SUMMARY
Transcription in eukaryotes produces a number of long noncoding RNAs (lncRNAs). Two of these, MALAT1 and Menβ, generate a tRNA-like small RNA in addition to the mature lncRNA. The stability of these tRNA-like small RNAs and bona fide tRNAs is monitored by the CCA-adding enzyme. Whereas CCA is added to stable tRNAs and tRNA-like transcripts, a second CCA repeat is added to certain unstable transcripts to initiate their degradation. Here, we characterize how these two scenarios are distinguished. Following the first CCA addition cycle, nucleotide binding to the active site triggers a clockwise screw motion, producing torque on the RNA. This ejects stable RNAs, whereas unstable RNAs are refolded while bound to the enzyme and subjected to a second CCA catalytic cycle. Intriguingly, with the CCA-adding enzyme acting as a molecular vise, the RNAs proofread themselves through differential responses to its interrogation between stable and unstable substrates.
INTRODUCTION
The CCA-adding enzyme adds the nucleotide triplet CCA to the 3′ end of all tRNAs (Deutscher, 1982), a step that is essential for tRNA aminoacylation (Sprinzl and Cramer, 1979) and correct tRNA positioning in the ribosome (Nissen et al., 2000). It is an intriguing member of the nucleotidyltransferase family, since it operates without a nucleic acid template and without translocation along the tRNA (Shi et al., 1998a). Instead, the tRNA 3′-end progressively refolds during synthesis and nucleotide selectivity switches from cytidine to adenosine after the addition of two C-nucleotides (Tomita et al., 2006; Xiong and Steitz, 2004). In addition to acting upon newly synthesized tRNA transcripts, the CCA-adding enzyme is able to accurately repair partially degraded tRNA 3′ ends. Interestingly, CCA-adding enzymes fall into two divergent classes with only the catalytic domains sharing significant homology. In class I enzymes (present in archaea), bound tRNA is involved in the selection of the correct nucleotides (Xiong and Steitz, 2004), where as in class II enzymes (in eubacteria and eukaryotes), proper nucleotides are selected by protein only (Li et al., 2002; Tomita et al., 2004).
Although the CCA-adding enzyme was long thought to terminate polymerization once a single CCA triplet had been added, recent work on tRNA-like small RNAs derived from long non-coding RNAs (lncRNAs) indicated this is not always the case (Sunwoo et al., 2009; Wilusz et al., 2011). Metastasis-associated long adenocarcinoma transcript 1 (MALAT1) and Menβ (also known as NEAT1) are well-characterized long, nuclear-retained non-coding transcripts that are involved in cancer progression (Ji et al., 2003; Lin et al., 2007) and paraspeckle formation (Clemson et al., 2009; Imamura et al., 2014; Sunwoo et al., 2009), respectively. In addition, both of these lncRNAs contain sequences that mimic the tRNA fold and are processed by the canonical tRNA biogenesis machinery to generate small RNAs (Wilusz et al., 2008; Wilusz et al., 2011). Just as for bona fide tRNAs, the trinucleotide CCA is post-transcriptionally added to the 3′-end of MALAT1-associated small cytoplasmic RNA (mascRNA), the tRNA-like transcript originating from the 3′-end of MALAT1 (Wilusz et al., 2008). In contrast, the Menβ tRNA-like small RNA was found to be subjected to CCACCA addition and efficiently degraded (Sunwoo et al., 2009; Wilusz et al., 2011). Unlike the acceptor stems of canonical tRNAs and mascRNA, the Menβ acceptor stem is destabilized through mismatches or wobble base pairs, somehow prompting the CCA-adding enzyme to repeat its catalytic cycle, thereby adding tandem CCA motifs (Wilusz et al., 2011). The CCACCA tail then serves as a degradation signal for the cellular RNA decay machinery.
It is now clear that CCA-adding enzymes from all three kingdoms of life survey the stability of their tRNA-type substrates and selectively add either CCA or CCACCA. For example, many bona fide tRNAs that are destabilized through mutations or the lack of proper modifications are subjected to CCACCA addition (Wilusz et al., 2011). The CCA-adding enzyme is thus not only critical for generating functional tRNAs, but also likely plays a universal and central role in tRNA and tRNA-like small RNA surveillance and quality control.
Here, we investigated how the CCA-adding enzyme distinguishes structurally stable from unstable RNAs so as to only mark unstable RNAs with CCACCA. We also unravel how a second CCA cycle can occur despite the enzyme possessing a unique mechanism that normally ensures a single cycle of CCA synthesis. We find that after the first CCA cycle, nucleotide binding to the active site induces the enzyme to apply torque on the RNA. A clockwise screw motion of the enzyme’s catalytic domain leads to RNA compression and overwinding. This causes unstable RNAs to extrude a distinctively positioned bulge from the acceptor stem while still bound to the enzyme. The bulge does not perturb the double-helical nature of the substrate and all other structural determinants near the active site remain in place. The catalytic mechanism is thus preserved between the first and second cycles of CCA addition (Pan et al., 2010; Tomita et al., 2006; Xiong and Steitz, 2004). In total, we find that tandem CCA addition is not the result of a modified enzymatic activity that is particular to unstable RNAs. Rather, it is a consequence of the natural activity of the CCA-adding enzyme on a substrate with increased conformational flexibility. By exploiting the versatility of RNA structure, the CCA-adding enzyme is able to trigger the degradation of potentially detrimental small RNAs and tRNAs.
RESULTS
Catalysis Is Crucial for the CCA-Adding Enzyme to Detect Unstable RNAs
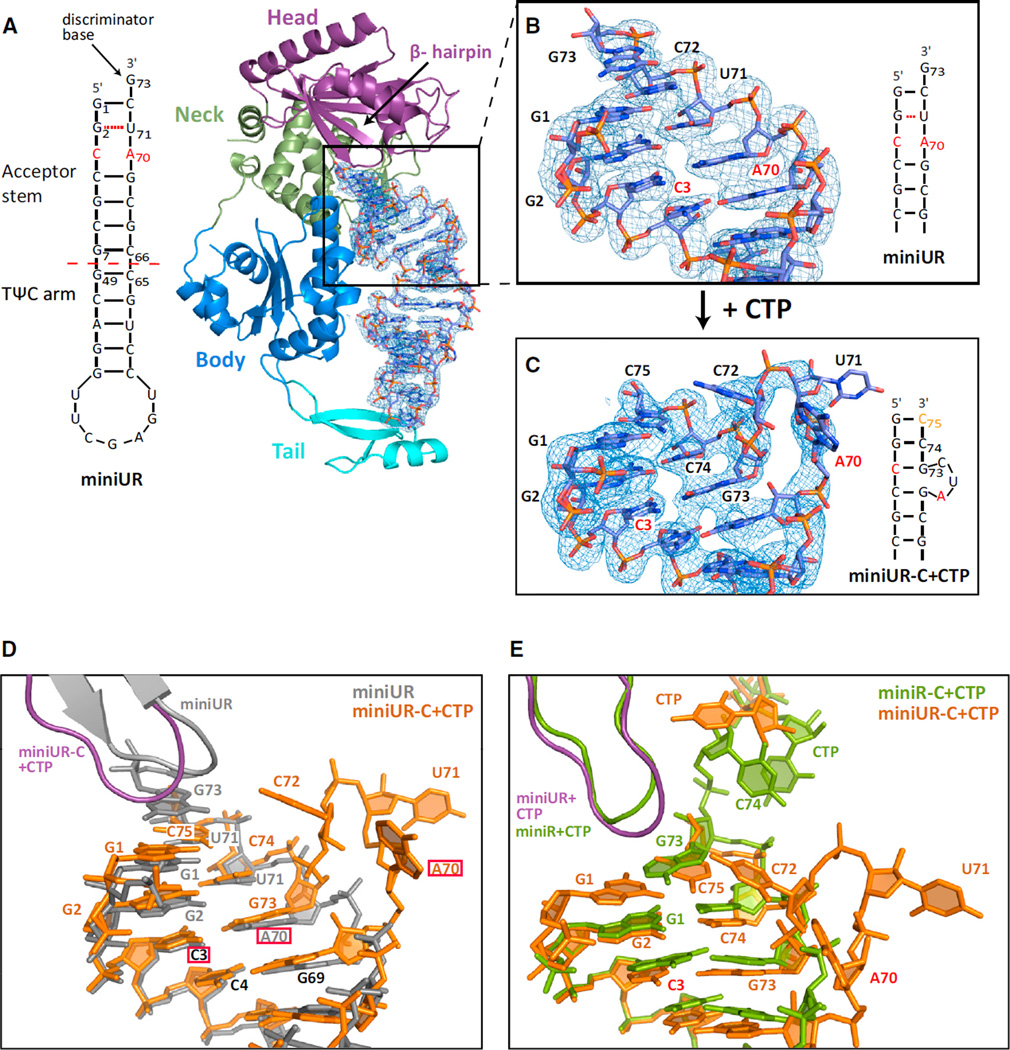
Certain full-length tRNAs that are hypomodified or contain mutations that destabilize their acceptor stems have previously been shown to be subjected to CCACCA addition (Wilusz et al., 2011). Using the CCA-adding enzyme from Archaeoglobus fulgidus (AfCCA), we found that tRNA minihelices, which contain only the acceptor stem and TΨC stem-loop, are also efficiently subjected to CCACCA addition when they have guanosines at the first and second positions as well as a destabilized acceptor stem by virtue of mismatches and G·U wobbles (Figure S1A available online). To understand how the CCA-adding enzyme determines whether a substrate is to receive CCA or CCACCA, we first used an arginyl-tRNATCG minihelix that contained a C-A mismatch (C3·A70) in its acceptor stem due to a mutation at position 70 (Figure 1A). This unstable tRNA minihelix (miniUR) was co-crystallized with AfCCA and the structure of the complex was determined by molecular replacement and refined to 2.95 Å resolution (Table S1).
