describe previously to 50–60% confluence (28). Cells were transfected with 1 μg of WNK1 single-guide (sg) RNA plasmid and 5 μl of Lipofectamine 2000/well. Cells were also transfected with pX330 empty vector as a control. Seventy-two hours posttransfection, the cells were harvested and genomic DNA was extracted (QuickExtract DNA extraction solution, Epicentre). A region of exon 1 of the WNK1 gene was amplified with genomic DNA-specific primers (forward primer, 5′-CCCTTCACGCCCTTTTCGTTCACGAATCC-3′; reverse primer, 5′-ACCTGCAGTTCACACCAGGCGACTTCC-3′). Homoduplex PCR products were denatured and rehybridized using stepdown annealing conditions to generate homo- and heteroduplexes. The mixture of duplexes was treated with T7E1 nuclease for 15 min at 37°C (New England Biolabs). The reaction was stopped using 1.5 μl of 0.25 M EDTA, and the products were analyzed on a 2% agarose gel.
Transfection and cell sorting.
HEK293T cells were cultured in six-well dishes to 70–80% confluence. Cells were cotransfected with 1 μg of WNK1 sgRNA plasmid plus 1 μg of pEGFP-N1 (Clontech) and 5 μl of Lipofectamine 2000/well. pEGFP-N1-derived enhanced green fluorescent protein (eGFP) was used as a fluorescent marker to sort transfected cells. Forty-eight hours posttransfection, cells were pelleted in PBS+ 2% FBS and sorted in 96-well plates using fluorescence-activated cell sorting (FACS) with a FACSAria II cell sorter (BD BioSciences). Single cells from two populations of GFP-expressing cells (high expression and medium expression) were expanded to obtain individual clones.
Clone validation.
Individual clones were lysed in detergent solution for 20 min at 4°C as described previously (8). The lysate was centrifuged at 16,000 g for 10 min, and 20 μg of supernatant was fractionated on 4–20% SDS-PAGE gels, transferred to nitrocellulose, and screened by immunoblotting with WNK1 antibodies. Genomic DNA was isolated from edited clones and nonedited HEK293T control cells as described above. Exon 1 of WNK1 was PCR amplified using the WNK1-specific PCR primers described above. The PCR products were A-tailed and cloned into pGEM-T Easy (Promega). Individually cloned amplicons were then analyzed by Sanger sequencing (GPCL). For imaging studies evaluating cellular morphology, cells were plated on Biocoat coverslips (BD), fixed for 30 min in 2% paraformaldehyde, and evaluated by differential interference contrast (DIC) microscopy using a Leica DM 6000 epifluorescence/DIC microscope equipped with a Retiga 400R digital imaging camera.
RT-PCR.
To detect the mRNA expression of endogenous WNK kinases in HEK293T cells, RNA was extracted from unedited cells using TRIzol (Life Technologies), and the RNA was reverse transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad). RT-PCR reactions for the four WNK kinases were carried out using the following primer sets: WNK1-forward: 5′- CGTCTGGAACACTTAAAACGTATCT-3′; WNK1-reverse: 5′- CACCAGCTTCTTAGAACTTTGATCT-3′ (43); WNK2-forward: 5′- ACGTCTATGCCTTTGGGATGT-3′; WNK2-reverse: 5′-GATCTCGTACCTTTCCTCCTT GT-3′ (14); WNK3-forward: 5′-ATTCAAGATAGCCCTGCACAAT-3′; WNK3-reverse: 5′-GTCAGAGGAATGGATCAGAAG-3′ (12); and WNK4-forward: 5′-TGCCTTGTCTATTCCACGGTCTG-3′; WNK4-reverse: 5′- CAGCTGCAATTTCTTCTGGGCTG-3′ (18).
Cell volume regulation studies.
Cell volume change was determined using calcein as a marker of intracellular water volume, as established previously (20). Briefly, cells on coverslips were incubated with 0.5 μM calcein-AM for 30 min at 37°C. The cells were placed in a heated (37°C) imaging chamber (Warner Instruments, Hamden, CT) on a Nikon Ti Eclipse inverted epifluorescence microscope equipped with perfect focus, a 40X Super Fluor oil-immersion objective lens, and a Princeton Instruments MicroMax CCD camera. Calcein fluorescence was monitored using a FITC filter set (excitation 480 nm, emission 535 nm, Chroma Technology, Rockingham, VT). Images were collected every 60 s with MetaFluor image-acquisition software (Molecular Devices, Sunnyvale, CA), and regions of interest (∼20–30 cells) were selected. Baseline drift resulting from photobleaching and dye leakage was corrected as described before (20). The fluorescence change was plotted as a function of the reciprocal of the relative osmotic pressure and the resulting calibration curve applied to all subsequent experiments as described before (20). The HEPES-buffered isotonic solution contained (in mM, pH 7.4) 100 NaCl, 5.4 KCl, 1.3 CaCl2, 0.8 MgSO4, 20 HEPES, 5.5 glucose, 0.4 NaHCO3. Anisosmotic solutions (250, 317, and 800 mosmol/kg H2O) were prepared by removal or addition of sorbitol to the above solution. Osmolarity was determined using an osmometer (Advanced Instruments, Norwood, MA).
Data analysis.
Quantification of Western blots was carried out using National Institutes of Health ImageJ software. Statistical analysis was performed using GraphPad Prism software. Measurements are presented as means ± SE. Comparisons between two groups were analyzed by Student's t-test. Multiple comparisons were performed by one-way ANOVA followed by either Dunnett's or Tukey's post hoc test. A base P value of <0.05 was considered statistically significant.
RESULTS
Generation of WNK1 knockout cell lines.
CRISPR/Cas systems are RNA-programmable host defense mechanisms in bacteria and archaea that degrade foreign nucleic acid (15). In 2013, multiple groups engineered the type II CRISPR system of S. pyogenes to edit genes in mammalian cells (6, 17, 23). This was accomplished by optimizing the S. pyogenes Cas9 enzyme for expression and localization in eukaryotic nuclei, and by fusing the essential noncoding RNA components of the CRISPR locus into a sgRNA whose 5′-end could be programmed with a 20-bp guide sequence for gene-specific targeting. While the 3′-end of the sgRNA associates with the Cas9 enzyme, the 5′ guide sequence will bind to its complementary genomic target through Watson-Crick base pairing. The target is always positioned next to a PAM. In S. pyogenes, the PAM corresponds to the sequence “NGG,” where N can be any nucleotide. When bound to this target site, the sgRNA-associated Cas9 enzyme generates a double stranded break near the PAM that undergoes imperfect repair by nonhomologous end joining (NHEJ). This results in the formation of insertions or deletions (“indels”) that create a frameshift mutation and premature termination of protein synthesis (30).
