Abstract
The forkhead box transcription factor FoxP3 controls the development and function of CD4+CD25+ regulatory T (Treg) cell. FoxP3 modulates gene expression in Treg cells by multiple epigenetic mechanisms that are not clearly defined. We identified FoxP3 interacting proteins in human T cells by co-IP/MS. We discovered that FoxP3 interacted with linker histone H1.5 via the leucine zipper (LZ) domain. Two independent IPEX patient-derived single residue mutations in the LZ of FoxP3 both abrogated its interaction with H1.5. Functionally, FoxP3 and H1.5 cooperatively repressed IL-2 expression in human T cells; and silencing of H1.5 expression inhibited the ability of FoxP3 to suppress IL-2 expression. We show that FoxP3 specifically enhanced H1.5 association at the IL-2 promoter, but reduce its association at the CTLA4 promoter, correlated with higher or lower histone acetylation of the respective promoters. Finally, silencing of H1.5 expression in human Treg cells impaired the Treg function to suppress target T cells. We conclude that FoxP3 interacts with H1.5 to alter its binding to target genes to modulate their expression and to program Treg function.
Introduction
Regulatory T (Treg) cells are required for the maintenance of self tolerance. The forkhead box transcription factor FoxP3 has been shown to be necessary for the Treg cell function. Scurfy mice deficient in FoxP3 function do not have CD4+CD25+ regulatory T cells and develop severe autoimmune lymphoproliferative disease1,2. Mutations in the foxp3 gene in humans also lead to immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome, characterized by multi-organ T cell infiltration and aggressive autoimmunity3,4. Furthermore, ectopic expression of FoxP3 confers a suppressive phenotype to CD4+CD25− T cells, characterized by decreased cytokine (IL-2, IL-4 and IFN-γ) production and ability to suppress proliferation of other T cells5-7.
FoxP3 is a member of the P subfamily of the forkhead (FKH) box transcription factors, which are characterized by a highly conserved winged-helix/FKH DNA binding domain8. The FoxP3 protein includes an N-terminal proline-rich (PRR) domain, a C2H2 zinc finger, a leucine zipper (LZ), and a C-terminal FKH DNA binding domain. The PRR domain of FoxP3 has been shown to be able to repress transcription by recruiting histone deacetylase containing complexes9,10. Mutations identified from IPEX patients within the FoxP3 gene show clusters of missense or in-frame deletion mutations within the leucine zipper and FKH domains, indicating the importance of these domains for FoxP3 function11. The leucine zipper region of FoxP3 is required for the function of FoxP3, as demonstrated by two independent mutations within the leucine zipper (Δ250K & Δ251E) derived from IPEX patients12. The FKH domain of FoxP3 has been shown to bind DNA and mediate repression of the IL-2 promoter in Treg cells13.
While IL-2 is important for Treg cell development and survival, Treg cells themselves do not produce IL-2 in response to TCR stimulation14,15. The repression of IL-2 in Treg cells has been linked directly to FoxP3 through several different mechanisms. FoxP3 has been proposed to bind NFAT and displace AP-1 binding, thereby inhibiting the transactivation of IL-2 by NFAT/AP113. Furthermore, FoxP3 is also shown to interact with the transcription factor AML1/Runx1 to lead to IL-2 repression and Treg suppressive function16. Additionally, FoxP3 interactions with chromatin remodeling complexes containing histone deacetylases have been implicated in IL-2 repression and Treg function9.
Linker histone H1 proteins have long been known to play an important role in the assembly of hetero-chromatin filaments associated with gene silencing. In mice and humans, 5 highly homologous linker histone isoforms (H1a to H1e or H1.1 to H1.5) have been identified, which are differentially regulated during development17. While histone H1 depletion alters global chromatin structure, it surprisingly affects expression of only a few specific genes18. Mice with mutations in individual histone H1a, H1c, H1d, and H1e genes have been generated, with only subtle phenotypes19. Mice with triple mutations in H1c, H1d, and H1e genes are embryonic lethal and also show defects in expression and DNA methylation of only a few specific genes20. Thus, specific histone H1 isoforms may function in coordination with tissue specific transcription factors to regulate specific genes during development and differentiation. Indeed, murine histone H1b is involved in cooperation with the muscle specific transcription repressor Msx1 to repress MyoD expression and muscle differentiation21.
We report here that the human linker histone H1.5 specifically binded FoxP3. The physical association of H1.5 with FoxP3 required the leucine zipper domain of FoxP3. Two independent IPEX patient derived FoxP3 mutants of single residues in the LZ of FoxP3 also abrogated its interaction with H1.5. We show that ectopic expression of FoxP3 and H1.5 synergistically repressed expression of IL-2 in human T cells, associated with chromatin modifications. Conversely, silencing of H1.5 expression in human T cells abrogated the ability of FoxP3 to repress IL-2 expression. FoxP3 interacted with H1.5 to specifically enhance its association at the IL-2 promoter, but reduce its association at the CTLA4 promoter, correlated with higher or lower acetylation of the respective promoters. Finally, silencing of H1.5 expression in human Treg cells impaired the Treg function to suppress target T cells. These findings suggest that interaction of linker histone H1.5 with FoxP3 plays an important role in FoxP3-mediated gene expression and in maintaining Treg cell functions by FoxP3.
