Abstract
Considerable evidence suggests that mitochondrial dysfunction occurs early in Alzheimer's disease, both in affected brain regions and in leukocytes, potentially precipitating neurodegeneration through increased oxidative stress. Epigenetic processes are emerging as a dynamic mechanism through which environmental signals may contribute to cellular changes, leading to neuropathology and disease. Until recently, little attention was given to the mitochondrial epigenome itself, as preliminary studies indicated an absence of DNA modifications. However, recent research has demonstrated that epigenetic changes to the mitochondrial genome do occur, potentially playing an important role in several disorders characterized by mitochondrial dysfunction. This review explores the potential role of mitochondrial epigenetic dysfunction in Alzheimer's disease etiology and discusses some technical issues pertinent to the study of these processes.
Keywords: 5-methylcytosine, 5-hydroxymethylcytosine, Alzheimer's disease, dementia, DNA methylation, epigenetics, heteroplasmy, mitochondria
Alzheimer's disease (AD) is a chronic, currently incurable, neurodegenerative disorder, accounting for more than 60% of dementia cases, with current estimates predicting more than 135 million dementia cases worldwide by 2050 [1]. The classic neuropathological hallmarks associated with AD include the formation of amyloid-β (Aβ) plaques and neurofibrillary tangles. These are suggested to play a role in the further development of other characteristics of the disease, such as disruption of calcium homeostasis, loss of connectivity, the generation of reactive oxidative species (ROS) and altered plasticity, ultimately leading to neurodegeneration [2,3,4,5,6]. Mito-chondrial dysfunction is a consistent feature of AD pathology in both the brain and white blood cells [7,8,9,10] although the molecular mechanism(s) mediating this phenomena are yet to be fully elucidated.
Mitochondrial dysfunction: a prominent feature of AD
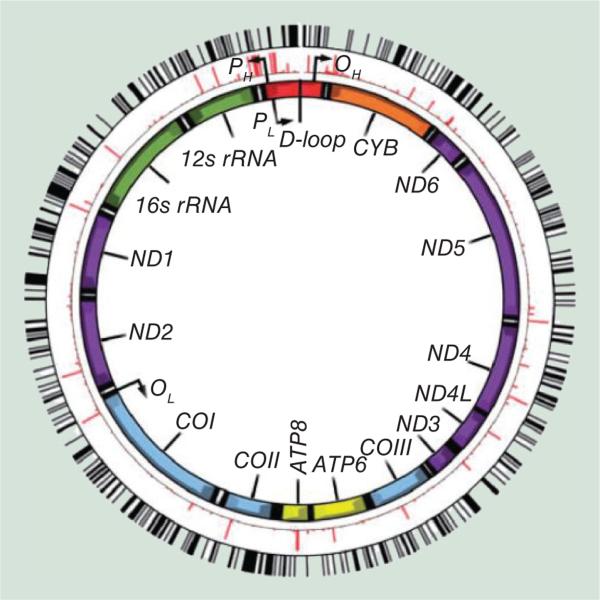
Being the site of ATP generation, mitochondria provide the cell with the energy required to properly function; as such they are often described as ‘the powerhouse of the cell’. Mitochondria are cylindrical organ-elles containing approximately 16.6 kb of DNA (mtDNA) [11], which is separate to the nuclear genome and inherited in a maternal, non-Mendelian fashion. The mitochondrial genome comprises 37 genes, 13 of which encode for polypeptides required for the electron transport chain (ETC) (Figure 1), in addition to two ribosomal RNAs and 22 transfer RNAs. The mitochondria play a vital role in a variety of key biological functions, including apoptosis via caspase dependent and independent mechanisms [12], the regulation of calcium homeostasis [13,14] and the production of reactive oxygen species (ROS) [15]. For these reasons, mitochondrial dysfunction has been implicated in the pathogenesis associated with AD [16,17] and forms the basis of the mitochondrial cascade hypothesis [18]. Proposed by Swerdlow et al., this hypothesis states that an individual's genetic code will determine their basal mitochondrial function and that, throughout aging, this function will decline due to a combination of genetic and environmental factors, determining an individual's time of disease onset [18].