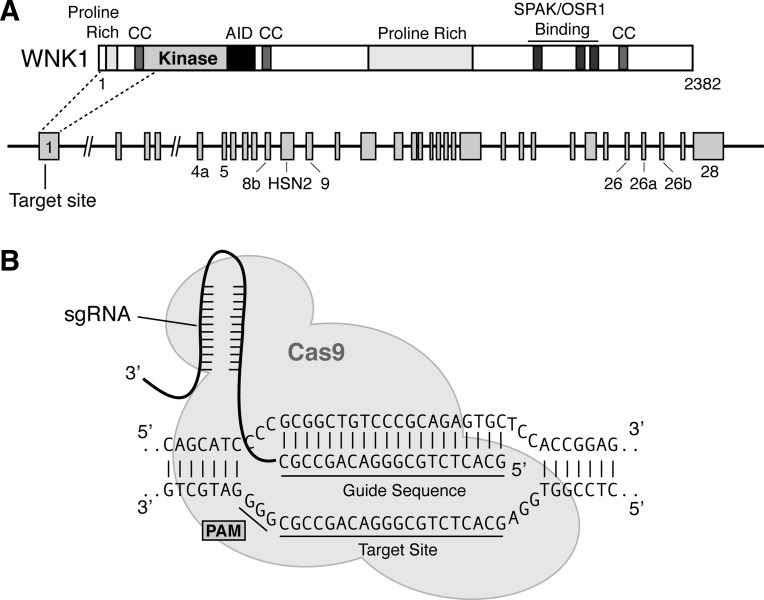
We first set out to identify an appropriate target site in WNK1 for gene editing. The site was selected from a bioinformatic database of 23-bp target-PAM sequences in the human exome that was filtered to minimize off-target cross-reactivity (23). We focused on exon 1 of WNK1 as a putative target site, because NHEJ-mediated frameshifts within this region could potentially terminate translation upstream of the N-terminal kinase domain, thereby eliminating kinase activity (Fig. 1A). From a list of several candidates, we chose a target sequence spanning bases 325–344 of the human WNK1 cDNA (CCDS8506.1). On the minus strand of the WNK1 gene, these nucleotides are positioned immediately 5′ to the trinucleotide PAM sequence “GGG” (Fig. 1B). Once identified, an oligo pair containing the guide sequence was cloned into pX330, a bicistronic vector that contains the Cas9 enzyme and the programmable chimeric sgRNA (6). For our initial attempt at generating a WNK knockout cell line, we elected to use HEK293T cells as the model system. We chose this cell line for two reasons. First, HEK239T cells are commonly used to edit genes with CRISPR/Cas technology, making them a well-established model to test the efficacy of RNA-guided endonucleases in deleting WNK kinase expression. Second, HEK293T cells are a frequently used model to study cation chloride cotransporter activity, trafficking, and regulation by the WNK-SPAK/OSR1 pathway.
Fig. 1.
With-No-Lysine (WNK) 1 single-guide (sg) RNA and target site. A, top: domain structure of full-length kinase active human WNK1 (also referred to as “L-WNK1”) (40). The locations of the kinase domain, 3 coiled coil-domains (CC), proline-rich domains, an autoinhibitory domain (AID), and C-terminal Ste20- and SPS1-related proline alanine-rich kinase (SPAK)/oxidative stress-responsive kinase 1 (OSR1) binding motifs are shown. Bottom: schematic representation of the WNK1 gene. The sgRNA target site is located at the 5′-end of exon 1, which encodes amino acids upstream of the kinase domain activation loop. B: close-up representation of the sgRNA target site. The 20-nt guide sequence comprising the 5′-end of the chimeric sgRNA is shown. This sequence pairs with the DNA target site (indicated on the bottom strand). Immediately 3′ to the target sequence is the trinucleotide protospacer adjacent motif (PAM; 5′-NGG). The Cas9 enzyme introduces a double stranded break (DSB) near the PAM, triggering imperfect nonhomologous end joining.
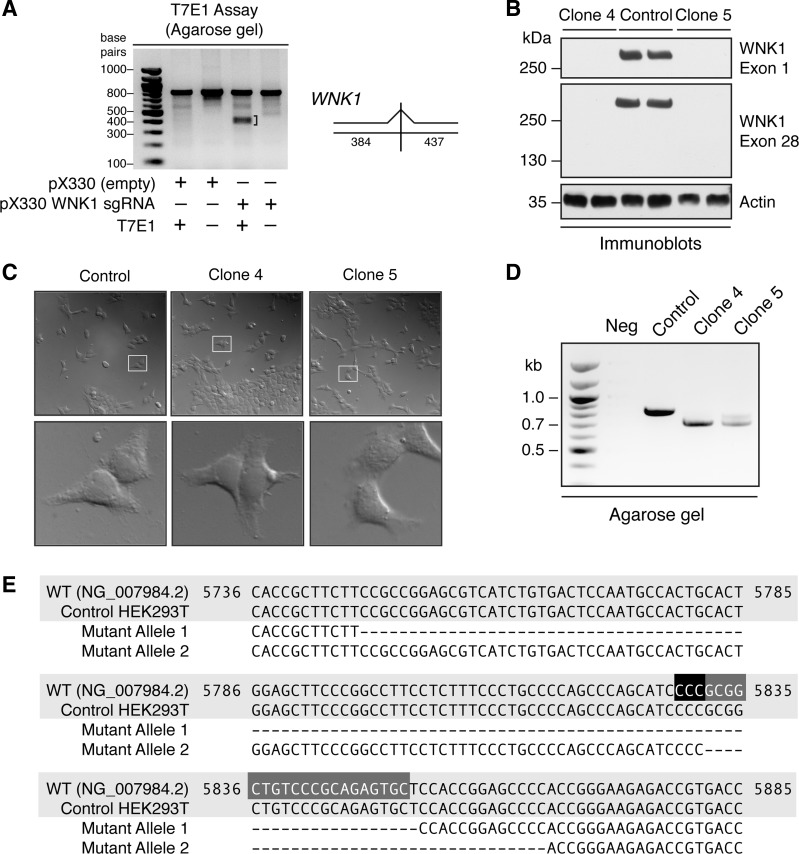
To test the effectiveness of the WNK1 sgRNA at triggering Cas9-mediated gene editing at the target site, HEK293T cells were transiently transfected with the pX330-WNK1 sgRNA plasmid, resulting in the generation of a mixed population of edited and nonedited cells. Seventy-two hours posttransfection, we extracted this heterogeneous pool of genomic DNA, amplified a genomic region containing the target site by PCR, denatured and reannealed the PCR amplicons in a thermal cycler to generate heteroduplex pairs, and subjected these rehybridized products to digestion with T7E1 nuclease. The T7E1 enzyme selectively recognizes and cleaves mismatched heteroduplexes harboring indels (5). Because CRISPR/Cas9 complexes trigger double stranded breaks and imperfect NHEJ near the PAM (6), we predicted that T7E1 digestion of mismatches in the target site should generate DNA fragments of ∼384–437 bp in size (Fig. 2A, right). Indeed, we found that in genomic DNA extracts from WNK1 sgRNA-transfected cells, the T7E1 enzyme cleaved exon 1 of WNK1 at a position targeted by the WNK1 sgRNA, resulting in the generation of lower molecular weight DNA fragments of ∼400 bp (Fig. 2A, left).
