Abstract
C57BL/6N inbred mice are used as the genetic background for producing knockout mice in large-scale projects worldwide; however, the genetic divergence among C57BL/6N-derived substrains has not been verified. Here, we identified novel single nucleotide polymorphisms (SNPs) specific to the C57BL/6NJ strain and selected useful SNPs for the genetic monitoring of C57BL/6N-derived substrains. Informative SNPs were selected from the public SNP database at the Wellcome Trust Sanger Institute by comparing sequence data from C57BL/6NJ and C57BL/6J mice. A total of 1,361 candidate SNPs from the SNP database could distinguish the C57BL/6NJ strain from 12 other inbred strains. We confirmed 277 C57BL/6NJ-specific SNPs including 10 nonsynonymous SNPs by direct sequencing, and selected 100 useful SNPs that cover all of the chromosomes except Y. Genotyping of 11 C57BL/6N-derived substrains at these 100 SNP loci demonstrated genetic differences among the substrains. This information will be useful for accurate genetic monitoring of mouse strains with a C57BL/6N-derived background.
Keywords: C57BL/6N, SNP, genetic background, inbred, substrain
Introduction
C57BL/6 is the best-known inbred mouse strain and has been used as the genetic background for spontaneous and induced mutations. To produce knockout mice, embryonic stem (ES) cells derived from 129 mouse substrains were used initially to manipulate the mouse genome [19]; however, these substrains were not suitable for most biomedical studies, especially in immunology, neurobiology, and physiology [4, 13, 28]. Backcrossing to C57BL/6 mice is carried out frequently to generate congenic strains to facilitate phenotypic analyses, but this procedure requires additional cost and time. In addition, the targeted locus from the original ES cell genome remains in the congenic mice and may confound the results of studies using these animals [6]. Therefore, ES cells with a pure C57BL/6 genetic background are more useful for the generation of knockout mice.
Recently, C57BL/6 mouse-derived ES cells were established in several laboratories. Importantly, ES cells derived from C57BL/6N mice maintained their pluripotency after homologous recombination [18, 25], and the methods used to generate germline-transmitting chimeric mice have been improved [5]. The International Knockout Mouse Consortium (IKMC) conducted large-scale mutagenesis to mutate all of the protein-coding genes in mice using gene trapping and targeting in ES cells [9, 22]. Since then, several mouse ES cells derived from the C57BL/6NTac strain have been used as standard ES cells for the production of mutant alleles [5, 18]. Moreover, the International Mouse Phenotyping Consortium has used the IKMC-targeted C57BL/6N ES cell clones to undertake the broad-based phenotyping of 20,000 mouse genes [2].
Since the 1950s, the C57BL/6 strain has diverged into several substrains, including two major groups, C57BL/6J and C57BL/6N. Currently, more than 20 inbred substrains derived from C57BL/6J and C57BL/6N mice have been established and distributed worldwide [1, 16]. The C57BL/6J strain has many specific single nucleotide polymorphisms (SNPs) that distinguish it from other inbred strains [17], and SNPs that can differentiate C57BL/6J substrains have also been identified [14, 32]. In addition, several phenotypic differences have been reported among C57BL/6J substrains [8, 20, 23, 24]. C57BL/6J-specific SNP information is useful for the genetic monitoring of mouse strains with a C57BL/6J-derived background and interpretation of phenotypic data.
At least, 11 C57BL/6N-derived substrains exist and are commercially available. However, genetic variation among C57BL/6N-derived substrains, including C57BL/6NTac, which was used to generate the IKMC ES cells has not yet been verified. Previously, C57BL/6J-specific SNPs detected by comparing the reference C57BL/6J sequence [15] with other inbred mouse strains have been reported [7, 17, 26, 27, 29, 31]; however, C57BL/6N was not included in these SNP data. Recently, the Wellcome Trust Sanger Institute (WTSI) published whole genome resequencing data of 17 key mouse inbred strains including C57BL/6NJ, which enabled us to identify C57BL/6NJ-specific SNPs through comparisons with other inbred strains [10, 30].
In this study, we searched for SNPs specific to the C57BL/6NJ strain using the resequence database of the WTSI. Moreover, in light of the branching history of C57BL/6N-derived substrains, we found variation in the number of accumulated C57BL/6NJ-specific SNPs among the C57BL/6N-derived substrains, which can be used to differentiate the substrains.
Materials and Methods
Animals
SNP genotyping was conducted in 11 C57BL/6N and 7 C57BL/6J-derived inbred substrains available from different breeders and holders around the world (Table 1). As for the C57BL/6NJ, C57BL/6By, C57BL/6ByJ, and C57BL/6JEiJ strains, genomic DNA from one animal of each strain was obtained from The Jackson Laboratory Mouse DNA Resources (stock #005304, #000663, # 001139, and #000924, respectively; Bar Harbor, ME). As for the other strains, live mice or frozen tissue from two animals of the C57BL/6N-derived substrain and one animal of the C57BL/6J-derived substrain were used, respectively. Genomic DNA was extracted from the tail tips or kidneys using an Autogen NA-2000 automatic nucleic acid isolation system (KURABO Industries Ltd., Osaka, Japan) and/or a DNeasy Blood & Tissue Kit (QIAGEN GmbH, Hilden, Germany). All animal experiments were conducted in accordance with the Regulations for Animal Experiments of RIKEN (October 1, 2003 Rule No. 129, last amendment on March 31, 2008 Rule No. 29). Our experimental protocols, including those involving animals (Exp10-002), were approved by the Animal Experiments Committee of the RIKEN Tsukuba Institute.
Table 1. C57BL/6 substrains investigated in this study.
| Substrain | Source | |
|---|---|---|
| C57BL/6N substrains | C57BL/6NJ | The Jackson Laboratory (Bar Harbor, MA, USA) |
| C57BL/6NCrSim | Simonsen Laboratories, Inc. (Gilroy, CA, USA) | |
| C57BL/6NTac | Taconic Farm Inc. (New York, NY, USA) | |
| C57BL/6NJcl | CLEA Japan Inc. (Tokyo, Japan) | |
| C57BL/6NSeac | Kyudo Co. Ltd. (Tosu, Japan) | |
| C57BL/6NCrlCrlj | Charls River Laboratories Japan, Inc. (Yokohama, Japan) | |
| C57BL/6NCrl | Charls River Laboratories International, Inc. (Wilmington, MA, USA) | |
| C57BL/6NHsd | Harlan Laboratories, Inc. (Indianapolus, IN, USA) | |
| C57BL/6NCrSlc | Japan SLC, Inc. (Hamamatsu, Japan) | |
| C57BL/6By | The Jackson Laboratory (Bar Harbor, MA, USA) | |
| C57BL/6ByJ | The Jackson Laboratory (Bar Harbor, MA, USA) | |
| C57BL/6J substrains | C57BL/6J | The Jackson Laboratory (Bar Harbor, MA, USA) via Charls River Laboratories Japan, Inc. (Yokohama, Japan) |
| C57BL/6JJcl | CLEA Japan Inc. (Tokyo, Japan) | |
| C57BL/6JJmsSlc | Japan SLC, Inc. (Hamamatsu, Japan) | |
| C57BL/6JEiJ | The Jackson Laboratory (Bar Harbor, MA, USA) | |
| C57BL/6JOlaHsd | Harlan Laboratories, Inc. (Indianapolus, IN, USA) | |
| C57BL/6JRccHsd | Harlan Laboratories, Inc. (Indianapolus, IN, USA) | |
| C57BL/6JBomTac | Taconic Farm Inc. (New York, NY, USA) | |
Nomenclatured strain names of each C57BL/6 substrain were in accordance with JAX® NOTES [16].
