Abstract
We present a novel partner-specific protein-protein interaction site prediction method called PAIRpred. Unlike most existing machine learning binding site prediction methods, PAIRpred uses information from both proteins in a protein complex to generate predict pairs of interacting residues from the two proteins. PAIRpred captures sequence and structure information about residue pairs through pairwise kernels that are used for training a support vector machine classifier. As a result, PAIRpred presents a more detailed model of protein binding, and offers state of the art accuracy in predicting binding sites at the protein level as well as inter-protein residue contacts at the complex level. We demonstrate PAIRpred's performance on Docking Benchmark 4.0 and recent CAPRI targets. We present a detailed performance analysis outlining the contribution of different sequence and structure features, together with a comparison to a variety of existing interface prediction techniques. We have also studied the impact of binding-associated conformational change on prediction accuracy and found PAIRpred to be more robust to such structural changes than existing schemes. As an illustration of potential applications of PAIRpred, we provide a case study in which PAIRpred is used to analyze the nature and specificity of the interface in the interaction of human ISG15 protein with NS1 protein from influenza A virus. Python code for PAIRpred is available at: http://combi.cs.colostate.edu/supplements/pairpred/.
Keywords: protein interface prediction, protein binding site prediction
1 Introduction
Proteins form the functional backbone of all living cells. They are involved in a variety of cellular functions and processes ranging from cell signaling and transportation to structural stability and gene expression control. Most protein functions are possible only through the interaction or binding of multiple proteins, and as a consequence, the study of protein binding is important in understanding protein function and disease mechanism as well as for drug design, discovery and effectiveness studies. The regions where binding occurs can be identified by analyzing the NMR or X-ray crystallography structures of bound protein complexes, or using biological assays such as mutagenesis experiments. However, these techniques are time consuming, difficult, and expensive to perform. As a result, computational methods for predicting the binding sites in protein-protein interactions are of great importance to help guide biological or structural biology e orts. However, the computational prediction of protein binding sites from their unbound structures or sequences is a complicated task owing to the large variety of physio-chemical phenomena involved in the process, not all of which are fully known or thoroughly understood [1]. For example, the conformation of two proteins in their bound state can be very different from their unbound configurations [2,3]. Furthermore, protein molecules are in a constant state of motion in which both the backbone and side chains of residues exhibit significant flexibility [4,5].
The problem of predicting binding regions in protein complexes from the unbound structures or sequences of the proteins involved in the complex has two flavors:
Partner-independent prediction: Given a protein A, find whether a residue a in the protein is involved in an interaction with any other protein.
Partner-specific prediction: Given proteins A and B, find whether a residue a in A interacts with residue b in B upon the formation of the complex A – B.
With this context in mind, we propose to differentiate between a binding site on a protein and the interface in a complex as follows: the region on a protein that is involved in an interaction with another protein is called its binding site, whereas, the group of interacting residues in a complex constitute the interface of the complex. Note that, given the interface of a complex, it is trivial to determine the binding region on each protein in the complex whereas the inverse is not. Thus, partner-independent predictors can only find protein binding sites whereas partner-specific predictors can provide information about both interfaces and the binding sites on the individual proteins.
A number of partner-independent methods have been proposed. However, in this paper, we focus on partner-specific prediction only. For reviews of partner-independent predictors, the interested reader is referred to [1,6–8].
Partner-specific predictions provide more information about the nature of the complex as they can tell which residues in one protein interact with which residues in the other protein. These more detailed predictions can then be used, for example, to enumerate the distinct binding modes of a protein and to find out whether a protein can bind two other proteins simultaneously or not. Furthermore, partner independent predictors ignore the fact that the binding propensity of a residue is dependent upon the nature and local environment of residues in its target protein. As a consequence, partner-specific interface predictors can be expected to be more accurate in comparison to binding site predictors, as it presents a more complete model of protein binding. This has been demonstrated by Ahmad et al. [9], and the results presented in this paper confirm these findings.
Existing techniques that can be used for the partner-specific prediction of interfaces can be divided in to three classes:
Docking methods
The objective of protein-protein docking methods is to predict the three dimensional structure of a macromolecular complex given the unbound structures of its constituent proteins. The docking problem has been extensively investigated using a large variety of strategies such as Fast Fourier Transform [10,11], geometric hashing [12, 13], Monte-Carlo search [14] and template matching [15]. Existing software for protein-protein docking include ZDOCK [11], HADDOCK [16] and RosettaDock [17]. For further details on different docking methods, the interested reader is referred to a recent review [13]. Docking methods typically produce a large number of putative complexes which are then ranked using a ranking criterion to identify the near-native structure. Once the predicted structure of a protein complex is available from docking, the binding interface can be easily recovered. However, docking solves a more general and complex problem than interface prediction as its primary objective is to construct the correct three dimensional structure of the protein complex. Docking methods are hampered by a lack of complete understanding of the factors involved in complex formation such as binding associated conformational changes [1]. As a consequence, docking methods do not fare well in cases with large conformational change. For example, ZDOCK is able to find near-native structures for 33 rigid-body complexes but for only two non-rigid body complexes in its top 10 predictions on a data set of 124 rigid and 52 non-rigid complexes [11]. Docking methods can benefit from binding site predictions as the correct identification of the interface of the complex can limit the degrees of conformational freedom in docking. Some machine learning schemes such as have been used for the scoring of docking conformations to predict which one is closest to the native structure [18, 19]. Some docking methods employ partner-independent predictors to accomplish this. However, partner-specific interface predictions can be expected to play a better role [20].
Template based methods
With the growth in the number of protein complexes in PDB, both template based interface predictors and template based docking schemes have attracted attention. In these methods, a protein complex is modeled using sequence or structural similarity to a known template protein complex. Template based methods can either use sequence or structural homology or interface similarity [21]. However, these methods are applicable only when template complexes or interfaces exist for a query complex. A recent comparison between template based and docking methods shows that both types of methods have comparable performance [22].
Machine learning methods
Direct prediction of interfaces using machine learning techniques is a relatively unexplored research area and the accuracy of existing methods in this category is low. Unlike template based methods, machine learning based techniques, depending upon the features used for the prediction, are also applicable in cases where template similarity of the query proteins to known interfaces cannot be established. In this work, we focus primarily on machine learning methods.
One of the first approaches to perform partner-specific binding site predictions is In-Site [23]. InSite models interactions at the motif or domain level, and predicts pairs of interacting motifs that best explain a given protein-protein interaction network. InSite does not use information about known interaction sites, and is limited by the richness of the motif library and its coverage across a given protein. Finally, it is more valuable to obtain binding site information at the residue level, as it allows for a more detailed understanding of the interaction.
Recently, Ahmad and Mizuguchi investigated the impact of performing partner-specific versus partner-independent binding site prediction [9]. Their analysis of the binding propensities of residue pairs in protein-protein interfaces clearly shows that the binding propensity of a residue is strongly dependent upon its partner in other proteins. On this basis, they hypothesized that considering residue pairs on interacting proteins in binding site prediction can improve performance and found out that this is in fact the case. Their neural network ensemble predictor (PPiPP) employed position specific scoring matrix and amino acid composition features. However, PPiPP's accuracy is low with area under the Receiver Operating Characteristic curve [24] of 72.9. The sequence-based nature of PPiPP allows the method to be applied to proteins for which only the sequence is known but at the same time it is unable to utilize the wealth of information contained in protein structures.