Figure 1. Catalysis Is Crucial to Detect Unstable RNAs.
(A) G70 of the arginyl-tRNATCG minihelix was mutated to A, resulting in the miniUR transcript that has significant structural instability within its acceptor stem. The archaeal CCA-adding enzyme is shown in cartoon representation with the head domain in purple, the neck domain in green, the body domain in blue and the tail domain in cyan. Bound RNA is shown in atom colors with oxygens in red, nitrogens in blue, phosphorous in orange and carbons in light blue. 2Fo-Fc density for the bound unstable RNA is contoured at 1 σ throughout Figure 1.
(B) Close-up view of the acceptor stem.
(C) Close-up view of the acceptor stem following CTP addition (miniUR-C+CTP).
(D) MiniUR before and after CTP addition. Nucleotides labeled in black have similar positions in both structures, the mutation is boxed in red.
(E) Comparison of the CCA-adding enzyme in complex with a wild-type tRNA minihelix (miniR) (PDB code 2DR5) (Tomita et al., 2006) or in complex with the unstable miniUR minihelix after CTP addition.
See also Figure S1.
The unstable minihelix is bound between the enzyme’s catalytic center, comprised of the head and neck domains, and its tail domain, which serves as a ruler to ensure that only tRNAs and tRNA-like transcripts are substrates for the enzyme (Figure 1A) (Tomita et al., 2004; Xiong and Steitz, 2004). As observed previously, the discriminator base (G73) at the RNA 3′-end extends the stacking of the minihelix and is not rotated out of the helical trajectory (Tomita et al., 2006). However, in contrast to Tomita et al. (2006), which described helical distortion in the middle of the minihelix, we find that the unstable tRNA minihelix perfectly mimics full-length tRNA with its acceptor and TΨC stems folding into a continuous A-type RNA helix. Furthermore, the TΨC loop is in the same conformation as in full-length tRNA, likely because two nucleotide fragments (derived from the mother liquor after nucleolytic cleavage) aid proper folding by mimicking nucleotides 17 and 18 of full-length arginyl-tRNATCG (Figure S1B). The C3·A70 mismatch is seamlessly incorporated into the RNA helix and adenine and cytosine face each other, albeit further apart, despite their hydrogen bonding incompatibility (Figure 1B). This larger distance is thus not sufficient to noticeably distort the bound minihelix and, therefore, unlikely to distort a full-length tRNA.
To understand at which step the enzyme is able to detect unstable RNAs, we added CTP prior to co-crystallization. The enzyme catalyzed the addition of two C-nucleotides to the arginyl-tRNATCG minihelix, but diffraction was poor. Nevertheless, we found that using a minihelix already containing the first C nucleotide (C74) improved diffraction to 3.2 Å (miniUR-C+CTP) (Table S1). In this case, the enzyme catalyzed the incorporation of one additional C-nucleotide, resulting in a minihelix ending in -CC. Strikingly, the addition of CTP resulted in a completely remodeled tRNA acceptor stem (Figure 1C and 1D). The original discriminator base G73 moved down by three positions to base pair with C3, resulting in a three-nucleotide bulge (A70-C72) protruding from the RNA helix. Apart from this bulge, which includes the originally mismatched (A70) and wobble (U71) bases, the RNA conformation was virtually unchanged. Two new base pairs G1:C75 and G2:C74 were formed, resulting in a blunt end close to the enzyme (Figure 1C). To determine whether full-length tRNA is able to form a bulge in the same position, we superimposed full-length tRNA (Protein Data Bank [PDB] code 1SZ1) (Xiong and Steitz, 2004) onto the miniUR-C+CTP structure (Figure S1C). The RNA bulge fits in the space between the enzyme and the anticodon arm, strongly suggesting that an unstable full-length tRNA can form an RNA bulge in the same position. Unfortunately, in the miniUR-C+CTP structure, the enzyme was captured in a catalytically impaired state, as the incoming nucleotide is not properly positioned in the active site. This resembles a complex of the enzyme with either nucleotide alone or with no RNA substrate (Figure S1D) (Tomita et al., 2006; Xiong et al., 2003). Further addition of ATP to our co-crystallization experiments did not yield an actively engaged complex. Although CTP addition results in a catalytically competent preinsertion complex when a stable substrate (denoted miniR) is used (PDB code 2DR5) (Tomita et al., 2006), we observed dramatic refolding of the RNA after CTP addition to an unstable substrate (Figure 1E). This clearly demonstrates that catalysis is necessary for the CCA-adding enzyme to detect unstable RNAs.
Clockwise RNA Overwinding and Compression upon Active Site Closure
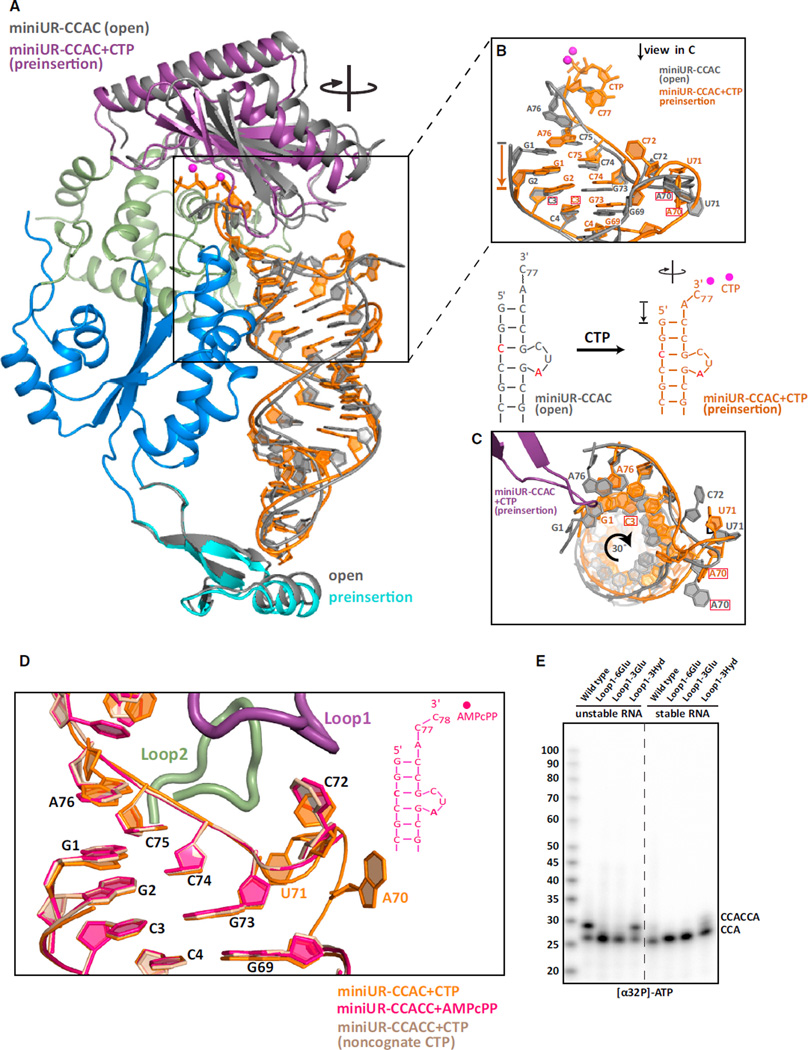
As we were unable to study the second CCA addition cycle by simply supplying nucleotides to the enzyme bound to unstable RNAs (Figures 1C and 1D), we incorporated CCAC at the 3′-end of our unstable model tRNA minihelix (miniUR-CCAC). This RNA was predicted to fold into a single species with a 3nt bulge (Zuker, 2003), reminiscent of our miniUR-C+CTP structure (Figure 1C). In the absence of CTP, triclinic, plate-like crystals formed, and intensity statistics confirmed pseudo-merohedral twinning, accounted for by twin refinement in PHENIX (Adams et al., 2010). Addition of CTP (miniUR-CCAC+CTP) eliminated twinning and the crystals diffracted to 3.15 Å (Table S1). The entire RNA is visible in both structures (Figures 2A and S2A). Prior to nucleotide binding (miniUR-CCAC/open complex), the CCA-adding enzyme adopts an open, inactive conformation consistent with what we observed for the miniUR complex (Figure 1A) and what is seen for stable RNAs (Tomita et al., 2006). Confirming the RNA folding prediction, a three nucleotide bulge (nucleotides 70–72) protrudes from the helix and does so in the same location as seen after CTP addition to the unstable RNA (miniUR-C+CTP) (Figures 1C and 1D).
