Abstract
The ongoing Ebola virus epidemic has presented numerous challenges with respect to control and treatment because there are no approved drugs or vaccines for the Ebola virus disease (EVD). Herein is proposed simple theoretical criterion for fast virtual screening of molecular libraries for candidate inhibitors of Ebola virus infection. We performed a repurposing screen of 6438 drugs from DrugBank using this criterion and selected 267 approved and 382 experimental drugs as candidates for treatment of EVD including 15 anti-malarial drugs and 32 antibiotics. An open source Web server allowing screening of molecular libraries for candidate drugs for treatment of EVD was also established.
Keywords: Ebola virus, drug candidates, entry inhibitors, virtual screening
Introduction
The current Ebola virus outbreak is one of the largest outbreaks of its kind in history and the first in West Africa. By January 14, 2015, a total of 21296 probable and confirmed cases, including 8429 deaths from Ebola virus disease (EVD), had been reported from five countries in West Africa - Guinea, Liberia, Nigeria, Senegal, and Sierra Leone ( http://apps.who.int/iris/bitstream/10665/148237/2/roadmapsitrep_14Jan2015_eng.pdf?ua=1). EVD with a high case-fatality rate of 40% and with currently no approved vaccine or therapy, represents a major public health threat.
In response to the current Ebola virus outbreak, the international community has urged for accelerated development of drugs against EVD but also has endorsed the clinical use of unregistered treatments for Ebola 1. Conventional time and the money consuming approach of drug development (> 10 years; > 2 billions $) does not meet the current urgent need for anti-Ebola drugs. Repurposing or repositioning of existing drugs could overcome some of these obstacles and help in the rapid discovery and development of therapeutics for EVD, although this approach does not negate the need for some preclinical studies and clinical trials for validation of the proposed indications. Recently, results of two large repurposing screenings of Food and Drug Administration (FDA)-approved drugs have been reported. In the first study, Madrid and co-workers performed in vitro and in vivo (in mice) screening of 1012 FDA-approved drugs and selected 24 candidate entry inhibitors for Ebola virus 2. In the second study, 53 inhibitors of Ebola virus infection with IC 50 < 10 µM and selectivity index SI > 10-fold have been identified by in vitro screening of 2816 FDA-approved drugs 3. In the same study, an additional 95 drugs which are active against Ebola virus infection with IC 50 > 10 µM and SI <10-fold were also reported.
Although in vitro and in vivo screening for repurposing/repositioning of existing drugs could significantly accelerate discovery of new drugs these approaches are time-consuming and costly for screening of large drug libraries. Recently, we proposed a novel approach for in silico screening of molecular libraries for drug candidates 4– 8. This approach, which uses the average quasi valence number (AQVN) and the electron-ion interaction potential (EIIP), parameters determining long-range interaction between biological molecules, might hold a key to overcoming some of these obstacles in experimental screening by significantly reducing the number of compounds which should be in vitro and in vivo tested 9.
Herein, 267 approved and 382 experimental drugs, selected by the EIIP/AQVN-based virtual screening of DrugBank ( http://www.drugbank.ca), have been proposed as candidate drugs for treatment of EVD. An open access portal allowing screening of molecular libraries for candidate drugs for treatment of EVD was established.
Material and methods
Molecular libraries
For screening of drugs for repurposing to select candidates for Ebola virus entry inhibitors, 1463 approved and 4975 experimental drugs from DrugBank ( http://www.drugbank.ca) were screened. For development of the predictive criterion used in this analysis, the learning set ( Dataset 1) encompassing 152 drugs which are selected as inhibitors of Ebola virus infection by in vitro and in vivo screening of 3828 FDA-approved drugs 2, 3, was established. As control data sets 45,010,644 compounds from PubChem ( http://www.ncbi.nlm.nih.gov/pccompound) and 49 Ebola virus entry inhibitors collected by data mining of literature and patents, were used. For screening of literature data the NCBI literature database PubMed ( http://www.ncbi.nlm.nih.gov/pubmed) was used. For search of patents and patent applications we used the Free Patent Online browser ( http://www.freepatentsonline.com).