RESULTS
FoxP3 Interacts Specifically with Linker Histone H1.5
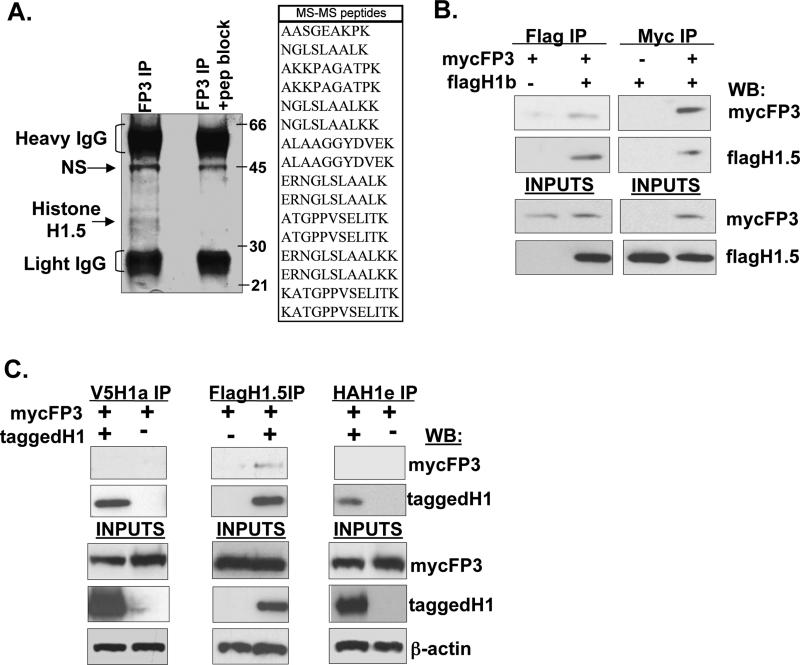
To investigate the molecular mechanisms by which FoxP3 functions in regulatory T cells, and to identify proteins that bind FoxP3, we expressed FoxP3 in a human CD4+ T cell line (Jurkat) with a retroviral vector. As expected, expression of FoxP3 in Jurkat cells inhibited IL-2 expression from the IL2 promoter following activation (Fig.S1). To search for FoxP3 interacting proteins in these cell, we performed immunoprecipitation (IP) with an antibody specific for FoxP3 22. Clarified lysates from Jurkat cells were IPed with an anti-FoxP3 antibody and resolved on an SDS−PAGE gel. Pre-blocking of the FoxP3 antibody with the specific FoxP3 peptide was used to identify specific FoxP3-interacting proteins that were pulled down by the IPed FoxP3 protein (Fig. 1A). Mass spectrometry revealed that one major FoxP3-associated protein was human linker histone H1.5, with a protein identification score of 269 and a total ion score of 169. The interaction was confirmed by co-IP, with either anti-FoxP3 or anti-H1.5 antibody, of coexpressed FoxP3 and H1.5 in 293 cells (Fig. 1B). In order to determine the specificity of the interaction of FoxP3 with H1.5, we performed co-immunoprecipitation of H1a, H1.5 (or H1b), or H1e proteins with FoxP3 (Fig. 1C). Only H1.5 was able to interact with FoxP3, suggesting that H1.5 may specifically play a role in the function of FoxP3 in programming regulatory T cells.
FIGURE 1. FoxP3 specifically interacts with linker histone H1.5.
(A) Cell extracts from Jurkat cells expressing FoxP3 (2-5x10e8 cells) were immuno-precipitated with anti-FoxP3 antibodies or anti-FoxP3 antibodies pre-blocked with the antigen peptide. Proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining of the gel. Positions of molecular weight markers are shown in kilodaltons (kd). The major bands that are specifically pulled down were identified by Mass spectrometry (MS). The peptides identified by MS from one major band (all matched to human H1.5) are listed in the table. (B) Nuclear extracts from 293T cells transfected with myc-FoxP3 and/or flag-H1.5 were immunoprecipitated with anti-myc or anti-flag antibodies, and immuno-blotted with the indicated antibodies. (C) Specificity of H1.5 binding to FoxP3. Nuclear extracts from 293T cells transfected with three different tagged-histone H1 isoforms (V5-H1a, Flag-H1.5, HA-H1e) and myc-FoxP3 were immunoprecipitated with the indicated antibodies and interactions were detected by immunoblotting. 10% inputs of the extract used for IPs are shown in the lower panels.
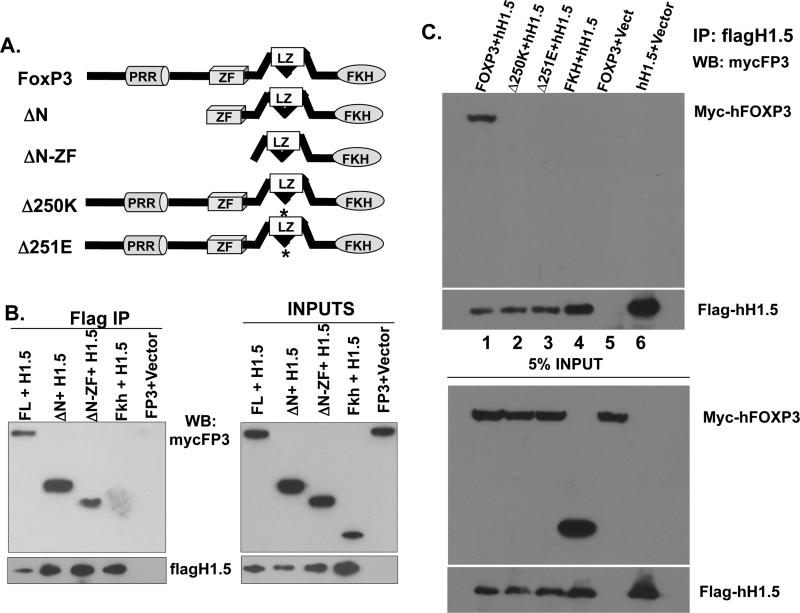
To delineate the domain of FoxP3 required for binding to H1.5, we performed co-IP of H1.5 with truncated mutants of FoxP3 (Fig. 2A). The N-terminus domains including PRR and the zinc finger regions of FoxP3 were dispensable for binding to H1.5, as the ΔN and ΔN-ZF FoxP3 proteins were able to interact with H1.5 (Fig. 2B). The FoxP3 C-terminus region containing the forkhead DNA binding domain (Fkh) was not sufficient for the interaction with H1.5, suggesting that the leucine zipper of FoxP3 is critical for the interaction. Importantly, two independent IPEX patients-derived mutations of a single amino acid deletion within the leucine zipper region of FoxP3 (del-251E or del-250K) abrogated FoxP3 binding to histone H1.5 (Fig. 2C). However, IPEX mutations in the FKH domain that abrogate FoxP3 binding to DNA did not alter the ability of FoxP3 to bind to H1.5 (data not shown). Therefore, the interaction between FoxP3 and H1.5 does not require DNA binding by FoxP3, but requires critical residues in the LZ domain that are correlated with the biological activity of FoxP3.
