Abstract
Human induced pluripotent stem cells (hiPSCs1–3) are useful in disease modeling and drug discovery, and they promise to provide a new generation of cell-based therapeutics. To date there has been no systematic evaluation of the most widely used techniques for generating integration-free hiPSCs. Here we compare Sendai-viral (SeV)4, episomal (Epi)5 and mRNA transfection mRNA6 methods using a number of criteria. All methods generated high-quality hiPSCs, but significant differences existed in aneuploidy rates, reprogramming efficiency, reliability and workload. We discuss the advantages and shortcomings of each approach, and present and review the results of a survey of a large number of human reprogramming laboratories on their independent experiences and preferences. Our analysis provides a valuable resource to inform the use of specific reprogramming methods for different laboratories and different applications, including clinical translation.
The goal of this analysis was to compare non-integrating reprogramming methods as they are being practiced now, using readily available and widely used reagents and kits. In SeV reprogramming4, Sendai-viral particles are used to transduce target cells with replication- competent RNAs that encode the original set of reprogramming factors (OCT4, SOX2, KLF4 and cMYC (together referred to as OSKM)). Here we used the Cytotune kit (Life Technologies). In Epi reprogramming5, prolonged reprogramming factor expression is achieved by Epstein-Barr virus–derived sequences that facilitate episomal plasmid DNA replication in dividing cells. Human episomal reprogramming was first realized by the Thomson laboratory7; here we use a more efficient method that employs the reprogramming factors OCT4, SOX2, KLF4, LMYC and LIN28A combined with P53 knock-down (shP53)5. In mRNA reprogramming6, cells are transfected with in vitro– transcribed mRNAs that encode OSKM, the additional reprogramming factor LIN28A and GFP. Several chemical measures are employed to limit activation of the innate immune system by foreign nucleic acids6, and, due to the very short half-life of mRNAs, daily transfections are required to induce hiPSCs. Here we used the mRNA reprogramming kit from Stemgent.
First, we looked at reprogramming efficiencies. Determining precise reprogramming efficiencies is complicated by differences between protocols in cell passaging regimes, cell plating efficiencies, as well as by variable somatic cell proliferation and transfection/transduction rates; nevertheless, the number of hiPSC colonies generated per somatic input cell is an important parameter. Of the three non-integrating methods, mRNA-based reprogramming was found to be the most efficient (Fig. 1a). The mean efficiency of successful reprogramming experiments was 2.1% for the mRNA method (n = 3 successfully reprogrammed samples), followed by SeV (0.077%) and Epi (0.013%) reprogramming; the differences in efficiencies (mRNA vs. Epi, mRNA vs. SeV, Epi vs. SeV) reached statistical significance (P < 0.05, Student’s t-test). For comparison, lentiviral (Lenti) reprogramming (with the reprogramming factors OSKM) generated colonies with an efficiency of 0.27% (n = 7). Efficiencies can be sample-dependent; however, the subset of samples that were successfully reprogrammed by all four methods (one neonatal (BJ) and two patient-derived lines (PS1, PS2)) showed the same trend and rank order (gray bars in Fig. 1a). Furthermore, our results are consistent with those reported by others5,6,8–11 (black bars in Fig. 1a).
Figure 1.
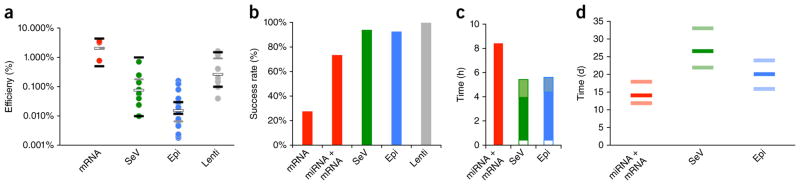
Performance comparison of non-integrating reprogramming methods. (a) Reprogramming efficiencies were calculated as the number of emerging hiPSC colonies per starting cell number; each dot represents the average efficiency of one sample. White bars indicate the mean efficiencies of successful experiments; black bars show the range of efficiencies reported in independent publications (see text); gray bars represent the method-specific means of the three samples that were reprogrammable by all methods. The number of biological replicates was n = 3 samples (a total of 11 biological replicates) for RNA, n = 9 (10) for SeV, n = 17 (25) for Epi and n = 9 (9) for Lenti. (b) Reprogramming success rates for human fibroblast reprogramming experiments using standard protocols. Only experiments yielding at least three colonies were counted as successes. The number of independent samples (biological replicates) was n = 11 for mRNA, 15 for miRNA + mRNA, 85 for SeV, 28 for Epi and 13 for Lenti. (c) Typical total hands-on workload (in hours) was assessed for the three non-integrating human fibroblast-reprogramming methods, until colonies have emerged and grown to a size large enough for picking. The Epi and SeV methods have higher starting cell requirements than the miRNA + mRNA method; the time required to perform the necessary additional somatic cell expansion is shown as a white box. Epi and SeV lines need to be tested for the absence of the reprogramming agents; shaded boxes indicate the workloads of nucleic acid isolation and analysis. (d) Skin fibroblast-reprogramming times (in days), from the first transfection and/or transduction until colonies are ready for picking. Bars show the observed range (light band) and average (dark band) for each method (n = 10 biological replicates for miRNA + mRNA, 44 for SeV and 12 for Epi). All pairwise comparisons reached statistical significance (Student’s t-test, P < 0.01).