To the best of our knowledge, no structure based machine learning scheme for partner-specific protein interface prediction exists in the literature. In this paper, we present a novel partner-specific SVM based interface predictor called PAIRpred (Partner-specific interacting residue predictor) that uses both sequence information and features computed from the unbound structures. We present performance analysis of PAIRpred at both the complex level, i.e., for the prediction of interfaces, and at the protein level, i.e., for the prediction of binding sites in the individual proteins in the complex using Docking Benchmark 4.0 [25] and independent test complexes from the CAPRI experiment [13]. At the protein level, we compare PAIRpred's performance to the binding site predictor PredUS [26] and the protein level results from PPiPP [9]. At the complex level, we compared against the docking method ZDOCK [11] and PPiPP [9]. We show that considering information about the binding partner of a protein enables more accurate prediction of its binding site. Furthermore, we also study the relation between PAIRpred's performance and the degree of binding associated conformational change.
2 Methods
2.1 Data and pre-processing
In the development of PAIRpred, we have used the protein-protein docking benchmark data set 3.0 (DBD 3.0) [25]. This data set has also been used in the performance analysis of PPiPP [9] and allows a direct performance comparison. DBD 3.0 contains 124 non-redundant complexes of pairs of proteins for which both the bound and unbound X-ray crystallography structures are known. The proteins structures in DBD 3.0 have resolutions better than 3.25 Å and a minimum sequence length of 30. No two complexes in DBD 3.0 share the same SCOP [27] family-family pair [28] and have sequence identity of more than 30% in both chains. Further testing was performed on version 4.0 of DBD which contains a total of 176 complexes including those already in DBD 3.0.
2.2 Interacting residue-pair definition
We define two residues belonging to two different proteins in a complex to be interacting if the distance between any two heavy atoms of those residues in the bound conformations of their proteins is less than or equal to 6.0 Å. All other residue pairs from the two proteins on that complex were taken as negative examples. Similar definitions have been used in previous studies (see [9] and references therein). Defining interacting residues in this way resulted in a total of about 11,500 positive examples in DBD 3.0, i.e., 93 interacting residue pairs per complex on average. The average number of residues pairs, in overall, for a complex is around 67,000.
2.3 Feature extraction
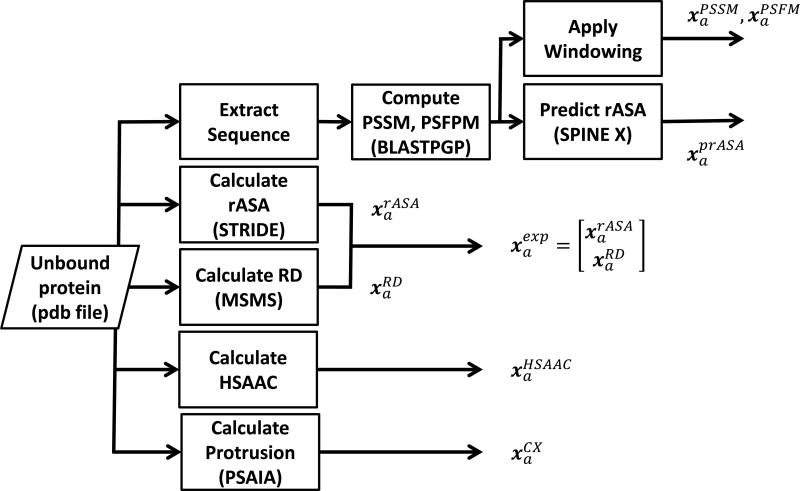
We extracted both sequence and structure features at the residue level from the unbound structure of each protein. When the three dimensional of proteins forming a complex is not available, PAIRpred can make predictions based on its sequence alone. We have used a number of existing programs and methods from the literature to extract features from protein sequences and structures (see Figure 1).
Figure 1.
Residue-level feature extraction in PAIRpred. The feature representation for residue a is denoted by xa. Different components of the feature representation are denoted by the superscript (e.g., indicates the relative accessible surface area for residue a). Each box also indicates the program used to extract a given set of features.
• Structure based features
The following features have been computed directly from the structure.
Relative Accessible Surface Area () The relative accessible surface area (rASA) from a given protein structure was computed using STRIDE [29].
Residue depth () Residue depth is defined as the minimum distance of a residue from the surface of the protein and has been computed using MSMS [30]. The residue depth values produced by MSMS were normalized to have the range from 0 to 1. and are combined to form a single surface exposure feature denoted by . We found that residue depth carries complimentary information to that in rASA for residue interaction prediction.
Half Sphere Amino Acid Composition () Hamelryck [31] found that the geometry and physiochemical characteristics of the regions in the direction of the side chain of a residue (called the ‘up’ direction) and in its opposite direction (called the ‘down’ direction) can be very different from each another. Based upon this observation, we computed a feature (called HSAAC) that captures the amino acid composition in the direction of the side chain of a residue and in the direction opposite to the side chain . The amino acid composition in a direction is defined as the number of times a particular amino acid occurs in that direction within a minimum atomic distance threshold of 8.0 Å from the residue of interest. Thus, HSAAC combines surface accessibility and amino acid composition within the neighborhood of a residue. These amino acid composition vectors in the two directions are then normalized to have unit norm to get and which are then concatenated to get . We utilized Biopython (Cock et al., 2009) to compute .
Protrusion Index () The protrusion index of a non-hydrogen atom is defined as the proportion of the volume of a sphere with a radius of 10.0 Å centered at that atom that is not filled with atoms [32]. The protrusion index has been calculated using PSAIA [33]. The protrusion index for single residue is a 6 dimensional vector comprising the mean, standard deviation, maximum and minimum of the protrusion values of all atoms in the residue along with the mean and standard deviation of the protrusion values of only its side chain atoms. Each element of this vector is normalized to have the range from 0 to 1.
• Sequence based features
We ran PSI-BLAST [34] against the non-redundant ‘nr’ database [35] to compute the Position Specific Scoring Matrix (PSSM) and the Position Specific Frequency Matrix (PSFM) for a given protein. The following sequence based features are then computed:
Profile Features (, ) In order to extract the profile features for a residue from the PSSM, we took the PSSM columns within a length 11 window centered at that residue. This 20 × 11 matrix is converted to a single 220 dimensional unit vector denoted by . is constructed in a similar manner from the PSFM.
Predicted Relative Accessible Surface Area () To determine whether predicted rASA can be used instead of the true rASA, we used SPINE X [36] to predict rASA using the PSI-BLAST data. The predicted rASA is denoted by prASA to emphasize the fact that it has been predicted from sequence.
2.4 Pairwise classification using SVMs
We model the interface prediction problem as a classification problem in which a classification example i is a pair of residues from two different proteins in a complex. Each example i is represented by ((ai, bi), yi), where (ai, bi) is a pair of residues and yi is the associated label, indicating whether the two residues interact (yi = +1) or not (yi = –1). Figure 2 illustrates this concept.
Figure 2.
Overview of PAIRpred. (i) Extract residue-level features from sequence and unbound structures (see Figure 1 for details). (ii) Construct pairwise kernel from the residue-level kernel Kr(ai, aj). (iii) Use the pairwise kernel to train the SVM and classify each residue pair in the query proteins.
As a classifier, we use a support vector machine (SVM) [37] trained over a set of N labeled examples. An SVM finds the optimal separating boundary between two classes by simultaneously maximizing the margin between them and minimizing the cost of misclassification over the training data. For an overview of SVMs in computational biology, the interested reader is referred to [38]. Due to its large-margin nature, an SVM can offer good accuracy over previously unseen examples during testing.