Fig. 2.
Generation and validation of WNK1 knockout (KO) cell lines. A: mismatch-specific endonuclease assay. Genomic PCR (gPCR) products spanning exon 1 of WNK1 were amplified from the template of a heterogeneous population of HEK293T cells transfected with px330-WNK1 guide. Rehybridization and treatment with T7E1 nuclease results in fallout bands of ∼380–430 bp, shown by a bracket. This is consistent with the predicted cleavage sizes of 384 and 437 bp. This fallout signal was not observed in gPCR amplicons from cells transfected with an empty px330 plasmid lacking the WNK1 guide. B: cell lysates (20 μg) of wild-type (WT) and clonal WNK1 KO isolates (clone 4 and clone 5), probed with exon 1- and exon 28-specific WNK1 antibodies. A parallel set of lysates (20 μg) was probed with an anti-actin antibody as a loading control. C: differential interference contrast (DIC) images of WT control and WNK1 KO clones. High-magnification images of areas bounded by white boxes are shown below the corresponding low-power image. D: after clone isolation, gPCR amplicons encompassing the modified locus of genomic DNA were run on a 1% agarose gel. The products from both WNK1 KO clones migrate faster than the unedited control. E: sequence alignment of bases 5736–5885 of exon 1 of the RefSeqGene human WNK1 sequence (NG_007984.2), a WT allele from control HEK293T cells, and two mutant alleles identified from each of the two WNK1 KO clones by genomic PCR. Mutant allele 1 was identified in both clones 4 and 5; mutant allele 2 was only isolated from clone 5. The complementary plus strand sequences corresponding to the 20-nt target and 3-nt PAM are highlighted in dark gray and black, respectively. See also Table 1.
Because the T7E1 screening assay suggested that the WNK1 sgRNA was functional, we next sought to generate clonal WNK1 knockout cell lines. We cotransfected a new batch of HEK293T cells with the px330-WNK1 sgRNA and a plasmid encoding eGFP and isolated transfected cells by FACS. The sorted cells were then plated as individual clones in 96-well plates and expanded. Eighteen clones were probed for WNK1 protein expression by Western blotting. From this initial screen, two clones (clones 4 and 5) were selected for further analysis. Both clones showed a complete absence of WNK1 expression compared with the unedited control, when probed with antibodies directed to the extreme N- and C-terminal ends of the protein (Fig. 2B). Despite the complete absence of WNK1 protein expression, DIC microscopy indicated that the cellular morphology of the two clones was grossly indistinguishable from wild-type controls (Fig. 2C).
To characterize the nature of gene editing in these cells, we PCR amplified exon 1 from genomic DNA extracted from the two individual clonal populations and analyzed the products on an agarose gel. We noted a substantial shift in the migration of the PCR products of both the knockout clones compared with the control DNA (Fig. 2D), indicating that the CRISPR/Cas9 machinery introduced deletions into the genomic sequence. To further confirm these findings, the PCR products were TA cloned and 12 clonal amplicons from each knockout cell line were sequenced. In clone 4, all 12 sequences exhibited a 105-bp deletion, which encompassed the target site+PAM (mutant allele 1; Fig. 2E and Table 1). This would be predicted to cause a frameshift mutation that generates a 91-amino acid truncated protein lacking kinase activity [p.Arg81Hisfs*10 per Human Genome Variation Society (HGVS) nomenclature (7); Table 1]. Sequence analysis of amplicons from clone 5 revealed a mixed population of mutated alleles. In 7 of the 12 amplicons, we detected the same 105-bp deletion that was present in clone 4. The other five amplicons revealed a second 34-bp deletion allele that also encompassed the target site+PAM (mutant allele 2; Fig. 2E and Table 1). The predicted consequence of this deletion is a frameshift mutation that generates five new amino acids and a premature stop codon downstream of alanine 109, also resulting in a kinase dead fragment (pAla109Thrfs*5; Table 1). Thus, in both clonal cell lines, we were unable to detect intact wild-type alleles, and all of the genomic DNA sequences that were identified contained deletions that created nonfunctional gene products. Taken together with the complete lack of detectable WNK1 protein expression by Western blotting with multiple WNK1-specific antibodies, these findings confirm the generation of two WNK1 knockout cell lines using the CRISPR/Cas9 system.
Table 1.
Captured genomic sequences from two WNK1 knockout clones
| Genomic Sequence Variant | Protein Mutation | No. of Amplicons Sequenced | |
|---|---|---|---|
| Clone 4 | |||
| Allele 1 | g.5749_5854del | p.Arg81Hisfs*10 | 12 of 12 |
| Clone 5 | |||
| Allele 1 | g.5749_5854del | p.Arg81Hisfs*10 | 7 of 12 |
| Allele 2 | g.5832_5865del | p.Ala109Thrfs*5 | 5 of 12 |
gPCR across the With-No-Lysine (WNK) 1 exon 1 target site was performed in clones 4 and 5, both of which did not exhibit WNK1 protein expression during screening by Western blotting. PCR products were TA cloned, and 12 clonal amplicons from each cell line was sent for Sanger sequencing. Two mutant alleles (allele 1 and allele 2) with deletions around the target site (Fig. 2E) were identified from these sequences. Genomic sequence variant, deletion mutation by HGVS nomenclature; protein mutation, translational consequence of deletion by HGVS nomenclature; no. of amplicons sequenced, details the number of PCR amplicons sent for sequencing with the identified mutation.
Deletion of WNK1 decreases WNK4 but not WNK3 expression.