Drug repurposing screen to identify active compounds that block Ebola entry
Specific recognition and targeting between interacting biological molecules at distances > 5Å are determined by the average AQVN and the EIIP 10, which are derived from the general model pseudopotential 11, 12. These parameters for organic molecules are determined by the following simple equations 10:
Where Z* is the average quasi-valence number (AQVN) determined by
where Z i is the valence number of the i-th atomic component, n i is the number of atoms of the i-th component, m is the number of atomic components in the molecule, and N is the total number of atoms. EIIP values calculated according to equation 1 and equation 2 are expressed in Rydberg units (Ry).
Among 3300 currently used molecular descriptors, AQVN and EIIP represent the unique physical properties which characterize the long-range interactions between biological molecules 10. Small molecules with similar AQVN and EIIP values interact with the common therapeutic target, which allow establish criterions for virtual screening of molecular libraries for compounds with similar therapeutic properties 4– 9. Here we develop the EIIP/AQVN-based criterion for virtual screening of molecular libraries for candidate drugs against Ebola virus infection.
Results and discussion
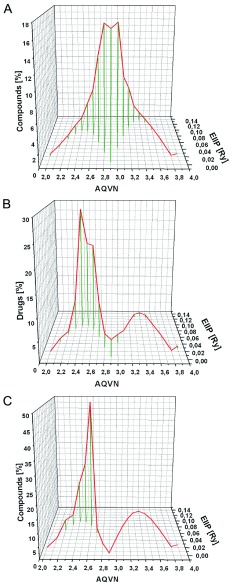
Previously, analyses of the EIIP/AQVN distribution of 45,010,644 compounds from the PubChem database ( http://www.ncbi.nlm.nih.gov/pccompound) revealed that 92.5% of presented compounds are homogenously distributed within EIIP and AQVN intervals (0.00 – 0.11 Ry) and (2.4 – 3.3), respectively). This domain of the EIIP/AQVN space, encompassing the majority of known chemical compounds, is referred to as the “basic EIIP/AQVN chemical space” (BCS) 6. Analysis of the molecular training set ( Dataset 1), encompassing 152 small molecule inhibitors of Ebola virus infection selected by in vitro screening of 3828 FDA approved drugs 2, 3, show that 79% of these compounds are placed within AQVN and EIIP region (2.3 – 2.7) and (0.0829 – 0.0954 Ry), respectively (“Ebola Virus Infection Inhibitors Space”, EVIIS). The AQVN region (2.36 – 2.54) and the EIIP region (0.0912 – 0.0924 Ry) form the part of EVIIS which encompasses 55.5% of all drugs from the learning set (core EVIIS, cEVIS). Literature data mining reveals 49 compounds with experimentally proved activity against Ebola virus infection ( Table 1) 13– 29. Most of these compounds 47 (95.9%) are placed within EVIIS ( Table 1). Of note is that EVIIS and cEVIIS domains contain only 14.6% and 6.5% of compounds from PubChem, respectively. This confirms high specificity of clustering of Ebola virus infection inhibitors within the EIIP/AQVN space. Comparison of distributions of Ebola virus infection inhibitors and compounds from PubMed is given in Figure 1.
Figure 1. Distribution of compounds according to their average quasivalence number (AQVN) and electron-ion interaction potential (EIIP) values.
( A) 45010644 compounds from the PubChem database ( http://www.ncbi.nlm.nih.gov/pccompound); ( B) FDA-approved drugs which are active against Ebola virus infection ( Dataset 1) 2, 3; ( C) Entry inhibitors of Ebola virus ( Table 1).
Table 1. Small-molecule entry inhibitors for Ebola virus.