FIGURE 2. FoxP3 specifically interacts with linker histone H1.5 via critical residues in the leucine zipper domain.
(A) Diagrams of the FoxP3 protein domain structure and deletion mutants31. FL=full length, ΔN=N-terminus deletion, ΔN-ZF= deleted N-terminus and zinc finger, Fkh=deleted N-terminus, zinc finger, and leucine zipper. Δ250K/Δ251E=mutant FoxP3 alleles isolated from IPEX patients. (B) Nuclear extracts from 293T cells transfected with flagH1.5 and mycFoxP3 deletion mutants were IPed with anti-flag antibodies and co-IPed mycFoxP3 proteins were detected by immunoblotting. (C) The amino acid residues 250K/251E in leucine zipper domain of FoxP3 are both critical for binding to H1.5, as well as for FoxP3 function. Nuclear extracts from 293T cells transfected with flagH1.5 and human FoxP3 (lane 1), Δ250K (lane 2) or Δ251E (lane 3), the Forkhead domain (lane 4), FoxP3 only (lane 5), and flagH1.5 only (lane 6). Flag-H1.5 was immuno-precipitated with anti-flag antibodies and Western blotted with the indicated antibodies. Inputs represent 5% of cell lysates used for each co-IP experiment.
FoxP3 and H1.5 functionally interact to repress IL2 expression in human T cells
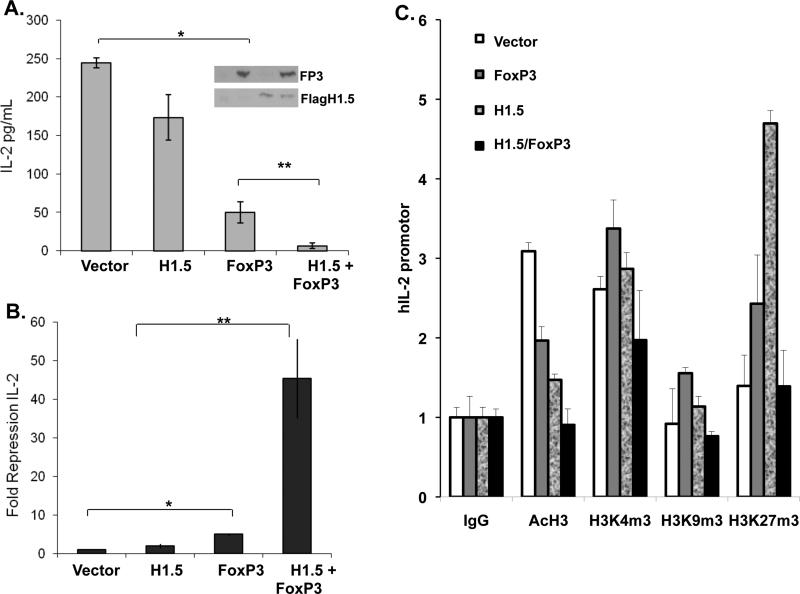
In order to determine the functional interaction of H1.5 and FoxP3, H1.5 and FoxP3 were ectopically expressed by retroviral transduction in Jurkat T cells. Co-expression of FoxP3 and H1.5 did not affect their relative expression (Fig. 3A, inset). The Jurkat T cells were stimulated and IL-2 expression was measured by IL-2 ELISA. Expression of FoxP3 alone in Jurkat T cells resulted in a 5-fold repression of IL-2 expression, while H1.5 expression alone caused low but not significant repression of IL-2 expression (Fig. 3A). Interestingly, co-expression of FoxP3 and H1.5 led to a 45-fold repression of IL-2 expression (Fig. 3B). When the chromatin structure at the IL2 locus was analyzed, we confirmed that histone acetylation was partially reduced by FoxP3 expression, but completely repressed by FoxP3/H1.5 coexpression. In addition, the relative trimethylation of H3 at K4, K9 and K27 positions was also differentially modulated by FoxP3 and H1.5 (Fig. 3C). Thus FoxP3 and H1.5 interact to cooperatively suppress IL-2 gene expression via epigenetic mechanisms in human T cells.
FIGURE 3. FoxP3 and H1.5 interact functionally to repress IL2 gene expression in T cells.