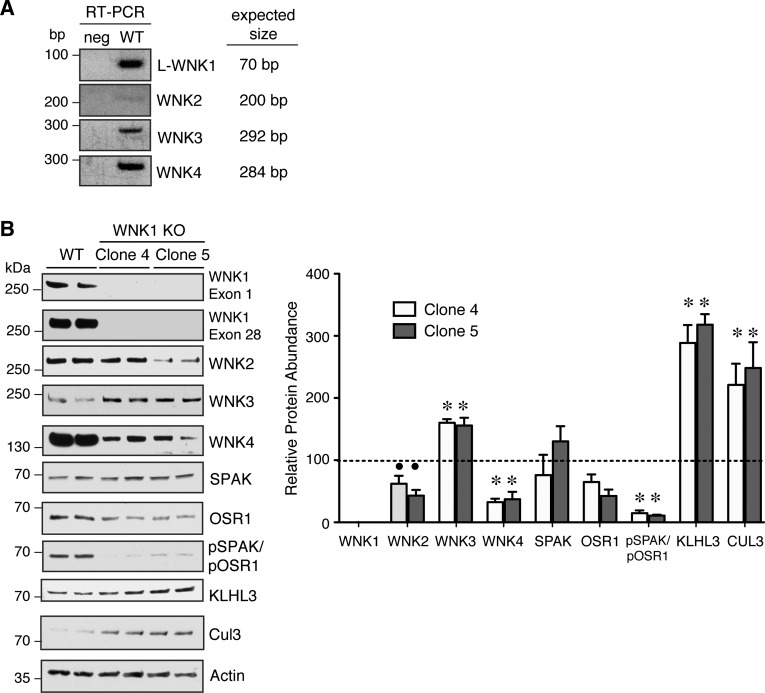
Several studies suggest that WNK kinases assemble and interact in macromolecular complexes (21, 40, 42, 52). Thus we set out to determine how the deletion of WNK1 protein expression in cells affects the abundance of the other WNK kinases and associated proteins in the pathway. We first performed RT-PCR studies to identify native WNK mRNA transcripts in wild-type HEK293T cells. These assays confirmed the expression of WNK1, WNK3, and WNK4 (Fig. 3A), but we were also able to detect WNK2, a brain-expressed kinase (33). When we probed wild-type and WNK1 knockout cells for WNK-SPAK/OSR1 protein expression, we found that WNK1 deletion strongly reduced the steady-state total abundance of WNK4 by ∼65% (Fig. 3B). WNK2 expression was also significantly reduced, although to a lesser degree. In contrast to WNK4 and WNK2, we observed an increase in WNK3 abundance. The levels of active phosphorylated SPAK and OSR1, detected with an antibody which recognizes phosphorylation of the C-terminal regulatory domain common to both kinases (34), were much lower in the knockouts compared with the wild-type, although the levels of total SPAK and OSR1 did not vary significantly. Collectively, these data confirm previous observations that WNK1 is a major determinant of SPAK and OSR1 activation status. The effects of WNK1 on downstream SPAK/OSR1 activity, however, may be due in part to its cooperative effects on the stability and activation status of other WNK kinases, such as WNK4.
Fig. 3.
Steady-state expression of the endogenous WNK-SPAK/OSR1 pathway in WT and WNK1 KO cells under isotonic conditions. A: agarose gel electrophoresis of RT-PCR products from WT HEK293T cells and template-free negative controls (neg). Predicted PCR amplicon sizes are provided to the right of each image. B: steady-state protein abundance of WNK-SPAK/OSR1 pathway components in WT and WNK1 KO clones 4 and 5. Total protein abundance of WNK2 and WNK4 were decreased in the WNK1 KO cell lines, while WNK3 expression was increased. SPAK/OSR1 activation was decreased in the WNK1 KO cells. The abundance of CUL3 and KLHL3 were similarly increased in WNK1 KOs; n = 4. ∙P < 0.05, *P < 0.001 by 1-way ANOVA, Dunnett's post hoc comparison with WT.
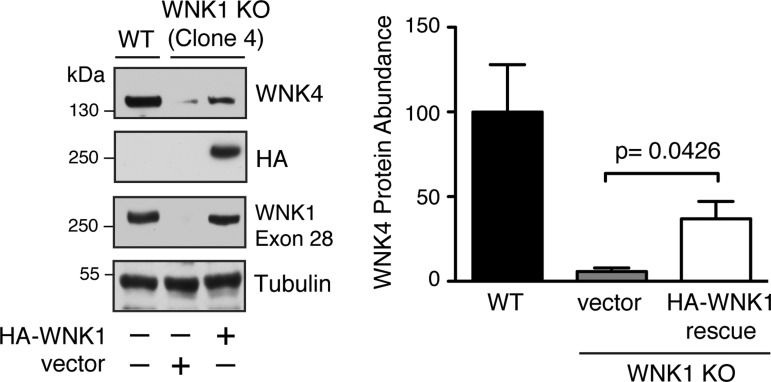
A major potential pitfall with the CRISPR/Cas9 system concerns the relatively short 20-bp length of the sgRNA guide sequence. Although Cas9-mediated gene editing only occurs at target sites localized immediately adjacent to a PAM (16), the short length of the guide increases the risk of cross-reactivity with target sites in different genes that share a similar sequence (19). Such off-target effects may lead to the misinterpretation of experimental results. To rule out the possibility that the reduced abundance of WNK4 observed in WNK1 knockout cells was due to an off-target effect, we performed a “rescue” experiment with heterologously expressed WNK1. For these studies, clone 4 WNK1 knockout cells were transiently transfected with full-length kinase active WNK1, and 48 h posttransfection we quantified WNK4 protein abundance by immunoblotting. Consistent with an on-target effect, reconstitution of WNK1 protein expression significantly increased WNK4 protein abundance (Fig. 4). These findings indicate that the reduced WNK4 abundance seen in WNK1 knockout cells is due to a WNK1-specific effect on WNK4 protein expression.
Fig. 4.
Heterologous expression of WNK1 rescues WNK4 protein abundance in WNK1 KO cells. Whole cell lysates from hemagglutinin (HA)-tagged L-WNK1-transfected clone 4 cells exhibit increased endogenous WNK4 expression by immunoblotting after 24 h; n = 3. P = 0.0426 by Student's t-test.
The E3 ubiquitin ligase CUL3 and its adaptor protein KLHL3 function in a protein complex that interacts with and degrades WNK1 and WNK4 (29, 45). A recent study provided evidence that the interaction between KLHL3-CUL3 and WNK4 can be regulated physiologically (36). We therefore wondered whether the absence of WNK1 protein expression upregulates the abundance of KLHL3-CUL3, providing a potential explanation for the reduction in WNK4 protein abundance. Consistent with this hypothesis, we observed a two- to threefold increase in the steady-state protein expression of KLHL3 and CUL3 in WNK1 knockout cells (Fig. 3B). Although additional studies will be needed to further evaluate the significance of this finding, the data suggest that WNK1 can regulate the stability of the KLHL3-CUL3 complex through feedback and that the abundance and activity of WNK4 may be influenced by this regulation.
WNK1 deletion reduces SPAK/OSR1 activation and impairs RVI.