| Compound | Formula | AQVN | EIIP [Ry] | Reference |
|---|---|---|---|---|
| Chloroquine | C18H26ClN3 | 2.375 | 0.0941 | 16 |
| Bafilomycin A1 | C35H58O9 | 2.471 | 0.0960 | 17 |
| Cytochalasin B | C29H37NO5 | 2.611 | 0.0810 | 17 |
| Cytochalasin D | C30H37NO6 | 2.676 | 0.0672 | 17 |
| Latruculion A | C22H31NO5S | 2.667 | 0.0693 | 17 |
| Jasplakinolide | C36H45BrN4O6 | 2.674 | 0.0676 | 18 |
| Clomiphene | C26H28ClNO | 2.526 | 0.0926 | 18 |
| Toremifene | C26H28ClNO | 2.526 | 0.0926 | 18 |
| Chlorpromazine | C17H19ClN2S | 2.600 | 0.0829 | 19 |
| Amiodarone | C25H29I2NO3 | 2.567 | 0.0880 | 20 |
| Dronedarone | C31H44N2O5S | 2.578 | 0.0864 | 20 |
| Verapamil | C27H38N2O4 | 2.535 | 0.0917 | 20 |
| Clomiphene | C26H28ClNO | 2.526 | 0.0926 | 21 |
| AY-9944 | C22H28Cl2N2 | 2.370 | 0.0938 | 21 |
| Ro 48-8071 | C23H27BrFNO2 | 2.505 | 0.0940 | 21 |
| U18666A | C25H41NO2 | 2.290 | 0.0849 | 21 |
| Terconazole | C26H31Cl2N5O3 | 2.687 | 0.0644 | 21 |
| Triparanol | C27H32ClNO2 | 2.508 | 0.0941 | 21 |
| Impramine | C19H24N2 | 2.444 | 0.0964 | 22 |
| 3.47 | C34H43N3O5 | 2.635 | 0.0763 | 23 |
| Cytochalasin B | C29H37NO5 | 2.611 | 0.0810 | 24 |
| Cytochalasin D | C30H37NO6 | 2.676 | 0.0672 | 24 |
| Latrunculin A | C22H31NO5S | 2.667 | 0.0693 | 24 |
| Jasplakinolide | C36H45BrN4O6 | 2.674 | 0.0676 | 24 |
| NSC62914 | C31H40O3 | 2.460 | 0.0962 | 25 |
| Compound 1 | C30H38N6O2 | 2.632 | 0.0770 | 26 |
| Compound 2 | C32H46N6 | 2.429 | 0.0963 | 26 |
| Compound 3 | C28H34N6O2 | 2.686 | 0.0635 | 26 |
| Compound 5 | C42H58N10O6 | 2.690 | 0.0635 | 27 |
| Compound 8a | C17H23N3O3 | 2.695 | 0.0621 | 27 |
| Compound 8b | C17H23N3O3 | 2.695 | 0.0621 | 27 |
| Compound 8y | C16H20BrNO2 | 2.610 | 0.0812 | 27 |
| Compound 15h | C15H20JN5O | 2.667 | 0.0693 | 27 |
| Compound 15k | C15H128Br3N5O | 2.667 | 0.0693 | 27 |
| Retinazone | C38H56Na3N5S2 | 2.385 | 0.0947 | 28 |
| Compound 7 | C17H12F4N2 | 2.467 | 0.0647 | 29 |
| Brincidofovir * | C27H52N3O7P | 2.467 | 0.0961 | 30 |
| Hit compound 3 | C25H35N3O2 | 2.494 | 0.0950 | 31 |
| Hit compound 3.1 | C21H24ClN3O2 | 2.667 | 0.0693 | 31 |
| Hit compound 3.2 | C20H29N3O2 | 2.518 | 0.0933 | 31 |
| Hit compound 3.3 | C30H35N3O2 | 2.556 | 0.0894 | 31 |
| Hit compound 3.4 | C25H32N4O3 | 2.656 | 0.0717 | 31 |
| Hit compound 3.5 | C20H23N3O2 | 2.708 | 0.0587 | 31 |
| Hit compound 3.6 | C25H33N3O2 | 2.540 | 0.09913 | 31 |
| Hit compound 3.7 | C22H27N3O3 | 2.691 | 0.0633 | 31 |
| Hit compound 3.18 | C26H37N3O2 | 2.471 | 0.0471 | 31 |
| Hit compound 3.48 | C34H43N3O5 | 2.635 | 0.0763 | 31 |
| Hit compound 3.105 | C34H40N6O2 | 2.658 | 0.0712 | 31 |
| NSC 62914 | C31H39O3 | 2.480 | 0.0957 | 32 |
*Experimental drug applied for treatment of Ebola patients in Liberia ( http://www.ox.ac.uk/news/2014-11-13-oxford-lead-trial-experimental-drug-ebola-patients)
AQVN: average quasivalence number; EIIP: electron-ion interaction potential
It was shown that Ebola virus glycoprotein (GP)-mediated entry and infection is subordinated with a membrane-trafficking event that translocates a GP binding partner to the cell surface, which depends on microtubules 30, 31. Consistently, microtubule inhibitors which block this trafficking process could decrease infection without interfering with the direct binding and translocation of the Ebola virus into cells. AQVN and EIIP values of microtubule modulators and transcription inhibitors with reported anti-Ebola virus activity are given in Table 2. As can be seen, all these compounds, which do not directly affect binding and internalization of Ebola virus, are located outside of EVIIS. This additionally confirms the specificity of the EVIS domain.