Figure 2. Clockwise Screw Motion and RNA Compression upon Active Site Closure Structural transitions upon active site closure.
(A) RNA and protein domains of the open complex (miniUR-CCAC) are shown in cartoon representation in gray. The preinsertion complex (miniUR-CCAC+CTP) is colored as in Figure 1A with miniUR-CCAC+CTP RNA, including the incoming CTP, in orange. Catalytic magnesium ions are in magenta. The clockwise screw motion of the head domain and RNA is indicated.
(B) Close-up view of the acceptor stem of miniUR-CCAC (gray) and miniUR-CCAC+CTP (orange). The A70 mutation is boxed in red. RNA compression is indicated by an arrow.
(C) Top-down view of (B) including the miniUR-CCAC+CTP β-hairpin (purple). Clockwise RNA rotation is indicated.
(D) Location of the RNA bulge in all three preinsertion complex structures of the second CCA cycle. Common nucleotides to all structures are numbered in black, unique ones follow the RNA color scheme. Loops 1 and 2 are shown for miniUR-CCAC+CTP only.
(E) In vitro CCA-addition assays were performed to assay the ability of AfCCA containing Loop 1 insertions to incorporate [α-32P] ATP onto stable or unstable (G70A) arginyl-tRNATCG minihelices ending in -C. The insertion of three or six glutamates abolished the ability of the CCA-adding enzyme to mark unstable RNAs for degradation by adding CCACCA.
CTP binding to the active site (miniUR-CCAC+CTP/preinsertion complex) leads to closure of the head domain over the RNA double helix (Figure 2A), a process that was previously likened to the closure of a clam shell (Tomita et al., 2006). This leads to compression of the RNA by an entire base-pair step such that G1 of the preinsertion complex is located in the same position as G2 of the open complex (miniUR-CCAC) (Figure 2B; Movie S1). The average helical rise from G1:C72 through C48:G64 decreases from 3.2 Å to 3 Å, accompanied by RNA overwinding of 3° per base-pair step (Movies S1 and S2). Unlike a clam shell, our data suggest that active site closure exhibits a clockwise screw motion of the entire head domain, leading to RNA overwinding (Figure 2C; Movie S2). A similar degree of overwinding was also observed when we reanalyzed active site closure over stable tRNA minihelices (Tomita et al., 2006). Although significant compression occurs in the acceptor stem, the TΨC arm of the minihelix is rather strikingly fixed and binds the tail domain of the enzyme in a similar manner before and after CTP addition (Figure 2A).
Upon comparing the location of the RNA bulges in the cytosine preinsertion complex (miniUR-CCAC+CTP, Figure 2) with two different adenosine preinsertion complexes (miniUR-CCACC+ AMPcPP and miniUR-CCACC+CTP, Figure S2B), we found that the RNA was present in by and large the same conformation regardless of how many nucleotides were added at the 3′ end (Figure 2D). However, the bulges exhibited varying degrees of disorder. A surface loop in the head domain (Loop1, residues 118–126) stabilizes nucleotide 72 (C72) through a weak hydrogen bond between N4 of C72 and the carbonyl oxygen of Lys124. A second surface loop (Loop2, residues 215–223) in the enzyme’s neck domain is oriented toward, but does not bind nucleotide 71. As Loop 1 only interacts with the RNA when it is present in the bulged conformation, we reasoned that inserting additional residues into Loop1 may sterically block bulge formation (thereby blocking CCACCA addition), but not affect the first CCA addition cycle. Three different insertions into Loop1 were tested: (1) insertion of six negatively charged glutamates that are predicted to repel the bulged RNA phosphates close to the enzyme, (2) insertion of three glutamates, and (3) insertion of three large hydrophobic residues (Phe-Leu-Trp) (Figure S2C). Although inserting the hydrophobic residues (Loop1-3Hyd) had no effect on tandem CCA addition, insertion of negatively charged glutamates (Loop1-6Glu and Loop1-3Glu) completely blocked the second CCA-addition cycle, whereas the first CCA triplet was readily added (Figure 2E). As our model arginyl-tRNATCG minihelix contains the wobble base pair G2:U72, which may disproportionally favor refolding, we confirmed that two additional minihelices lacking this wobble pair gave identical results (Figure S2D).
We conclude that bulge formation is critical for tandem CCA addition and RNA surveillance of unstable RNAs. Furthermore, RNA compression and overwinding, induced by the head domain acting as a “molecular screwdriver,” appears to be a way for the CCA-adding enzyme to select for proper substrates by challenging the stability of the substrates.
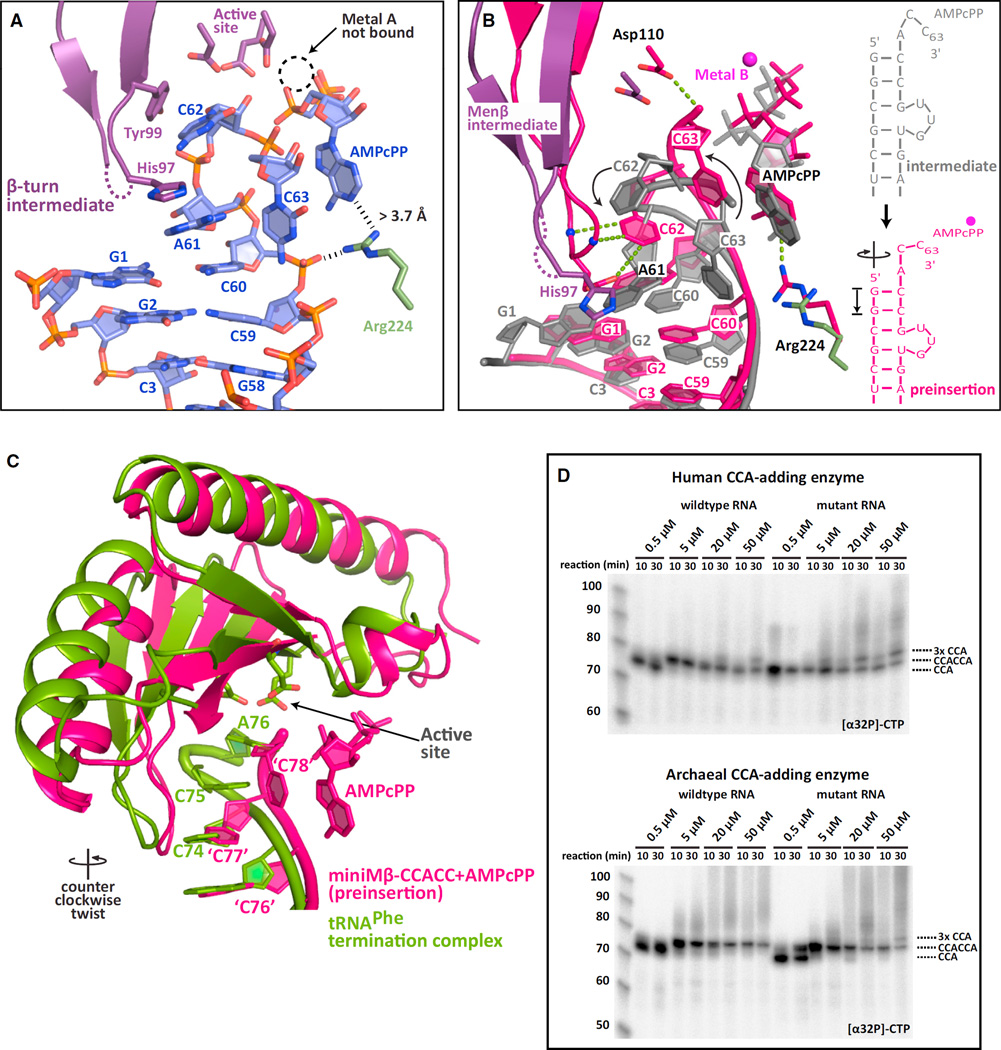
Menp τPNA-like Small RNA Behaves Like an Unstable tRNA and Is Held onto the CCA-Adding Enzyme through Ionic “Tweezers”
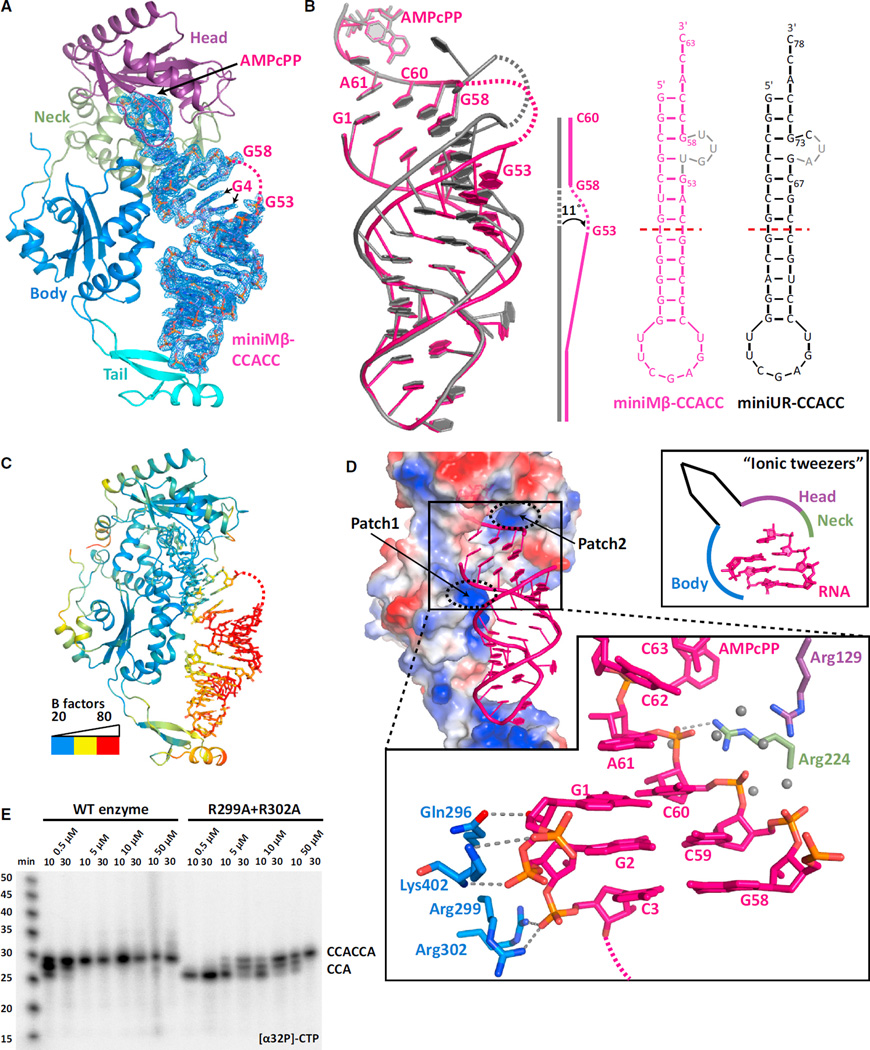