In silico selection of informative SNPs for C57BL/6NJ
Informative SNPs were selected from the public SNP database at the Mouse Genome Project, WTSI (http://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1303), by comparing the sequence data of C57BL/6NJ with that of the C57BL/6J reference strain. SNPs marked with a “high confidence” call on the database were extracted, and then candidate C57BL/6NJ-specific SNPs were selected through a comparison with sequence data from the 12 other inbred mouse strains in the database (129P2/OlaHsd, 129S1/SvImJ, 129S5/SvEvBrd, A/J, AKR/J, BALB/cJ, C3H/HeJ, CBA/J, DBA/2J, LP/J, NOD/ShiLtJ, and NZO/HlLtJ).
Experimental confirmation of SNPs and genotyping
To confirm whether the in silico-selected C57BL/6NJ-specific candidate SNPs were present in mouse DNA samples, the SNP loci of the C57BL/6NJ and C57BL/6J strains were genotyped by PCR and direct sequencing. Target regions containing the candidate SNPs were amplified by PCR with flanking primers designed by using BatchPrimer3 v1.0 (probes.pw.usda.gov/batchprimer3/index.html). PCR was performed using a QIAGEN multiplex PCR Kit (QIAGEN GmbH) according to the manufacturer’s protocol. The PCR products were electrophoresed and separated on an E-Gel CloneWell 0.8% SYBR Safe gel using an E-Gel iBase Power system (both from Life Technologies, Carlsbad, CA). Sequencing reactions were performed in a DNA Engine® and Dyad™ PTC-220 Peltier Thermal Cycler (Bio-Rad, Laboratories, Inc., Hercules, CA) using an ABI BigDye® Terminator v3.1 Cycle Sequencing Kit with AmpliTaq DNA polymerase (Life Technologies), following the protocols supplied by the manufacturers. Single-pass sequencing was performed on each template primer. The fluorescently labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and then subjected to electrophoresis in an ABI 3730 × l sequencer (Life Technologies). After the SNPs were confirmed in both strains, the other C57BL/6 substrains were genotyped by the same method. The flanking primers used for each SNP typing are listed in Table 4.
Table 4. Position of the 100 selected SNP loci and their flanking primer sequence.
| Locus No. |
dbSNP ID |
Chromo some |
Position (bp) (GRCm38) |
C57BL/6J allele |
C57BL/6NJ allele |
Flanking primer sequence (5’ to 3’) | |
|---|---|---|---|---|---|---|---|
| Forward primer | Reverse primer | ||||||
| 1 | rs246236360 | 1 | 11,996,705 | C/C | T/T | ACCCCCTGAACCTTCAATTC | TTTCCCATGGAATTCTGCTC |
| 2 | rs246490354 | 1 | 14,344,561 | T/T | A/A | GGGGAAGAGTGGGATGACTA | TGGTCAGCATATTCCAGTGC |
| 3 | rs212521754 | 1 | 16,968,405 | C/C | A/A | GCAACGAAGGAAATTGAAGC | TGTTGAGGCATGTCCCTTTT |
| 4 | rs227394849 | 1 | 19,544,960 | A/A | T/T | GGGCAGAACTTCCTTTTCCT | TCTCACCTGAGTCCCTGGAT |
| 5 | rs232920323 | 1 | 21,639,642 | C/C | G/G | GAAATAGCACAGGTCCATCAAA | CCCAGCAGACAAGAGACAAA |
| 6 | rs213024334 | 1 | 30,167,141 | C/C | T/T | GGAATCTCAACCTAAGCAGCA | TGGAAAATTGAAGCAACCAG |
| 7 | rs244794780 | 1 | 40,107,883 | T/T | A/A | TCTGTTGCTCTCCAGCATTG | CTACACCCTGGCCTGACACT |
| 8 | rs260670033 | 1 | 50,146,448 | C/C | T/T | AGCAGAAATGCCAAAATGCT | TCAGACCCAAAAGGACATGC |
| 9 | rs249907793 | 1 | 61,950,812 | C/C | A/A | CCAGTGGGTTAAGTGGGATT | ATCAAATGGGGTGGCATTTA |
| 10 | rs265151779 | 1 | 83,182,637 | G/G | A/A | ATTCTGCTAACTGCGGAGGA | TTGTTCACCCTCTTCCCCTA |
| 11 | rs229124202 | 1 | 89,861,338 | G/G | T/T | GGAAGGCAGATCACCAACTC | TCTATGGTGGCCCTAGGATG |
| 12 | rs237656339 | 1 | 99,547,673 | G/G | A/A | TGGCTCCTGACATCTTTCCT | GCTCCTGGATCGGCATATTA |
| 13 | rs223540754 | 1 | 110,024,886 | G/G | C/C | GACCAAATGCCTTGAAAATGA | CTCTCCCCATCCCTTTTCTT |
| 14 | rs259683638 | 1 | 119,116,297 | C/C | T/T | AGGTCTTGGGCTCTTTAGGG | ACTTGCTGGCTGACTCCTTC |
| 15 | rs229911289 | 1 | 132,980,179 | T/T | A/A | TTTTATTTCCTCCGCATTGG | ACTCGGGAACACACAAGCTC |
| 16 | rs215622703 | 1 | 142,008,378 | C/C | A/A | TTTTTGTGTTGGCCAAGGAT | CCTTTCTCTTCAGAGGGGTTTT |
| 17 | rs239017398 | 1 | 154,474,620 | C/C | T/T | CAGATCCCGGCTCAATTTTA | AGCTCATTAGCCTGGCATGT |