In order to use an example in training or classification, a classifier needs the feature representation for the pair of residues in that example. However, it is easier and computationally more efficient to extract features for a single residue in a given protein than for a pair of residues from two different proteins. Thus, we would like to be able to use the residue level features directly to generate predictions at the residue pair level. This is where our use of SVMs for classification offers an advantage in comparison to other classifiers. Unlike other classifiers, such as the neural networks employed in PPiPP, SVMs can operate without requiring the explicit feature representation of an example by using a kernel function [38]. A kernel function is, in essence, a dot product that measures the degree of similarity between two examples. In this work, we employ pairwise kernels of the form K((a, b), (a′, b′)) which can directly score the similarity between examples (a, b) and (a′, b′) by comparing the feature representations of individual residues in these examples. The pairwise kernel eliminates the need of constructing an explicit feature representation of each example because the scoring function of the SVM can be expressed only in terms of this pairwise kernel as: . In this scoring function, the values of αi are obtained through training.
One of the interesting features of using pairwise kernels in the SVM is that these kernels can themselves be built from kernels over individual residues. Such residue level kernels, denoted by Kr (a, b), compare the explicit feature representations of residues a and b to score the degree of similarity between them. The problem of constructing pairwise kernels from kernels over individual objects has been studied in the machine learning and bioinformatics communities [39–42]. We constructed the pairwise kernel Kpw for our SVM as the additive combination of one or more of the following pairwise kernels from the literature:
Here, Ktppk is the tensor product pairwise kernel (TPPK) proposed by Ben-Hur and Noble [39]. TPPK detects high similarity between examples (a, b) and (a′, b′) if a, expressed in terms of its feature representation, is similar to one of the residues in (a′, b′) and b is also similar to the other residue in the other example. It can be shown that the feature space of TPPK consists of products of features of the underlying residue kernel Kr.
Kmlpk((a, b), (a′, b′)) is the metric learning pairwise kernel (MLPK) [40]. If the feature representation of a residue a is given by ϕ(a), then the MLPK kernel can be written as:
This shows that the MLPK is a homogeneous polynomial kernel of degree 2 between pairs after mapping a pair (a, b) to the vector Φmlpk((a, b)) = ϕ(a) – ϕ(b). Vert et al. have shown that MLPK performs slightly better than TPPK for predicting protein-protein interactions and that their additive combination performs better than either of the kernels [40].
Given the feature space representation ϕ(a) of a residue a, the direct sum pairwise kernel can be written as [41,43]:
This shows that the sum kernel uses the underlying feature map Φsum((a, b)) = ϕ(a) + ϕ(b).
We found that the simple kernel Ksum performed better than both TPPK and MLPK for our problem. However, the additive combination of the three kernels performed better than any of the individual kernels (see the results section for more details). Finally, each pairwise kernel Kpw is normalized as for use in the SVM.
To produce a pairwise prediction for an example (a, b), PPiPP [9] concatenates the feature representation of the two residues in the example in both orders and . In comparison to PPiPP, our pairwise kernel based approach is computationally more efficient as it requires no duplication of the data. Moreover, pairwise kernels in our formulation directly model the inter-dependencies within individual feature components.
The residue kernel Kr used in constructing the pairwise kernel in PAIRpred is itself an unweighted summation of one or more of the following kernels, which are computed using the features described in section 2.3:
In the above equations, g(a, b; γ) = exp(–γ∥a – b∥2) is the Gaussian kernel. The parameter γ in the Gaussian kernel controls the decay of the exponential function. If γ is set too high or too low, the exponential function can saturate at 0 or 1 which will inhibit effective learning from the training data. We chose the values of these parameters so that, for the majority of non-identical input vectors for a kernel, the similarity score does not saturate and maintains good dynamic range. This heuristic is inspired by the literature about parameter selection in radial basis function neural networks [44]. Once chosen in this manner, these parameters were not changed to optimize accuracy. The selected values of these parameters are as follows: γPSSM = γPSFM = γHSAAC = 0.5, γCX = 1.0 and γexp = γprASA = 3.0. As discussed in the results section, these values give good performance over test data. Training and classification has been performed using the SVM implementation in the machine learning library PyML [45].
2.5 Post-processing
A binding site or interface is a collection of spatially neighboring residues whose binding propensities are correlated. Keeping this in mind, we smoothed the prediction score for a pair of residues by averaging prediction scores within their local neighborhoods through the following post-processing step:
| (1) |
where fAB((a, b)) is the raw PAIRpred discriminant score from the trained SVM and N(r) is the set of the 10 neighboring residues of residue r on the same protein including r itself. Thus the post-processed scores is the sum of the averages of the prediction scores of a residue on one protein with a set of residues on the other protein. As discussed in the results section, this simple post-processing scheme improves the prediction performance significantly.
2.6 Performance evaluation
Performance evaluation was carried out in two stages. In the first stage we compared different kernel designs, and residue-level features using five-fold cross-validation at the complex level. In this cross-validation procedure, examples from all complexes in our data set were divided into 5 folds such that all examples from a complex are found in exactly one fold. To reduce computational time during model selection, the 5 fold cross-validation was done using a class-size balanced sample from DBD 3.0 in which the number of randomly chosen negative examples for a complex is equal to the number of positive examples in it. For each fold, the value of the parameter C that controls the cost of misclassification over training data in the SVM was selected by performing a similar nested 5-fold cross-validation. The value of C was selected from {0.1, 1.0, 10.0, 100.0}. The classification function values and the known true labels of the examples were used to compute the Receiver Operating Characteristic (ROC) curve for each complex. The average of the area (expressed as a percentage) under the ROC curve for all complexes, labeled as AUC, has been used as the performance statistic for selecting the optimal model.
In the second stage of performance evaluation, we performed a leave-one-complex-out cross-validation analysis with the optimal kernel design selected in the first stage. In this cross-validation procedure, a classifier is trained on a balanced set of examples extracted from all but one of the complexes, and testing is performed on all pairs of residues from the left-out complex. This evaluation protocol is identical to the one used for PPiPP [9] and allows a direct and fair comparison between the two methods. The average area (expressed as percentage) under the ROC curves for all complexes (AUC) is used as a performance metric as it allows a quantitative comparison with other interface prediction methods. However, AUC scores are not easy to interpret in this setting. In cases with highly unbalanced data with a big difference in the number of positive and negative test examples as we have here, AUC can give a false impression of accuracy. For these reasons we propose a measure of accuracy that is specifically designed for this domain. Our measure, which we call RFPP (rank of the first positive prediction), is defined as follows: RFPP(p) = q, if p % of the complexes tested have at least one true positive interacting residue pair among the top q predictions. Thus, an ideal classifier will have RFPP(100) = 1, i.e., in every complex, the top scoring prediction from the classifier belongs to the interface. In comparison to an ROC curve, this measure is more informative for the biologist as it tells us directly how often the top ranking predictions can be expected to correspond to known interactions.
We also evaluate the performance of PAIRpred for binding site prediction at the single protein level (i.e., binding site prediction) and compare it to existing partner-independent methods. Pairwise predictions of interacting residues at the complex level (from Equation (1)) are converted into predictions at the protein level for each protein as follows: and . AUC scores for an individual protein can then be easily computed.