Upon comparing cytosine selection and incorporation during the first and second CCA cycle, we noticed a strict conservation of the catalytic mechanism between the two cycles (Figure S3A). Since the same holds true for adenosine incorporation (Figure S2B), we were surprised to observe a kinked minihelix in the adenosine preinsertion complex of AfCCA bound to a human Menβ-derived minihelix (miniMβ-CCACC+AMPcPP) (Figures 3A and S3B). Despite diffraction to 2.6 Å (Table S1), the entire 3nt RNA bulge and the adjacent base U54 are invisible. In addition, G4 shows two distinct conformations (Figures 3A and 3B) at the center of a clear 11° kink in the helical axis of the Menβ minihelix (Figure 3B). Nevertheless, the two ends of the Menβ minihelix, the three 5′-terminal “acceptor-stem” base pairs and the “TΨC-arm” base pairs, superimpose perfectly with the equivalent features of the unstable arginyl-tRNATCG minihelix structure (miniUR-CCACC+AMPcPP) (Figure 3B). Since the kink essentially uncouples the acceptor stem from the TΨC arm, it seemed that the tail domain may not actually be anchoring the RNA on the enzyme, as has been previously proposed (Cho and Weiner, 2004; Tomita et al., 2006), during the second cycle of CCA addition.
Figure 3. Unstable RNA Is Held onto the CCA-Adding Enzyme through Ionic “Tweezers”.
(A) Structure of the adenosine preinsertion complex of the Menβ tRNA-like small RNA (miniMβ-CCACC+AMPcPP), as in Figure 1A. The disordered RNA bulge is dashed and colored in pink.
(B) Comparison between the miniMβ-CCACC+AMPcPP and miniUR-CCACC+AMPcPP RNA structures. Both RNAs are schematically diagrammed on the right with disordered nucleotides in gray. The kink in miniMβ-CCACC RNA is sketched.
(C) Temperature factor distribution of miniMβ-CCACC+AMPcPP. Low temperature factors of 20 Å2 are in blue, intermediate values in yellow, and values above 80 Å2 in red.
(D) Electrostatic surface potential of the archaeal CCA-adding enzyme. Blue depicts positively charged, white neutral, and red negatively charged areas. The two positively charged patches holding the acceptor stem are indicated. MiniMβ RNA is in cartoon and colored in pink. The close-up view details the ionic interactions that glue the top of the acceptor stem to the enzyme. A schematic of the “ionic tweezers” is shown.
(E) In vitro CCA-addition assays using wild-type or mutant AfCCA and the unstable (G70A) arginyl-tRNATCG minihelix (miniUR) substrate. Weakening the strong interactions between the RNA 5′-end and the enzyme impaired the second CCA-addition cycle, whereas the first cycle was unaltered.
See also Figure S3.
To gain insights into whether a kinked Menβ tRNA-like substrate conforms to the normal CCA-adding enzyme mechanism, we plotted the temperature-factor distribution of the entire mini-Mβ-CCACC+AMPcPP complex (Figure 3C and S3C). Strikingly, the most stable region of the entire complex is the active site cleft and the adjacent top part of the acceptor stem. The tail domain and the TΨC arm, on the other hand, are highly mobile. Analogous results were obtained for canonical tRNA minihelices undergoing the second CCA cycle (miniUR-CCACC+AMPcPP and miniUR-CCACC+CTP), which all exhibit identical crystal packing (Figure S3D). In contrast, the tail domain and the TΨC arm are both stable during the first CCA addition cycle (Figure S3E). Therefore, it appears that the tail domain does not serve as the major RNA anchor during the second CCA addition cycle.
Why then is the top of the acceptor stem so stable? Upon calculating the electrostatic surface potential of the CCA-adding enzyme, we noticed two positively charged surface patches binding the top of the acceptor stem (Figure 3D). Patch1, consisting of body domain residues Gln296, Arg299, Arg302, and Lys402, binds the 5′-end of Menβ via direct ionic interactions to bridging RNA phosphates. Patch2 is comprised of Arg129 from the head domain and the catalytically essential Arg224 from the neck domain. Interestingly, most contacts on this 3′-side are water-mediated and, therefore, weaker than on the 5′-side (Figure 3D). To address whether those ionic interactions keep the unstable RNA in place for the second CCA-addition cycle, we mutated both Arg299 and Arg302 to alanine and tested enzymatic activity in vitro (Figure 3E). While the wild-type enzyme was able to extend the unstable arginyl-tRNATCG minihelix to CCACCA at a nucleotide concentration of 0.5–5 µM, the mutant enzyme required 10–100× higher concentration before switching from CCA to CCACCA addition (Figure 3E). These results are consistent with Patches1+2 forming “ionic tweezers” (Figure 3D, schematic) that ensure the second CCA cycle proceeds efficiently.
Apart from a kink in the acceptor stem helix, we conclude that tandem CCA addition proceeds in a highly similar fashion for canonical tRNAs and tRNA-like small RNAs. Furthermore, it is clear that the tail domain of the CCA-adding enzyme has two separate functions. First, it serves as a ruler to ensure selective binding of tRNA-type substrates. Second, it allows torque to be applied on the helix upon active site closure and thereby contributes to the discrimination between stable and unstable RNA substrates.
On-Enzyme RNA Refolding Permits the Second CCA Addition Cycle
As the CCA-adding enzyme is renowned for its precision in terminating synthesis after a single CCA triplet, we were eager to determine how it transitions from the first to the second cycle of CCA addition. For unstable RNAs, we hypothesized that this transition may occur in one of two ways: (1) the RNA may dissociate from the enzyme after the first round of catalysis and rebind in the bulged conformation to initiate the second cycle or, alternatively, (2) the RNA may refold while bound to the enzyme.