| 18 | rs214254072 | 1 | 161,859,644 | C/C | T/T | GTTTGCTCCTCCCCCTACTC | TGTATGAAAATGTGCAGGTTCTG |
| 19 | rs255914894 | 1 | 172,611,934 | G/G | A/A | ATGCCGGTGTACCTTCAGAG | CCCCAGTAACCATTCTCCTG |
| 20 | rs222303818 | 1 | 179,503,532 | G/G | A/A | CTGCCCATACTCCTGTCCAT | AGGGCCTGGTACTGAGAACA |
| 21 | rs262282675 | 1 | 188,434,376 | A/A | T/T | ACCCTTTGATGGTTCCCATT | AATTTTGCTAGGCCCATGAAT |
| 22 | rs251979693 | 2 | 11,214,185 | C/C | T/T | CCCCACATTTGCTTATCCAG | GCCAATTGTGAGGAATGCTT |
| 23 | rs230600693 | 2 | 21,681,174 | G/G | T/T | GGTCCAGCATTATTGGCATT | GTGATCCCATCTGCCATCTT |
| 24 | rs242780245 | 2 | 30,188,489 | A/A | C/C | CCACTGTCACCAGCACATTC | CCACCACTCCTCTCCGAATA |
| 25 | rs228546410 | 2 | 41,205,764 | T/T | A/A | ATGCCCACAATGCAAACATA | TAGCCCCTCTGACTGTCCAC |
| 26 | rs254996546 | 2 | 51,969,852 | C/C | T/T | CCGTGACCAAGTATGCACAG | GCTGAGGGTTCAGTTGTGGT |
| 27 | rs256541267 | 2 | 70,251,451 | C/C | T/T | CTGAAGAAAGGCCTGTTTGG | CGAATTCAATGCTGCCAATA |
| 28 | rs248280077 | 2 | 80,873,138 | C/C | T/T | TGTGCCGATTCCTCTAGCTT | CTGCACCAATTAGCAGCAA |
| 29 | rs214356625 | 2 | 96,674,180 | G/G | T/T | GTGTATGCCCCCAACCTTTA | CCAGTGATTGCATTTCACCTT |
| 30 | rs224344563 | 2 | 102,710,505 | A/A | T/T | CAGGACAGGAGAGGGTCAAG | ATCCCAGGCCATAGGATTTT |
| 31 | rs251933504 | 2 | 112,966,408 | T/T | C/C | GCTCGGTCTGAAAGGTCAAC | GGAAGCAAGAGCTTGGAAGA |
| 32 | rs258508221 | 2 | 122,708,738 | T/T | A/A | ACTTTGTGCCTTTTGCAACC | GAGGGGGATCCAAGGATAAG |
| 33 | rs255014110 | 2 | 132,432,999 | C/C | T/T | CAGCACAGATGGTTTCATGG | AGATGCACAAGTGGCTCTGA |
| 34 | rs242413924 | 2 | 140,793,056 | T/T | G/G | AAATTTTCTTCCCCACAGCA | GTGAGCCAGTACAGGGGAGA |
| 35 | rs217443774 | 2 | 152,781,403 | A/A | G/G | CTCTTCTTCCTGCCCTTCCT | AGCCATTGAGTGAGGTGCTT |
| 36 | rs253212197 | 2 | 164,813,748 | C/C | T/T | CTGAACTGCAACCCTCATCA | AGTGTAGCCCTCCCTGTCCT |
| 37 | rs213376233 | 2 | 170,240,435 | G/G | A/A | GCTAAGTGGTCTTGGGATGC | GCCACCACACACAGCTAATTT |
| 38 | rs264719247 | 2 | 180,149,012 | A/A | G/G | TTCTGCTAGGCTTCCTGGTG | CTCCCTCACAACAGGCTCAT |
| 39 | rs221521392 | 2 | 181,868,891 | C/C | T/T | TCTTTTTGCCTCTTGTTGGAA | CGTGCTTGTGAGCTCTCTGA |
| 40 | rs256520809 | 3 | 8,498,163 | G/G | A/A | TGGCAGAAGTTTGTTTCAGG | CCATCTGGGGCTGAATACTT |
| 41 | rs214801792 | 3 | 32,773,111 | G/G | C/C | TGCTGGGGTAGTTTTCCACT | GGAGGGAGTCAGGTGCAATA |
| 42 | rs222821429 | 3 | 66,305,330 | C/C | T/T | CACCCAACACCCACAGAGTA | TGTCTTTTCAAAGGGCCAGA |
| 43 | rs243656799 | 3 | 72,616,062 | G/G | A/A | CCCATTGGACACGAAAACTT | CACTGCTGCTCATTGGTTCA |
| 44 | rs262827930 | 3 | 109,597,274 | A/A | T/T | GGCAGTTTGGCCTGTAGGTA | CTTTACTGGCTTGCCTCACC |
| 45 | rs254145219 | 3 | 147,657,255 | C/C | T/T | CAGCAGGATATGCGTCCTCT | GCTTCCCCTCCCATAATTTC |
| 46 | rs219227155 | 4 | 19,328,298 | T/T | C/C | ACACAAGAACTGGCACATGG | TTGGGACCTGTCAGCCTATC |
| 47 | rs235104023 | 4 | 56,463,984 | C/C | T/T | AGCAGTTGGTGTGTTTGCTG | CCCCCATTGCTTTGTGTCTA |
| 48 | rs261879287 | 4 | 104,973,294 | T/T | G/G | GACGAGGGAAAATGAGTGGA | CAAATGGCATGTTCGTTTGA |
| 49 | rs256724446 | 5 | 35,701,259 | G/G | A/A | ATTCATTCCTGACCCATCCA | CTTCCTCAATTCCCCTCCAT |
| 50 | rs260260338 | 5 | 80,026,465 | G/G | A/A | TGGGGAAAGAATGTGCCTAC | TTGGTCCAACATCAAACTACCTT |
| 51 | rs217297994 | 5 | 117,118,668 | G/G | A/A | CCAAGGAGCAGCCCTACTAA | CAACTCCTGGTCAACGCTCT |
| 52 | rs221990668 | 5 | 150,224,989 | G/G | T/T | CTTGTAGAACCCAGGCCATC | GTCCCCACCCATTACATCAG |
| 53 | rs257294810 | 6 | 39,971,164 | G/G | A/A | GCATTCAGCTCTCCTTCCTG | GAGACCTGGGCACAATGACT |
| 54 | rs224069095 | 6 | 74,169,211 | C/C | A/A | CTCATCATGACACAAGGAGCA | CATGTGTGGCCCCTAGTTCT |
| 55 | rs37540455 | 6 | 113,159,679 | G/G | A/A | ATTCCTGGCCAGCCTTAGAT | TGTTGGTGAGAGTCCTTCCA |
| 56 | rs217544076 | 6 | 144,513,005 | G/G | C/C | CACACATCCATCTGCCTCTG | GCAGCCGGAGTATTAGCAAG |
| 57 | rs212452109 | 7 | 16,595,985 | A/A | T/T | GAGTTCAATCCCTGGGACAA | GTGTCACTGTGGCTGGCTTA |
| 58 | rs224103578 | 7 | 53,390,545 | C/C | T/T | GGAGGGAATGTGTCAGTAACG | CCTGACCTCAGTGTGCAGAA |
| 59 | rs243575509 | 7 | 102,973,309 | C/C | T/T | TCATCACAGGAGGGAAGAGG | GGCTATCTGTCGTCCTTTGC |