3 Results and Discussion
3.1 Comparison of residue and pairwise representations
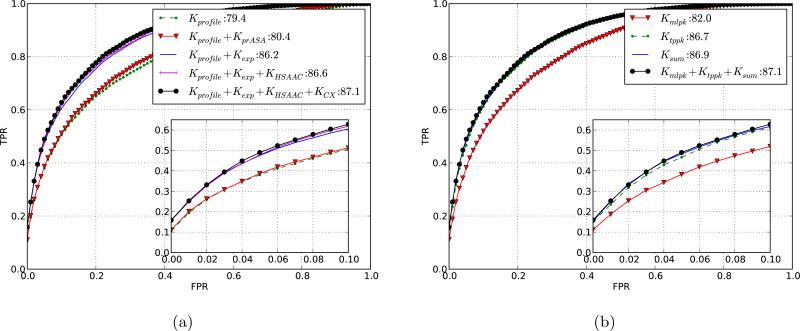
We analyzed and compared different feature representations and pairwise kernel formulations in order to see the contribution of different features towards prediction accuracy and the impact of pairwise kernel design. Figure 3 shows the complex-wise averaged ROC curve for different feature and kernel combinations. In order to compare different feature representations, we chose to use Kpw = Kmlpk + Ktppk + Ksum as the pairwise kernel. Our first step was to analyze the accuracy when our method is restricted to using sequence-based features only, which include sequence profile and relative accessible surface area predicted from sequence. As shown in figure 3, profile features alone give an AUC of 79.4, and adding the predicted rASA (i.e., Kr = Kprofile +KprASA) increases the AUC to 80.4. For the profile-based features we found that the combination of PSSM and PSFM features performed slightly better than either of the two alone (results not shown).
Figure 3.
Selecting the optimal sequence/structure representation by comparing ROC curves for different kernel designs. Shown are the averaged ROC curves computed using 5-fold cross-validation over complexes in DBD 3.0. The inset shows the true positive rate (TPR) vs. false positive rate (FPR) for up to first 10 % false positives. The legend shows the AUC scores for the different kernels used. (a) Results for different residue kernels Kr using the pairwise kernel Kpw = Kmlpk + Ktppk + Ksum. The curves illustrate the increase in performance as additional structural information is added to the sequence-based kernel. Recall that Kprofile is the PSI-BLAST profile kernel; KprASA uses predicted rASA; Kexp is the residue exposure kernel; KHSAAC is the half-sphere exposure kernel; KCX uses protrusion-index features. (b) Results for different pairwise kernels Kpw with residue kernel Kr = Kprofile + Kexp + KHSAAC + KCX.
The addition of structure-based features provides a big boost in performance: the combination of true surface accessibility features (Kexp) with the profile features (Kprofile) gives an AUC of 86.2 compared to 79.4 for the profile-based features alone and 80.4 using the combination of profile and predicted rASA features. Such an improvement is to be expected because most of the residues involved in the interaction have high surface accessibility. However, the use of predicted rASA did not result in such a big increase. This is because the protein-wise averaged correlation between predicted and true rASA values for binding residues is low (r = 0.56, against r = 0.76 for non-interacting residues). Thus, the use of a better sequence based predictor of surface accessibility can help improve the accuracy of the sequence based predictions in future.
Addition of HSAAC and protrusion index based features (KHSAAC + KCX) improves the accuracy of the method even further (AUC of 87.1). For the rest of the analyses in the paper we have used Kr = Kprofile + Kexp + KHSAAC + KCX.
The choice of a pairwise kernel has a strong influence on accuracy. Figure 3 shows the ROC curves for different pairwise kernel formulations. Kmlpk, Ktppk, and Ksum produce AUC scores of 82.0, 86.7, and 86.9 respectively, while adding all three provides an AUC score of 87.1. For the rest of the analyses in the paper we have used Kpw = Kmlpk + Ktppk + Ksum.
In order to test the variability of the results, the cross-validation procedure in model selection was repeated 5 times with change both in the randomly selected negative examples and the membership of complexes in different folds. We then evaluated the mean and standard deviation of different cross-validation runs. The maximum standard deviation in the AUC scores for any kernel combination was 0.2. This shows that these results are robust to changes in the training data.
3.2 Prediction using residue exposure alone
As discussed above, residue exposure features result in a big improvement in accuracy. To explore the contribution of the residue exposure features (rASA, reside depth, and mean protrusion), we computed the sum of the residue exposure of the two residues in each example. Using this combination as a ranking criterion we computed the AUC score for each complex. This naïve way of classification yields some interesting results. The average AUC scores for all complexes from rASA, reside depth (RD) and the mean protrusion value are 71.9, 69.4 and 71.2 respectively. These results are only marginally inferior to the leave-one-complex-out cross-validation results from PPiPP (AUC = 72.9) [9]. Since rASA and RD are both measures of the surface accessibility of a residue, the AUC values for these features clearly reflect the known fact that surface residues are more likely to participate in protein-protein interactions. The AUC score of the protrusion index shows that the residues that interact have few atoms around them. This includes surface atoms, and especially those atoms on the surface that lie in cavities or protrude out from their local neighborhood. The protrusion index captures more local shape information than rASA and the two can be complementary to one another. The fact that pairwise summation of surface exposure features provides good results explains why the pairwise sum kernel Ksum was able to perform better than the other two pairwise kernels.
The same ranking criterion over the relative accessible surface area predicted from sequence using SPINE X gives an AUC of only 0.56. This clearly shows that these predictions need to be more accurate to be effective in finding interfaces in protein complexes.
3.3 Results for leave-one-complex-out cross-validation
For comparison with other methods we used the optimal kernel combination found through kernel evaluation (section 3.1) and recomputed its performance using the leave-one-complex-out cross-validation protocol detailed in Section 2.6. Results of this analysis are reported in table I. The AUC scores for interface prediction, averaged across the 123 complexes in DBD 3.0, for sequence and structure kernels are 80.9 and 87.3, respectively. It is interesting to note that these scores from leave-one-complex-out cross-validation over all examples are very close to those obtained with the balanced sample. Evaluation over all the 176 complexes in DBD 4.0 gives an AUC score of 87.0 with the structure kernel.
Table I.
PAIRpred and PPiPP performance. We compare the performance of PAIRpred and PPiPP [9] using Area Under the ROC Curve (AUC) and the rank of the first positive prediction (RFPP). RFPP(p) indicates that p percent of the proteins achieve that level of performance. For example, on DBD 4.0 without post processing, the second PAIRpred prediction is part of the interface for 10% of the complexes. PAIRpred results are provided for two residue kernels: the sequence-based kernel, and for the kernel that uses all the features computed from sequence and structure.
| Dataset | Method | RFPP (p) | AUC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 10% | 25% | 50% | 75% | 90% | Complex | Protein | |||
| DBD 3.0 (124 complexes) | PPiPP | 9 | 19 | 78 | 297 | 760 | 72.9 | 66.1 | |
| PAIRPred | |||||||||
| Kr = Kprofile + KprASA | No post-processing | 2 | 13 | 68 | 257 | 804 | 80.9 | 70.8 | |
| Kr = Kprofile + Kexp + KHSAAC + KCX | No post-processing | 1 | 5 | 22 | 89 | 282 | 87.3 | 73.4 | |
| With post-processing | 1 | 3 | 16 | 103 | 272 | 88.7 | 77.0 | ||
| DBD 4.0 (176 complexes) | Kr = Kprofile + Kexp + KHSAAC + KCX | No post-processing | 2 | 6 | 19 | 75 | 340 | 87.0 | 73.1 |
| With post-processing | 1 | 3 | 18 | 101 | 282 | 87.8 | 75.4 | ||
At the protein level, the AUC scores of PAIRpred for sequence and structure kernels for DBD 3.0 are 70.8 and 77.0 (with post-processing), respectively. It can also be noted that post-processing increases the performance of the method. This is particularly true at the protein level.