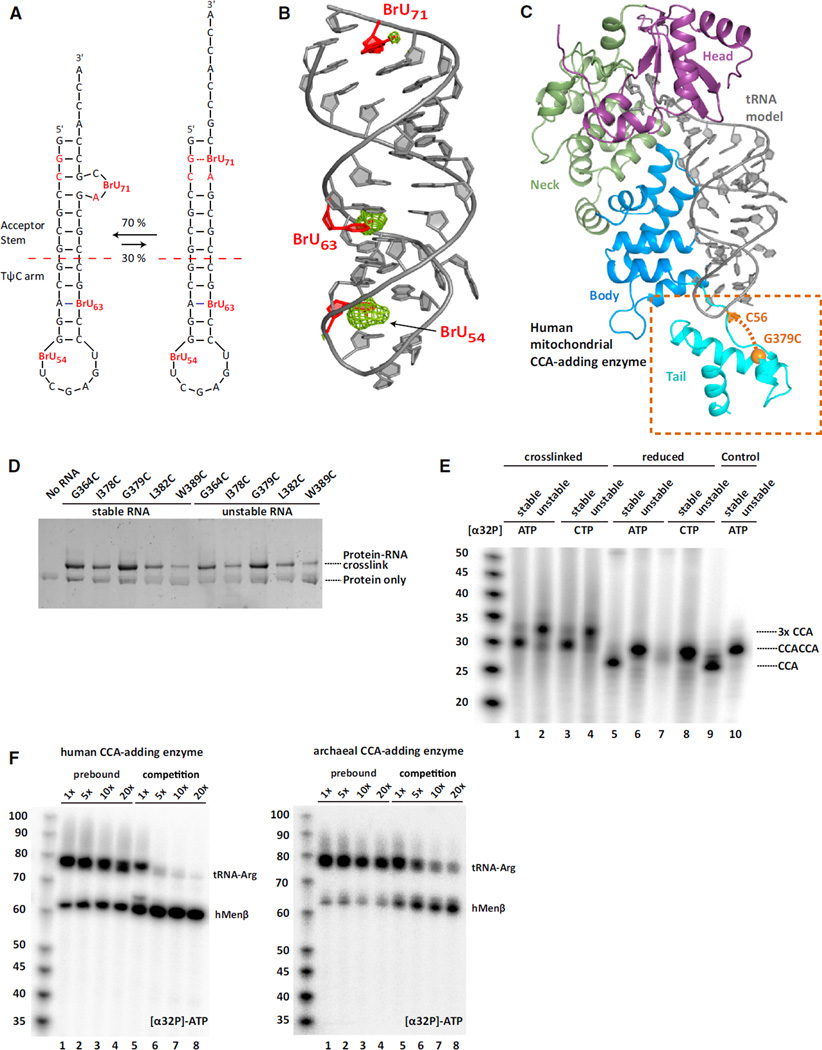
Initial support for the latter model came from determining the structure of AfCCA in complex with the mutant arginyl-tRNATCG minhelix ending in CCACCA that was bromouridine (BrU)-labeled at nucleotides 54, 63, and 71(miniUR-CCACCA) (Figure 4A). Two strong anomalous signals were detected at the expected positions for BrU54 and BrU63 (Figure 4A). Surprisingly, we also observed a weak anomalous peak for BrU71 near the top of the acceptor stem. This result was unexpected since U71 falls within the location of the bulge, which was disordered in this structure. The only reasonable explanation is the presence of two alternative RNA conformations, one representing the RNA prior, and one after refolding. Indeed, we were able to refine the non-refolded state of the RNA to an occupancy of 30%, thereby providing an explanation for the weak anomalous signal. The remaining 70% is disordered in the bulge. Considering that the electron density for the tRNA 5′-end was unambiguous in this structure (as in all other structures of this study) and that the tRNA 5′-end is tightly bound by the enzyme (Figures 3C–3E), we hypothesized that the tRNA 3′-end refolded independently of the 5′ -end to allow addition of the second CCA.
Figure 4. On-Enzyme RNA Refolding Permits the Transition from the First to the Second CCA Cycle.
(A) The two interchangeably bound RNA conformations observed in bromouridine (BrU)-labeled miniUR-CCACCA co-crystals. BrU and RNA instabilities are highlighted in red.
(B) The double-helical part of the non-refolded RNA from (A) is shown in cartoon representation and colored in gray. BrU are in red. The anomalous difference Fourier map is colored in green and contoured at 3.5 σ.
(C) Structure of the full-length human mitochondrial CCA-adding enzyme. Protein domains are colored according to the archaeal enzyme in Figure 1A. The newly resolved tail domain is highlighted. A tRNA minihelix is modeled according to PDB code 1VFG (Tomita et al., 2004). An ethylthio-purine crosslinker was incorporated at tRNA position C56 and crosslinked to the CCA-adding enzyme carrying a G379C mutation.
(D) Silver-stained SDS-PAGE of the crosslinked human CCA-adding enzyme and the ethylthio-purine-derivatized minihelices. Crosslinked complexes show retarded gel mobility, with the G379C and G364C mutants showing the highest crosslinking efficiency.
(E) In vitro CCA addition assays with crosslinked complexes. The wild-type and G70A mutant arginyl-tRNATCG minihelices are denoted as stable and unstable, respectively. Reactions 2–5 were under oxidizing, whereas reactions 6–10 were under reducing conditions. Radioactive nucleotides are indicated above the lanes. Unstable minihelices that had never been crosslinked were used as controls (reactions 9–10).
(F) In vitro competition experiments. In lanes 1–4, full-length arginyl-tRNATCG (G70A) was pre-bound to the human and archaeal CCA-adding enzyme in the presence of 2 µM CTP before adding cold ATP supplemented with [α32P]-ATP and varying amounts of the hMENβ transcript (up to a 20-fold excess). In lanes 5–8, both RNAs were added simultaneously along with ATP. An equal amount of arginyl-tRNATCG was added in all lanes, whereas hMENβ amounts varied as denoted at the top. Pre-bound RNA was efficiently extended to CCACCA irrespective of added excess RNA, indicating that the bound RNA refolds “on-enzyme.” When added simultaneously, both RNAs compete for binding to the CCA-adding enzyme.
See also Figure S4.
To test this model, we used two different approaches: first by covalently tethering the RNA to the enzyme, and second using a competition experiment. For the first, we were inspired by earlier work on DNA (Huang et al., 1998) and we set out to reversibly crosslink the CCA-adding enzyme to the unstable tRNA using a site-specific link. We reasoned that if tandem CCA addition proceeded efficiently when the RNA substrate and CCA-adding enzyme were covalently joined, refolding most likely occurred “on-enzyme.”
To engineer a covalent complex with a site-specific linkage, a purine nucleoside analog bearing a short linker terminating in a free thiol was incorporated at position 56 of the TΨC arm of stable (WT) and unstable (mutant) arginyl-tRNATCG minihelices (Figure S4A) (Peacock et al., 2011). The TΨC arm seemed to be the best location for not interfering with catalysis, as much of the rest of the minihelix is compressed prior to catalysis (this study and Tomita et al., 2006). Likewise, Asp351 and Arg344 of the AfCCA tail domain are located closest to the crosslinkable nucleotide and were therefore replaced with cysteines to allow a protein-RNA disulfide crosslink to form, a prediction we confirmed experimentally (Figure S4B).
Since tRNA surveillance through CCACCA addition is conserved across all kingdoms of life (Wilusz et al., 2011), we also wanted to include class II CCA-adding enzymes. Given a lack of sufficient structural information on class II enzymes, the structure of the full-length human mitochondrial CCA-adding enzyme was determined to 1.9 Å (Figure S4C). Although the tail domain was disordered in Augustin et al. (2003), it is clearly resolved in our structure, and we were able to construct a model of a tRNA-bound complex (Figure 4C). Based on this model and allowing for conformational changes in the protein, several cysteine mutants were tested for their ability to form disulfide bonds with the crosslinkable tRNA. Two mutants, G364C and G379C, crosslinked efficiently to both stable and unstable tRNA minihelices (Figure 4D).
We next purified these crosslinked complexes to test whether RNA refolding occurs on the enzyme in a standard CCA addition assay (Shi et al., 1998b). Reactions were performed in the presence of 2 µM cold ATP and CTP, combined with either radioactive ATP or CTP to measure A or C incorporation, respectively. Since the minihelices used ended in -CC (miniUR-CC), further C incorporation served as a sensitive readout of refolding, as it should only occur during the second CCA cycle, after the substrate has successfully completed the first cycle. A stable tRNA minihelix (miniR-CC) was used as a control as it should terminate synthesis after the first CCA cycle and thus not incorporate CTP.
When we measured radioactive ATP incorporation onto the 3′-end of stable and unstable human complexes, we detected a lower molecular weight product for the stable tRNA complex and a higher molecular weight product for the unstable tRNA complex, as would be expected for CCA versus CCACCA addition (Figure 4E, lanes 1 and 2). Surprisingly, similar results were obtained using radioactive CTP (Figure 4E, lanes 3 and 4), despite our expectation not to detect CTP incorporation into stable tRNAs ending in -CC. Upon reversing the crosslink by adding 20 mM DTT prior to performing the CCA addition assay, CTP incorporation was detected only onto the unstable tRNA, indicating that the stable tRNA now completed only the first CCA cycle (Figure 4E, lanes 7 and 8). Interestingly, all RNA products were smaller under reducing (Figure 4E, lanes 5–8) versus oxidizing conditions (Figure 4E, lanes 1–4), with the products under reducing conditions running at the same sizes as never-cross-linked reaction controls (Figure 4E, lanes 9 and 10). We therefore conclude that crosslinking the human CCA-adding enzyme to its RNA substrate resulted in tandem CCA addition to stable RNAs and three CCA triplets being added to unstable RNAs. Similar results were obtained with the crosslinked archaeal enzyme, although the difference between stable and unstable RNA was less pronounced (Figure S4D). We should note that upon cross-linking the substrate to either enzyme and increasing the nucleotide concentration to 50 mM, both enzyme classes behaved like “oligo-CCA”-adding enzymes and added extended CCA tails to both stable and unstable RNAs, with unstable RNAs harboring longer tails on average (Figure S4E). This “oligo-CCA” addition is not observed in vivo (Wilusz et al., 2011) since refolding and CCA addition rates are much higher for crosslinked complexes compared to those of the uncrosslinked versions.