| 60 | rs229340185 | 7 | 140,821,590 | A/A | T/T | CTTCAGGCCCTTCACGAGTA | GATTCCTATTGGCTGGCTTG |
| 61 | rs263791105 | 8 | 22,903,742 | G/G | A/A | CGAATGTGCATTTGTCATCC | GCCCTTCCAACTACCTCACA |
| 62 | rs255341040 | 8 | 58,790,625 | G/G | A/A | GGCACTGTTTATCTTGGGAAC | TGCCAAACAGCACTCAGAAG |
| 63 | rs239219835 | 8 | 79,117,401 | G/G | A/A | TAAATGGCCCGAATTCACAT | TGTGCACCTTCCTTTGTTCA |
| 64 | rs256624163 | 8 | 94,046,068 | G/G | A/A | TCAGAGCCCACAGAAAAAGG | CCATGGGTTTCACACATTCA |
| 65 | rs211750147 | 8 | 118,442,679 | G/G | A/A | TCGGGGGCTTAATTTCTCTT | TGCCTAGACCTGGATTTGGT |
| 66 | rs52003732 | 9 | 10,125,248 | T/T | C/C | TTCTCCCCTCTGTGAGCAAG | TTGCCACCACCTCAAAAACT |
| 67 | rs214490504 | 9 | 60,662,109 | G/G | A/A | TCATCCCGGAACATAAATGG | AGTCTCGCCAATACGACTGC |
| 68 | rs243500146 | 9 | 116,160,235 | C/C | T/T | CAGAAGGATCCTGGACTTGC | CTCTTATCTCCCCGCCAGA |
| 69 | rs51123066 | 10 | 11,070,460 | G/G | T/T | CAAGGCCCCTGTAAATCCTT | CGAGGGCCTAGCATATTTCA |
| 70 | rs219489973 | 10 | 41,944,745 | G/G | A/A | CCTCGGCTAACTTCAAGCAC | GCCTGTGCGCTTACTTTGAT |
| 71 | rs213583872 | 10 | 49,357,252 | G/G | T/T | GTTGCACAGGCTGAGAATGA | CCCAAATGAATTGCAAAGGT |
| 72 | rs246274290 | 10 | 88,091,833 | T/T | C/C | CACAGAACACAGGCTCCAAA | GCCAACATGGTCGGTAGACT |
| 73 | rs223857079 | 11 | 12,253,003 | A/A | T/T | CTGGTTTGGAGGTGAGCATT | AAAAGCTCCGGAAGGTGAAT |
| 74 | rs240617401 | 11 | 46,222,615 | G/G | A/A | TGACCCCCAATCACACATTA | GCCAGCTTATCCATCTGCAC |
| 75 | rs231656457 | 12 | 29,886,947 | T/T | G/G | TCTTGGTTAAGGTGGCAAGG | ATTCACAAATGTCGGCATCA |
| 76 | rs217422777 | 12 | 70,772,479 | C/C | T/T | CCGGGAAAAACATACACACC | ACCCTGCTCTCCTTGACAAA |
| 77 | rs221345442 | 12 | 97,702,669 | A/A | T/T | TCCTCTCACCCGTATCTGCT | GCGTCTTCAGAGACCTTCTCA |
| 78 | rs226310424 | 13 | 41,494,375 | G/G | C/C | CATCTCCATGGTGCTCGATA | TCCACAGTTCAGCCAAAGG |
| 79 | rs230596409 | 13 | 64,921,972 | C/C | T/T | GGTGTTGACCATGAGCCTTC | CTGGGGTGAGCTTAGGTCTG |
| 80 | rs251507217 | 13 | 101,112,155 | A/A | T/T | CCCTGTACCGTCCAATCATC | TTCTCCCCACCTCTGATGTC |
| 81 | rs242991609 | 13 | 119,477,808 | C/C | A/A | TTTTGGCTGTGCAATTCTTG | CACACAGAGGTCGCCTATCA |
| 82 | rs265193270 | 14 | 39,164,780 | C/C | T/T | GGCCATCTCATCAGTGCATA | AGGCTGACATGGTTTTGAGC |
| 83 | rs235428682 | 14 | 75,727,727 | G/G | A/A | ACATCTCCAGCTTCCAGACC | GAGGCGGTGACTATGAAGGA |
| 84 | rs222607275 | 14 | 117,850,332 | G/G | A/A | TTGTGGTTTCAGGAATGTCG | GGCAAACTTCTTGCCTCAGA |
| 85 | rs243245803 | 15 | 22,748,238 | T/T | G/G | TGTTTCCTGAGAGGGTTTGC | TTTGGGAAAGACAAGGGAGA |
| 86 | rs243400512 | 15 | 55,816,925 | C/C | T/T | TCAGAGGCTGAAGTGACAGC | GGGCAGTCTGTCTGTGGAAG |
| 87 | rs231321125 | 15 | 97,760,563 | C/C | G/G | CTCTCACGAGGACATGAGCA | GGCTCCCCAGTAAAACATGA |
| 88 | rs230243864 | 16 | 20,458,800 | C/C | T/T | TGGGGGCTTATCTTGTTCAC | ACTTAACCACAAGCCCAGGA |
| 89 | rs240948896 | 16 | 61,450,798 | G/G | A/A | TGGAGCATGACAAGGAATCA | TTTGCAAAATCCATGATTGG |
| 90 | rs240067957 | 17 | 40,854,409 | C/C | T/T | TGCTCATGGTAAATGCTGGA | TCAGCACTCAGGTGATTTCC |
| 91 | rs259144033 | 17 | 69,131,609 | C/C | T/T | CATGCACACGGCAGTAGAAG | CAGAGGTGGAACCAGGAAGA |
| 92 | rs225963780 | 18 | 22,530,101 | G/G | T/T | AGCGGTATGCTTGCTTTGAT | ACAAGGGCCAAATATTGCTG |
| 93 | rs214638331 | 18 | 41,344,993 | T/T | C/C | CTGCCAGATAAGCCACCAAT | TCACCAATGACAGAGCAAAAA |
| 94 | rs255789242 | 18 | 59,519,801 | C/C | T/T | TTCCCCTAGCTTGGAAACCT | TCTTTCCTGGAGTTGCCCTA |
| 95 | rs263687961 | 18 | 90,448,757 | A/A | G/G | TTCCCATTGTGGTCATTGAA | TGAGCTAAATTTGGAGCAAGC |
| 96 | rs232414357 | 19 | 23,329,888 | C/C | T/T | CAGCCCTCCCCTTTATCTTC | GTATGCCCCTGTTGGGTCTA |
| 97 | rs230656170 | 19 | 40,364,531 | G/G | A/A | AGCGCTCGCTTTGACATAAT | TGGGACAGGAGGAGGTTACA |
| 98 | rs246037535 | X | 84,805,631 | C/C | T/T | CCCTAGGGCAACATGGTAAA | CATTCCGTGCAAATGAGATG |
| 99 | rs266019057 | X | 112,095,948 | A/A | G/G | GGTGGCAGAGATGGAAACAT | CTGTCTTGCTTGGTCGCTAA |
| 100 | rs212226666 | X | 157,445,480 | T/T | A/A | TGCACTTGCACATCCTACAG | GGGGTTTGGGTTTTCATTTT |
SNP: single nucleotide polymorphism.