3.4 Comparison with PPiPP and ZDOCK
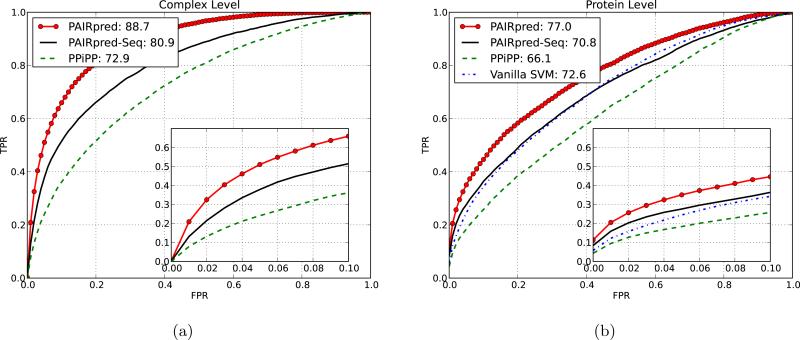
PPiPP [9] is a recently proposed sequence based method for partner-specific predictions that uses an ensemble of neural networks trained with a more elaborate version of our profile representation with different window sizes [9]. Table I shows the results of leave-one-complex-out cross-validation for DBD 3.0 using PPiPP. Even with the sequence features alone, PAIRpred gives better AUC and RFPP scores than PPiPP. As shown in figure 4, PAIRpred's performance at the complex level (i.e., for interface prediction) is superior to PPiPP not only in overall AUC but also in the number of true positives within the first 10% false positives.
Figure 4.
Comparison between PAIRpred, PPiPP [9] and vanilla SVM at the complex and protein level predictions on DBD 3.0. PAIRpred-seq refers to PAIRpred based only on sequence features.
PPiPP offers better accuracy than other published sequence based methods for binding site prediction such as PSIVER and SPPIDER (results given in [9]). PAIRpred's performance at the protein level (i.e., for binding site prediction) is also superior to PPiPP using either sequence features alone or in conjunction with protein structure (see Table I and Figure 4).
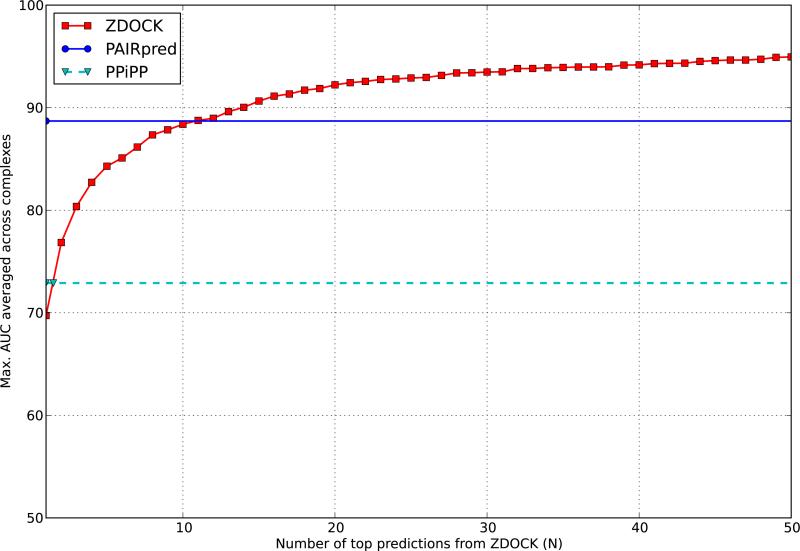
We also compared PAIRpred with the docking method ZDOCK [11] over the 176 complexes in DBD 4.0. For this purpose, we have used, for each complex, the top 2000 predictions in the 15-degrees sampling data available online for ZDOCK v. 3.02. For each ZDOCK prediction for a complex, we computed the pairwise minimum inter-atomic distance between all residues of the two proteins in the predicted complex. The inverse of this distance was used as a ranking criterion in the evaluation of the AUC score at the complex level. The AUC score of a ZDOCK prediction tells us how good that prediction is at identifying the known interface in the complex and is directly comparable to the AUC scores given earlier for PAIRpred and PPiPP. For a given complex, we computed the maximum AUC score in the top N ZDOCK predictions and then averaged these scores across all complexes for a given value of N to obtain the results shown in Figure 5. These results show that PAIRpred is better than the best of the top 11 ZDOCK predictions. The AUC score of the top prediction by ZDOCK is roughly comparable to that of PPiPP.
Figure 5.
The maximum AUC from top N ZDOCK predictions in comparison to PAIRpred and PPiPP [9].
3.5 Comparison with partner-independent predictions
In order to test the hypothesis that a partner-specific predictor can perform better than partner-independent predictors, we developed an SVM based binding site predictor (referred to as vanilla SVM) using the same structural features as in PAIRpred and compared its leave-one-protein-out cross validation performance to the PAIRpred results at the protein level. Figure 4 shows the ROC curve for vanilla SVM which gives an AUC score of 72.6. PAIRpred's performs much better than the vanilla SVM. This clearly shows that partner-specific predictors can offer superior performance in comparison to partner-independent ones even when the same residue level features are used. Moreover, PAIRpred's AUC score of 70.8 with the sequence features alone is only marginally inferior to vanilla SVM even though the latter employs structure based features. As a matter of fact, PAIRpred with sequence features alone gives better true positive rates than the vanilla SVM consistently for false positive rates less than 0.4.
At the protein level, PAIRpred's performance using structure based features can be roughly contrasted to PredUS [26], a recently published structure based binding site predictor. PredUS performs better than other similar predictors available in the literature and gives an AUC score of 73.9 over 188 chains in DBD 3.0. It must be noted that a direct comparison between the performance of the two methods is not possible because of differences in their evaluation data sets, interface definitions, and cross-validation protocols. PAIRpred's performance with structure features can be expected to be equal or slightly better than that of PredUS as PAIRpred gives an AUC score of 77.0 over 248 proteins in DBD 3.0.
3.6 Spatial proximity of PAIRpred predictions
In order to see whether the top predictions by PAIRpred are spatially close, we compared pairwise distances between residues in our top predictions with a random sample of residues. More specifically, we computed the pairwise distances among the top 20 residue predictions from PAIRpred for each protein and also between the remaining pairs of residues from each protein. The average of the pairwise distances in the top predictions is 15.6 Å and 20.1 Å for the remaining pairs. These distances are significantly different (with a p-value of 4.7 × 10–25 using the Wilcoxon Rank Sum test on all complexes in DBD 4.0). This indicates the top PAIRpred predictions exhibit spatial clustering.
Furthermore, we found that the difference between the mean pairwise distances across the top predictions and the remaining residues in a protein is inversely correlated with the its AUC (correlation coefficient of −0.49, 2 tailed p-value of 1.1×10–21). Thus, this difference in distances is a rough indication of the quality of prediction.