For the competition experiment, we reasoned that if the RNA refolded between the two CCA-addition cycles while bound to the enzyme, a pre-bound tRNA would not be competed off the enzyme by another RNA when transitioning from the first to the second CCA cycle. We, therefore, pre-bound unstable full-length arginyl-tRNATCG (G70A) ending in its discriminator base to either the human or the archaeal CCA-adding enzyme in the presence of CTP only. After having incorporated two C nucleotides, we expected the enzymes to remain bound to their substrate RNAs, awaiting ATP to complete the first CCA cycle. ATP was then added simultaneously with an excess of full-length human Menβ tRNA-like small RNA already ending in CCA, and product formation was monitored. Consistent with the tRNA refolding on enzyme, pre-binding of unstable arginyl-tRNATCG to both classes of enzymes rendered the enzymes “immune” toward excess RNA and resulted in similar amounts of mature arginyl-tRNATCG product, irrespective of the competing Menβ concentration (Figure 4F, lanes 1–4). In contrast, when both arginyl-tRNATCG and Menβ substrates ending in -CC were added at the same time, final products were predominantly synthesized for Menβ, the substrate present in excess (Figure 4F, lanes 5–8). We note that human Menβ is a better substrate for the human CCA-adding enzyme compared to the archaeal enzyme (Figure 4F). Also, when the archaeal reaction scheme was inverted such that human Menβ was pre-bound and arginyl-tRNATCG served as the competitor, we observed a less pronounced trend, since Menβ is not as good a substrate as arginyl-tRNATCG (Figure S4F).
Taken together, it is clear that tandem CCA addition does not involve RNA dissociation. Instead, RNA refolding takes place “on-enzyme.”
Active Site Closure Gauges RNA Stability and Triggers Refolding
What triggers “on-enzyme” RNA refolding to enable the second CCA cycle? Termination of the first CCA cycle, which precedes refolding, has previously proven difficult to study. It is, however, known that in the termination state, the entire CCA triplet stacks onto the acceptor stem and abuts against the β sheets containing the active site residues, thereby blocking the addition of further nucleotides (Xiong and Steitz, 2004).
Insight regarding how refolding is triggered was gained from a 2.7 Å structure of a monoclinic crystal of the Menβ tRNA-like small RNA adenosine preinsertion complex (miniMβ-CCACC+ AMPcPP-i). Most of the RNA bound in the active site pocket of this complex is ordered, despite the pocket not being entirely closed (Figures 5A and S5A). As expected for a substrate ending in CCACC, the tRNA-like transcript is bound in its refolded state, ready for the second CCA cycle to be completed with the addition of an A. Nucleotides C59 and C60 (equivalent to C74 and C75 for tRNAs) form strong base pairs to G1 and G2. A61 (A76 for tRNAs), the last A of the first CCA triplet, is in a similar position to a discriminator base. However, its base edge projects toward a β-hairpin that has been previously shown to be critical for catalysis and proofreading (Cho et al., 2005). This A also stacks with His97, thereby displacing Asp96 and Ala95. C62 (C77 for tRNAs), the first C of the second cycle, packs against Tyr99, whereas C63 (C78) folds back onto the 3′-end of the acceptor stem, lying orthogonal to it and tucking against Arg129 (Figures 5A and S5A). Although electron density for the C63 base is weak, its 3′-hydroxyl group is clearly not in a position to attack the α-phosphate of the incoming nucleotide. Interestingly, the incoming AMPcPP is also not properly locked into its binding pocket as its base edge is not recognized by Arg224, nor do the three phosphate groups coordinate metal ion A.
Figure 5. Active Site Closure Gauges RNA Stability and Triggers Refolding.
(A) The intermediate adenosine preinsertion complex of Menβ (miniMβ-CCACC+AMPcPP-i) shown as in Figure 1A. Residues contributed by the head domain are in atom colors with carbons in purple. The disordered residues Ala95 and Asp96 are dashed in purple. Arg224 from the neck domain is in atom colors with carbons in green. A dashed circle highlights the missing Metal B. Improper positioning of the incoming nucleotide is shown with black dashes.
(B) Superposition of the adenosine intermediate and preinsertion complexes (miniMβ-CCACC+AMPcPP). The intermediate state RNA and AMPcPP are in gray, while the intermediate β-hairpin and Asp110 are in purple. Intermediate Arg224 is in green. The entire preinsertion complex (miniMβ-CCACC+AMPcPP) is in pink. Three proofreading interactions, the AMPcPP base edge recognition and the Asp110 general base contact to the 3′-hydroxyl are shown with green dashed lines for the preinsertion complex, as is Metal B bound to AMPcPP.
(C) Comparison of the miniMβ-CCACC+AMPcPP complex and the termination complex of the first CCA cycle (PDB code 1SZ1) (Xiong and Steitz, 2004). The incoming AMPcPP is depicted for Menβ. Canonical tRNA numbering is used for Menβ to simplify comparison. The active site residues that the RNA stacks against in the termination complex are shown as sticks. The counter-clockwise twist between the Menβ preinsertion complex and the tRNA termination complex is indicated.
(D) In vitro CCA addition assays with full-length stable and unstable (G70A) arginyl-tRNATCG and either the human or the archaeal CCA-adding enzyme. Cold nucleotide concentrations (ATP+CTP) were increased in the presence of radioactive CTP. CCA tail length is indicated.
When we compared this structure to the proper adenosine preinsertion complexes of Menβ (miniMβ-CCACC+AMPcPP, Figure 3A) and unstable tRNA (miniUR-CCACC, Figure S2B), we realized that the observed state represents a functional intermediate between the open (inactive) and the closed preinsertion complexes. The bound small RNA exhibits an intermediate degree of compression and the β-hairpin displays an intermediate degree of closure (Figure S5B). Similar to our observations for unstable tRNA (Figure 2), the transition from the intermediate to the preinsertion complex revealed a coordinated clockwise screw motion of the head domain, resulting in RNA overwinding and compression of the acceptor stem by an entire base pair step (Figure 5B; Movies S3 and S4). The head domain pushed the protruding nucleotide A61 of the intermediate state back into its proper position to stack onto the 3′-end of the acceptor helix, unstacking it from His97. The β-hairpin is now in a proper conformation to proofread C62, which undergoes a 90° rotation, whereas C63 reorients its 3′-hydroxyl toward Asp110, which serves as a general base during adenosine incorporation (Pan et al., 2010). The incoming AMPcPP moves to its proper preinsertion position, interacting with Arg224 and coordinating Metal B along with the catalytic residues Glu59 and Asp61. Interestingly, only the Menβ preinsertion complex shows a kinked helix, likely induced by complete active site closure (Figures 3A and Figure S5C).
The apparent correlation between active site closure and RNA compression indicates that apart from being essential for catalysis, active site closure is the way by which the stability of the RNA is assessed. Following the first CCA addition, the torque applied on the RNA double helix essentially gauges weak spots in the duplex and causes refolding of the RNA in the form of extrusion of a bulge.
Nucleotide Binding Is Critical for Triggering RNA Substrate Refolding
Three lines of evidence suggested that nucleotide binding is critical to trigger refolding. First, our ability to isolate an intermediate complex is likely attributed to the presence of a sub-optimally positioned nucleotide in the active site pocket. Proper binding of the triphosphate moiety locks the enzyme in the catalytically active preinsertion state. Second, the termination state after the first CCA cycle (PDB 1SZ1) (Xiong and Steitz, 2004) has sufficient space for an incoming nucleotide, which could start off the second CCA-addition cycle (Figure 5C). Finally, since active site closure is accompanied by a clockwise rotation, we reasoned that the termination complex is formed from the adenosine pre-insertion complex by a counter-clockwise motion of the head domain, which appears to be the case (Figure 5C). This movement is essential to reset the enzyme prior to a new cycle of CCA addition.