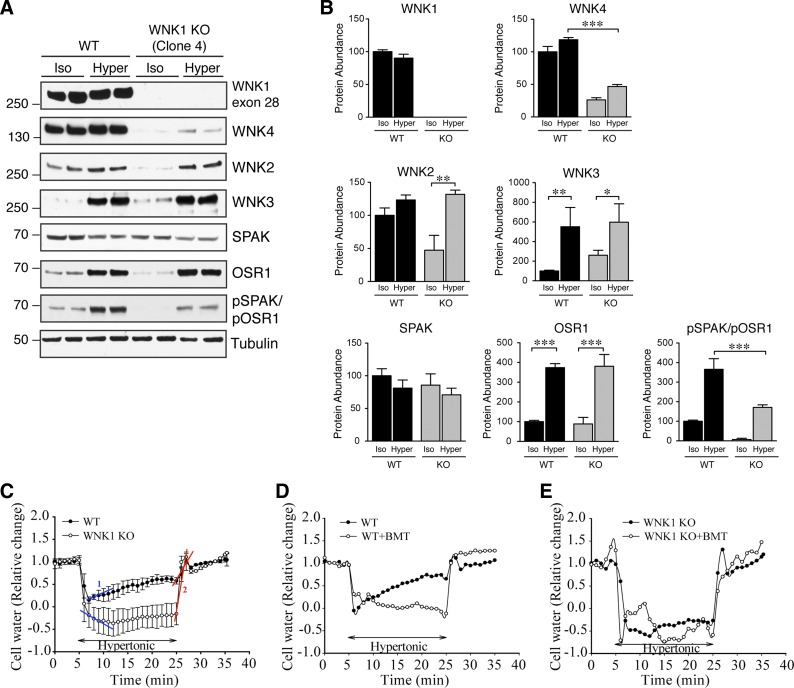
Hyperosmotic stress activates WNK1 by multisite phosphorylation (50, 53). To test the effect of hypertonicity on WNK1-dependent activation of SPAK/OSR1, we monitored the abundance and phosphorylation status of several components of the network after subjecting wild-type and WNK1 knockout cells to a hypertonic solution containing sorbitol. We elected to treat cells with 0.5 M sorbitol (800 mosmol/kgH2O) for 30 min, a previously reported hypertonic stimulus that activates the WNK-SPAK/OSR1 pathway (53). Consistent with previous observations, exposure of wild-type cells to 800 mosmol/kgH2O sorbitol solution reduced the electrophoretic mobility of WNK1 (Fig. 5A), a phenomenon that has been attributed to enhanced WNK1 phosphorylation and activation (53). Moreover, hypertonic stress did not significantly reverse the dramatic inhibitory effect of WNK1 deletion on WNK4 expression. In contrast, although WNK2 protein abundance was lower in WNK1 knockout cells under unstressed isotonic conditions, its protein abundance recovered dramatically during hypertonic stress, such that its expression was indistinguishable from WNK2 in sorbitol-treated wild-type cells (Fig. 5, A and B). The abundance of WNK3 was also significantly increased by either WNK1 knockout (Figs. 5, A and B, and 3B; compare WNK3 signal in WT and knockout cells under isotonic conditions) or hypertonicity.
Fig. 5.
Effect of hyperosmotic stress on WNK-SPAK/OSR1 pathway expression and cell volume regulation in WT and WNK1 KO. A: whole cell lysates (20 μg) from WT and WNK1 KO clone 4 cells were probed for WNK-SPAK/OSR1 pathway components by immunoblotting following a 30-min exposure to isotonic control media or media containing 0.5 M sorbitol. B: quantification of immunoblots for WNK kinases in A. In WNK1 KO cells, the abundance of WNK2, WNK3, and OSR1 increased significantly following exposure to hypertonicity, approximating their protein levels in WT cells subjected to the same condition. In contrast, the abundance of WNK4 and phosphorylated SPAK/OSR1 was lower in sorbitol-exposed WNK1 KO cells compared with WT; n = 4 for all samples. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA, Tukey's multiple comparison post hoc test. C: average changes in relative cell volume were determined in WT and WNK1 KO cells (clone 4) with the calcein method (20). Regulatory volume increase (RVI) after a 20-min exposure to 0.5 M sorbitol in HEPES buffer (800 mosmol/kgH2O) was detected in WNK1 WT cells. Each tracing represents measurements taken in 60 cells across 3 separate experiments. D: effects of bumetanide (BMT; 10 μM) on RVI. WT+BMT tracing (○) represents measurements taken from 20 cells across a single experiment. E: lack of RVI in WNK1 KO cells. WNK1 KO+BMT tracing (○) represents measurements taken from 20 cells across a single experiment.
We also evaluated the effect of hypertonic stress on downstream SPAK/OSR1 expression and activity. Unexpectedly, and despite their structural homology, sorbitol treatment had different effects on total SPAK and OSR1 protein abundance. In wild-type cells, OSR1 expression was dramatically increased by sorbitol hypertonicity, while SPAK expression did not change significantly (Fig. 5, A and B). The net result of these effects, however, was a significant increase in net SPAK/OSR1 phosphorylation status. WNK1 knockout cells exhibited similar trends in SPAK and OSR1 total protein abundance. Thus WNK1 knockout cells exhibited an overall increase in SPAK/OSR1 phosphorylation in response to hypertonicity; however, relative to wild-type cells, this phosphorylation was 50% reduced (Fig. 5, A and B; compare pSPAK/pOSR1 signal under hypertonic conditions in WT and WNK1 knockout cells).
Previous studies have suggested that WNK1 plays an important role in the cellular response to hypertonic stress. Following hypertonicity-induced shrinkage, cells recover their volume by stimulating NKCC1-mediated Na-K-2Cl cotransport, and by inhibiting potassium and chloride efflux via K-Cl cotransporters (KCCs) (13). This process, termed RVI, is directly linked to cotransporter phosphorylation status (13). WNK1 is believed to be an important mediator of this process, since it triggers SPAK/OSR1-mediated stimulatory phosphorylation of NKCC1 while simultaneously influencing the phosphorylation of KCCs at key inhibitory residues that block its activity (9, 27, 32, 44). To definitively test this hypothesis, we used a previously validated cell volume measurement assay with the fluorescent dye calcein (20) to determine whether deletion of WNK1 affects RVI in HEK293T cells during osmotic challenge. WNK1 wild-type and knockout cells were exposed to a 20-min hypertonic stress with 0.5 M sorbitol. WNK1 WT cells exhibited an immediate sharp drop in cell volume, followed by a slow volume increase (10.7 ± 3%/min), representing RVI (Fig. 5C, slope 1). At the end of the 20-min hypertonic stress, WNK1 WT cells recovered to 64% of the initial volume (63.5 ± 7.0). In contrast, WNK1 KO cells underwent more severe cell shrinkage and failed to show any RVI (Fig. 5C), with a recovery rate of −1.6 ± 0.3%/min. Both WNK1 WT and KO cells recovered their original volume upon returning to the isosmotic solution (Fig. 5C, slope 2). Inhibiting NKCC1 activity with bumetanide blocked the RVI response in WNK1 wild-type cells (Fig. 5D). However, bumetanide did not exhibit any effects in WNK1 knockout cells (Fig. 5E). Taken together, these data further suggest that WNK1 deletion weakens the RVI response, which is in part likely due to impaired SPAK/OSR1-NKCC1 activation.