3.7 Effects of conformational change
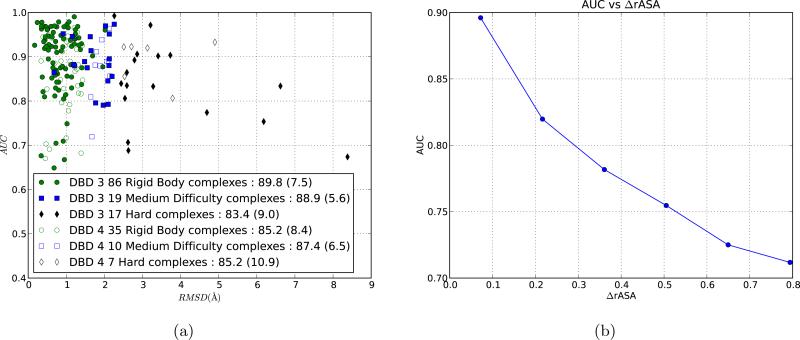
Proteins can undergo significant conformation change upon binding as buried residues can become exposed and vice versa. In order to observe the effects of the degree of conformational change on the accuracy of PAIRpred, we plotted the AUC of a complex against the root mean square deviation (RMSD) between the bound and the unbound states over the interface residue for in the complex. A large RMSD value for a complex corresponds to a large binding-associated conformation change. Figure 6 shows that the accuracy decreases with increase in conformational change. This effect was also observed for PPiPP. However, PAIRpred performs much better than PPiPP for complexes with large conformational change. Based on the degree of conformational change, the complexes in the docking benchmark datasets have been divided into three categories: rigid body, medium difficulty and hard. Figure 6 shows the prediction performance across complexes in these categories. As expected, PAIRpred performs better for rigid body complexes in comparison to the other two categories that involve larger conformational changes.
Figure 6.
Effect of conformational change on PAIRpred performance. (a) AUC vs. RMSD for each complex in DBD 3.0 and the 53 new complexes in DBD 4.0 using leave-one-complex-out cross-validation. The legend shows mean AUC and standard deviation (within paranthesis) of complexes in each category for each data set. (b) Relationship between AUC and the change in rASA (ΔrASA). Residue pairs were binned into groups based on their ΔrASA and the AUC score was computed for all the residues within each bin using our 5-fold cross-validation scheme on DBD 3.0 complexes.
We investigated the effects of conformational change on PAIRpred performance at the residue level as well. As we had access to both the bound and the unbound states of each protein, we were able to calculate the absolute difference in rASA for a residue between the two states of the protein. A large difference is indicative of a large conformational change in the environment around that residue. For a pair of residues we define the degree of conformational change as the sum of the changes in the individual residues, and denote it as ΔrASA(a, b). AUC exhibits a high negative correlation (see figure 6) with ΔrASA (correlation coefficient of −0.97, p-value of 1.5 × 10–3). AUC vs. change in residue depth shows a similar trend. This demonstrates the inherent difficulty of predicting residue-residue interactions in protein complexes that undergo a large conformational change. This difficulty is exacerbated by the fact that there is only a small amount of training data (24 complexes in DBD 4.0) available for such cases. Furthermore, the standard deviation of AUC scores for complexes from the hard category in DBD 4.0 shown in figure 6 is much larger in comparison to other categories. This suggests that effective handling of complexes with large conformational change requires a larger number of training examples with this property.
3.8 Evaluation on CAPRI targets
In order to further analyze the performance of PAIRpred, we tested it on nine recent targets from the Critical Assessment of Protein Interactions (CAPRI) experiment [13]. We used all heteromeric protein complexes published after 2007 for which both the bound and unbound X-ray crystallography structures are available. For this task, PAIRpred was trained using DBD 4.0, and results of this analysis are reported in Table II. This table shows that PAIRpred is able to predict the interface with good accuracy for most targets. For seven out of these nine targets, the top 15 PAIRpred predictions contain at least one true positive. It is interesting to note that even for complexes involving large conformational changes, such as 3BX1 and 2WPT, the first true positive lies within the top 10 predictions. PAIRpred does not perform well on two targets: 3FM8 and 2VDU. These targets have proven to be very challenging for docking methods as well: only 1% and 4% of the models predicted by docking methods in CAPRI have an acceptable complex structure for 3FM8 and 2VDU, respectively [46].
Table II.
PAIRpred evaluation results on recent heteromeric protein complex targets from CAPRI with both bound and unbound X-ray structures available. The degree of conformational change for the ligand and receptor proteins has been measured as the backbone root mean square deviation. The maximum sequence identity of a protein from the CAPRI set to any protein in DBD 4.0 has been calculated using local sequence alignment. The AUC score for each complex and the rank of the first positive prediction (RFPP) is reported.
| Complex ID in PDB | Target ID in CAPRI | Ligand Backbone RMSD (Å) | Receptor Backbone RMSD (Å) | Max. Seq Id. of ligand to DBD4 | Max. Seq Id. of receptor to DBD4 | AUC | RFPP |
|---|---|---|---|---|---|---|---|
| 4G9S | T58 | 0.3 | 0.7 | 28 % | 27% | 89.7 | 4 |
| 4EEF | T56 | 0.7 | 0.5 | 27 % | 29 % | 76.3 | 1 |
| 3R2X | T50 | 0.5 | 0.6 | 29 % | 26 % | 90.3 | 15 |
| 3U43 | T47 | 0.9 | 1.5 | 60 % | 55 % | 88.9 | 2 |
| 2WPT | T41 | 2.0 | 0.7 | 62 % | 66 % | 85.8 | 1 |
| 3E8L | T40 | 0.2 | 0.4 | 100 % | 28 % | 92.1 | 9 |
| 3FM8 | T39 | 0.0 | 1.6 | 28 % | 25 % | 79.6 | 71 |
| 3BX1 | T32 | 2.0 | 0.4 | 30 % | 56 % | 89.7 | 10 |
| 2VDU | T29 | 1.1 | 0.4 | 28 % | 27 % | 82.9 | 302 |
3.9 Application to Human ISG15-Influenza A NS1 interaction
Due to its partner-specific nature and state of the art accuracy, PAIRpred can be used to study the nature and mechanics of an interface beyond what is possible with partner-independent predictors. In this section, we demonstrate PAIRpred's capabilities beyond the simple prediction of an interface by using the interaction between ISG15 protein in human and mouse and NS1 protein from Influenza A virus as a case study.
The influenza B virus is known to infect only human and non-human primates and the cause of this specific behavior have been investigated in [47] through a study of the bound and unbound structures of NS1 protein from the virus and the ISG15 protein in humans and other species. We have used PAIRpred to study the binding between these two proteins and compare the findings from this computational analysis to the results published in [47].
We first predicted the interface of the complex from the unbound structures of the two proteins using both PPiPP and PAIRpred and used the known interface to compare the performance of the two methods. The unbound PDB structures of NS1 and ISG15 are available as 1XEQ [48] and 1Z2M [49]. The complex structure (PDB ID: 3SDL) has two chains each of NS1 and ISG15 [50]. There is no significant conformational change in NS1 upon binding to ISG15 with only a disorder to order change in a short C-terminal polypeptide sequence. ISG15 undergoes modest conformational change upon binding NS1 with a backbone RMSD of 1.05 Å. We obtained the predictions from the unbound proteins by training PAIRpred on DBD 3.0 to allow for a comparison with PPiPP, and used structure-based features. This complex is not a part of training sets of PAIRpred or PPiPP. The AUC scores for PPiPP and PAIRpred for this complex are 67.2 and 92.4, respectively. The first true positive detected by PAIRpred is the top-most prediction, whereas the first true positive detected by PPiPP occurs at rank 174. PAIRpred is able to find more than half of the interacting residue pairs within its top 100 predictions (see Figure 7). The predictions correspond very closely to the interactions discussed in [47]. We also compared the interface prediction performance of PAIRpred to that of ZDOCK for this complex by using the inverse of the inter-residue distance from ZDOCK predictions as a ranking criterion as described in Section 3.4. It was found that the AUC score from PAIRpred is better than the best of the top 13 ZDOCK predictions for this complex.