As nucleotide binding is critical to trigger refolding, we hypothesized that increasing the nucleotide concentrations in standard CCA-addition assays (Shi et al., 1998b) may lower the threshold of instability needed in a tRNA acceptor stem for CCACCA addition to occur. Using a nucleotide concentration of 0.5 µM, we recapitulated what is seen in vivo for full-length wild-type or mutant arginyl tRNA (Figure 5D). However, at nucleotide concentrations above 5 µM, the wild-type tRNA was converted into a tandem CCA target by both classes of CCA-adding enzymes (Figure 5D). Similar results were obtained for minihelix substrates (Figure S5D). At nucleotide concentrations above 20 µM, we detected even further extension of the CCA tails, reminiscent of the results obtained with crosslinked complexes (Figure 4E). Most surprisingly, both enzyme classes converted stable tRNAs into CCACCA targets at identical nucleotide concentrations, which was unexpected since tRNA approaches their respective active sites from entirely different directions (Xiong et al., 2003). Whereas the extruded RNA bulge tucks against the protein for class I enzymes (Figure 2), it almost certainly protrudes into the solvent in class II enzymes (Figure S5E).
Additional tRNAs were subsequently tested to determine whether sequence elements within the tRNA may affect the ability of nucleotide concentration to regulate CCA versus CCACCA choice. As in our model arginyl-tRNATCG substrate, wild-type cysteinyl-tRNAGCA contains a wobble base pair in its acceptor stem and was converted into a tandem CCA target at a nucleotide concentration of 5 µM (Figure S5F). On the other hand, wild-type tRNAs with acceptor stems containing only Watson-Crick base pairs required nucleotide concentrations to be at least ten times higher (50 µM) before they were converted into CCACCA targets (Figure S5G and S5H).
We suggest that the choice between CCA versus CCACCA addition is made when a nucleotide binds to the active site in the termination state following the first CCA addition cycle. For stable substrates, clockwise active site closure triggered by nucleotide binding leads to RNA dissociation as the energetic cost of breaking multiple Watson-Crick base pairs is too high. Unstable RNAs, however, will refold “on-enzyme” and a second CCA triplet will be added. Elevated nucleotide concentrations shift the reaction equilibrium toward tandem CCA addition with the actual concentration wherein this switch occurs dependent on acceptor stem stability. Elevated nucleotide concentrations might favor a closed active site, which would trap even transiently refolded stable substrates after the first CCA cycle on the enzyme by adding the first C of the second cycle.
RNAs with Longer Bulges Are Not Readily Accommodated by the CCA-Adding Enzyme
Why, however, is CCA addition limited to a tandem triplet in vivo? Although extended tails longer than CCACCA are efficiently added under certain conditions in vitro (higher nucleotide concentrations or when the RNA is covalently bound to the enzyme), they are only rarely added in vivo (Wilusz et al., 2011, 2012). To understand why, we carried out fragment-based assembly of RNA with full atom refinement (FARFAR), a ROSETTA-based de novo RNA prediction algorithm (Das and Baker, 2007; Das et al., 2010). Using the miniUR-CCAC+CTP complex as a template (Figure 2), 5,000 structures were calculated assuming either a 3nt (for tandem CCA) or a 6nt bulge (for three CCAs). As expected, models containing a 6nt bulge deviated more significantly from the template structure and none formed a continuous helix (Figure S6A). In contrast, about a third of the models with a 3nt bulge exhibited a continuous helix (Figure S6B).
We posit that in order to apply torque onto the acceptor stem to trigger refolding, a continuous helix would be necessary. From these calculations, it appears that the reason that three CCAs are not accommodated is because the resulting RNA molecules are unable to form continuous helices.
DISCUSSION
Model of Tandem CCA-Addition
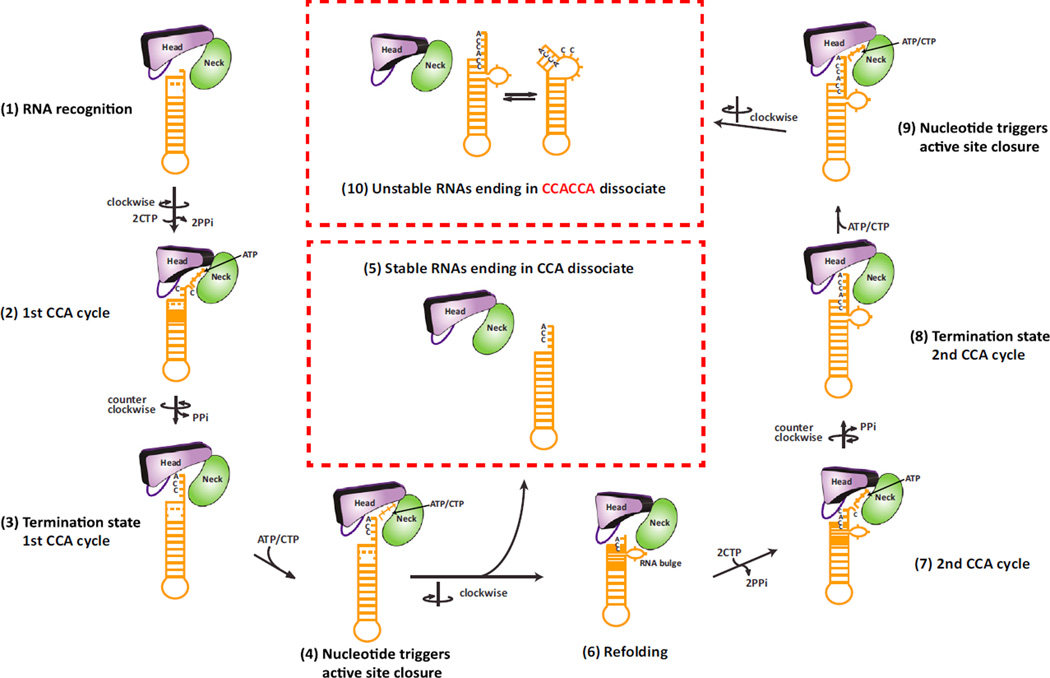
tRNA surveillance through tandem CCA addition is a universally conserved mechanism by which the cell distinguishes between stable and certain unstable tRNAs and tRNA-like transcripts (Wilusz et al., 2011). While stable tRNAs receive only CCA, unstable tRNAs beginning with GG are marked with CCACCA with near 100% efficiency and rapidly degraded (Wilusz et al., 2011). We show that the structural flexibility of RNA controls the choice between CCA versus CCACCA addition and propose the following model for tandem CCA addition (Figure 6): (1–3) a proper tRNA-type substrate is first recognized by the CCA-adding enzyme. The discriminator base is inserted between the head and neck domains, while the TΨC loop interacts with the enzyme’s tail domain (Figure 1A). The first CCA addition cycle then proceeds as has been established (Pan et al., 2010; Tomita et al., 2006; Xiong and Steitz, 2004). Following the addition of the terminal A, pyrophosphate is released and the head domain rotates counter-clockwise to allow continuous stacking of the CCA triplet against the head domain (Figure 5C). (4) Nucleotide binding at this stage induces the clockwise closure of the active site with torque applied on the RNA duplex (Figures 2 and 5). This “interrogation” of the RNA by the enzyme results in two possible outcomes: (5) if the bound RNA is stable, it will dissociate from the enzyme since breaking multiple stable Watson-Crick base pairs for RNA refolding is unfavorable; (6) however, if the RNA is unstable, the screw motion will trigger on-enzyme refolding of the RNA 3′-end, in the form of extrusion of a bulge, while the 5′-end and the TΨC loop remain bound to the enzyme (Figure 4). Refolding thus leads to a structure that is reminiscent of a misaligned zipper with the bulged nucleotides protruding from an otherwise continuous RNA helix (Figures 2 and 3). This bulged substrate is stabilized by two new base pairs that form between the Cs added during the first CCA cycle and the 5′-terminal Gs and further stabilized by ionic interactions to the enzyme (Figure 3) as well as interactions between the bulge and loop 1 of the head domain. These interactions help lower the energy for the refolded conformation. (7–8) Once the active site is closed over the refolded RNA, the second cycle of CCA addition proceeds analogous to the first. Upon termination, the second CCA triplet again stacks against the head domain following a counter-clockwise rotation. (9) Nucleotide binding could then, in principle, result in yet another round of RNA refolding (10). However, RNA refolding at this stage would result in a 6nt bulge, a structure that is unable to accommodate a continuous helix, which is required for the RNA to remain bound to the enzyme (Figure S6). Therefore, the screw motion for active site closure and torque applied on the RNA following the second CCA-addition cycle induces dissociation of the RNA carrying CCACCA at its 3′ end.
Figure 6. Model of RNA Surveillance by the CCA-Adding Enzyme.
The head and neck domains of the CCA-adding enzyme are shown as abstract bodies colored in purple and green, respectively, with the β-hairpin as an oval.
Bound unstable RNA is shown in orange with its compression state changing during the CCA-addition cycles. Steps 1–10 are described in the text.
See also Figure S6.
Proofreading Unstable RNAs through Their Structural Versatility
Proofreading is an important feature of all polymerases. As DNA and RNA polymerases scan along a template strand while synthesizing a product strand, they recognize misincorporated nucleotides or erroneous templates either before or immediately after nucleotide incorporation (Kunkel and Bebenek, 2000; Sydow and Cramer, 2009). These transcriptional obstacles are usually removed through endonucleolytic cleavage before nucleotide synthesis resumes.