Next, we considered the success rates, defined as the percentage of samples for which at least three hiPSC colonies emerged (Fig. 1b). In our hands, the Lenti (100% success rate), Epi (93%) and SeV (94%) methods very reliably generated multiple hiPSC colonies. In contrast, with the mRNA method, the success rate was significantly lower (27%, P < 0.001, Fisher’s exact test). Failures did not appear to be due to reduced mRNA transfection efficiencies (GFP expression); rather, they were associated with massive cell death and detachment. Furthermore, whereas skin fibroblast samples BJ, PS1 and PS2 were readily reprogrammed using all methods, two other patient skin samples (PS3, PS4) that could be reprogrammed using Epi and SeV methods failed with the mRNA method, strongly suggesting that these failures were method-specific and sample-dependent. When we used a modified protocol that employed transfection of microRNAs (miRNAs) (miRNA Booster Kit, Stemgent) and mRNAs, the success rate improved significantly, to 73% overall (P < 0.05) and to 100% for samples refractory to reprogramming by mRNA alone (n = 4). The mean reprogramming efficiency of miRNA + mRNA reprogramming was 0.19% for the 11 fibroblast samples that were reprogrammable with this method.
To allow us to directly compare the workload of generating hiPSCs with the three non-integrating methods, we measured the hands-on time required, including reagent, media and feeder cell preparations, from initial seeding of the target somatic cells to the picking of hiPSC colonies (Fig. 1c,d). The SeV method demanded the least amount of work, consuming 3.5 h of hands-on time until colonies were ready for picking around day 26. Epi reprogramming consumed about 4 h, with colonies large enough for picking appearing around day 20, and the miRNA + mRNA method required about 8 h, although colonies were ready to be picked around day 14. SeV and Epi reprogramming required a larger starting cell number and that more clones be expanded and tested for the loss of the reprogramming agents (see below), adding to the workload (Fig. 1c).
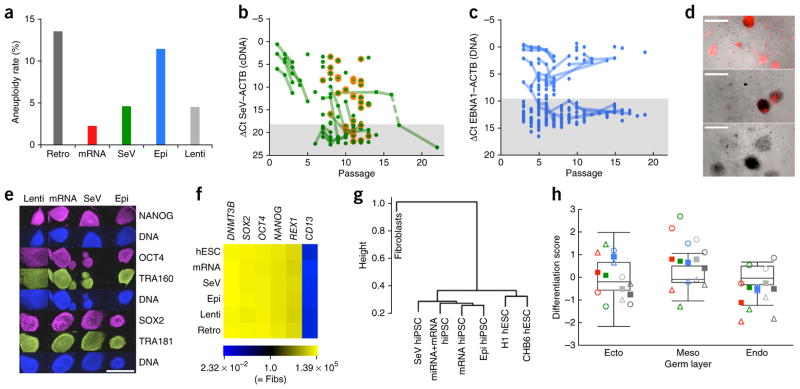
Next, we examined karyotypes of the hiPSC lines derived by the different methods (Fig. 2a and Supplementary Fig. 1a). Among 470 qualifying hiPSC karyotypic analyses, 42 (8.9%) were aneuploid. Retroviral (Retro) hiPSCs had the highest aneuploidy rate (13.5%), and RNA-hiPSCs, the lowest (2.3%). Lower aneuploidy rates were also seen with Lenti (4.5%) and SeV (4.6%) reprogramming, whereas the rate in Epi-hiPSCs was 11.5%. Statistical significance (Fisher’s exact test, P < 0.05) was achieved for the differences between RNA versus Retro, SeV versus Epi and SeV versus Retro.
Figure 2.
Comparison of the genetic integrity of hiPSCs derived by different reprogramming methods. (a) Aneuploidy rates of low-passage (P < 30) hiPSC lines derived by different reprogramming methods. The data include only standard reprogramming methods and the results from the lowest available passage for each hiPSC line. All observed abnormal karyotypes are listed in Supplementary Figure 1a. Donor age did not contribute to the increased rate of aneuploidy among Epi hiPSCs (data not shown). n = 192 (Retro), 44 (mRNA), 151 (SeV), 61 (Epi), 22 (Lenti). (b) Quantification of SeV (Cytotune) RNA in SeV hiPSC lines at different passages by TaqMan RT-QPCR analysis. Ct values for ACTB were subtracted from those for SeV. Lines connect data points that represent the same stem cell line over time. A 5-day heat treatment (39 °C) was performed between collections of the samples connected by the dashed line. The shaded area marks the detection limit (3 s.d. removed from the mean of the SeV-negative controls (n = 10)). Orange circles highlight erythroblast-derived SeV hiPSC samples showing a slightly delayed loss of SeV RNA. Cytotune2 SeV RNA loss kinetics are shown in Supplementary Figure 2a. (c) Quantification of EBNA1 DNA in Epi hiPSC lines at different passages by TaqMan QPCR analysis. Ct values for RNAseP (single-copy control gene locus) were subtracted from those for EBNA1. Lines connect data points that represent the same stem cell line over time. The shaded area marks the detection limit (3 s.d. removed from the mean of the EBNA1-negative controls (n = 41)). See also Supplementary Figure 2b. (d) Epi hiPSCs (passage 3) generated with a modified OCT4-p53 plasmid containing a H2B-mKO2 cassette for fluorescent labeling of hiPSCs that have retained this plasmid (shown are examples of lines containing varying amounts of H2B-mKO2; some dead show auto-fluorescence). Scale bars, 1,000 μm. (e) Immunofluorescence analysis of hiPSC lines representing the indicated reprogramming methods. Scale bar, 1,000 μm. (f) Quantitative