Figure 1. The structure of the mitochondrial genome showing genes encoded by the mitochondria.
3358 mtDNA genetic variants shown in red and black lines highlight the predicted CpG sites relative to mutations that define the mitochondrial haplogroup. PH and PL represent the heavy and light strand promoter regions and OH and OL represent the origins of heavy-strand and light-strand replication, respectively. Reproduced with permission from [19].
Mitochondrial-encoded ETC gene expression has been shown to be altered in both early and late stages of AD, with decreased expression of complex I and increased expression of complexes III and IV [7]. Increased expression of mitochondrial-encoded ETC complex genes has also been associated with aging, with increased expression of complexes I, III, IV and V in 12- and 18-month mice compared with 2-month mice, which was accompanied by increased oxidative damage [20]. However, decreased expression of these genes was seen in older, 24-month-old mice, suggesting an initial compensatory upregulation of proteins in the ETC, which failed as aging continued. Further evidence for a role of mitochondria in AD pathogenesis comes from a study demonstrating increased levels of mitochondrial gene expression and oxidative damage in a transgenic amyloid precursor protein (APP) mutant mouse model of AD [21]. In addition, various components of the mitochondrial permeability transition pore (mPTP), which acts as a voltage-dependent channel regulating mitochondrial membrane permeability, have been shown to interact with Aβ in various murine models of AD. For example, one recent study found that, in APP transgenic mice, Aβ acts to upregulate VDAC1, a component of the mPTP, leading to mPTP blockade [22]. Interestingly, this study also reports that VDAC1 may interact with hyperphosphorylated tau, suggesting another mechanism of mitochondrial dysfunction. An earlier study found that Aβ present in mitochondria interacts with CypD, another component of the mPTP, in cortical samples from postmortem AD patients and mAPP transgenic mice [23]. In the mouse model, this was shown to lead to increased ROS production and neuronal cell death. Taken together, this illustrates how mitochondrial-encoded gene expression is altered in AD, a variety of mechanisms by which Aβ interacts with mitochondria in AD and how mitochondrial dysfunction can lead to changes associated with AD, thus highlighting the need for continued research into the field.
Epigenetics & AD
Given the high heritability estimates for AD [24], considerable effort has focused on understanding the role of genetic variation in disease etiology, although more recently it has been hypothesized that epigenetic dys-function may also be important [25]. A number of studies have shown reduced global levels of the DNA modifications 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) in AD brain [26,27,28,29], with only a handful of studies having looked at changes occurring at specific loci (reviewed in [25]). Recent methodological advances in microarray and genomic sequencing technologies have enabled researchers to undertake epigenome-wide association studies in AD brain, identifying several consistent differentially methylated regions associated with disease [30,31,32]. Many of these differentially methylated regions are tissue specific, restricted to regions of the brain associated with AD pathology and correlate strongly with quantitative measures of neuro-pathology. As such, a strong case is being built for a role of epigenetics in the etiology of AD.
Epigenetic regulation of the mitochondrial genome
Although hypotheses about the importance of mtDNA modifications are by no means recent, research in this area has been marred by contradictory results since the 1970s [33-36]. The confirmation in 2011 of both 5-mC and 5-hmC occurring in mtDNA prompted a resurgence of interest in mitochondrial epigenomics [37]. The mitochondrial epigenome has some notable differences compared with the nuclear epigenome, and an overview of the mitochondrial genome, including its CpG sites, can be seen in Figure 1. Unlike the nuclear genome, the mitochondrial genome does not contain classical CpG islands [37], and is not associated with chromatin; instead it is structurally organized by nucleoids [38,39]. As a result, mtDNA is not associated with histone proteins and relies on transcription factors such as mitochondrial transcription factor A (TFAM) to mediate compaction [40]. Histone modifications do not therefore play a direct role in regulating mitochondrial gene expression, highlighting the potential importance of DNA modifications in the regulation of mitochondrial function [41]. Evidence suggests that mtDNA methylation largely influences mtDNA structure and replication and is affected by factors that influence nucleoid compaction and DNA methyltransferase (DNMT) binding [42]. It has been shown that different areas of mtDNA are packaged differently and that a depletion of the nucleoid protein ATAD3 can reduce mtDNA methylation, resulting in an open circular state mitochondrial genome, although evidence for an effect of TFAM on mtDNA methylation was inconclusive [42].