DISCUSSION
In this study, we used CRISPR/Cas9 technology to knock out WNK1 gene expression in HEK293T cells, a model commonly used to study WNK-SPAK/OSR1 regulation of cation chloride cotransport. Our findings indicate that an sgRNA programmed to a target site in exon 1 of the WNK1 gene triggers sequence-specific Cas9-mediated NHEJ, resulting in the total ablation of WNK1 protein expression. Using two separate clones, we found that WNK4 expression is dependent on the presence of WNK1. In addition, WNK1 deletion attenuated the normal RVI response to hypertonicity, implicating WNK1 as an essential regulator of cell volume. The methodology presented here offers a simple and broadly applicable framework for generating validated knockout cell lines. Our workflow is adapted from previously published methods (30) and can be divided into four phases: 1) guide sequence design and sgRNA construction, 2) sgRNA screening, 3) clone isolation, and 4) clone screening and validation. In our experience, this straightforward approach can be used to generate gene-edited clones within a matter of weeks. The technique also provides certain distinct advantages, since it permits the study of endogenous genes that are resistant to conventional RNAi, or are embryonically lethal when knocked out in vivo. In theory, the approach could be applied to any cell culture model that can tolerate FACS and can receive cDNA via transfection or viral transduction. Thus, the gene editing workflow presented here could potentially be adapted to a variety of immortalized renal cell types, including tubular epithelial cells, podocytes, pericytes, or other cells relevant to kidney physiology.
RNA programmable gene editing is a rapidly evolving technology with a growing number of applications (15). Although we used the technique to knock out a single gene in this study, CRISPR/Cas9 nucleases can also be harnessed for multiplex genome engineering (6, 23). Thus sgRNAs targeting distinct sites within the genome can be coexpressed to knock out multiple genes simultaneously. This approach can potentially be used to rapidly eliminate redundancy in signaling pathways. For example, the multiplexing approach could be applied to the WNK-SPAK/OSR1 pathway to genetically delete several or all of the WNK kinases, or perhaps both SPAK and OSR1. This may help define the physiological roles of specific network components, or identify new kinases that phosphorylate cation chloride cotransporters independently of SPAK/OSR1. This multiplexing technique has been applied to the manipulation of genes in cultured cells and has also been used to knock out multiple genes from mice (47). Equally intriguing, CRISPR/Cas systems can also knock in point mutations or a new sequence into genes. In this case, a donor DNA template containing the desired sequence modification is introduced into cells with the Cas9 enzyme and sgRNA, triggering gene editing through homology-directed repair (23). Other than the requisite introduction of this donor template, the workflow for generating mutant clones is analogous to that outlined in this study.
The expanding armamentarium of CRISPR/Cas-related applications is quickly pushing it to the forefront of currently available tools for in vitro and in vivo gene modification. However, the system has important caveats that should be considered. The first and most significant of these is the potential for CRISPR to trigger off-target gene disruption (19). The short 20-nt guide sequence that directs Cas9 to a given target site may cross-react with other sequences present in the genome. It is therefore critically important to design guide sequences with high target specificity. Off-target cross-reactivity occurs at sites that pair with the 3′-most 13 bp of the guide sequence, termed the “seed sequence” (16, 23). In this study, we chose a WNK1 guide sequence that binds perfectly to a single 23-bp target site (including the NGG PAM) in the human exome but does not pair with unwanted target sites that harbor a matching seed sequence (23). Although this in silico approach helps minimize off-target cleavage, databases such as the one used in our study are still in the process of being optimized (23). Moreover, single base pair mismatches within the seed sequence may not completely abrogate off-target binding (16), and in silico reference sequences may not be a perfect match to the genome of an immortalized cell line. Thus it is essential to carry out supplemental experiments verifying that the ablation of a target gene of interest is responsible for a specific physiological effect. In our study, we used two approaches to test the specificity of WNK1 deletion on WNK4 abundance. First, we surveyed the WNK-SPAK/OSR1 pathway in two genetically distinct knockout cell lines that completely lack WNK1 protein expression and confirmed that all of the known major components of the signaling pathway were similarly affected. During this analysis, we observed that both clones exhibited increased expression of the KLHL3/CUL3 E3 ligase complex, suggesting a potential mechanism that might decrease WNK4 protein abundance. Second, we performed a rescue experiment in which exogenous WNK1 was transiently transfected into knockout cells and WNK4 expression was compared with vector transfected knockouts and wild-type controls. These studies confirmed that reconstituting WNK1 back into the network restored WNK4 abundance. Although we did not pursue other validation studies, an alternative approach would be to evaluate additional WNK1 knockout clones generated through the usage of a second sgRNA targeting a different area of the WNK1 gene. Because this nonoverlapping sgRNA should generate a different complement of off-target events, similar experimental results would be more likely to be the consequence of target gene knockout. Finally, deep sequencing of knockout cell lines would be a comprehensive gold standard to guide study interpretation. Although such studies can be costly, it seems advisable to pursue such characterization if multiple genes are going to be knocked out, or if edited cells will be used to generate large “unbiased” mRNA or proteomic datasets.
Another important issue that should be considered is the possibility that endogenous protein expression may drift during cell passage. In our hands, we found that the endogenous expression of certain components of the WNK-SPAK/OSR1 signaling pathway in HEK cells, particularly WNK3, can change over multiple cell passages. More specifically, the increased WNK3 expression noted in the knockouts (Figs. 3 and 5) was lost over multiple passages, and the WNK3 protein levels drifted down to that of wild-type cells (data not shown). Thus comparisons between knockout and control cells with significant discrepancies in passage number may not necessarily be reflective of target gene ablation. For this reason, we recommend analyzing control and knockout cell lines within a tight passage window and performing side-by-side comparisons with multiple genotypically distinct knockout clones.