RT-PCR analysis (passage 10–15) of fibroblast and pluripotency marker gene expression in parental fibroblasts and hiPSCs, derived by the indicated methods. Heat map representation of the average fold-induction and fold-repression (compared to fibroblasts) of GAPDH-normalized expression levels of three hiPSC lines per method (three technical replicates per line). Additional genes are shown in Supplementary Figure 3. (g) Hierarchical cluster analysis of CpG genomic DNA methylation levels of partially methylated domains in method-specific hiPSC (passage 11–21) and control cells (Supplementary Fig. 4). (h) Comparison of the scorecard differentiation propensity of method-specific hiPSC lines (passage 15–27). Boxplots show the distribution of differentiation propensities for the pluripotent reference set (hESCs); circles = BJ-derived hiPSCs, triangles = PS1-derived hiPSCs; solid squares = method-specific score averages; for each germ layer, method-specific hiPSC scorecard data are shown (from left to right) for mRNA (red), SeV (green), Epi (blue), Lenti (light gray) and Retro hiPSCs (dark gray). None of the pairwise scorecard-score comparisons (mRNA vs. Epi, mRNA vs. SeV, Epi vs. SeV) reached statistical significance (Student’s t-test, P > 0.5).
Several studies have reported small genomic alterations in hiPSC lines, including copy number variations (CNVs) and coding mutations, possibly reflecting a mutagenic effect of the reprogramming process itself12,13. We conducted a small-scale genome-wide array comparative genomic hybridization (aCGH) analysis of representative patient-derived Lenti, SeV, Epi and RNA hiPSC lines and found the majority of CNVs preexisting and the frequency of possible de novo aberrations uniformly low across all methods (Supplementary Fig. 1b).
Next, we tested how quickly the exogenous reprogramming agents were lost during hiPSC expansion. Because modified mRNAs have a short half-life and cannot integrate or replicate, we focused on SeV and Epi hiPSC samples. Loss of SeV RNA was independent of sample type except for erythroblast-derived SeV hiPSCs, which showed a somewhat delayed loss of SeV RNA (Fig. 2b). We observed a passage-dependent decrease in the number of hiPSC lines retaining SeV RNA, from 100% at passage 1–5, to around 53.8% at passage 6–8 (66.7% including erythroblast hiPSC samples) and down to 21.2% (34.3% including erythroblast hiPSC samples) at passage 9–11.
EBNA1 DNA was detected in ~39.1% of Epi hiPSCs analyzed before passage 6. This rate decreased, but slowly, to 37.9% at passages 6–8 and 33.3% at passage 9–11 (Fig. 2c), a frequency identical to the one observed by Okita et al.5. Most lines either showed an increase or a decrease in EBNA1 DNA levels, resulting in a distinct separation into EBNA1 DNAnegative and EBNA1 DNAhigh lines by passage 10. PCR analyses revealed that O4-shP53 plasmid sequences were retained in 13/14 higher-passage DNAhigh lines (Supplementary Fig. 2b). A few of the O4-shP53 plasmid-positive clones also contained the LIN28A-LMYC plasmid, whereas sequences derived from the SOX2-KLF4 encoding were not detected in this sample set. Retention of episomal plasmid sequences in rare hiPSCs that may have a growth advantage is a potential concern that necessitates continued vigilance. To address this issue, we developed a fluorescent protein (H2B-mKO2)–tagged version of the OCT4-shP53 episomal plasmid that facilitates identification of plasmid-retaining colonies (Fig. 2d).
We also looked at marker expression in mRNA-, Epi-, SeV-hiPSCs and in Retro- and Lenti-hiPSCs. Independent of the reprogramming method, all hiPSC lines expressed markers of undifferentiated and completely reprogrammed human pluripotent stem cells, such as TRA160, NANOG, SSEA4, TRA181, OCT4, DNMT3B, SOX2, REX1, LIN28, UTF1 and CDH1, and downregulated the fibroblast-expressed gene ANPEP. Expression of these markers was indistinguishable between human embryonic stem cell (hESC) and hiPSC lines, whether assessed by immunofluorescence (Fig. 2e) or QPCR (Fig. 2f and Supplementary Fig. 3).
When we extended our analysis to genes previously reported as differentially expressed between hESC and hiPSC lines (TCERG1L, FAM19A5 (refs. 14,15) and MEG3/RIAN16), we found that they were indeed differentially expressed, but only in some of the hiPSC lines. Notably, all reprogramming methods performed quite similarly, with a small fraction of all method-specific sets of the lines showing the ‘correct’ hESC-like gene expression levels for the three variable genes (Supplementary Fig. 3). Our observations are in agreement with reports showing that aberrant methylation of the TCERG1L gene locus17 or aberrant expression of Meg3/Meg8 (ref. 16) is observed only in some iPSC lines.