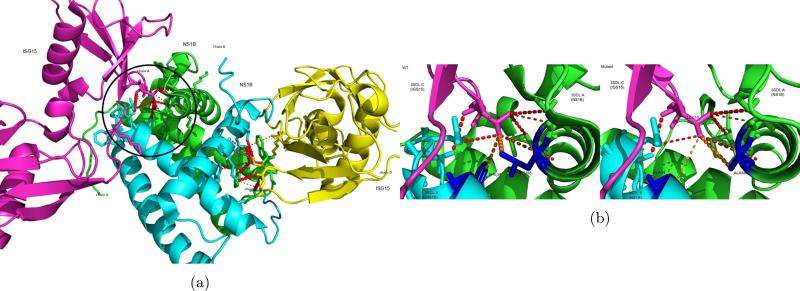
Figure 7.
(a) PAIRpred predictions for human ISG15 and influenza B NS1 mapped onto the 3D structure of the complex (PDB ID: 3SDL). The red dotted lines indicate the true positives in the top predictions with the width of the line proportional to the prediction score. The circled area is expanded in (b). (b) Results of in silico mutagenesis. The blue residues were changed to alanines. Notice the change in the prediction score (indicated by the width of the orange dotted lines) for the mutated residues between the wild-type (left) and the mutant (right).
Next we used, PAIRpred predictions in order to identify the residues that are crucial for binding. Specifically, we conducted an in silico mutagenesis experiment in which we changed the NS1: L88 residue involved in our top prediction (ISG15: L10, NS1: L88) to an alanine. We also recapitulated one mutagenesis experiments reported in (Guan et al., 2011) which involved changing NS1: F34 (which also interacts with ISG15: L10) to an alanine. The (ISG15: L10, NS1: F34) interaction is originally ranked 8th in PAIRPRed predictions for this complex. We obtained the predicted structure after the mutations using I-TASSER (Roy et al., 2010). In comparison to the wild-type predictions for (ISG15: L10, NS1: L88) and (ISG15: L10, NS1: F34), we observed a decrease of 25% and 53% in prediction scores for L88 and F34 mutations in NS1, respectively (see Figure 7). The prediction scores for other interacting residues were essentially unchanged. These results indicate that both these residues are, as experimentally determined in [47], very important for this interaction.
As stated earlier, NS1 binds specifically to ISG15 from human and non-human primates and does not bind to mouse ISG15. Guan et al. [47] attribute this binding specificity to residues 47-52 and 76-80 in the sequence alignment of ISG15s from these three species. We obtained the unbound structure of mouse ISG15 using I-TASSER. We then compared the PAIRpred prediction scores for (human ISG15,NS1) complex to those from the (mouse ISG15, NS1) interaction. This comparison allowed us to identify the ISG15 residues that are interacting in (human ISG15, NS1) complex but undergo a large decrease in their prediction scores in the (mouse ISG15, NS1) interaction. These locations (in order of decreasing magnitude of change in predictions scores) are 76, 77, 72, 74 and 49. This strengthens the claim made in [47].
These analyses clearly demonstrate the usefullness of partner-specific predictions generated from PAIRpred as the mutagenesis studies explained above cannot be performed with conventional partner-independent predictors.
3.10 Using PAIRpred
PAIRpred has been implemented in Python and its architecture allows future extensions to include additional residue-level features or pairwise kernels. Complete implementation of PAIRpred, together with the pre-trained classifier, can be downloaded at http://combi.cs.colostate.edu/supplements/pairpred/. PAIRpred users need to supply the sequences in FASTA format or, when available, the PDB format structure files as input. PAIRpred then automatically extracts features from these files and produces predictions using a pre-trained SVM. Users also have the option of training the classifier on their own data sets. PAIRpred generates its prediction for a complex as a text file which contains the pairwise interaction scores for each pair of residues from the two query proteins. This pairwise prediction file can then be used to generate protein-level binding site predictions through scripts available as part of the PAIRpred package. PAIRpred implementation also provides PyMOL scripts for visualizing top PAIRpred predictions both at the complex and protein levels as shown in Figure 7.
4 Conclusions
We have presented a new method for predicting the interface of a protein complex called PAIRpred that offers state-of-the-art accuracy for both interface and binding site prediction. The proposed scheme is able to make accurate predictions using either sequence information alone or in conjunction with structure-based features. There are very few machine learning based methods that perform partner-specific prediction of interactions, and PAIRpred provides a large improvement over the recently published PPiPP method. We investigated the merit of sequence and structure-based features and found that using structure provides a big improvement in performance. Furthermore, the analysis of the accuracy of PAIRpred shows much better scaling of performance with respect to the degree of conformational change upon complex formation in comparison to PPiPP. However, there is still plenty of room for improvement, especially for complexes that exhibit a large degree of conformational change upon binding. In the future we plan on adding features to capture shape complementarity between binding interfaces, information about correlated mutations [51,52], protein flexibility and predictors of degree of conformational change [53] in order to improve the predictions even further. Moreover, PAIRpred can potentially improve the accuracy of docking methods if used as a filter or by direct incorporation into the energy function [6].
Acknowledgements
The authors would like to thank Shandar Ahmad for his correspondence. Fayyaz Minhas is supported by a grant from the Fulbright scholarship program of the U.S. State Department and the Higher Education Commission (HEC) of Pakistan. Brian Geiss is supported by NIH (NIAID) grant U54 AI065357.
Contributor Information
Fayyaz ul Amir Afsar Minhas, Department of Computer Science Colorado State University Fort Collins, Colorado 80523, USA fayyazafsar@gmail.com.
Brian J. Geiss, Department of Microbiology, Immunology and Pathology Colorado State University Fort Collins, Colorado 80523, USA brian.geiss@colostate.edu
Asa Ben-Hur, Department of Computer Science Colorado State University Fort Collins, Colorado 80523, USA asa@cs.colostate.edu.