(A-B) FoxP3 and H1.5 cooperate to synergistically repress IL-2 expression in human T cells. (A) Jurkat T cells stably expressing flag-H1.5 were re-transduced with control vector or FoxP3-expressing vector (>95% with each or both vectors). Inset shows Western blot of FoxP3 and flagH1.5 expression. Transduced cells were stimulated with PMA and Ionomycin, and IL-2 expression was measured by IL-2 ELISA in culture supernatants in triplicates. Shown are IL2 levels normalized to total live cell numbers in each sample. Error bars indicate standard deviation. (B) Summary of relative repression of IL-2 from four independent experiments. Error bars indicate standard error, p-values are determined by two-tailed students’ t-test. *, p<0.05, **, P<0.005. (C) ChIP assays were performed to assess histone modifications (Acetylation of H3/H4, tri-methylation of H3 K4, K9 and K27) at the IL2 promoter. Results are quantified by real-time PCR and normalized to binding by control IgG antibody. Error bars indicate standard deviation. * = p<0.05.
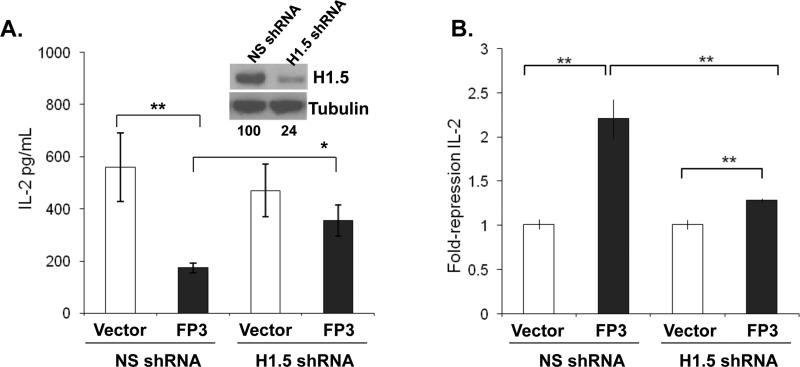
To genetically prove the role of H1.5 in FoxP3-mediated suppression of IL-2 gene expression, we silenced expression of endogenous H1.5 gene and tested its effect on FoxP3's ability to repress IL-2 expression in T cells. The H1.5 shRNA construct was able to reduce expression of the flag-tagged H1.5 by 75% in co-transfected cells (Fig. 4A, inset). Human primary CD4+ T cells were co-transfected with a non-specific shRNA or an H1.5 specific shRNA plasmid in combination with either a control vector or a FoxP3 expression vector. The transfected cells were activated with anti-CD3/CD28, and IL-2 expression was measured by ELISA. Silencing of H1.5 expression by shRNA alone did not significantly affect IL-2 production. However, the activity of FoxP3 to repress IL-2 expression was severely impaired in CD4+ T cells with reduced expression of H1.5 (Fig. 4). When the IL2 promoter (-226 to + 45)-luciferase gene was co-transfected with FoxP3 and H1.5 (Fig. S2A) or with FoxP3 and shRNA-H1.5 (Fig. S2B), similar inhibition results by FoxP3 and H1.5 as the endogenous IL2 gene were observed. Thus, FoxP3 requires H1.5 to repress IL-2 expression in human CD4+ T cells.
FIGURE 4. H1.5 is required for FoxP3-mediated IL-2 repression in human T cells.
(A) Expression of H1.5 is specifically knocked down by the H1.5 shRNA construct. H1.5 levels are determined by Western blot analysis of flag-H1.5 in 293T cells cotransfected with NS (non-specific) or H1.5-specific shRNA vector. Relative H1.5 expression normalized to tubulin protein is shown under the panels (inset). Primary human CD4+ T cells were transfected by Amaxa nucleofection with either the control NS-shRNA or H1.5-specific shRNA plasmids with or without FoxP3. Cells in triplicate wells were stimulated with anti-CD3 and anti-CD28 antibody for 48h and culture supernatants were analyzed by IL-2 ELISA. Error bars indicate standard deviation. (B) Relative repression (fold) of IL-2 in primary CD4+ T cells from two independent experiments is summarized. * = p<0.05, ** = p<0.005.