To assess the degree of epigenetic reprogramming, we analyzed genomic regions that are partially methylated in fibroblasts and strongly methylated in hESCs and hiPSCs, the partially methylated domains (PMDs14). Genomic DNA was isolated from the parental sample (BJ) and the method-specific, reprogramming factor-free hiPSCs. We failed to observe meaningful method-specific differences, either at the level of individual PMDs or in hierarchical cluster analysis of global DNA methylation (Fig. 2g and Supplementary Fig. 4). Furthermore, all hiPSCs showed methylation patterns that were very similar to those of two hESC lines and distinct from those of the parental cells.
To evaluate their pluripotency, we differentiated Lenti, Retro, as well as reprogramming factor-free SeV, mRNA and Epi hiPSCs derived from two samples (BJ, PS1) as embryoid bodies and determined each cell line’s differentiation propensity for the three germ layers using the scorecard approach18. All lines scored as pluripotent, with differentiation score averages well within the non-outlier range of the human pluripotent reference set and with no apparent trends specific to the reprogramming method (Fig. 2h).
Finally, to allow us to compare our observations with those of other human cell reprogramming laboratories, we designed an online survey (Supplementary Fig. 5). The survey was distributed to a set of hiPSC core facilities and research laboratories. We received 55 responses from laboratories representing 12 countries and an estimated combined experience of >1,450 human somatic cell reprogramming experiments. The most commonly attempted methods for human fibroblast reprogramming were Lenti (38 laboratories), Retro (33) and SeV (35), followed by RNA (22) and Epi (21) methods (Table 1). All laboratories were able to successfully use all these methods on fibroblasts, with the exception of RNA reprogramming, which could not be established in 41% of laboratories. When we focused on just the responses of the most experienced laboratories (>20 reprogramming samples processed), a similarly high fraction (47%) reported an inability to generate RNA-hiPSCs, suggesting that the difficulties associated with the RNA method are not merely due to a lack of sufficient reprogramming expertise. The survey responses allowed us to calculate method ‘adoption’ and ‘rejection’ rates specifically for those laboratories that were able to establish the method. Very high method-adoption rates were reported for SeV, whereas the rates for Retro and/or Lenti methods were low, indicating that most laboratories eventually switched to non-integrating techniques. Most of the reported experience with fibroblast reprogramming extended to blood cells, with a few notable exceptions. None of the laboratories reported successful RNA reprogramming of hematopoietic cells, and although Retro reprogramming of blood cells was seldom if ever practiced, Lenti reprogramming was more popular with blood cells than with fibroblasts.
Table 1.
Reprogramming survey results
| Survey results | Laboratories | Method | Tried (no. of labs) | Success (% labs) | Success (no. of labs) | Rejected (%) | Adopted (%) |
|---|---|---|---|---|---|---|---|
| Skin fibroblast reprogramming | All | RNA | 22 | 59 | 13 | 38 | 46 |
| SeV | 35 | 97 | 34 | 18 | 62 | ||
| Epi | 21 | 100 | 21 | 48 | 33 | ||
| Lenti | 38 | 95 | 36 | 64 | 19 | ||
| Retro | 33 | 97 | 32 | 75 | 9 | ||
| Experienced | RNA | 15 | 53 | 8 | 50 | 25 | |
| SeV | 21 | 95 | 20 | 18 | 62 | ||
| Epi | 12 | 100 | 12 | 58 | 42 | ||
| Lenti | 25 | 100 | 25 | 68 | 20 | ||
| Retro | 21 | 100 | 21 | 86 | 5 | ||
|
| |||||||
| Blood reprogramming | All | RNA | 4 | 0 | 0 | n/a | n/a |
| SeV | 19 | 95 | 18 | 11 | 61 | ||
| Epi | 11 | 82 | 9 | 33 | 44 | ||
| Lenti | 15 | 100 | 15 | 40 | 40 | ||
| Retro | 6 | 67 | 4 | 75 | 0 | ||
| Experienced | RNA | 3 | 0 | 0 | n/a | n/a | |
| SeV | 13 | 100 | 13 | 15 | 54 | ||
| Epi | 8 | 88 | 7 | 29 | 57 | ||
| Lenti | 13 | 100 | 13 | 38 | 46 | ||
| Retro | 6 | 67 | 4 | 75 | 0 | ||
Experienced refers to laboratories that have successfully reprogrammed at least 20 human cell samples; Tried to the number of laboratories that reported having attempted to reprogram somatic cells using the indicated method (RNA refers to the mRNA method with or without miRNA use); Success to the number and percentage of these laboratories that were able to derive hiPSC lines with the method. The survey also included questions about the method-specific usage frequencies. Rejected refers to the % of successful laboratories reporting usage frequencies of ≤10% (≤10% of samples reprogrammed using this method); Adopted refers to usage frequencies of ≥60%. The survey questionnaire is provided as Supplementary Figure 5.
In brief, we detected no substantial method-specific differences in marker expression levels or patterns, developmental potential, DNA methylation or CNV loads of euploid lines. It is possible that more detailed studies will reveal method-specific differences, such as in the potential to form specific cell types. For example, the potential of miPSCs was recently shown to depend on the factors used for reprogramming19. Nevertheless, our conclusion that no overt method-specific differences exist echoes findings by other groups who performed less comprehensive cross-comparisons of smaller sets of reprogramming methods8,12.