The studies presented here highlight the importance of the WNK-SPAK/OSR1 pathway in cell volume regulation. We found that in wild-type cells, sorbitol-induced hypertonic stress shifted the electrophoretic mobility of WNK1. Previous work indicates that this alteration in mobility correlates with enhanced phosphorylation and activation, suggesting a role for WNK1 in SPAK/OSR1 activation by hypertonicity (53). Consistent with this, WNK1 knockout cells exposed to sorbitol hypertonicity exhibited reduced SPAK/OSR1 regulatory domain phosphorylation and a blunted RVI response. These results demonstrate that WNK1 plays a critically important role in cell volume regulation, either acting by itself, or via coregulation with WNK4, whose expression was strongly downregulated in WNK1 knockout cells. Despite the absence of significant WNK1 and WNK4 expression during hypertonicity, SPAK/OSR1 phosphorylation was not completely abolished. This suggests that other kinases could phosphorylate SPAK and OSR1 upon osmotic stress. WNK3 and WNK2 were detectable during sorbitol hypertonicity; thus, these other WNKs could mediate this process. Alternatively, there are WNK-independent mechanisms that can selectively activate OSR1, whose total protein abundance was strongly increased by hypertonicity. One such activation mechanism could be via the recently described P13K-mTORC2-OSR1 pathway (35).
The WNK1 knockout cell lines have also started to provide new information on the behavior of endogenously expressed WNK kinases in cells expressing a functional WNK-SPAK/OSR1 network. Our data indicate that WNK1 influences the total protein abundance of other WNK kinases. Specifically, WNK1 knockout cells exhibited a strong decrease in the abundance of WNK4, and to a lesser degree WNK2. This could be due to changes in the composition of WNK complexes, as previous studies have suggested that the WNKs can heterooligomerize via their C-terminal coiled-coil domains (42). Alternatively, WNK1 might regulate proteins that destabilize the other WNK kinases. Indeed, we found that WNK1 deletion increases the abundance of the KLHL3/CUL3 complex, a known negative regulator of WNK4 abundance (37). This suggests that WNK1 may control WNK4 ubiquitylation and degradation through feedback inhibition of KLHL3/CUL3. Although the mechanism by which WNK1 deletion downregulates WNK4 abundance will require further study, our data clearly show that selectively knocking out one WNK kinase can dramatically influence the abundance of others. This has important implications for the correct interpretation of knockdown and heterologous overexpression studies that implicate a specific WNK kinase in a particular physiological response. Most of these types of studies have not evaluated the effects of WNK overexpression on other endogenous components of the pathway. Yet, for most of the studies conducted in heterologous expression systems such as Xenopus oocytes or HEK293 cells, this unmonitored endogenous network is co-opted for at least some of the intermediate signaling stream that triggers downstream cation chloride cotransporter activation. To fully ascertain the role of specific WNKs in signal transduction, it seems that future studies would benefit from the development of gene-edited cell lines that contain solitary WNK kinases, or a “WNKless” cell line in which specific WNK-dependent signaling cascades can be reconstituted by rescue.
In conclusion, we report a simple approach that uses programmable CRISPR/Cas9 nucleases to generate knockout cell lines. The new WNK1 knockout model generated via this methodology highlights new relationships between WNK1, WNK4, and other associated proteins and confirms an essential role for WNK1 in NKCC1-dependent cell volume recovery. Thus our data demonstrate how this new and powerful genome-editing tool can be used to dissect and analyze WNK signaling networks.
GRANTS
The study was supported by National Institute of Health Grants R01DK098145 (to A. R. Subramanya), P30DK79307 (Pittsburgh Center for Kidney Research), R01 NS038118, and NS075995 (to D. Sun), and R01DK55881 (to E. J. Weinman), a Mid-Level Career Development Award from the US Department of Veterans Affairs (to A. R. Subramanya), and funds from a US Department of Veterans Affairs Middleton Award (to E. J. Weinman).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: A.R., J.H.G., G.B., G.P., E.J.W., D.S., and A.R.S. provided conception and design of research; A.R., J.H.G., G.B., B.F.D., and G.P. performed experiments; A.R., J.H.G., G.B., G.P., D.S., and A.R.S. analyzed data; A.R., J.H.G., G.B., G.P., D.S., and A.R.S. interpreted results of experiments; A.R., J.H.G., G.B., G.P., D.S., and A.R.S. prepared figures; A.R., G.B., D.S., and A.R.S. drafted manuscript; A.R., G.B., E.J.W., D.S., and A.R.S. edited and revised manuscript; A.R., J.H.G., G.B., B.F.D., G.P., E.J.W., D.S., and A.R.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Erich Wilkerson for FACS sorting and Deborah Steplock and Alexandra Socovich for excellent technical assistance. We are grateful to Drs. John Aach (Department of Genetics, Harvard Medical School, Boston, MA), Mike Butterworth, Adam Kwiatkowski, and Anatalia Labilloy for helpful discussions.
REFERENCES
- 1.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol 8: 348–357, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Valimaki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castaneda-Bueno M, Cervantes-Perez LG, Vazquez N, Uribe N, Kantesaria S, Morla L, Bobadilla NA, Doucet A, Alessi DR, Gamba G. Activation of the renal Na+:Cl− cotransporter by angiotensin II is a WNK4-dependent process. Proc Natl Acad Sci USA 109: 7929–7934, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavez-Canales M, Zhang C, Soukaseum C, Moreno E, Pacheco-Alvarez D, Vidal-Petiot E, Castaneda-Bueno M, Vazquez N, Rojas-Vega L, Meermeier NP, Rogers S, Jeunemaitre X, Yang CL, Ellison DH, Gamba G, Hadchouel J. WNK-SPAK-NCC cascade revisited: WNK1 stimulates the activity of the Na-Cl cotransporter via SPAK, an effect antagonized by WNK4. Hypertension 64: 1047–1053, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232, 2013. [DOI] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15: 7–12, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly BF, Needham PG, Snyder AC, Roy A, Khadem S, Brodsky JL, Subramanya AR. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem 288: 13124–13135, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-Cl−1 cotransporters is modulated by the interaction of two kinases: Ste20-related proline-alanine-rich kinase and WNK4. Am J Physiol Cell Physiol 290: C134–C142, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Ginns SM, Knepper MA, Ecelbarger CA, Terris J, He X, Coleman RA, Wade JB. Immunolocalization of the secretory isoform of Na-K-Cl cotransporter in rat renal intercalated cells. J Am Soc Nephrol 7: 2533–2542, 1996. [DOI] [PubMed] [Google Scholar]
- 12.Glover M, Zuber AM, O'Shaughnessy KM. Renal and brain isoforms of WNK3 have opposite effects on NCCT expression. J Am Soc Nephrol 20: 1314–1322, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev 89: 193–277, 2009. [DOI] [PubMed] [Google Scholar]
