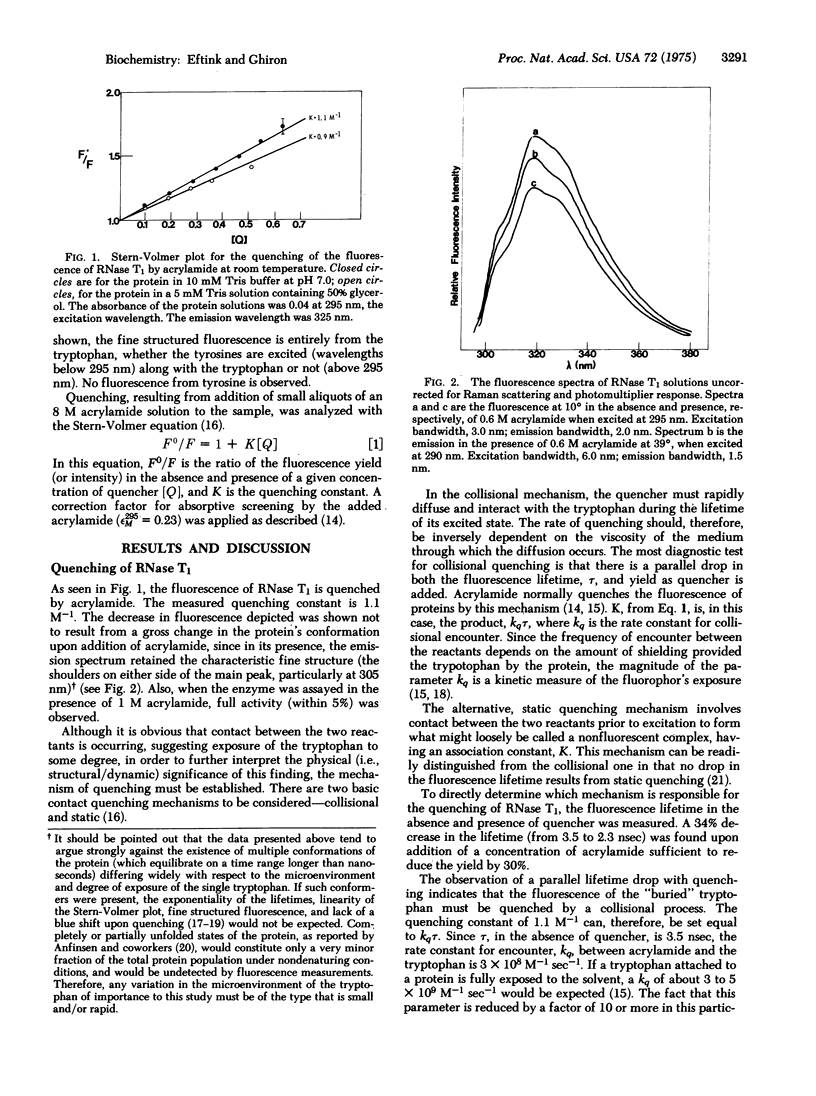
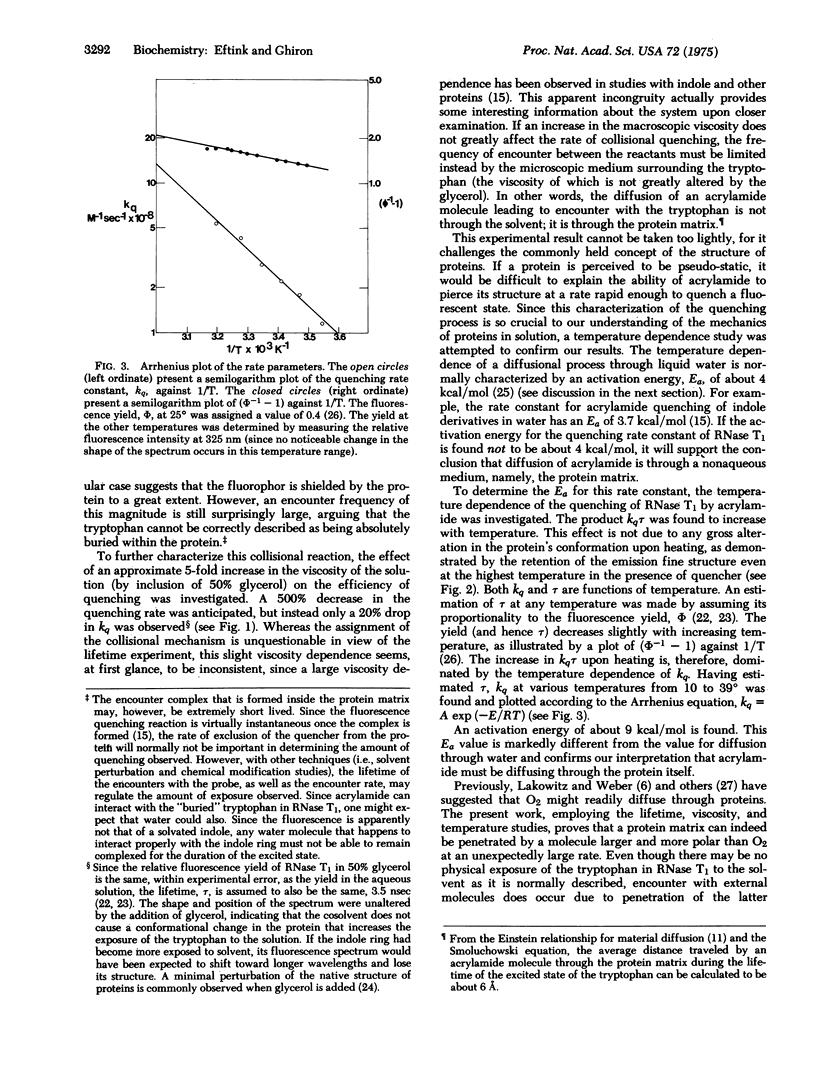
Abstract
The fluorescence of the supposedly buried tryptophan in ribonuclease T1 has been found to be collisionally quenched by acrylamide with a rate constant of 3 X 10(8) M--1 sec--1. Only a slight decrease in the quenching rate is observed upon a 5-fold increase in the viscosity of the solution. For this to be the case, the diffusion of the quencher must be limited by the protein matrix. To explain the process of diffusion through this complex material, the formation of "holes" in the lattice of a protein due to nanosecond fluctuations must be invoked. Thus, the dynamic character of a protein molecule is revealed. The quenching rate constant has an activation energy of 9 kcal/mol which can be used to characterize the nature of the cohesive forces in the microenvironment about the indole ring. The mechanical properties of a portion of a protein matrix can, therefore, be described as one would for a fluid.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allerhand A., Doddrell D., Glushko V., Cochran D. W., Wenkert E., Lawson P. J., Gurd F. R. Conformation and segmental motion of native and denatured ribonuclease A in solution. Application of natural-abundance carbon-13 partially relaxed Fourier transform nuclear magnetic resonance. J Am Chem Soc. 1971 Jan 27;93(2):544–546. doi: 10.1021/ja00731a053. [DOI] [PubMed] [Google Scholar]
- Badley R. A., Teale F. W. Resonance energy transfer in pepsin conjugates. J Mol Biol. 1969 Aug 28;44(1):71–88. doi: 10.1016/0022-2836(69)90405-7. [DOI] [PubMed] [Google Scholar]
- Brand L., Gohlke J. R. Nanosecond time-resolved fluorescence spectra of a protein-dye complex. J Biol Chem. 1971 Apr 10;246(7):2317–2319. [PubMed] [Google Scholar]
- Burstein E. A., Vedenkina N. S., Ivkova M. N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem Photobiol. 1973 Oct;18(4):263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Downer N. W., Teitelbaum H. Hydrogen exchange. Annu Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- Furie B., Schechter A. N., Sachs D. H., Anfinsen C. B. An immunological approach to the conformational equilibrium of staphylococcal nuclease. J Mol Biol. 1975 Mar 15;92(4):497–506. doi: 10.1016/0022-2836(75)90305-8. [DOI] [PubMed] [Google Scholar]