SeV reprogramming was efficient and highly reliable, with a low workload and a complete absence of viral sequences in most lines at higher passages. The shortcomings of SeV reprogramming include the dependence on one commercial vendor (making it, for example, difficult to test other reprogramming factors), the relatively slow clearance of SeV RNA and the current lack of clinical-grade SeV for reprogramming. The Cytotune SeV reprogramming kit has achieved a high acceptance rate for skin fibroblast and blood cell reprogramming; we share this enthusiasm and highly recommend SeV reprogramming to laboratories that do not focus on generating clinical-grade hiPSCs.
The main advantages of the RNA method are the speed of colony emergence, high efficiency, a complete absence of integration, a very low aneuploidy rate and a low donor cell requirement (typically 50,000 cells, but as few as 1,000 human fibroblasts can be reprogrammed; data not shown). Minor issues are the increased workload and the need for a tissue culture incubator with O2 control. More significantly, RNA reprogramming was successful only with a small fraction of the samples; we and others8 frequently experienced difficulties when attempting to reprogram primary fibroblasts obtained from patient skin biopsies. Problems with implementing this technique were also reported in our survey, and to date there have been no reports of successful generation of RNA-hiPSCs from blood cells. One promising approach for improving RNA-based reprogramming is the inclusion of pluripotency-inducing miRNAs (Fig. 1b). Others have reported increased RNA reprogramming efficiencies and accelerated colony emergence by fusing Oct4 to a heterologous transactivation domain10. The conventional RNA method can therefore be useful for easy-to-reprogram fibroblast samples, but needs to be further optimized to overcome the reprogramming resistance and excessive cell death observed with many patient samples.
Key advantages of Epi reprogramming are the high reliability of hiPSC generation from fibroblast and blood samples (CD34+ and peripheral blood mononuclear cells (PBMNCs))20,21; see also Supplementary Figs. 6 and 7) and the quick loss of the reprogramming agent (relative to SeV). However, Epi reprogramming does raise concerns regarding the genetic integrity of the resulting hiPSC lines due to the use of a TP53 short hairpin RNA (shRNA) cassette. Notably, we did not observe an increase in CNVs, and the rate of aneuploidies, although elevated relative to SeV, RNA and Lenti hiPSCs, was still lower than what we observed with Retro hiPSCs. To further boost Epi reprogramming efficiencies, several groups have successfully employed small molecules22,23, increased levels of EBNA1 (ref. 21), or used additional or modified reprogramming factors (such as BCL-XL24 or OCT4-VP16 (ref. 25)).
Although xeno-free procedures have been described for all major reprogramming methods8,10,23,26–28, Epi reprogramming seems particularly well-suited for clinical translation because it is integration-free, works reliably with patient fibroblasts and blood cells, and is based on a very simple reagent (plasmid DNA) that can easily be generated using current good manufacturing practice (cGMP)- compatible processes. SeV, on the other hand, is currently not available commercially as a cGMP-grade reagent for reprogramming, although Sendai virus has been used as a vehicle in human vaccination studies. Another important consideration are the restrictions imposed by the respective licensors or vendors. In general, the commercial kits are restricted to educational use and noncommercial or nonclinical in vitro research; however, additional rights to commercial or clinical use can be acquired (Supplementary Table 1).
In summary, the choice of reprogramming method will depend on each laboratory’s particular requirements (Table 2). The field of pluripotency induction continues to develop at a rapid pace, as evidenced by the recent publication of a gene-free, small molecule–based method29, as well as the identification of novel pathways that can be manipulated to augment the efficiency and completeness of reprogramming30. The focus of technology development efforts should now move toward leveraging these findings to generate improved methods for clinical translation of human pluripotent stem cells.
Table 2.
Summary of the critical features and characteristics of the RNA, SeV, Epi and Lenti reprogramming methods
| Feature | RNA Stemgent | SeV Life Technologies | Epi Yamanaka/Addgene | Lenti Mostoslavsky/BU |
|---|---|---|---|---|
| Efficiency (fibroblasts) | ~1% | ~0.1% | ~0.01% | ~0.5% |
| Reliability (fibroblasts) | <50% (miRNA + mRNA: >70%) | >90% | >90% | >90% |
| Reprogramming workload | High | Low | Low | Very high (excision) |
| Aneuploidy rate | 2.3% | 4.6% | 11.5% | 4.5% |
| Input cell requirement (fibroblasts) | ~50 k | ~500 k | ~500–800 k | ~100 k |
| Time to colony emergence | Fast | Moderate | Moderate | Moderate |
| Adoption rate (fibroblasts) | 46% | 62% | 33% | 19% |
| Efficiency (blood) | Not yet reported | High | High | High |
| Reliability (blood) | Not yet reported | High | High | High |
| Adoption rate (blood) | Not yet reported | 61% | 44% | 40% |
| Special equipment needs | 5% O2 incubator | None | Transfection device, and O2 incubator (blood) | None |
| Lines free of reprogramming agents by passage 5 | 100% | 0% (Cytotune 1) | 60.9% | 0% |
| Lines free of reprogramming agents by passage 9–11 | 100% | 78.8% | 66.7% | 0% |
| Xeno-free cGMP-like reprogramming | (Ref. 10) | (Ref. 25) | (Refs. 8,27) see also Supplementary Figure 7 | (Ref. 26) |
ONLINE METHODS
Cells
HDF-f fibroblasts were obtained from ScienceCell, and NuFF and BJ fibroblasts were obtained from Stemgent and ATCC. Inactivated MEF feeder cells were obtained from Millipore, and patient fibroblasts were obtained from Coriell Institute and from skin biopsies procured under IRB-approved protocols.