- 14.Hong C, Moorefield KS, Jun P, Aldape KD, Kharbanda S, Phillips HS, Costello JF. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci USA 104: 10974–10979, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife 2: e00471, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahle KT, Gimenez I, Hassan H, Wilson FH, Wong RD, Forbush B, Aronson PS, Lifton RP. WNK4 regulates apical and basolateral Cl- flux in extrarenal epithelia. Proc Natl Acad Sci USA 101: 2064–2069, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32: 677–683, 2014. [DOI] [PubMed] [Google Scholar]
- 20.Lenart B, Kintner DB, Shull GE, Sun D. Na-K-Cl cotransporter-mediated intracellular Na+ accumulation affects Ca2+ signaling in astrocytes in an in vitro ischemic model. J Neurosci 24: 9585–9597, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH. Properties of WNK1 and implications for other family members. J Biol Chem 2005. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Schreck C, Coleman RA, Wade JB, Hernandez Y, Zavilowitz B, Warth R, Kleyman TR, Satlin LM. Role of NKCC in BK channel-mediated net K+ secretion in the CCD. Am J Physiol Renal Physiol 301: F1088–F1097, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 339: 823–826, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markadieu N, Delpire E. Physiology and pathophysiology of SLC12A1/2 transporters. Pflügers Arch 466: 91–105, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick JA, Ellison DH. The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev 91: 177–219, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick JA, Yang CL, Zhang C, Davidge B, Blankenstein KI, Terker AS, Yarbrough B, Meermeier NP, Park HJ, McCully B, West M, Borschewski A, Himmerkus N, Bleich M, Bachmann S, Mutig K, Argaiz ER, Gamba G, Singer JD, Ellison DH. Hyperkalemic hypertension-associated cullin 3 promotes WNK signaling by degrading KLHL3. J Clin Invest 124: 4723–4736, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem 280: 42685–42693, 2005. [DOI] [PubMed] [Google Scholar]
- 28.Needham PG, Mikoluk K, Dhakarwal P, Khadem S, Snyder AC, Subramanya AR, Brodsky JL. The thiazide-sensitive NaCl cotransporter is targeted for chaperone-dependent endoplasmic reticulum-associated degradation. J Biol Chem 286: 43611–43621, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T. The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451: 111–122, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson C, Alessi DR. The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121: 3293–3304, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Rinehart J, Maksimova YD, Tanis JE, Stone KL, Hodson CA, Zhang J, Risinger M, Pan W, Wu D, Colangelo CM, Forbush B, Joiner CH, Gulcicek EE, Gallagher PG, Lifton RP. Sites of regulated phosphorylation that control K-Cl cotransporter activity. Cell 138: 525–536, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinehart J, Vazquez N, Kahle KT, Hodson CA, Ring AM, Gulcicek EE, Louvi A, Bobadilla NA, Gamba G, Lifton RP. WNK2 kinase is a novel regulator of essential neuronal cation-chloride cotransporters. J Biol Chem 286: 30171–30180, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saritas T, Borschewski A, McCormick JA, Paliege A, Dathe C, Uchida S, Terker A, Himmerkus N, Bleich M, Demaretz S, Laghmani K, Delpire E, Ellison DH, Bachmann S, Mutig K. SPAK differentially mediates vasopressin effects on sodium cotransporters. J Am Soc Nephrol 24: 407–418, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sengupta S, Lorente-Rodriguez A, Earnest S, Stippec S, Guo X, Trudgian DC, Mirzaei H, Cobb MH. Regulation of OSR1 and the sodium, potassium, two chloride cotransporter by convergent signals. Proc Natl Acad Sci USA 110: 18826–18831, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata S, Arroyo JP, Castaneda-Bueno M, Puthumana J, Zhang J, Uchida S, Stone KL, Lam TT, Lifton RP. Angiotensin II signaling via protein kinase C phosphorylates Kelch-like 3, preventing WNK4 degradation. Proc Natl Acad Sci USA 111: 15556–15561, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci USA 110: 7838–7843, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanya AR, Ellison DH. Distal convoluted tubule. Clin J Am Soc Nephrol 9: 2147–2163, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanya AR, Liu J, Ellison DH, Wade JB, Welling PA. WNK4 diverts the thiazide-sensitive NaCl cotransporter to the lysosome and stimulates AP-3 interaction. J Biol Chem 284: 18471–18480, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanya AR, Yang CL, Zhu X, Ellison DH. Dominant-negative regulation of WNK1 by its kidney-specific kinase-defective isoform. Am J Physiol Renal Physiol 290: F619–F624, 2006. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi D, Mori T, Nomura N, Khan MZ, Araki Y, Zeniya M, Sohara E, Rai T, Sasaki S, Uchida S. WNK4 is the major WNK positively regulating NCC in the mouse kidney. Biosci Reps 34: e00107, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thastrup JO, Rafiqi FH, Vitari AC, Pozo-Guisado E, Deak M, Mehellou Y, Alessi DR. SPAK/OSR1 regulate NKCC1 and WNK activity: analysis of WNK isoform interactions and activation by T-loop trans-autophosphorylation. Biochem J 441: 325–337, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal-Petiot E, Cheval L, Faugeroux J, Malard T, Doucet A, Jeunemaitre X, Hadchouel J. A new methodology for quantification of alternatively spliced exons reveals a highly tissue-specific expression pattern of WNK1 isoforms. PLoS One 7: e37751, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391: 17–24, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, Chiga M, Kikuchi E, Nomura N, Mori Y, Matsuo H, Murata T, Nomura S, Asano T, Kawaguchi H, Nonoyama S, Rai T, Sasaki S, Uchida S. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep 3: 858–868, 2013. [DOI] [PubMed] [Google Scholar]
- 46.Wall SM, Weinstein AM. Cortical distal nephron Cl− transport in volume homeostasis and blood pressure regulation. Am J Physiol Renal Physiol 305: F427–F438, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welling PA, Chang YP, Delpire E, Wade JB. Multigene kinase network, kidney transport, and salt in essential hypertension. Kidney Int 77: 1063–1069, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J, Yoon J, Yang SS, Lin SH, Huang CL. WNK1 protein kinase regulates embryonic cardiovascular development through the OSR1 signaling cascade. J Biol Chem 288: 8566–8574, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275: 16795–16801, 2000. [DOI] [PubMed] [Google Scholar]
- 51.Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH. Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277: 48456–48462, 2002. [DOI] [PubMed] [Google Scholar]
- 52.Yang CL, Zhu X, Ellison DH. The thiazide-sensitive Na-Cl cotransporter is regulated by a WNK kinase signaling complex. J Clin Invest 117: 3403–3411, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, Alessi DR. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol 176: 89–100, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, Gupta A, Hansen G, Hu Y, Huang W, Jaing C, Key BW Jr, Kipp P, Kohlhauff B, Ma ZQ, Markesich D, Payne R, Potter DG, Qian N, Shaw J, Schrick J, Shi ZZ, Sparks MJ, Van Sligtenhorst I, Vogel P, Walke W, Xu N, Zhu Q, Person C, and Sands AT. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci USA 100: 14109–14114, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]