Table 2. Viral transcription inhibitors and microtubule modulators with anti-Ebola virus activity.
| Compound | Formula | AQVN | EIIP [Ry] |
|---|---|---|---|
| Viral transcription inhibitors | |||
| BCX4430 | C11H15N5O3 | 3.000 | 0.0439 |
| Favipiravir | C5H4FN3O2 | 3.467 | 0.1304 |
| C-c3Ado | C12H16N4O3 | 2.914 | 0.0112 |
| c3Nep | C12H14N4O3 | 3.030 | 0.0552 |
| “D-like” 1’-6’-isoneplanocin | C11H12N5O3 | 3.194 | 0.1076 |
| “L-like” 1’-6’-isoneplanocin | C11H12N5O3 | 3.194 | 0.1076 |
| CMLDBU3402 | C30H26BrN3O7 | 3.045 | 0.1343 |
| Microtubule modulators | |||
| Vinblastine | C13H8Cl2N2O4 | 3.310 | 0.0130 |
| Vinorelbine | C45H54N4O8 | 2.721 | 0.0552 |
| Vincristine | C46H56N4O10 | 2.759 | 0.0439 |
| Colchicine | C22H25NO6 | 2.852 | 0.0121 |
| Nocodazole | C14H11N3O3S | 3.312 | 0.1298 |
| Mebendazole | C16H13N3O3 | 3.143 | 0.0934 |
| Albendazole | C12H15N3O2S | 2.909 | 0.0092 |
In further analysis we used EVIIS as a filter for virtual screening for candidate Ebola virus infection inhibitors. In Dataset 2 622 approved and 1089 experimental drugs in Dataset 3 selected by EVIIS screening of 6532 drugs from DrugBank are reported. Using cEVIIS, we located 267 approved and 382 experimental drugs. This small molecular library represents a source of candidate drugs for treatment of Ebola virus disease (EVD), which can be further experimentally tested.
AQVN: average quasivalence number; EIIP: electron-ion interaction potential
AQVN: average quasivalence number; EIIP: electron-ion interaction potential
Madrid and co-workers selected 24 drugs by in vitro screening of 1012 FDA-approved drugs, which are effective against Ebola virus infection 2. They also showed that among these compounds, four antimalarial drugs (chloroquine, hydroxychloroquine, amodiaquine and aminoquinoline-13) also are effective against Ebola virus infection in vivo 2. Among 53 compounds which effectively inhibit Ebola virus infection in vitro, which Kouznetsova and co-workers selected from 2816 approved drugs, are also three anti-malarial drugs (mefloquione, chloroquine, amodiaquine) 3. It was also suggested that application of chloroquine for prevention of virus transmission should be considered because this compound significantly inhibits Ebola virus infection 13. Our analysis showed that 15 of 22 approved ant-malarial drugs ( http://en.wikipedia.org/wiki/Antimalarial_medication) are located in EVIIS ( Table 3). Six 2-alkylquinolines have been also included in this study. This chemical series is promising as some derivatives exhibited antiviral activity such as 2PQ, and 2QQ 32, 33 antimalarial activity such as 2PQ and 2PentQ2 34, antileishmanial activity such as 2PQ 35, 36 and neurotrophin-like activity on dopaminergic neurons such as 2QI15 37. These compounds exhibit some advantages in regard to their chemical synthesis with few steps and good yields as well as their chemical stability in tropical conditions of storage. Their combined effects against virus and Leishmania parasites suggested they could be an advantage for the treatment of Leishmania/HIV co-infections and they were considered as attractive enough to enter the pipeline of DNDi on 2010.
All these data strongly suggest that this class of drugs should be further investigated as a promising source of therapeutics for treatment of EVD. Anti-malarial drugs with dual activity should be of special interest because malaria represents the highest health-related disease in African countries with EVD.