SeV reprogramming
SeV reprogramming was performed using the Cytotune reprogramming kit (Lifetech) following the manufacturer’s protocol. In brief: 80% confluent fibroblasts grown in fibroblast medium (10% FCS (Gemini), 1% L-glutamine, 1% sodium pyruvate and 1% MEM-NEAA in high-glucose DMEM (Lifetech)) in a 6-well plate were transduced with each of the four viruses at a multiplicity of infection (MOI) of 3. Cells were fed every other day, and 50 k, 100 k and 200 k cells were replated on day 7 onto 0.1% gelatin-coated, 10-cm dishes containing CF1 MEFs (Globalstem). Medium was switched to hESC medium (7 μl/liter 2ME (Sigma), 20% KOSR, 2× L-Glutamine, 1× MEM-NEAA, 10 ng/ml bFGF, in DMEM/F12 (Lifetech)). Colonies were picked by mechanical dissection, were transferred to fresh feeders, and were expanded using mechanical or enzymatic (Collagenase IV, Lifetech) clump passaging methods31. Reprogramming efficiencies were determined by TRA-1-60 immunocytochemistry as described32.
mRNA reprogramming
On day −2, irradiated human fibroblast feeder cells (NuFFs (Stemgent), 250 k per well) were plated in NuFF culture medium (Stemgent). On day −1, 50 k target cells were passaged onto the NuFF feeder layer in fibroblast medium. Two hours before each transfection, the culture was pretreated with B18R (300 ng/ml; Stemgent). A total of 1.2 μg mRNA was transfected with 6 μl Lipofectamine RNAiMAX (Life Technologies). After 4 h, transfection medium was exchanged with Pluriton Medium. After the first transfection, cells were cultured under low (physiological) oxygen conditions (5%). Daily mRNA transfections were performed through day 16. From day 6, reprogramming cultures were fed with NuFF-conditioned Pluriton Medium supplemented with B18R and Pluriton Supplement (Stemgent).
miRNA + mRNA reprogramming
50 k fibroblasts were plated onto a Matrigel (BD)-coated, 6-well tissue culture plate at a density of 50 k cells per well on day −1. Two hours before the first transfection on day 0, cells were pretreated with B18R (300 ng/ml; Stemgent) in NuFF-conditioned Pluriton Medium (Stemgent). The microRNA reprogramming cocktail was transfected with 4 μl Stemfect Transfection Reagent (Stemgent). After the first transfection, cells were cultured under low (physiological) oxygen condition (5%). On days 1 to 3, cells were transfected with mRNA reprogramming cocktail using Stemfect. On day 4, cells were transfected with both microRNA and mRNA reprogramming cocktails. Daily mRNA transfections were done from days 5 through 12. Cells were cultured in NuFF-Conditioned Pluriton Medium until colonies were mechanically picked on day 14.
Epi reprogramming
80% confluent fibroblasts in a T25 flask (500–800k cells) were nucleofected with 1 μg of each of the three episomal reprogramming plasmids (Addgene plasmids #27077, #27078, #27080) using program P22 and then plated onto a 6-well plate in fibroblast medium. Upon reaching confluency around day 6, the cells were split onto two 10-cm plates, coated with 0.1% gelatin and MEFs using fibroblast medium. Beginning the following day, the cells were fed daily with hESC medium until colonies were ready for picking. For Xeno-free reprogramming of PBMNCs, see Supplementary Methods.
Retro/Lenti reprogramming
The human STEMCCA lentiviral plasmid was kindly provided by Gustavo Mostoslavsky, and viral particles were generated in 293T cells using standard procedures33. One day after plating 100 k fibroblasts per 6-well plate, STEMCCA virus was added for 24 h at an MOI of 5 in 2 ml fresh medium with 2 μl of 10 mg/ml protamine sulfate in H2O. Cells were fed until they reached 80% confluency when they were split 1:6 and 1:36 onto 0.1% gelatin-coated, 10-cm dishes with MEF in fibroblast medium. From 24 h after plating the cells were fed daily with hESC medium until colonies were ready for picking. Retro reprogramming was performed as previously described34.
Statistical analysis
For reprogramming efficiency experiments, the method-specific efficiencies were calculated as the geometric means of the reprogramming efficiencies of the successfully reprogrammed sample sets. P values were calculated using the unpaired two-tailed Student’s t-test (efficiencies) or Fisher’s exact test (success rates, aneuploidies).
Characterization of hiPSC lines
For aneuploidy rate analyses, we only counted karyotype results from low-passage samples (P < 30), and excluded lines generated using nonstandard protocols for which our samples size was very low (e.g., feeder- or xeno-free), and disease samples where the disease was known to affect genetic integrity (e.g., Fanconi Anemia). For each of the non-integrating methods, karyotypes from at least 12 different donor fibroblast samples were analyzed. Karyotyping analyses were performed by Cell Line Genetics or the Tuft’s Cytogenetics facility. Array-CGH CNV analysis was performed using the StemArray platform (Cell Line genetics) with samples at P15-16. Immunofluorescence analysis was performed as described34–36. For immunofluorescence, gene expression, epigenetic and in vitro differentiation analyses, we only used Epi and SeV hiPSCs in which the exogenous reprogramming agents could no longer be detected.