FoxP3 modulates H1.5 association at the IL-2 and the CTLA4 promoters
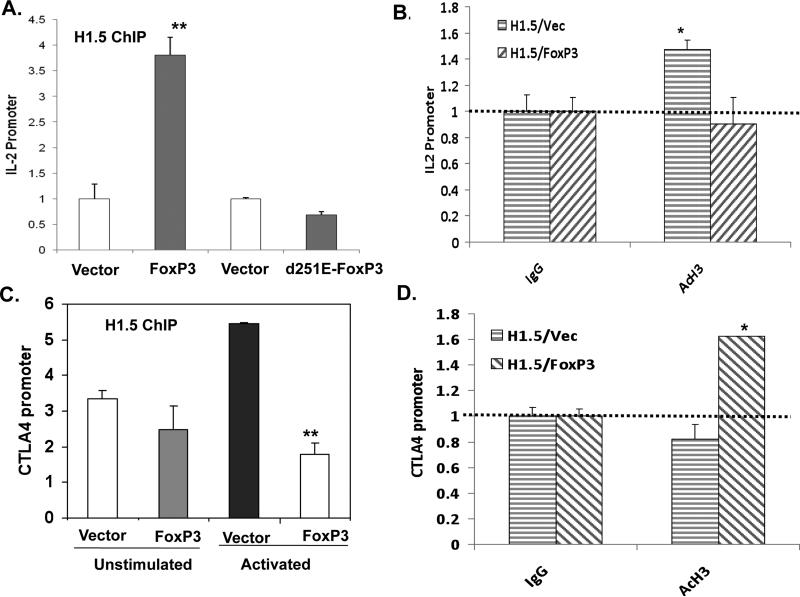
In order to determine whether FoxP3 alters the binding of H1.5 at the FoxP3 target genes in human T cells, we performed Chromatin ImmunoPrecipitation (ChIP) analysis of flag-tagged H1.5 and FoxP3 in human T cells. FoxP3 binds to the IL-2 proximal promoter in Jurkat T cells with or without exogenously expressed H1.5 (data not shown and 13). We show that binding of H1.5 at the IL-2 promoter was increased when FoxP3 was expressed in both unstimulated and stimulated Jurkat T cells (Fig. 5A and data not shown). When the IPEX patient-derived FoxP3 mutant (del-251E), which abrogated FoxP3 binding to H1.5 (Fig. 2C), was tested by ChIP, it also failed to enhance binding of H1.5 to the IL-2 promoter (Fig. 5A). This increase in FoxP3-mediated H1.5 binding is associated with the repression of IL-2 gene expression and with reduced histone acetylation level of the IL-2 promoter (Fig. 5B and DH-JBC). When the FoxP3-enhanced CTLA4 gene was analyzed, FoxP3 significantly reduced the H1.5 association at the CTLA4 promoter (Fig. 5C), consistent with its enhanced expression and elevated histone acetylation level (Fig. 5D). In sum, our data support a model that FoxP3 modulates gene expression in T cells by modulating H1.5 binding at distinct target promoters.
FIGURE 5. FoxP3 specifically modulates H1.5 association and histone modification at the IL-2 or CTLA4 promoter.
Jurkat T cells stably expressing flag-H1.5 were transduced with control vector or FoxP3-expressing vector. ChIP assays were performed with anti-flag antibody to assess H1.5 binding to the target promoters. The distal region of IL-2 gene showed only background binding (data not shown). ChIP assay was also performed with anti-AcH3/4 mAb to measure relative acetylation of histones H3 andH4 at the target promoters. Results are quantified by real-time PCR and normalized to binding by control IgG antibody. Error bars indicate standard deviation. * = p<0.05, **=p<0.01. (A) αFlagH1.5 ChIP at the IL-2 promoter in FlagH1.5-Jurkat cells transduced with wild type or del-251E IPEX mutant FoxP3. (B) Relative acetylation of the IL2 promoter was measured by anti-AcH3/4 ChIP. (C) αFlagH1.5 ChIP at the CTLA4 promoter in FlagH1.5-Jurkat cells transduced with control vector or FoxP3. Both un-stimulated and activated cells were analyzed by ChIP. (D) Relative acetylation of the CTLA4 promoter was measured by anti-AcH3/4 ChIP.
H1.5, similar to FoxP3, is required for the suppression activity of Treg cells
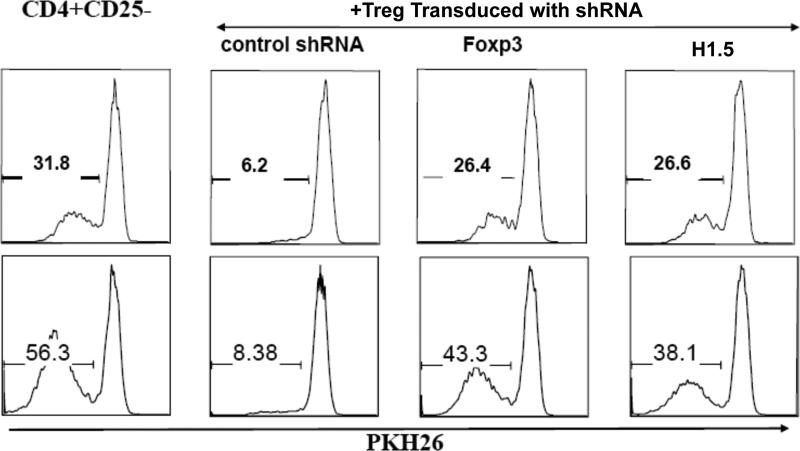
The human CD4+ T cell subset (CD4+ CD45RA+CD25+) with Treg cell potential (TNreg) has been recently reported to become fully functional Treg cells after activation and expansion, with further induction of FoxP3 expression and suppression activity 23. To demonstrate that H1.5 is critical in the FoxP3-induced maturation of Treg cells with suppression activity, we investigated the effect of silencing endogenous H1.5 expression in human TNreg cells on their ability to suppress proliferation of CD4 T cells. Human TNreg cells were purified (>99% by FACS) and transduced with GFP-lentiviral vectors expressing non-specific shRNA (control), FoxP3-shRNA or H1.5-shRNA. The TNreg cells were then activated with monocyte derived dendritic cells (mDC) pulsed with Staphylococcal Enterotoxin B (SEB), and expanded to derive mature Treg cells 23. Transduced GFP+ Treg cells were then purified and used as Treg effectors in the suppression assay with CD4+CD25- T cells labeled with PKH26 as target cells. Proliferation of the target cells was examined by measuring PKH26 dilution at 4 or 5 days post stimulation (Fig. 6). Treg cells transduced with control shRNA vector efficiently suppressed target cell proliferation. Silencing of FoxP3 expression in Treg cells significantly reduced the suppression activity of Treg cells as recently reported 23. When expression of H1.5 was silenced in Treg cells, the suppression activity of Treg cells was also similarly reduced. Therefore, H1.5 is required for the FoxP3-induced suppression activity in Treg cells.