References
- 1.Ezkurdia I, Bartoli L, Fariselli P, Casadio R, Valencia A, Tress ML. Progress and challenges in predicting protein-protein interaction sites. Briefings in Bioinformatics. 2009 May;10:233–246. doi: 10.1093/bib/bbp021. [DOI] [PubMed] [Google Scholar]
- 2.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chemical reviews. 2008 Apr.108:1225–1244. doi: 10.1021/cr040409x. PMID: 18355092. [DOI] [PubMed] [Google Scholar]
- 3.Changeux J-P, Edelstein S. Conformational selection or induced-fit? 50 years of debate resolved. F1000 Biology Reports. 2011 Sept.3 doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunasekaran K, Nussinov R. How different are structurally flexible and rigid binding sites? sequence and structural features discriminating proteins that do and do not undergo conformational change upon ligand binding. Journal of Molecular Biology. 2007 Jan.365:257–273. doi: 10.1016/j.jmb.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 5.Wass MN, David A, Sternberg MJ. Challenges for the prediction of macro-molecular interactions. Current Opinion in Structural Biology. 2011 Jun;21:382–390. doi: 10.1016/j.sbi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H-X, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007 Sept.23:2203–2209. doi: 10.1093/bioinformatics/btm323. PMID: 17586545. [DOI] [PubMed] [Google Scholar]
- 7.Ofran Y. Computational Protein-Protein Interaction. CRC Press; 2009. Prediction of protein interaction sites; pp. 167–184. [Google Scholar]
- 8.Leis S, Schneider S, Zacharias M. In silico prediction of binding sites on proteins. Current Medicinal Chemistry. 2010;2010;(17):1550–1562. doi: 10.2174/092986710790979944. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad S, Mizuguchi K. Partner-aware prediction of interacting residues in protein-protein complexes from sequence data. PLoS ONE. 2011 Dec.6 doi: 10.1371/journal.pone.0029104. PMID: 22194998 PMCID: 3237601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proceedings of the National Academy of Sciences. 1992 Mar.89:2195–2199. doi: 10.1073/pnas.89.6.2195. PMID: 1549581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011 Sept.6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duhovny D, Nussinov R, Wolfson HJ. Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI) Notes in Computer Science 2452. Springer Verlag; 2002. Efficient Unbound Docking of Rigid Molecules; pp. 185–200. [Google Scholar]
- 13.Janin J. Identification of Ligand Binding Site and Protein-Protein Interaction Area (I. Roterman-Konieczna, ed.), no. 8 in Focus on Structural Biology. Springer Netherlands; Jan. 2013. Docking predictions of protein-protein interactions and their assessment: The CAPRI experiment; pp. 87–104. [Google Scholar]
- 14.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 15.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proceedings of the National Academy of Sciences. 2012 May; doi: 10.1073/pnas.1200678109. PMID: 22645367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries SJ, van Dijk ADJ, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AMJJ. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007 Dec.69:726–733. doi: 10.1002/prot.21723. PMID: 17803234. [DOI] [PubMed] [Google Scholar]
- 17.Schueler-Furman O, Wang C, Baker D. Progress in proteinprotein docking: Atomic resolution predictions in the CAPRI experiment using RosettaDock with an improved treatment of side-chain flexibility. Proteins: Structure, Function, and Bioinformatics. 2005 Aug;60(2):187–194. doi: 10.1002/prot.20556. [DOI] [PubMed] [Google Scholar]
- 18.Bordner AJ, Gorin AA. Protein docking using surface matching and supervised machine learning. Proteins: Structure, Function, and Bioinformatics. 2007;68(2):488502. doi: 10.1002/prot.21406. [DOI] [PubMed] [Google Scholar]
- 19.Martin O, Schomburg D. Efficient comprehensive scoring of docked protein complexes using probabilistic support vector machines. Proteins: Structure, Function, and Bioinformatics. 2008;70(4):13671378. doi: 10.1002/prot.21603. [DOI] [PubMed] [Google Scholar]
- 20.Huang B, Schroeder M. Using protein binding site prediction to improve protein docking. Gene. 2008 Oct.422:14–21. doi: 10.1016/j.gene.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Tuncbag N, Gursoy A, Keskin O. Prediction of proteinprotein interactions: unifying evolution and structure at protein interfaces. Physical Biology. 2011 Jun;8:035006. doi: 10.1088/1478-3975/8/3/035006. [DOI] [PubMed] [Google Scholar]
- 22.Vreven T, Hwang H, Pierce BG, Weng Z. Evaluating template-based and template-free protein-protein complex structure prediction. Briefings in Bioinformatics. 2013 Jul; doi: 10.1093/bib/bbt047. PMID: 23818491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Segal E, Ben-Hur A, Li Q-R, Vidal M, Koller D. InSite: a computational method for identifying protein-protein interaction binding sites on a proteome-wide scale. Genome Biology. 2007;8(9):R192. doi: 10.1186/gb-2007-8-9-r192. PMID: 17868464 PMCID: 2375030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CD, Davis HT. Receiver operating characteristics curves and related decision measures: A tutorial. Chemometrics and Intelligent Laboratory Systems. 2006 Jan.80:24–38. [Google Scholar]
- 25.Hwang H, Vreven T, Janin J, Weng Z. Protein-protein docking benchmark version 4.0. Proteins: Structure, Function, and Bioinformatics. 2010;78(15):31113114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang QC, Deng L, Fisher M, Guan J, Honig B, Petrey D. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Research. 2011 May;39:W283–W287. doi: 10.1093/nar/gkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic acids research. 2004 Jan.32:D226–229. doi: 10.1093/nar/gkh039. PMID: 14681400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins: Structure, Function, and Bioinformatics. 2008;73(3):705709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins: Structure, Function, and Bioinformatics. 1995;23:566579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 30.Sanner M, Olson A, Spehner J-C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38(3):305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 31.Hamelryck T. An amino acid has two sides: A new 2D measure provides a different view of solvent exposure. Proteins: Structure, Function, and Bioinformatics. 2005;59(1):3848. doi: 10.1002/prot.20379. [DOI] [PubMed] [Google Scholar]
- 32.Pintar A, Carugo O, Pongor S. CX, an algorithm that identifies protruding atoms in proteins. Bioinformatics (Oxford, England) 2002 Jul;18:980–984. doi: 10.1093/bioinformatics/18.7.980. PMID: 12117796. [DOI] [PubMed] [Google Scholar]
- 33.Mihel J, iki M, Tomi S, Jeren B, Vlahoviek K. PSAIA protein structure and interaction analyzer. BMC Structural Biology. 2008 Apr.8:21. doi: 10.1186/1472-6807-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Sch er AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997 Sept.25:3389–3402. doi: 10.1093/nar/25.17.3389. PMID: 9254694 PMCID: 146917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research. 2004 Jul;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faraggi E, Zhang T, Yang Y, Kurgan L, Zhou Y. SPINE x: Improving protein secondary structure prediction by multistep learning coupled with prediction of solvent accessible surface area and backbone torsion angles. Journal of Computational Chemistry. 2012 Jan.33:259–267. doi: 10.1002/jcc.21968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995:273297. [Google Scholar]
- 38.Ben-Hur A, Ong CS, Sonnenburg S, Schlkopf B, Rtsch G. Support vector machines and kernels for computational biology. PLoS Comput Biol. 2008 Oct.4:e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Hur A, Noble WS. Kernel methods for predicting protein-protein interactions. Bioinformatics. 2005 Jun;21:i38–i46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- 40.Vert J-P, Qiu J, Noble W. A new pairwise kernel for biological network inference with support vector machines. BMC Bioinformatics. 2007 Dec.8:S8. doi: 10.1186/1471-2105-8-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar-hillel A, Weinshall D. Boosting margin based distance functions for clustering. In Proceedings of the Twenty-First International Conference on Machine Learning. 2004:393400. [Google Scholar]
- 42.Brunner C, Fischer A, Luig K, Thies T. Pairwise support vector machines and their application to large scale problems. Journal of Machine Learning Research. 2012 Aug.13:22792292. [Google Scholar]
- 43.Brunner C, Fischer A, Luig K, Thies T. Pairwise kernels, support vector machines, and the application to large scale problems. tech. rep., Technische Universitat Dresden Institut fur Numerische Mathematik. 2011 [Google Scholar]
- 44.Haykin SS. Neural networks a comprehensive foundation. Prentice Hall. (2nd ed.) 1999 [Google Scholar]
- 45.Ben-Hur A. PyML: machine learning using python. 2012 Dec. http://pyml.sourceforge.net/
- 46.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins: Structure, Function, and Bioinformatics. 2010;78(15):30733084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 47.Guan R, Ma L-C, Leonard PG, Amer BR, Sridharan H, Zhao C, Krug RM, Montelione GT. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza b virus. Proceedings of the National Academy of Sciences. 2011 Aug.108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin C, Khan JA, Swapna GVT, Ertekin A, Krug RM, Tong L, Montelione GT. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza a and b viruses. Journal of Biological Chemistry. 2007 May;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- 49.Narasimhan J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. Journal of Biological Chemistry. 2005 Jun;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 50.Guan R, Ma L-C, Leonard PG, Amer BR, Sridharan H, Zhao C, Krug RM, Montelione GT. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza b virus. Proceedings of the National Academy of Sciences. 2011 Aug.108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008 Feb.24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 52.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE. 2011 Dec.6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marsh J, Teichmann S. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure. 2011 Jun;19:859–867. doi: 10.1016/j.str.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]