In contrast, the CCA-adding enzyme uses proofreading on two different levels. First, it is able to add the nucleotides CCA in a template-independent manner to the 3′ ends of all tRNAs and tRNA-like molecules without translocating along its substrate (Pan et al., 2010; Toh et al., 2008; Tomita et al., 2006; Xiong and Steitz, 2004). Second, the enzyme exploits the structural versatility of unstable RNAs to mark them for degradation, even though the mutation or instability is never directly recognized by the active site of the enzyme. The enzyme simply sticks to its substrate requirements and catalytic principles regardless of the substrate. If a continuous A-form RNA helix of proper length is bound between the head and tail domains, and an unpaired discriminator base can be properly positioned in the active site, then the CCA-adding enzyme will add a CCA triplet to the RNA 3′-end. Rather than dissociating after the first CCA cycle, unstable RNAs are refolded in response to the regular screw motion of the enzyme during active site closure. CCA-addition terminates once the RNA substrate is unable to remain bound to the enzyme. We speculate that tandem CCA addition is the unavoidable consequence of CCA addition to a large subset of unstable RNAs beginning with GG, since refolding is triggered during catalysis. As it is driven by the RNA substrate itself and is beneficial for removing erroneous tRNAs, this mechanism is conserved across both classes of CCA-adding enzymes.
We suggest that RNA compression after completion of the first CCA cycle, which challenges RNA stability and triggers refolding of unstable RNAs, is an example of proof-reading via an energy relay mechanism, first proposed by Hopfield (1980). RNA compression as seen in the dynamic intermediate state of the enzyme is at a branch point, characteristic of these types of mechanisms. The RNA substrate at this branch point either “resists” and falls off the enzyme when it contains a stable acceptor stem, or “buckles,” extruding a bulge, and then receives another CCA when the acceptor stem is weakened due to a mismatch. The compression of the RNA provides the energy relay for proofreading in this case.
Our study elucidates how non-coding RNAs (ncRNAs) that utilize the tRNA fold are either marked for rapid degradation or stabilized by the addition of a single CCA triplet. Since bona fide tRNAs possess very long half lives (Kanerva and Mäenpää, 1981), we speculate that CCA-addition is a way for non-coding RNAs (ncRNAs) to increase their stability. Interestingly, while the Menβ tRNA-like small RNA is rapidly degraded due to CCACCA addition in humans and mice, the Old World Monkey homolog is stable and thus a CCA target (Wilusz et al., 2011). In the future, it will be instructive to study the functional significance of these differences. Furthermore, the surveillance mechanism presented here is conserved across all kingdoms of life and across different classes of RNA. This underlines the centrality of RNA structure in regulating transcript stability and function (Ding et al., 2014; Rouskin et al., 2014; Wan et al., 2014) and calls for a more detailed search into the structural elements of lncRNAs.
EXPERIMENTAL PROCEDURES
Structures of the Archaeal CCA-Adding Enzyme in Complex with Unstable RNAs
The A. fulgidus CCA-adding enzyme (AfCCA) was expressed in Escherichia coli and all RNAs used for crystallography were purchased from Dharmacon. Preinsertion complexes were obtained by co-crystallization with AMPcPP or CTP. Most reaction steps crystallized in 16%–24% PEG-3350 and 0.2–0.27 M sodium-potassium tartrate. Diffraction data were collected at beamline X25 of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. All structures were solved by molecular replacement using PHASER (McCoy et al., 2007) and refined with PHENIX (Adams et al., 2010). For details, see Extended Experimental Procedures.
Structure of the Full-Length Human CCA-Adding Enzyme
The human mitochondrial CCA-adding enzyme was expressed in E. coli and crystallized in 2.3 M ammonium sulfate and 0.5 M tri-sodium citrate pH 5.8. Its structure was solved by molecular replacement and refined with PHENIX (Adams et al., 2010). For details, see Extended Experimental Procedures.
Crosslinking Experiments, CCA-Addition Assays
A 2-amino-(S-trityl-ethylthio)-purine phosphoramidite was synthesized as described (Peacock et al., 2011). Crosslinkable RNA was prepared at ChemGenes. All in vitro assays were carried out with crosslinked and soluble components as described (Shi et al., 1998b). For details, see Extended Experimental Procedures.
Competition Experiments
To study the effect of RNA pre-binding, full-length arginyl tRNATCG (G70A) ending in its discriminator base was pre-incubated in the presence of cold CTP only. Cold ATP spiked with [α32P] ATP was added simultaneously with equimolar or excess amounts of full-length human Menβ already carrying CCA. Reactions were allowed to proceed for 2 min for AfCCA and 30 s for the human CCA-adding enzyme. As a control experiment, both RNAs were added simultaneously along with ATP. For details, see Extended Experimental Procedures.
Rosetta-Based RNA Modeling
De novo RNA folding was carried out using Rosetta 3.5 (http://www.rosettacommons.org) (Das et al., 2010). Detailed procedure in Extended Experimental Procedures.
Supplementary Material
Highlights.
The CCA-adding enzyme interrogates substrates like a nucleotide-driven molecular vise
Unstable RNAs refold “on-enzyme” after completion of the first CCA cycle
An RNA bulge permits addition of a second CCA triplet
RNAs proofread themselves through differential responses to the CCA-adding process
ACKNOWLEDGMENTS
We thank Annie Héroux for help with data collection at beamline X25 at the National Synchrotron Light Source, which is supported by the Department of Energy, Office of Basic Energy Sciences, Andrei Laikhter from ChemGenes for RNA synthesis, Alan Weiner for the AfCCA expression plasmid, and Phillip Sharp and members of the L.J. laboratory for discussions and advice. We thank J. Jansen for comments on the manuscript. C.-D.K. was supported by a Postdoctoral Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. Early stages of this research were performed at MIT by J.E.W. supported in part by NIH grants R01-GM34277 and R01-CA133404 to P.S. This work was supported by NIH grants R00-GM104166 (to J.E.W), R01-GM61115 (to P.A.B), the Robertson Research Fund of Cold Spring Harbor Laboratory and the Cold Spring Harbor Laboratory Women in Science Award (to L.J.). L.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors of the archaeal CCA-adding enzyme complexes have been deposited in the Protein Data Bank (PDB) under accession codes 4X4N, 4X4O, 4X4P, 4X4Q, 4X4R, 4X4S, 4X4T, 4X4U, and 4X4V. The full-length human mitochondrial CCA-adding enzyme has been deposited under the accession code 4X4W.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, four movies, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.01.005.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HD, Weiner AM. A single catalytically active subunit in the multimeric Sulfolobus shibatae CCA-adding enzyme can carry out all three steps of CCA addition. J. Biol. Chem. 2004;279:40130–40136. doi: 10.1074/jbc.M405518200. [DOI] [PubMed] [Google Scholar]
- Cho HD, Verlinde CL, Weiner AM. Archaeal CCA-adding enzymes: central role of a highly conserved beta-turn motif in RNA polymerization without translocation. J. Biol. Chem. 2005;280:9555–9566. doi: 10.1074/jbc.M412594200. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. tRNA nucleotidyltransferase. Enzymes. 1982;15:183–215. [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. The energy relay: a proofreading scheme based on dynamic cooperativity and lacking all characteristic symptoms of kinetic proofreading in DNA replication and protein synthesis. Proc. Natl. Acad. Sci. USA. 1980;77:5248–5252. doi: 10.1073/pnas.77.9.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to para-speckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kanerva PA, Mäenpää PH. Codon-specific serine transfer ribonucleic acid degradation in avian liver during vitellogenin induction. Acta Chem. Scand. B, Org. Chem. Biochem. 1981;35:379–385. doi: 10.3891/acta.chem.scand.35b-0379. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Li F, Xiong Y, Wang J, Cho HD, Tomita K, Weiner AM, Steitz TA. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111:815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large non-coding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Pan B, Xiong Y, Steitz TA. How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA. Science. 2010;330:937–940. doi: 10.1126/science.1194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock H, Bachu R, Beal PA. Covalent stabilization of a small molecule-RNA complex. Bioorg. Med. Chem. Lett. 2011;21:5002–5005. doi: 10.1016/j.bmcl.2011.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genomewide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Maizels N, Weiner AM. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J. 1998a;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Weiner AM, Maizels N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA. 1998b;4:276–284. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Cramer F. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1979;22:1–69. doi: 10.1016/s0079-6603(08)60798-9. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow JF, Cramer P. RNA polymerase fidelity and transcripstional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Toh Y, Numata T, Watanabe K, Takeshita D, Nureki O, Tomita K. Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme. EMBO J. 2008;27:1944–1952. doi: 10.1038/emboj.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishitani R, Fukai S, Nureki O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature. 2006;443:956–960. doi: 10.1038/nature05204. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Li F, Wang J, Weiner AM, Steitz TA. Crystal structures of anarchaeal classI CCA-adding enzyme and its nucleotide complexes. Mol. Cell. 2003;12:1165–1172. doi: 10.1016/s1097-2765(03)00440-4. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.