FIGURE 6. H1.5 is required for FoxP3-induced Treg suppression activity.
CD4+CD25+CD45RA+ (TNreg) cells were sorted from human blood (>99% purity), activated with SEB-pulsed DC, and transduced with shRNA expressing lentiviral-GFP vectors. Cells were then expanded, and sorted based on GFP expression. Transduced GFP+ cells were used as Treg effectors in a suppression assay, with CD4+CD25- T cells labeled with PKH26 as target cells. Treg cells and target cells were mixed at 1:8, and stimulated with mDC (1:30) pulsed with SEB (1 pg/ml). Proliferation of the target cells were examined 4 (top) or 5 (bottom) days post stimulation. Numbers indicate relative proliferation (PKH26 dilution). Similar results are observed from two independent experiments at multiple conditions.
DISCUSSION
In this study we demonstrate a novel mechanism by which FoxP3 modulates gene expression in regulatory T cells. FoxP3 specifically interacted with the linker histone H1.5. Co-expression of H1.5 and FoxP3 lead to synergistic repression of IL-2 expression in human T cells. Conversely, knockdown of H1.5 expression in human T cells inhibited the ability of FoxP3 to repress IL-2 expression. In addition, FoxP3 interacted with H1.5 to specifically enhance its association at the IL-2 promoter, but reduce its association at the CTLA4 promoter, correlated with higher or lower histone acetylation of the respective promoters. Finally, knockdown of either FoxP3 or H1.5 expression in human TNreg cells inhibited the ability of Treg cells to suppress target T cells, indicating that the interaction between FoxP3 and linker histone H1.5 is important for FoxP3 function in human regulatory T cells.
The FoxP3-H1.5 interaction required the leucine zipper domain of FoxP3, suggesting that dimerization of FoxP3 with itself or other proteins may be necessary for H1.5 binding. Importantly, the human IPEX mutations with the 251E or the 250K amino acid deletion in the LZ region also both impaired its binding to H1.5, suggesting that H1.5 binding by FoxP3 plays an important role in FoxP3 function. It has been reported that region of FoxP3 between the leucine zipper and forkhead domain (AA278-336) is critical for binding to the transcription factor RUNX/AML116. It is possible that interaction with RUNX/AML1 may affect FoxP3 binding with H1.5, but it is not clear if the 251E or the 250K FoxP3 mutant is still able to bind RUNX/AML1. While previous studies have shown that the FoxP2/3 forkhead domain binds DNA and interacts with NFAT13, our studies demonstrated that the FoxP3 DNA binding activity is not required for the physical interaction with H1.5, as shown with the IPEX mutations within the forkhead region of FoxP3 (Fig. S2). However, it is clear that this FoxP3-H1.5 interaction is critical for IL-2 repression by FoxP3 and likely may modulate the expression of a group of target genes that are modulated by FoxP3 in regulatory T cells24. It will be of interest to identify genes/loci whose association with H1.5 is modulated by FoxP3 in T cells.
We propose that FoxP3 binds to the IL-2 promoter and recruits H1.5, leading to its repression. Two independent IPEX patients-derived FoxP3 mutants with a single deletion in the LZ domain also failed to recruit H1.5 to the IL2 promoter, suggesting that the binding of FoxP3 with H1.5 may play a critical role in the FoxP3 activity to reprogram Treg cells. Interestingly, the muscle specific transcription repressor Msx1 also specifically interacts with murine H1b (homolog of human H1.5) to regulate mouse muscle cell differentiation. Msx1 binds and recruits H1b to the MyoD gene to repress its expression and muscle differentiation21. For target genes whose expression is enhanced by FoxP3 in Treg cells, FoxP3 reduced H1.5 binding to the CTLA4 promoter in activated T cells, via unclear mechanisms (Fig. 5C). Therefore, linker histone H1.5 plays a special role in modulating gene transcription in T cells through association with transcription factors such as FoxP3. However, the H1.5 association at the CD25 promoter was not altered by FoxP3. Instead, T cell activation significantly reduced H1.5 association at CD25 promoter (unpublished results). FoxP3 may therefore enhance target gene expression via multiple mechanisms.
While expression of H1.5/H1b decreases during differentiation in most tissues, H1.5/H1b expression is maintained at high levels in lymphocytes and lymphoid organs such as thymus and spleen25. It is likely that H1b is preferentially involved in lymphocyte differentiation and functions. To date, however, the H1b gene has not been knocked out in mice, which may prove to have a specific phenotype due to its expression and interactions with tissue specific transcription factors. On the other hand, mice with mutations in one or two of the linker H1 histone genes have developed normally, with no significant differences in the total H1-to-nucleosome core H2-4 histone stoichiometry, indicating that H1 subtypes are able to compensate for each other during mouse development18,19. It may be necessary to generate conditional mutant mice with inducible deletion of H1b gene in order to observe its effect in specific tissues or cell types.