Results
From the SNP database, 1,361 informative SNPs on all of the chromosomes except Y were screened, and the SNPs were able to distinguish C57BL/6NJ from the other 12 inbred strains in silico (Table 2). No informative SNPs on chromosome Y were found in the database. Among the informative SNPs, 486 candidate SNPs from chromosome 1–X were selected to include approximately 1 SNP locus per 10 Mb, and the genotypes of the selected SNP loci were examined by direct sequencing of C57BL/6J and C57BL/6NJ DNA samples. As a result, 315 SNP loci were sequenced successfully and 277 SNPs were confirmed to be specific to C57BL/6NJ. The genotypes of the remaining 38 SNP loci were not consistent with the data in the SNP database. According to the Ensembl Mouse Genome Server (www.ensembl.org/Mus_musculus/Info/Index), 10 of the 277 SNPs were nonsynonymous variant SNPs, such as missense or stop-gain variants, which were predicted to affect the amino acid sequence of the protein (Table 3).
Table 2. Confirmation of C57BL/6NJ-specific SNPs by direct sequencing of PCR products.
| Chromosome | Number of SNPs selected |
Number of SNPs sequenced |
Direct sequencing succeeded |
Confirmed | Data discrepancy |
|---|---|---|---|---|---|
| 1 | 91 | 91 | 67 | 60 | 7 |
| 2 | 98 | 98 | 67 | 61 | 6 |
| 3 | 89 | 45 | 28 | 25 | 3 |
| 4 | 74 | 18 | 8 | 8 | 0 |
| 5 | 64 | 16 | 13 | 11 | 2 |
| 6 | 71 | 48 | 21 | 10 | 11 |
| 7 | 57 | 16 | 9 | 9 | 0 |
| 8 | 82 | 13 | 9 | 8 | 1 |
| 9 | 68 | 12 | 8 | 8 | 0 |
| 10 | 61 | 14 | 10 | 8 | 2 |
| 11 | 50 | 11 | 8 | 8 | 0 |
| 12 | 80 | 12 | 7 | 7 | 0 |
| 13 | 85 | 13 | 9 | 8 | 1 |
| 14 | 54 | 12 | 9 | 8 | 1 |
| 15 | 59 | 11 | 8 | 8 | 0 |
| 16 | 69 | 11 | 7 | 7 | 0 |
| 17 | 62 | 10 | 4 | 4 | 0 |
| 18 | 67 | 10 | 8 | 8 | 0 |
| 19 | 20 | 9 | 5 | 4 | 1 |
| X | 60 | 16 | 10 | 7 | 3 |
| Total | 1361 | 486 | 315 | 277 | 38 |
SNP: single nucleotide polymorphism.
Table 3. Status of 10 nonsynonymous variant SNPs.
| dbSNP ID | Chromo some |
Position (bp) (GRCm38) |
Linked gene | C57BL/6J allele |
C57BL/6NJ allele |
Variant type of C57BL/6NJ allele |
|---|---|---|---|---|---|---|
| rs262569844 | 5 | 89,775,351 | Adamts3 | C/C | T/T | Missense (Val199Ile) |
| rs229712565 | 5 | 112,762,721 | Myo18b | C/C | T/T | Missense (Arg1935His) |
| rs243575509 | 7 | 102,973,309 | Olfr577 | C/C | T/T | Missense (Val228Ile) |
| rs246274290 | 10 | 88,091,833 | Pmch | T/T | C/C | Missense (Ile132Thr) |
| rs240617401 | 11 | 46,222,615 | Cyfip2 | G/G | A/A | Missense (Ser968Phe) |
| rs238893157 | 11 | 90,480,671 | Stxbp4 | C/C | T/T | Missense (Ala535Thr) |
| rs242991609 | 13 | 119,477,808 | 4833420G17Rik | C/C | A/A | Missense (Thr484Lys) |
| rs248157600 | 14 | 70,586,204 | Fam160b2 | G/G | T/T | Missense (Ser575Arg) |
| rs246033409 | 15 | 11,336,383 | Adamts12 | G/G | T/T | Missense (Cys1518Phe) |
| rs230596409 | 13 | 64,921,972 | Spata31 | C/C | T/T | Stop gained (Arg645Ter) |
Information of the variant type was obtained from the Ensembl Mouse Genome Server.
SNP: single nucleotide polymorphism.
Next, 100 SNPs for genetic monitoring of C57BL/6N-derived substrains were selected from the 277 SNPs to include 1 SNP locus per 10–40 Mb to cover all of the chromosomes except Y (Table 4), and these SNPs were genotyped in the other 10 C57BL/6N and 6 C57BL/6J-derived substrains. All C57BL/6N and C57BL/6J-derived substrains were homozygous for these 100 SNP loci. In addition, when two samples from the same strain were genotyped in the C57BL/6N-derived substrains, they were found to be completely identical. The genotyping results for the C57BL/6J-derived substrains were consistent with the C57BL/6J reference sequence in the database. SNP genotyping demonstrated variation in the number of C57BL/6NJ-specific SNPs among the C57BL/6N-derived substrains (Table 5 ). Fourteen SNPs at Locus Nos. 10, 13, 14, 22, 30, 42, 44, 45, 54, 58, 63, 68, 84, and 93 were shared commonly, while the remaining 86 SNPs were only shared partly among the 11 C57BL/6N-derived substrains.
Table 5. SNPs among C57BL/6 substrains.