DNMTs are a family of enzymes that catalyze the removal of a methyl group from methyl donors such as S-adenosylmethionine (SAM) for addition to the 5-position of cytosine. Recently, a DNMT isoform, mitochondrial DNMT1 (mtDNMT1), has been found to contain a mitochondrial targeting sequence allowing it to bind to the D-loop of the mitochondrial genome, which contains the promoter sites for both the light and heavy strand of mtDNA and can therefore influence mitochondrial gene expression by altering transcriptional activity [37]. Furthermore, it has been suggested that the presence of these methyltransferases in mitochondria may be tissue specific. Although Shock et al., did not observe mitochondrial localization of DNMT3a in the two cell lines they investigated, a later paper has found that DNMT3a is present, and in higher levels than mtDNMT1, in the mitochondria of motor neurons [43]. This study also demonstrated significantly higher global levels of both mitochondrial DNMT3a and 5-mC in amyotrophic lateral sclerosis (ALS) motor neurons in vivo, suggesting a potential role for mtDNA methylation in motor neurons. DNMT1 and DNMT3b have also been observed in the mitochondria, with their inactivation reducing methylation at CpG sites [44].
Recently, it has been debated whether 5-hmC is just an intermediary product of the demethylation process of 5-mC to cytosine or could represent an independent epigenetic mark [45]. Growing evidence now suggests that 5-hmC could be a mark in its own right, produced from the conversion of 5-mC by TET1, TET2 and TET3 [46], with both TET1 and TET2 being present in the mitochondria [44]. Taken together with the presence of 5-hmC in the mitochondrial D-Loop [37], this strengthens the evidence suggesting that demethylation pathways are not only important in nuclear epigenetics, but may also play a role in the mitochondria. Furthermore, recent evidence suggests that 5-mC and 5-hmC exist stably within mtDNA at cytosines not preceding a guanine base, suggesting a role for non-CpG methylation in mtDNA [47]. Further, CpG and non-CpG methylation has been observed in the mitochondrial D-loop at conserved regions associated with DNARNA hybrid formation during transcription, suggesting that DNA methylation in mitochondria shares similarities with plants and fungi and that this methylation may play a role in regulating mtDNA transcription and replication in a cell-type-specific fashion [44].
MtDNA modifications in disease
Despite little being known about the physiological impact of variation in mtDNA methylation, some recent studies have shown that it may be associated with a variety of diseases. The majority of studies have focused on diseases where mitochondrial dysfunction is known to be prevalent, for example in cancer, which has been previously linked with mitochondrial dysfunction [48] and more recently in Down's syndrome, where mitochondrial abnormalities have also been reported [49]. Particularly, for the purpose of this review, mitochondrial dysfunction and mtDNA methylation aberrations in Down's syndrome cells (see Table 1) are interesting given that these patients have an increased likelihood of presenting with AD-like phenotypes throughout aging [50,51] due to possessing an extra copy of APP. An overview of studies of mtDNA epigenetics in disease is given in Table 1.
Table 1.
An overview of current studies of mitochondrial epigenetics in disease.