Among 3828 FDA-approved drugs screened for anti-Ebola activity were six antibiotics which inhibit Ebola virus infection (azthromycin, erythromycin, spiramycin, dirithromycin, maduramicin, charitromycin) 2, 3. All these antibiotics are within EVIIS and four of them are in cEVIIS. Analysis of 184 approved antibiotics ( Dataset 4) showed that only 32 (17.4%) have AQVN and EIIP values in EVIIS, and that 11 of them are located within cEVIIS. Previously we reported domains of AQVN and EIIP which characterize different classes of antibiotics ( Table 4) 6. According to these data, among antibiotics some macrolides, pleuromutilins and aminoglycosides have the highest chance for inhibition of Ebola virus infection. Of note is that five of six antibiotics with experimentally proved activity against Ebola virus infection (azthromycin, erythromycin, spiramycin, dirithromycin, charitromycin) are macrolides. Antibiotics representing candidate Ebola virus infection inhibitors selected by EIIP/AQVN criterion are given in Table 5.
Table 3. Approved anti-malarial drugs selected as candidate drugs for EVD.
| Compound | Formula | AQVN | EIIP [Ry] |
|---|---|---|---|
| Quinine | C20H24N2O2 | 2.625 | 0.0784 |
| Chloroquinine | C18H26ClN3 | 2.375 | 0.0941 |
| Amodiquinine | C20H22ClN3O | 2.638 | 0.0756 |
| Proguanil | C11H16ClN5 | 2.606 | 0.0819 |
| Mefloquine | C17H16F6N2O | 2.524 | 0.0928 |
| Primaquine | C15H21NO3 | 2.600 | 0.0829 |
| Halofantrine | C26H30Cl2F3NO | 2.381 | 0.0945 |
| Clindamycin | C18H33ClN2O5S | 2.533 | 0.0919 |
| Artemether | C16H26O5 | 2.553 | 0.0897 |
| Piperaquine | C29H32Cl2N6 | 2.609 | 0.0814 |
| Artemotil | C17H28O5 | 2.520 | 0.0931 |
| Dihydroartemisin | C15H24O5 | 2.591 | 0.0844 |
| Quinidine | C20H24N2O2 | 2.625 | 0.0784 |
| Cinchonidine | C19H22N2O | 2.591 | 0.0844 |
| Artemisin | C15H22O5 | 2.667 | 0.0693 |
Table 4. AQVN and EIIP range of different antibiotics classes 6.
| Antibiotic class | AQVN | EIIP [Ry] |
|---|---|---|
| Penicillins | 2.975 - 3.180 | 0.035 - 0.124 |
| Cephalosporins | 3.071 - 3.473 | 0.070 - 0.130 |
| Carbapenems & Penems | 2.973 - 3.059 | 0.022 - 0.066 |
| Monobactams | 3.166 - 3.581 | 0.100 - 0.134 |
| Quinolines | 2.760 - 3.060 | 0.003 - 0.065 |
| Aminoglycosides | 2.552 - 2.820 | 0.024 - 0.084 |
| Tetracyclines | 2.933 - 3.111 | 0.018 - 0.084 |
| Macrolides | 2.467 - 2.630 | 0.077 - 0.096 |
| Pleuromutilins | 2.395 - 2.473 | 0.095 - 0.096 |
| Nitrofurans | 3.652 - 3.826 | 0.010 - 0.086 |
AQVN: average quasivalence number; EIIP: electron-ion interaction potential
Previous, we determined AQVN and EIIP domains characterizing different classes of anti-HIV drugs 4– 9. As can be seen in Table 6, the EIIP/AQVN domain of CCR5 HIV entry inhibitors is within EVIIS, and domains of CXCR4 HIV entry inhibitors and HIV protease inhibitors partially overlaps EVIIS. The EIIP/AQVN domains of other classes of anti-HIV agents are located outside EVIIS. This indicates that some HIV entry inhibitors and HIV protease inhibitors could also be effective drugs against Ebola virus infection.