Scorecard analysis
Stem cell colonies were harvested as small clumps and differentiated in vitro as embryoid bodies (EBs) in hESC medium without bFGF. RNA was isolated from day 15 EBs (P15-27) and Nanostring RNA expression profiles were obtained for 500 marker genes, normalized and analyzed as described37. All sample differentiation propensities fell within the normal, non-outlier range of the reference set of 18 hESC lines18.
DNA methylation analysis methodology
Bisulfite sequencing libraries were prepared using TruSeq DNA library preparation kits (Illumina) with some modifications to the manufacturer’s protocol. Genomic DNA from each sample (P11-21 for hiPSCs) was purified using the MasterPure DNA purification kit (Epicentre) based on a protocol by the manufacturer. 1.5 μg of genomic DNA for all samples and lambda phage genomic DNA as bisulfite conversion control were sheared for 100 s using Covaris sonicator with the following setting: duty cycle = 10%, intensity = 5, burst per second = 200. End repair, the addition of A tail and adaptor ligation to library fragments were performed following the Truseq protocol. Libraries were size selected to get 400- to 500-bp fragments by cutting them from 2% agarose gel (Bio-Rad-certified low Range Ultra Agarose) and extracted using Qiagen MinElute Gel Extraction Kit. Libraries were bisulfite-converted using Zymo EZ DNA Methylation Gold kit and then amplified using KAPA HiFi HotStart Uracil+ Ready Mix (Kapa Biosystems) for 11 cycles. 100-bp paired-end HiSeq2000 sequencing reads were aligned by BSmooth bisulfite alignment pipeline (version 0.7.1)38 as previously described in detail39. Briefly, reads were aligned by Bowtie2 (version 2.0.1) against human genome (hg19) as well as the lambda phage genome. After alignment, methylation measurements for each CpG were extracted from aligned reads. We filtered out measurements with mapping quality <20 or nucleotide base quality on cytosine position <10, and we also removed measurements from the 5′-most ten nucleotides of both mates. Methylation level of CpGs was smoothed by bsseq package from BSmooth using a window size of 200 CpGs or 10 kb, whichever is larger. For hierarchical cluster analysis of methylomes, we first calculated average methylation levels of each 2-kb sliding window along every chromosome for all samples. Then all sliding window methylation profiles were used for hierarchical cluster using pair-wise Pearson correlation distance and Ward’s method by ‘hclust’ function of R package. The coordinates of PMDs (partially methylated domains) were obtained from Lister et al.40. Methylation levels of all samples used in this study were plotted in PMDs and their 5-kb flanking regions based on smoothed methylation level.
EBNA DNA and SeV RNA TaqMan assays
Genomic DNA was prepared using Qiagen DNAeasy or Viagen Direct PCR Lysis reagent with Proteinase K. Genomic DNA samples were subjected to 2-color TaqMan analysis using a custom-designed EBNA-specific assay (TGTCTG ACAGCGACCATGAAG, FAM/NFQ- GATCAATAGACATCTTTATTAG ACGACGCT, TCTGCACTCCCTGTATTCACTG) and a VIC-labeled, human genomic DNA copy-number reference assay (RNAseP, Lifetech) with TaqMan Genotyping Master Mix (Liftech) on a Bio-Rad C1000 CFX96 system (95 °C for 10 min; 40 cycles, 95 °C for 15 s, 60 °C for 60 s) in white PCR plates (Bio-Rad). Thresholds were set to 3,000 relative fluorescence units and results were plotted as ΔCt. SeV hiPSC line RNA was reverse-transcribed and 0.25 μl cDNA were subjected to TaqMan gene expression analysis using probes Mr04269880_mr (SeV) and Hs01060665_g1 (ACTB). Thresholds were set to 1 k RLUs and results were plotted as ΔCt. Retained plasmids were identified using primers O4p53-F (ATCCGACGCCGCCATCTCTA), O4p53-R (AGTCGGCCCAAACAAGGCTT); UL-F (GGCAGCAGCAGTT GCAGAA), UL-R (GTGCAGCAGCTGAGGCTCGT); and klf4f2 (GACA CTGCGTCAAGCAGGTGC), Klf4r2 (AAGTCCAGGTCCAGGAGATCG).
Gene expression
QPCR analyses were performed on Trizol (Lifetech) or RNAeasy (Qiagen) extracted RNA (hiPSC harvested at passage 10–15) following reverse-transcription (High Capacity cDNA Reverse Transcription kit, Life Technologies) using iQ SYBR Green (Bio-Rad). PCR primers are as described36. Results were normalized to GAPDH RNA levels (SYBR Green assays) or to the geometric mean of ACTB, UBC and TBP mRNA levels (TaqMan assays).
TaqMan gene expression assay
The following TaqMan assays were used: MEG3 (Hs01087966_m1); FAM19A5 (Hs01015862_m1); LIN28A (Hs00702808_s1); NANOG (Hs04260366_g1); ZFP42 (Hs01938187_s1); UTF1 (Hs00864535_s1); POUF51 (Hs01895061_u1); TCERG1 (Hs00545416_m1); TERT (Hs00972656_m1); CDH1 (Hs01023894_m1); ACTB (Hs01060665_g1); TBP (Hs00427620_m1); UBC (Hs01871556_s1).