Discussion
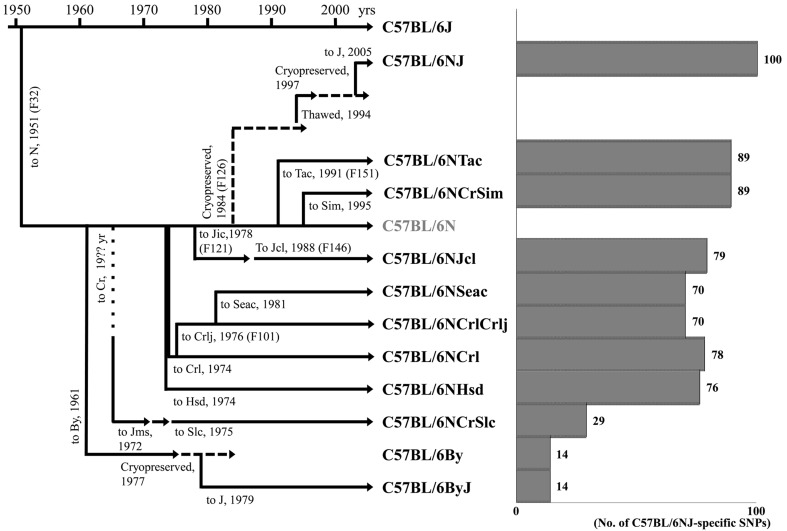
The relationship between the genealogy of the C57BL/6N-derived substrains used in this study and the number of the 100 C57BL/6NJ-specific SNPs in the substrains is summarized in Fig. 1. Milestones in the establishment of the strains were obtained from the product catalogs of the breeders and previous reports [14, 16]. The C57BL/6 strain was separated from the C57BL parental strain at the end of the 1940s and introduced to The Jackson Laboratory. A few years later, C57BL/6 mice were sent to the National Institutes of Health (NIH; Bethesda, MD) from The Jackson Laboratory, and the C57BL/6N mice were separated into the other C57BL/6N substrains, including the C57BL/6By strain, at different times. C57BL/6NCrSim mice from Simonsen Laboratories (Gilroy, CA) were derived from C57BL/6N mice at the NIH in 1995. C57BL/6NTac mice from Taconic Farms (New York, NY) were derived from the NIH Animal Genetic Resource at F151 in 1991. C57BL/6NJ mice were derived from embryos cryopreserved at the NIH in 1984. C57BL/6NJcl mice were introduced to the Central Institute for Experimental Animals (Kawasaki, Japan) from the NIH at F121 in 1978, and then transferred to CLEA Japan (Tokyo, Japan) at F146 in 1988. Charles River Laboratories (Wilmington, MA) obtained C57BL/6N mice from the NIH in 1974. The C57BL/6NCrl mice were further transferred to Charles River Laboratories Japan (Yokohama, Japan) at F101 in 1976, and since then, the mice have been distributed as C57BL/6NCrlCrlj. C57BL/6NSeac mice were introduced to Kyudo (Tosu, Japan) from the Charles River Laboratory Japan in 1981. C57BL/6NHsd mice were derived from a nucleus colony at the NIH in 1974. C57BL/6NCrSlc mice were introduced to the Institute of Medical Science, The University of Tokyo (Tokyo, Japan) in 1972 by Mr. Samuel M. Poiey, and then the mice were transferred to Japan SLC (Hamamatsu, Japan) in 1975. C57BL/6By (C57BL/6ByJ) mice were derived from the breeding stocks of Dr. Donald Bailey at the NIH in 1961.
Fig. 1.
Genealogy of C57BL/6N-derived substrains (left) and a bar graph for the number of C57BL/6NJ-specific SNPs (single nucleotide polymorphisms) (right). Abbreviations in the genealogy: N: National Institutes of Health; By: Dr. Donald Bailey; J: The Jackson Laboratory; Cr: National Cancer Institute; Jms: Institute of Medical Science, The University of Tokyo; Slc: Japan SLC; Crl: Charles River Laboratories; Crlj: Charles River Laboratories Japan; Seac: Kyudo; Hsd: Harlan Laboratories; Jic: Central Institute for Experimental Animals; Jcl: CLEA Japan; Sim: Simonsen Laboratories; Tac: Taconic Farm. Doted line: the year of introduction was unknown. Broken line: the strain was stored as frozen embryos. The C57BL/6NHsd and C57BL/6NCrl strains were derived independently from the NIH on unknown dates in 1974. On the right (bar graph), the number of C57BL/6NJ-specific SNPs of each C57BL/6N-derived substrain is indicated as a solid bar. The number of SNPs is also indicated at the right side of each bar.
The number of C57BL/6NJ-specific SNPs in each C57BL/6N-derived substrain was well correlated with its branching date from the original C57BL/6N strain (Fig. 1). Each C57BL/6N-derived substrain, except C57BL/6NTac and C57BL/6NCrSim, could only share C57BL/6NJ-specific SNPs that had arisen before its branching from the original C57BL/6N strain. The oldest C57BL/6By and C57BL/6ByJ strains may well preserve the genotype of C57BL/6N in approximately 1961 and only share 14 SNPs with other later substrains. As for the C57BL/6NTac and C57BL/6NCrSim strains, they only shared C57BL/6NJ-specific SNPs that had arisen before the C57BL/6NJ mice branched from the original C57BL/6N strain in 1984. Eleven SNPs that were detected only in the C57BL/6NJ strain were assumed to have arisen in C57BL/6NJ after it branched from the C57BL/6N strain in 1984. In other words, we could determine retrospectively when the C57BL/6NJ-specific SNPs had arisen by comparing the branching history and accumulated C57BL/6NJ-specific SNPs in different C57BL/6N-derived substrains. Similar observations have been reported previously in C57BL/6J-derived substrains [14, 32].
The C57BL/6NCrlCrlj and C57BL/6NSeac substrains that branched directly from C57BL/6NCrl and C57BL/6NCrlCrlj, respectively, but not directly from the original C57BL/6N, had only 70 C57BL/6NJ-specific SNPs, which is less than that of C57BL/6NCrl (Fig. 1). This corresponds to the number of discontinuous distribution patterns of SNPs seen in Table 5. One possible explanation for the discontinuous distribution of SNPs is that the 9 SNP loci, Nos. 20, 32, 38, 46, 48, 52, 66, 70, and 72 (Table 5), were heterozygous just before the branching of the C57BL/6NHsd and C57BL/6NCrl strains in 1974. Then, the SNPs were independently fixed to either C57BL/6NJ type or C57BL/6J type in the subsequent inbreeding of each substrain. Finally, 6 SNP loci (Nos. 20, 38, 46, 48, 52 and 66) of C57BL/6NHsd and 8 SNP loci (Nos. 32, 38, 46, 48, 52, 66, 70 and 72) of C57BL/6NCrl were thought to be fixed to C57BL/6NJ type, while the 9 SNP loci of C57BL/6NCrlCrlj and C57BL/6NSeac were fixed to C57BL/6J type. Although there was no significant difference in the pattern of the 100 selected SNPs between C57BL/6By and C57BL/6ByJ, and C57BL/6NTac and C57BL/6NCrSim, informative SNPs that can be used to discriminate these substrains can be found by genotyping additional candidate SNPs in the future.
In the selected 277 SNPs, 10 nonsynonymous SNPs were identified that were predicted to affect the amino acid sequence of the protein (Table 3). According to the Ensembl Mouse Genome Server, 4 of these 10 SNP variants, which occurred in the Myo18b, Olfr577, Cyfip2, and Adamts12 genes, were predicted to affect adversely each protein’s function. Notably, the Ser968Phe variant of Cyfip2 in C57BL/6N mice was reported recently by Kumar et al. [11]; this mutation destabilizes CYFIP2 protein and leads to acute and sensitized cocaine-response phenotypes. Although the C57BL/6N-derived substrains are closely related strains, such genetic differences would undoubtedly affect the phenotypes in various studies and interact with targeted mutations. Other phenotypic differences in behavior have been also reported among several C57BL/6N-derived substrains [3, 12]. As for the C57BL/6J and C57BL/6N strains, a relationship between genome variation and phenotypic changes has also been demonstrated [21]. In the future, advanced phenotypic analyses among the C57BL/6N-derived substrains will reveal novel functions of the genome variations detected among these substrains.