Table 5. Antibiotics selected as candidate drugs for EVD.
| Antibiotics | Formula | AQVN | EIIP [Ry] |
|---|---|---|---|
| Tiamulin | C 28H 47NO 4S | 2.395 | 0.095 |
| Retapamulin | C 30H 47NO 4S | 2.434 | 0.096 |
| Valnemulin | C 31H 52N 2O 5S | 2.440 | 0.096 |
| Azithromycin | C 38H 72N 2O 12 | 2.468 | 0.096 |
| BC-3205 | C 32H 51N 2O 5S | 2.472 | 0.096 |
| Dirithromycin | C 42H 78N 2O 14 | 2.500 | 0.095 |
| Clarithromycin | C 38H 69NO 13 | 2.512 | 0.094 |
| Surfactin | C 53H 93N 7O 13 | 2.518 | 0.093 |
| Erythromycin | C 37H 67NO 13 | 2.525 | 0.093 |
| Clindamycin | C 18H 33ClN 2O 5S | 2.533 | 0.092 |
| Roxithromycin | C 41H 76N 2O 15 | 2.537 | 0.092 |
| Oleandomycin | C 35H 61NO 12 | 2.550 | 0.090 |
| Gentamicin | C 21H 43N 5O 7 | 2.553 | 0.090 |
| Spiramycin | C 43H 74N 2O 14 | 2.556 | 0.089 |
| Mupirocin | C 26H 44O 9 | 2.557 | 0.089 |
| Lincomycin | C 18H 34N 2O 6S | 2.590 | 0.085 |
| Netilmicin | C 21H 41N 5O 7 | 2.595 | 0.084 |
| Astromicin | C 17H 35N 5O 6 | 2.603 | 0.082 |
| Tylosin | C 46H 77NO 17 | 2.610 | 0.081 |
| Kitasamycin | C 35H 59NO 13 | 2.611 | 0.081 |
| Josamycin | C 42H 69NO 15 | 2.614 | 0.080 |
| Telithromycin | C 43H 65N 5O 10 | 2.618 | 0.080 |
| Telithromycin | C 43H 65N 5O 10 | 2.618 | 0.080 |
| Verdamicin | C 20H 39N 5O 7 | 2.620 | 0.080 |
| Midecamycin | C 41H 67NO 15 | 2.629 | 0.078 |
| Troleandomycin | C 41H 67NO 15 | 2.629 | 0.078 |
| Sisomicin | C 19H 37N 5O 7 | 2.647 | 0.074 |
| Cethromycin | C 42H 59N 3O 10 | 2.649 | 0.073 |
| Carbomycin A | C 42H 67NO 16 | 2.667 | 0.069 |
| Dibekacin | C 18H 37N 5O 8 | 2.676 | 0.067 |
| Echinocandin B | C 52H 81N 7O 16 | 2.692 | 0.063 |
| Rifabutin | C 46H 62N 4O 11 | 2.699 | 0.061 |
Table 6. AQVN and EIIP range of anti-HIV drugs 6.
| Target | AQVN | EIIP [Ry] |
|---|---|---|
| CXCR4 | 2.16 - 2.53 | 0.062 - 0.096 |
| CCR5 | 2.42 - 2.63 | 0.079 - 0.099 |
| PI | 2.61 - 2.78 | 0.040 - 0.080 |
| NRTI/NtRTI | 2.92 - 3.20 | 0.040 - 0.100 |
| INI | 3.00 - 3.20 | 0.044 - 0.116 |
| Anti-HIV flavonoids | 3.34 - 3.59 | 0.110 - 0.135 |
In conclusion, the presented results show that the EIIP/AQVN criterion can be used as an efficient filter in virtual screening of molecular libraries for candidate inhibitors of Ebola virus infection. Approved ( Dataset 2) and experimental drugs ( Dataset 3), anti-malarial drugs ( Table 3) and antibiotics ( Table 5) selected by this criterion represents a valuable source of candidate therapeutics for treatment of EVD, some of which are already approved by FDA for treatment of other diseases which can be repurposed for use in EVD. We hope that these data, obtained by an in silico drug repurposing screen, will accelerate discovery of drugs for treatment of EVD, which are necessary in this ongoing emergency situation caused by the current unprecedented Ebola virus outbreak. To enable other researchers working on online EIIP/AQVN-based screening of different sources of small molecules for candidate Ebola drugs, we established an open web server ( http://www.biomedconsulting.info/ebola_screen.php).
Data availability
The virtual screen for candidate inhibitors of EBOLA virus infection web tool is available at: http://www.biomedconsulting.info/tools/ebolascreen.php. An archived version can be accessed at: http://www.webcitation.org/6Vxtuojgx 38
F1000Research: Dataset 1. FDA-approved drugs which are active against Ebola virus infection 2, 3, 10.5256/f1000research.6110.d42876 39
F1000Research: Dataset 2. Approved and experimental drugs selected as candidate for treatment of EVD, 10.5256/f1000research.6110.d42877 40
F1000Research: Dataset 3. Experimental drugs selected as candidate for treatment of EVD, 10.5256/f1000research.6110.d42878 41
F1000Research: Dataset 4. Approved antibiotics screened for candidate anti-Ebola drugs, 10.5256/f1000research.6110.d42879 42
Acknowledgments
The authors would like to gratefully acknowledge networking support by the COST Action CM1307.