Survey
An online questionnaire (https://www.surveymonkey.com, see Supplementary Fig. 3) was prepared and sent to hiPSC core facilities and laboratories using a list curated by S. D’Souza from hESC/iPSC Shared Resource Facility of the Icahn School of Medicine at Mount Sinai, New York. The survey was open from 11/15 to 12/09/2014. Replies lacking crucial information (reprogramming experience) were excluded.
Supplementary Material
Acknowledgments
We would like to thank B. Hamilton (Stemgent) for miRNA reprogramming agents and protocols, G. Mostoslavsky (Boston University) for human STEMCCA lentiviral plasmid constructs, M. Armant (Boston Children’s Hospital) for MRC5 and hCD34+ cells, G. MacLean (Boston Children’s Hospital) for advice on episomal blood reprogramming and S. D’Souza (Icahn School of Medicine at Mount Sinai) for a list of contacts for human cell reprogramming laboratories. This work was supported in part by grants R01HL75737, U01HL107440, UO1-HL100001, U01HL87402 and U01HL100408 (National Heart, Lung, and Blood Institute (NHLBI) Progenitor Cell Biology Consortium), R24DK092760 (the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)), PO1NS066888 (the National Institute of Neurological Disorders and Stroke (NINDS)), by the SMA Foundation, the Jerome Le Jeune Foundation, an EMBO postdoctoral fellowship (E.B.) and the Harvard Stem Cell Institute.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
T.M.S., L.D., C.A.C., A.M., L.L.R., A.P.F., L.I.Z. and G.Q.D. contributed to the conception and design of the study; T.M.S., L.D. and G.Q.D. wrote the manuscript; T.R.B., S.E., K.C., A.C., A.D., A.E., K.F., M.G., D.G., J.M., P.M., M.G. and B.B. performed reprogramming and cell culture experiments; A.D., N.J., X.L. and A.P.F. performed epigenetics analyses, T.M.S., K.F., D.G., P.M., L.D., T.R.B. and S.E. performed PCR assays; M.S.L., E.B., A.B.C.C. and D.D. provided cell samples and reprogramming data; A.M.T. and A.M. performed the scorecard analysis; T.M.S. performed statistical analyses; T.M.S. and L.D. performed the survey.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 4.Fusaki N, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad, Ser B, Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 6.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goh PA, et al. A systematic evaluation of integration free reprogramming methods for deriving clinically relevant patient specific induced pluripotent stem (iPS) cells. PLoS ONE. 2013;8:e81622. doi: 10.1371/journal.pone.0081622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieu PT, et al. Generation of induced pluripotent stem cells with CytoTune, a non-integrating Sendai virus. Methods Mol Biol. 2013;997:45–56. doi: 10.1007/978-1-62703-348-0_5. [DOI] [PubMed] [Google Scholar]
- 10.Warren L, Ni Y, Wang J, Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci Rep. 2012;2:657. doi: 10.1038/srep00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somers A. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz S, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey BW, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Koyanagi-Aoi M, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci USA. 2013;110:20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock C, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buganim Y, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park TS, et al. Growth factor-activated stem cell circuits and stromal signals cooperatively accelerate non-integrated iPSC reprogramming of human myeloid progenitors. PLoS ONE. 2012;7:e42838. doi: 10.1371/journal.pone.0042838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okita K, et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Chau KF, Vodyanik MA, Jiang J, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS ONE. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su RJ, et al. Efficient generation of integration-free ips cells from human adult peripheral blood using BCL-XL together with Yamanaka factors. PLoS ONE. 2013;8:e64496. doi: 10.1371/journal.pone.0064496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011;12:373–378. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacArthur CC, et al. Generation of human-induced pluripotent stem cells by a nonintegrating RNA Sendai virus vector in feeder-free or xeno-free conditions. Stem Cells Int. 2012;2012:564612. doi: 10.1155/2012/564612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awe JP, et al. Generation and characterization of transgene-free human induced pluripotent stem cells and conversion to putative clinical-grade status. Stem Cell Res Ther. 2013;4:87. doi: 10.1186/scrt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa M, et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou P, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartung O, Huo H, Daley GQ, Schlaeger TM. Clump passaging and expansion of human embryonic and induced pluripotent stem cells on mouse embryonic fibroblast feeder cells. Curr Protoc Stem Cell Biol. 2010;14:1C.10. doi: 10.1002/9780470151808.sc01c10s14. [DOI] [PubMed] [Google Scholar]
- 32.Manos PD, Ratanasirintrawoot S, Loewer S, Daley GQ, Schlaeger TM. Live-cell immunofluorescence staining of human pluripotent stem cells. Curr Protoc Stem Cell Biol. 2011;19:1C.12. doi: 10.1002/9780470151808.sc01c12s19. [DOI] [PubMed] [Google Scholar]
- 33.Somers A. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 35.Miller JD, Schlaeger TM. Generation of induced pluripotent stem cell lines from human fibroblasts via retroviral gene transfer. Methods Mol Biol. 2011;767:55–65. doi: 10.1007/978-1-61779-201-4_5. [DOI] [PubMed] [Google Scholar]
- 36.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 37.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen KD, et al. From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.