- Ghiron C. A., Longworth J. W., Ramachandran N. Triplet-triplet energy transfer in alpha-trypsin. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3703–3706. doi: 10.1073/pnas.70.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. Fast relaxation processes inn a protein revealed by the decay kinetics of tryptophan fluorescence. Biochemistry. 1974 Dec 3;13(25):5170–5178. doi: 10.1021/bi00722a019. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal Biochem. 1974 Jun;59(2):583–598. doi: 10.1016/0003-2697(74)90312-1. [DOI] [PubMed] [Google Scholar]
- Irie M. Studies on the state of tryptophan residue in ribonuclease T1 and carboxymethyl ribonuclease T1. J Biochem. 1970 Jul;68(1):31–37. [PubMed] [Google Scholar]
- Lakowicz J. R., Weber G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 1973 Oct 9;12(21):4171–4179. doi: 10.1021/bi00745a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S. S. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971 Aug 17;10(17):3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- Likhtenshtein G. I., Grebenshchikov Y. B., Avilova T. V. An investigation of the microrelief and conformational mobility of proteins by the ESR method. Mol Biol. 1972 Jan-Feb;6(1):52–60. [PubMed] [Google Scholar]
- Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9(10):1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- Pennock B. E., Schwan H. P. Further observations on the electrical properties of hemoglobin-bound water. J Phys Chem. 1969 Aug;73(8):2600–2610. doi: 10.1021/j100842a024. [DOI] [PubMed] [Google Scholar]
- TAKAHASHI K. The structure and function of ribonuclease T1. II. Further purification and amino acid composition of ribonuclease T1. J Biochem. 1962 Feb;51:95–108. doi: 10.1093/oxfordjournals.jbchem.a127515. [DOI] [PubMed] [Google Scholar]
- Weinryb I., Steiner R. F. The luminescence of tryptophan and phenylalanine derivatives. Biochemistry. 1968 Jul;7(7):2488–2495. doi: 10.1021/bi00847a007. [DOI] [PubMed] [Google Scholar]
- Wishnia A. On the thermodynamic basis of induced fit. Specific alkane binding to proteins. Biochemistry. 1969 Dec;8(12):5070–5075. doi: 10.1021/bi00840a059. [DOI] [PubMed] [Google Scholar]