FoxP3 has been recently described to be associated with a chromatin remodeling complex including TIP60, HDAC7, and members of the SWI/SNF family9,26. We describe here another mechanism of FoxP3 transcriptional modulation through modulation of linker histone H1.5 binding to alter the chromatin structure at FoxP3-responsive genes. Our results do not preclude the idea that FoxP3 may form a large complex with H1.5, other transcription factors, and chromatin remodeling complexes. Indeed, another human histone H1 isoform, H1.4, has recently been reported to specifically interact with the malignant brain tumor protein L3MBTL1 to compact chromatin and repress expression of E2F regulated genes27. FoxP3 may similarly alter the chromatin structures at distinct gene loci to coordinate Treg maturation and functions. Additionally, the reciprocal binding of PARP-1 and Histone H1 has been shown to affect gene expression outcomes28. Thus, it is likely that the combination of these factors is required for FoxP3 function. Different classes of FoxP3 regulated genes may use different combinations of mechanisms to specifically up- or down-modulate their expression during Treg development or in response to Treg activation. Our findings on the functional interaction of FoxP3 with linker histone H1.5 have important implications in understanding the transcriptional program of Treg cells. The findings will also help to elucidate how linker histones modulate expression of specific genes through interactions with specific transcription factors. Finally, as single residues in the LZ domain of FoxP3 are critical for H1.5 binding and FoxP3 function, the interaction between FoxP3 and H1.5 may provide a novel target to modulate Treg development and functions.
Materials and Methods
Plasmids, antibodies, and cell lines
FoxP3 was expressed from the HSPG retrovirus vector 29. Deletion mutants of FoxP3 were generated by PCR amplification to specifically remove the N-terminus (aa 1-189), the N-terminus with the Zinc finger motif (aa1-241), the N-terminus including the zinc finger and the leucine zipper (aa 1-337). Human H1.5 was PCR amplified from genomic Jurkat cell DNA and cloned into the pHITflag vector. pcDNA-V5-H1a and pcDNA-HA-H1d were generous gifts from Dr. Corey Abate-Shen21. The Δ251E and Δ250K point mutants were introduced into FoxP3 by site-directed mutagenesis and verified by DNA sequencing. The NS shRNA plasmid (pLL5.0NS) was a kind gift from Dr. Jim Bear30. The non-specific shRNA was generated by using the target loop sequence GATCGACTTACGACGTTAT. The H1.5 shRNA was purchased from Open Biosystems in the pGINZeo vector, using the H1.5 target loop sequence TAGTGAAGCCACAGATGTA. Jurkat cells were maintained in RPMI 1640 (Gibco BRL), 293T cells were maintained in DMEM and human primary T cells were maintained in Iscoves MEM, supplemented with 10% FBS (Sigma), 2mM L-glutamine, 100U/ml penicillin, 100U/ml streptomycin. For T cell activation, PMA (Sigma, 25ng/ml) and ionomycin (Sigma, 1uM), mouse anti-CD3 mouse, anti-CD28 (BD Biosciences), and goat anti-mouse IgG (Caltag) were used. FoxP3 antibodies have been previously described22. The peptide from the N-terminus of murine FoxP3 (CLLGTRGSGGPFQGRDLRSGAH) was used to immunize rabbits for production of polyclonal antibodies. Affinity purification of the antibodies was performed with a SulfoLink Kit (Pierce Biotechology, Rockford, IL). The mouse anti-myc (9E10) monoclonal supernatant was kindly provided by Dr. Y. Xiong (UNC, NC). The mouse anti-Flag M2 (Sigma) and the rabbit anti-histone H1.5 from Abcam were used.
Retrovirus and lentiviral vector production and transduction
293T cells were cotransfected with retroviral vector, VSV-g, and Gag-Pol containing plasmids by calcium phosphate, as previously described29. For retroviral transduction, Jurkat cells were spinoculated with retrovirus to >90% efficiency as measured by GFP expression31. For VSV-g pseudotyped HIV-derived lentiviral vector production, 293T cells were transfected with Effectene (Qiagen) transfection reagent per manufacturer's instructions. 48 hour post transfection supernatants were used to infect Jurkat cells by spinoculation.
Immunoprecipitations
Whole cell extracts were made from FoxP3 transduced Jurkat cells using modified RIPA extraction buffer. Extracts (40mg) were immunoprecipitated using affinity purified anti-FoxP3 (Li195) antibodies in PBS, 10uM NaVO4 , 10uM DTT, 1uM PMSF, and 1X protease inhibitor cocktail (Roche). FoxP3 antibodies were pre-incubated for 1h with the Li195 peptide, prior to immunoprecipitation for the peptide specificity control. Immune complexes were precipitated by addition of Protein A agarose beads (Pierce). Complexes were eluted by boiling in SDS-loading dye buffer, resolved by SDS-PAGE and visualized with Coomassie Brilliant Blue stain. Protein bands were cut from the gel and sequence analysis was performed by the UNC Proteomics Core Facility by reverse phase HPLC tandem mass spectrometry (LC/MS). Co-immunoprecipitation experiments were performed using nuclear extracts of 293T cells transfected (Effectene, Qiagen) with flag-tagged H1b and myc-tagged-FoxP3, using either mouse anti-myc (9E10) hybridoma supernantant or mouse anti-flag M2 (Sigma) antibodies. Proteins were analyzed by immunoblotting and detection with ECL plus western blotting detection (Amersham).
Chromatin immunoprecipitation assays (ChIPs)
Jurkat T cells were retrovirallytransduced with control or flag-H1b expressing virus, followed by a second transduction 48h later with control or FoxP3 expressing virus. Sonicated chromatin from the Jurkat cells was immunoprecipitated with control IgG (IgG) or anti-myc (9E10) (Santa Cruz Biotechnologies) or anti-flagM2 (Sigma) by chromatin immunoprecipitation assay kit perthe manufacturer's instruction (Upstate, Millipore), as previously reported31. Sampleswere purified and subject to real-time qPCR analysis. IL-2 promoter was analyzed withSybergreen reagents using the following primers: 5’-CAC CTA AGT GTG TGG GCT AAT GTA AC-3’ and 5’-CTG ATG ACT CTT TGG AAT TTC TTT AAA CC-3’. This amplicon spans –226 to –133 in the human IL-2 promoter and spans the NFκB and AP-1 sites. The relative mycFP3 or flagH1b binding of IL-2 promoter is expressed with ratios of ChIP-specific signal divided by signals from control 10% input chromatins, then normalized to non-specific IgG antibody binding.