Funding Statement
This work was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grant no. 173001).
v1; ref status: indexed
References
- 1. Enserink M: Infectious diseases. Debate erupts on ‘repurposed’ drugs for Ebola. Science. 2014;345(6198):718–9. 10.1126/science.345.6198.718 [DOI] [PubMed] [Google Scholar]
- 2. Madrid PB, Chopra S, Manger ID, et al. : A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One. 2013;8(4):e60579. 10.1371/journal.pone.0060579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kouznetsova J, Sun W, Martínez-Romero C, et al. : Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microb Infect. 2014;3:e84 10.1038/emi.2014.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veljkovic V, Veljkovic N, Este J, et al. : Application of the EIIP/ISM bioinformatics concept in development of new drugs. Curr Med Chem. 2007;14(4):441–53. 10.2174/092986707779941014 [DOI] [PubMed] [Google Scholar]
- 5. Veljkovic V, Mouscadet JF, Veljkovic N, et al. : Simple criterion for selection of flavonoid compounds with anti-HIV activity. Bioorg Medic Chem Lett. 2007;17(5):1226–32. 10.1016/j.bmcl.2006.12.029 [DOI] [PubMed] [Google Scholar]
- 6. Veljkovic N, Glisic S, Perovic V, et al. : The role of long-range intermolecular interactions in discovery of new drugs. Exp Opin Drug Disc. 2011;6(12):1263–70. 10.1517/17460441.2012.638280 [DOI] [PubMed] [Google Scholar]
- 7. Maga G, Veljkovic N, Crespan E, et al. : New in silico and conventional in vitro approaches to advance HIV drug discovery and design. Exp Opin Drug Discov. 2013;8(1):83–92. 10.1517/17460441.2013.741118 [DOI] [PubMed] [Google Scholar]
- 8. Veljkovic N, Glisic S, Prljic J, et al. : Simple and general criterion for “ in silico” screening of candidate HIV drugs. Curr Pharm Biotechnol. 2013;14(5):561–9. 10.2174/138920101405131111105301 [DOI] [PubMed] [Google Scholar]
- 9. Tintori C, Veljkovic N, Veljkovic V, et al. : Computational studies of the interaction between the HIV-1 integrase tetramer and the cofactor LEDGF/p75: insights from molecular dynamics simulations and the informational spectrum method. Proteins. 2010;78(16):3396–408. 10.1002/prot.22847 [DOI] [PubMed] [Google Scholar]
- 10. Veljkovic V: A theoretical approach to preselection of carcinogens and chemical carcinogenesis. New York: Gordon & Breach1980. Reference Source [Google Scholar]
- 11. Veljkovic V, Slavic I: Simple general-model pseudopotential. Phys Rev Lett. 1972;29:105–7. 10.1103/PhysRevLett.29.105 [DOI] [Google Scholar]
- 12. Veljkovic V: The dependence of the Fermi energy on the atomic number. Phys Lett. 1973;45A(1):41–2. 10.1016/0375-9601(73)90497-0 [DOI] [Google Scholar]
- 13. Wool-Lewis RJ, Bates P: Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72(4):3155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yonezawa A, Cavrois M, Greene WC: Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79(2):918–26. 10.1128/JVI.79.2.918-926.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansen LM, Brannan JM, Delos SE, et al. : FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med. 2013;5(190):190ra79. 10.1126/scitranslmed.3005471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhattacharyya S, Warfield KL, Ruthel G, et al. : Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology. 2010;401(1):18–28. 10.1016/j.virol.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gehring G, Rohrmann K, Atenchong N, et al. : The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J Antimicrob Chemother. 2014;69(8):2123–31. 10.1093/jac/dku091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shoemaker CJ, Schornberg KL, Delos SE, et al. : Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PloS One. 2013;8(2):e56265. 10.1371/journal.pone.0056265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carette JE, Raaben M, Wong AC, et al. : Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477(7364):340–3. 10.1038/nature10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Côté M, Misasi J, Ren T, et al. : Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477(7364):344–8. 10.1038/nature10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yonezawa A, Cavrois M, Greene WC: Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79(2):918–26. 10.1128/JVI.79.2.918-926.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panchal RG, Reid SP, Tran JP, et al. : Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antiviral Res. 2012;93(1):23–9. 10.1016/j.antiviral.2011.10.011 [DOI] [PubMed] [Google Scholar]