All of the SNPs detected in this report, including several nonsynonymous SNPs, will also be instrumental in the accurate identification of the C57BL/6N-derived substrain status of mutant mice and avoid the incorrect interpretation of data due to background effects. Furthermore, the selected SNP markers will be useful for quickly producing congenic strains to move a targeted mutation from one C57BL/6N-derived substrain to another substrain. Hence, our findings will serve as useful markers for the accurate and sophisticated genetic monitoring of C57BL/6N-derived congenic strains.
Acknowledgments
This work was supported by a Grand-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant Number 23700518) to M.K. We are grateful to Mr. Ikuo Miura and Drs. Shigeharu Wakana and Kuniya Abe at the RIKEN BioResource Center for their valuable advice. We also thank Dr. Naomi Nakagata, Kumamoto University for kindly providing the information about the origin of C57BL/6NSeac. The RIKEN BioResource Center is participating in the National BioResource Project (NBRP) by the MEXT, and is designated as a central core facility of the NBRP for mouse resources in Japan.
References
- 1.Bailey D.W.1978. Source of subline divergence and their relative importance for sublines of six major inbred strains of mice. pp. 197–215. In: Origins of inbred mice (Morse III, H.C. ed.), Academic Press, Bethesda, Maryland. [Google Scholar]
- 2.Brown S.D., Moore M.W.2012. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23: 632–640. doi: 10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang H.Y., Mitzner W., Watson J.2012. Variation in airway responsiveness of male C57BL/6 mice from 5 vendors. J. Am. Assoc. Lab. Anim. Sci. 51: 401–406. [PMC free article] [PubMed] [Google Scholar]
- 4.Crawley J.N., Belknap J.K., Collins A., Crabbe J.C., Frankel W., Henderson N., Hitzemann R.J., Maxson S.C., Miner L.L., Silva A.J., Wehner J.M., Wynshaw-Boris A., Paylor R.1997. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology (Berl.) 132: 107–124. doi: 10.1007/s002130050327 [DOI] [PubMed] [Google Scholar]
- 5.Gertsenstein M., Nutter L.M., Reid T., Pereira M., Stanford W.L., Rossant J., Nagy A.2010. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE 5: e11260. doi: 10.1371/journal.pone.0011260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlai R.1996. Gene-targeting studies of mammalian behavior: is it the mutation or the background genotype? Trends Neurosci. 19: 177–181. doi: 10.1016/S0166-2236(96)20020-7 [DOI] [PubMed] [Google Scholar]
- 7.Grupe A., Germer S., Usuka J., Aud D., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G.2001. In silico mapping of complex disease-related traits in mice. Science 292: 1915–1918. doi: 10.1126/science.1058889 [DOI] [PubMed] [Google Scholar]
- 8.Huang T.T., Naeemuddin M., Elchuri S., Yamaguchi M., Kozy H.M., Carlson E.J., Epstein C.J.2006. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum. Mol. Genet. 15: 1187–1194. doi: 10.1093/hmg/ddl034 [DOI] [PubMed] [Google Scholar]
- 9.International Mouse Knockout Consortium, Collins F.S., Rossant J., Wurst W.2007. A mouse for all reasons. Cell 128: 9–13. doi: 10.1016/j.cell.2006.12.018 [DOI] [PubMed] [Google Scholar]
- 10.Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., Furlotte N.A., Eskin E., Nellåker C., Whitley H., Cleak J., Janowitz D., Hernandez-Pliego P., Edwards A., Belgard T.G., Oliver P.L., McIntyre R.E., Bhomra A., Nicod J., Gan X., Yuan W., van der Weyden L., Steward C.A., Bala S., Stalker J., Mott R., Durbin R., Jackson I.J., Czechanski A., Guerra-Assunção J.A., Donahue L.R., Reinholdt L.G., Payseur B.A., Ponting C.P., Birney E., Flint J., Adams D.J.2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. doi: 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar V., Kim K., Joseph C., Kourrich S., Yoo S.H., Huang H.C., Vitaterna M.H., de Villena F.P., Churchill G., Bonci A., Takahashi J.S.2013. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342: 1508–1512. doi: 10.1126/science.1245503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuo N., Takao K., Nakanishi K., Yamasaki N., Tanda K., Miyakawa T.2010. Behavioral profiles of three C57BL/6 substrains. Front. Behav. Neurosci. 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McVicar D.W., Winkler-Pickett R., Taylor L.S., Makrigiannis A., Bennett M., Anderson S.K., Ortaldo J.R.2002. Aberrant DAP12 signaling in the 129 strain of mice: implications for the analysis of gene-targeted mice. J. Immunol. 169: 1721–1728. doi: 10.4049/jimmunol.169.4.1721 [DOI] [PubMed] [Google Scholar]
- 14.Mekada K., Abe K., Murakami A., Nakamura S., Nakata H., Moriwaki K., Obata Y., Yoshiki A.2009. Genetic differences among C57BL/6 substrains. Exp. Anim. 58: 141–149. doi: 10.1538/expanim.58.141 [DOI] [PubMed] [Google Scholar]
- 15.Mouse Genome Sequence Consortium, Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Antonarakis S.E., Attwood J., Baertsch R., Bailey J., Barlow K., Beck S., Berry E., Birren B., Bloom T., Bork P., Botcherby M., Bray N., Brent M.R., Brown D.G., Brown S.D., Bult C., Burton J., Butler J., Campbell R.D., Carninci P., Cawley S., Chiaromonte F., Chinwalla A.T., Church D.M., Clamp M., Clee, C., Collins F.S., Cook L.L., Copley C., Coulson A., Couronne O., Cuff J., Curwen V., Cutts T., Daly M., David R., Davies J., Delehaunty K.D., Deri J., Dermitzakis E.T., Dewey C., Dickens N.J., Diekhans M., Dodge S., Dubchak I., Dunn D.M., Eddy S.R., Elnitski L., Emes R.D., Eswara P., Eyras E., Felsenfeld A., Fewell G.A., Flicek P., Foley K., Frankel W.N., Fulton L.A., Fulton R.S., Furey T.S., Gage D., Gibbs R.A., Glusman G., Gnerre S., Goldman N., Goodstadt L., Grafham D., Graves T.A., Green E.D., Gregory S., Guigó R., Guyer M., Hardison R.C., Haussler D., Hayashizaki Y., Hillier L.W., Hinrichs A., Hlavina W., Holzer T., Hsu F., Hua A., Hubbard T., Hunt A., Jackson I., Jaffe D.B., Johnson L.S., Jones M., Jones T.A., Joy A., Kamal M., Karlsson E.K., Karolchik D., Kasprzyk A., Kawai J., Keibler E., Kells C., Kent W.J., Kirby A., Kolbe D.L., Korf I., Kucherlapati R.S., Kulbokas E.J., Kulp D., Landers T., Leger J.P., Leonard S., Letunic I., Levine R., Li J., Li M., Lloyd C., Lucas S., Ma B., Maglott D.R., Mardis