Primary human T-cell purification and transfection
CD4+ primary T cells were purified from Ficoll-separated PBMC (peripheral blood mononuclear cell) by MACS human CD4+ T-cell isolation kit (>90% purity, Miltenyi-Biotech, Auburn, CA). Purified primary CD4+ T cells were transfected by using the Amaxa nucleofector kit (Amaxa Biosystems, Gaithersburg, MD). Briefly, 10 × 106 unstimulated CD4+ T cells were transfected with 2 μg of plasmid DNA (~50% efficiency by GFP expression), and cultured for 24h before activation.
IL-2 ELISA
Jurkat T cells were activated by PMA (25ng/ml) and ionomycin (1uM) for 16h. Media supernatants were harvested and analyzed by the human IL-2 ELISA kit (BD Biosciences), as per manufacturer's instructions. Primary CD4+ T cells were activated by anti-CD3/CD28 monoclonal antibody cross-linked by plate-bound anti-mouse IgG as previously described. Briefly, purified cells are stained with anti-CD3 (0.5 μg/ml) and anti-CD28 (2 μg/ml) and incubated on plated-bound goat anti-mouse IgG. Cells were activated for 72h, and media supernatants were harvested for IL-2 ELISA. Four independent experiments (each in triplicates) are carried out.
Isolation, expansion and lentiviral transduction of TNreg cells
were performed as recently reported23. Briefly, naive and TNreg cells were sorted from human blood as described before23. Cells were activated with monocyte derived dentric cells (DC) and Staphylococcal Enterotoxin B (SEB), and grown for 5 days before infected by shRNA expressing virus supernatant. Preparation of the control and Foxp3 shRNA viruses has been described23. Cells were then expanded for another week, sorted based on GFP expression and GFP+ cells were used as effectors in a suppression assay.
Treg suppression assay
Naive CD4+ T cells were labeled with PKH26 (Invitrogen), and used as target cells. Target cells and different effector cells were mixed at a ratio 1:4 or 1:8, and stimulated with DC (1:30) and SEB (10~0.1pg/ml). Proliferation of the target cells were examined 4 or 5 days post stimulation23.
Statistical Analysis
For statistical analysis, a two-tailed Student t test is employed where p < 0.05 is considered significant.
Supplementary Material
Acknowledgment
We thank Drs. Yi Zhang, Art Skoultchi, Jenny Ting, Yue Xiong and members of the Su lab for critical reading and/or discussion of the manuscript. The project was supported by grants from NIH (AI048407 & AI077454 to L. S., AI065303 to D. U.) and UNC training grant (106693-39-RFMT to S.M.C.).
Footnotes
The authors declare no competing financial interests.
References
- 1.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 2.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 3.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 4.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 5.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 6.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 7.Yagi H, et al. Crucial role of FOXP3 in the development and function of human CD25+CD4+ regulatory T cells. Int Immunol. 2004;16:1643–1656. doi: 10.1093/intimm/dxh165. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- 9.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 11.Campbell DJ, Ziegler SF. FOXP3 modifies the phenotypic and functional properties of regulatory T cells. Nat Rev Immunol. 2007;7:305–310. doi: 10.1038/nri2061. [DOI] [PubMed] [Google Scholar]
- 12.Chae WJ, Henegariu O, Lee SK, Bothwell AL. The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc Natl Acad Sci U S A. 2006;103:9631–9636. doi: 10.1073/pnas.0600225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 14.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 15.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 17.Doenecke D, et al. Histones: genetic diversity and tissue-specific gene expression. Histochem Cell Biol. 1997;107:1–10. doi: 10.1007/s004180050083. [DOI] [PubMed] [Google Scholar]
- 18.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Q, Su H, Knudsen G, Helms W, Su L. Delayed functional maturation of natural regulatory T cells in the medulla of postnatal thymus: role of TSLP. BMC Immunol. 2006;7:6. doi: 10.1186/1471-2172-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antons AK, et al. Naive Precursors of Human Regulatory T Cells Require FoxP3 for Suppression and Are Susceptible to HIV Infection. J Immunol. 2008;180:764–773. doi: 10.4049/jimmunol.180.2.764. [DOI] [PubMed] [Google Scholar]
- 24.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T- cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lennox RW, Cohen LH. The histone H1 complements of dividing and nondividing cells of the mouse. J Biol Chem. 1983;258:262–268. [PubMed] [Google Scholar]
- 26.Li B, et al. FOXP3 ensembles in T-cell regulation. Immunol Rev. 2006;212:99–113. doi: 10.1111/j.0105-2896.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 27.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 28.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 29.Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral protein. Nat Biotechnol. 2003;21:1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitriol EA, Uetrecht AC, Shen F, Jacobson K, Bear JE. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci U S A. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes D, Knudsen G, Mackey-Cushman S, Su L. FoxP3 enhances HIV-1 gene expression by modulating NFkappaB occupancy at the long terminal repeat in human T cells. J Biol Chem. 2007;282:15973–15980. doi: 10.1074/jbc.M702051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.