- 23. Selaković Z, Opsenica D, Eaton B, et al. : A limited structural modification results in a significantly more efficacious diazachrysene-based filovirus inhibitor. Viruses. 2012;4(8):1279–88. 10.3390/v4081279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yermolina MV, Wang J, Caffrey M, et al. : Discovery, synthesis, and biological evaluation of a novel group of selective inhibitors of filoviral entry. J Med Chem. 2011;54(3):765–81. 10.1021/jm1008715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kesel AJ, Huang Z, Murray MG, et al. : Retinazone inhibits certain blood-borne human viruses including Ebola virus Zaire. Antivir Chem Chemother. 2014;23(5):197–215. 10.3851/IMP2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basu A, Li B, Mills DM, et al. : Identification of a small-molecule entry inhibitor for filoviruses. J Virol. 2011;85(7):3106–19. 10.1128/JVI.01456-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bishop BM: Potential and emerging treatment options for Ebola virus disease. Ann Pharmacother. 2015;49(2):196–206. 10.1177/1060028014561227 [DOI] [PubMed] [Google Scholar]
- 28. Cunninham J, Lee K, Ren T, et al. : Small molecules inhibitors of Ebola and Lassa fever viruses.2014. Reference Source [Google Scholar]
- 29. Panchal RG, Reid SP, Tran JP, et al. : Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antiviral Res. 2012;93(1):23–9. 10.1016/j.antiviral.2011.10.011 [DOI] [PubMed] [Google Scholar]
- 30. Dube D, Schornberg KL, Shoemaker CJ, et al. : Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci USA. 2010;107(38):16637–42. 10.1073/pnas.1008509107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yonezawa A, Cavrois M, Greene WC: Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79(2):918–26. 10.1128/JVI.79.2.918-926.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fakhfakh MA, Fournet A, Prina E, et al. : Synthesis and biological evaluation of substituted quinolines: potential treatment of protozoal and retroviral co-infections. Bioorg Med Chem. 2003;11(23):5013–23. 10.1016/j.bmc.2003.09.007 [DOI] [PubMed] [Google Scholar]
- 33. Fournet A, Mahieux R, Fakhfakh MA, et al. : Substituted quinolines induce inhibition of proliferation of HTLV-1 infected cells. Bioorg Med Chem Lett. 2003;13(5):891–4. 10.1016/S0960-894X(02)01085-5 [DOI] [PubMed] [Google Scholar]
- 34. Gantier JC, Fournet A, Munos MH, et al. : The effect of some 2-substituted quinolines isolated from Galipea longiflora on Plasmodium vinckei petteri infected mice. Planta Med. 1996;62(3):285–6. 10.1055/s-2006-957883 [DOI] [PubMed] [Google Scholar]
- 35. Campos Vieira N, Vacus J, Fournet A, et al. : Antileishmanial activity of a formulation of 2-n-propylquinoline by oral route in mice model. Parasite. 2011;18(4):333–6. 10.1051/parasite/2011184333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakayama H, Loiseau PM, Bories C, et al. : Efficacy of orally administered 2-substituted quinolines in experimental murine cutaneous and visceral leishmaniases. Antimicrob Agents Chemother. 2005;49(12):4950–6. 10.1128/AAC.49.12.4950-4956.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmidt F, Champy P, Séon-Méniel B, et al. : Chemicals possessing a neurotrophin-like activity on dopaminergic neurons in primary culture. PLoS One. 2009;4(7):e6215. 10.1371/journal.pone.0006215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perovic VR: Virtual screen for candidate inhibitors of EBOLA virus infection. F1000Res. 2015. Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Veljkovic V, Loiseau PM, Figadère B, et al. : Dataset 1 in: Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veljkovic V, Loiseau PM, Figadère B, et al. : Dataset 2 in: Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veljkovic V, Loiseau PM, Figadère B, et al. : Dataset 3 in: Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Veljkovic V, Loiseau PM, Figadère B, et al. : Dataset 4 in: Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]