E.R., Matthews L., Mauceli E., Mayer J.H., McCarthy M., McCombie W.R., McLaren S., McLay K., McPherson J.D., Meldrim J., Meredith B., Mesirov J.P., Miller W., Miner T.L., Mongin E., Montgomery K.T., Morgan M., Mott R., Mullikin J.C., Muzny D.M., Nash W.E., Nelson J.O., Nhan M.N., Nicol R., Ning Z., Nusbaum C., O’Connor M.J., Okazaki Y., Oliver K., Overton-Larty E., Pachter L., Parra G., Pepin K.H., Peterson J., Pevzner P., Plumb R., Pohl C.S., Poliakov A., Ponce T.C., Ponting C.P., Potter S., Quail M., Reymond A., Roe B.A., Roskin K.M., Rubin E.M., Rust A.G., Santos R., Sapojnikov V., Schultz B., Schultz J., Schwartz M.S., Schwartz S., Scott C., Seaman S., Searle S., Sharpe T., Sheridan A., Shownkeen R., Sims S., Singer J.B., Slater G., Smit A., Smith D.R., Spencer B., Stabenau A., Stange-Thomann N., Sugnet C., Suyama M., Tesler G., Thompson J., Torrents D., Trevaskis E., Tromp J., Ucla C., Ureta-Vidal A., Vinson J.P., Von Niederhausern A.C., Wade C.M., Wall M., Weber R.J., Weiss R.B., Wendl M.C., West A.P., Wetterstrand K., Wheeler R., Whelan S., Wierzbowski J., Willey D., Williams S., Wilson R.K., Winter E., Worley K.C., Wyman D., Yang S., Yang S.P., Zdobnov E.M., Zody M.C., Lander E.S.2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- 16.New resource illustrates divergence of C57BL/6 laboratory mouse substrains JAX® NOTES Issue 512, Winter 2008. http://jaxmice.jax.org/jaxnotes/512/512s.html. [Google Scholar]
- 17.Petkov P.M., Ding Y., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Scott V.E., Wiles M.V.2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14: 1806–1811. doi: 10.1101/gr.2825804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettitt S.J., Liang Q., Rairdan X.Y., Moran J.L., Prosser H.M., Beier D.R., Lloyd K.C., Bradley A., Skarnes W.C.2009. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods 6: 493–495. doi: 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seong E., Saunders T.L., Stewart C.L., Burmeister M.2004. To knockout in 129 or in C57BL/6: that is the question. Trends Genet. 20: 59–62. doi: 10.1016/j.tig.2003.12.006 [DOI] [PubMed] [Google Scholar]
- 20.Siegmund A., Langnaese K., Wotjak C.T.2005. Differences in extinction of conditioned fear in C57BL/6 substrains are unrelated to expression of alpha-synuclein. Behav. Brain Res. 157: 291–298. doi: 10.1016/j.bbr.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 21.Simon M.M., Greenaway S., White J.K., Fuchs H., Gailus-Durner V., Wells S., Sorg T., Wong K., Bedu E., Cartwright E.J., Dacquin R., Djebali S., Estabel J., Graw J., Ingham N.J., Jackson I.J., Lengeling A., Mandillo S., Marvel J., Meziane H., Preitner F., Puk O., Roux M., Adams D.J., Atkins S., Ayadi A., Becker L., Blake A., Brooker D., Cater H., Champy M.F., Combe R., Danecek P., di Fenza A., Gates H., Gerdin A.K., Golini E., Hancock J.M., Hans W., Hölter S.M., Hough T., Jurdic P., Keane T.M., Morgan H., Müller W., Neff F., Nicholson G., Pasche B., Roberson L.A., Rozman J., Sanderson M., Santos L., Selloum M., Shannon C., Southwell A., Tocchini-Valentini G.P., Vancollie V.E., Westerberg H., Wurst W., Zi M., Yalcin B., Ramirez-Solis R., Steel K.P., Mallon A.M., de Angelis M.H., Herault Y., Brown S.D.2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14: R82. doi: 10.1186/gb-2013-14-7-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P.J., Stewart A.F., Bradley A.2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. doi: 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sluyter F., Marican C.C., Crusio W.E.1999. Further phenotypical characterisation of two substrains of C57BL/6J inbred mice differing by a spontaneous single-gene mutation. Behav. Brain Res. 98: 39–43. doi: 10.1016/S0166-4328(98)00049-7 [DOI] [PubMed] [Google Scholar]
- 24.Specht C.G., Schoepfer R.2001. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2: 11. doi: 10.1186/1471-2202-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanimoto Y., Iijima S., Hasegawa Y., Suzuki Y., Daitoku Y., Mizuno S., Ishige T., Kudo T., Takahashi S., Kunita S., Sugiyama F., Yagami K.2008. Embryonic stem cells derived from C57BL/6J and C57BL/6N mice. Comp. Med. 58: 347–352. [PMC free article] [PubMed] [Google Scholar]
- 26.Tsang S., Sun Z., Luke B., Stewart C., Lum N., Gregory M., Wu X., Subleski M., Jenkins N.A., Copeland N.G., Munroe D.J.2005. A comprehensive SNP-based genetic analysis of inbred mouse strains. Mamm. Genome 16: 476–480. doi: 10.1007/s00335-005-0001-7 [DOI] [PubMed] [Google Scholar]
- 27.Wade C.M., Kulbokas E.J., 3rd, Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J.2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574–578. doi: 10.1038/nature01252 [DOI] [PubMed] [Google Scholar]
- 28.Wahlsten D., Ozaki H.S., Livy D.1992. Deficient corpus callosum in hybrids between ddN and three other abnormal mouse strains. Neurosci. Lett. 136: 99–101. doi: 10.1016/0304-3940(92)90657-S [DOI] [PubMed] [Google Scholar]
- 29.Wiltshire T., Pletcher M.T., Batalov S., Barnes S.W., Tarantino L.M., Cooke M.P., Wu H., Smylie K., Santrosyan A., Copeland N.G., Jenkins N.A., Kalush F., Mural R.J., Glynne R.J., Kay S.A., Adams M.D., Fletcher C.F.2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA 100: 3380–3385. doi: 10.1073/pnas.0130101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yalcin B., Wong K., Agam A., Goodson M., Keane T.M., Gan X., Nellåker C., Goodstadt L., Nicod J., Bhomra A., Hernandez-Pliego P., Whitley H., Cleak J., Dutton R., Janowitz D., Mott R., Adams D.J., Flint J.2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. doi: 10.1038/nature10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Hunter K.W., Gandolph M., Rowe W.L., Finney R.P., Kelley J.M., Edmonson M., Buetow K.H.2005. A high-resolution multistrain haplotype analysis of laboratory mouse genome reveals three distinctive genetic variation patterns. Genome Res. 15: 241–249. doi: 10.1101/gr.2901705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zurita E., Chagoyen M., Cantero M., Alonso R., González-Neira A., López-Jiménez A., López-Moreno J.A., Landel C.P., Benítez J., Pazos F., Montoliu L.2011. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 20: 481–489. doi: 10.1007/s11248-010-9403-8 [DOI] [PubMed] [Google Scholar]