| Research question | Techniques | Main findings | Ref. |
|---|---|---|---|
| The effect of different environmental exposures (metal-rich particulate matter, air benzene levels and traffic-derived elemental carbon levels) on mitochondria | Pyrosequencing; qRT-PCR | Increased exposure to particulate matter increases MT-RNR1 and MT-TF gene methylation. Increased MT-RNR1 methylation is associated with a significant increase in mtDNA copy number | [52] |
| The effect of mtDNA methylation in the mitochondrial D-loop on gene expression in colorectal cancer cells | Methylation-specific PCR; western blotting | An increased level of demethylated sites in the D-loop of tumor cells is strongly associated with increased MT-ND2 expression and mtDNA copy number | [48] |
| The effect of methylation in the D-loop, MT-ND6 and MT-CO1 on disease progression in SS and NASH | Methylation-specific PCR; qRT-PCR | Increased MT-ND6 methylation and decreased MT-ND6 protein levels in NASH compared with SS. Physical activity reduced MT-ND6 methylation in NASH | [53] |
| The effect of decreased SAM on mtDNA methylation in Down's syndrome lymphoblastoid cells | LC-ESI-MS; LC-MS/MS | Decreased SAM availability in Down's syndrome lymphoblastoid cells reduce methyl uptake to mitochondria and lead to mtDNA hypomethylation | [54] |
| The tissue specificity of DNMTs and 5-mC in the mitochondria in relation to ALS models | IF; pyrosequencing | Increased methylation at six cytosine sites in the 16S rRNA gene in the spinal cord of an ALS mouse cell line. Reduced levels of mtDNMT3a protein in skeletal muscle and spinal cord early in disease | [55] |
| The effect of mtDNA methylation on mtDNA copy number in gastric cancer | qRT-PCR; pyrosequencing | Reduced mtDNA copy number levels in late clinicopathological stages. Demethylation of mtDNA increases mtDNA copy number | [56] |
ALS: Amyotrophic lateral sclerosis; IF: Immunofluorescence; LC–ESI-MS/MS: Liquid chromatography–electrospray ionization tandem mass spectrometry; LC–MS: Liquid chromatography mass spectrometry; mtDNA: Mitochondrial DNA; NASH: Nonalcoholic steatohepatitis; qRT-PCR: Quantitative real-time PCR; SAM: S-Adenosylmethionine; SS: Simple steatosis.
MtDNA modifications: evidence for a role in AD & aging
Until recently, the role of mtDNA modifications in AD has been largely ignored, despite the evidence that mitochondrial dysfunction is involved in AD [18] and that ncDNA methylation differences are associated with the disease [30,31]. At a global level, an initial dot blot study showed some evidence for increased mitochondrial 5-hmC in AD superior temporal gyrus tissue, although definitive conclusions could not be drawn given the small number of samples used [57]. Mitochondrial DNA modifications in the brain have been shown to be associated with aging, with global mtDNA 5-hmC levels reduced in the frontal cortex of aged mice and specifically decreased 5-hmC levels being found in the regulatory D-Loop, as well as in two genes encoding ETC complex I polypeptides (MT-ND2 and MT-ND5) [58]. Aging was not only found to be associated with overall decreased mtDNA 5-hmC levels but also with increased cortical expression of the mitochondrial ETC genes MT-ND2, MT-ND4, MT-ND4L, MT-ND5 and MT-ND6 [58]. A post-mortem study of frontal cortex described differential mtDNA gene expression of these genes, and other mitochondrial-encoded genes, in both early-and late-stage AD [7]. Taken together, these findings illustrate that alterations in mitochondrial-encoded genes do occur with aging and in age-related diseases, yet without further studies, the exact role of mtDNA methylation on mitochondrial gene expression in these instances remains uncertain.
Two genomes are better than one: interactions between the nuclear & mitochondrial genomes
As research into the field of mitochondrial epigenetics gains momentum, studies have focused on a potential trans-acting role of mtDNA in the epigenetic regulation of ncDNA, whereby covalent modifications across the mtDNA genome may affect not only the expression of a gene in cis, but also have trans-acting effects on the transcription of genes in the nuclear genome. Evidence for this is provided by cybrid models, which combine the nuclear genome of one source with the mitochon drial genome of another in an attempt to determine the functional role of the mtDNA. Using restriction landmark genomic scanning and Rho0 cells, a form of cybrid cell line designed for investigating mtDNA depletion, one study found that mtDNA depletion significantly altered DNA methylation at CpG islands in nuclear-encoded genes [59], indicating that there are functional interactions between the two genomes. Re-introduction of wild-type mtDNA restored DNA methylation levels, at some restriction landmark genomic scanning spots, suggesting that, at least for some genes, mitochondria may play a role in nuclear DNA methylation. This is corroborated by a recent study demonstrating that mitochondrial haplotype variation can affect ncDNA methylation, with mtDNA haplotype J exhibiting higher global DNA methylation levels, reduced ATP and overexpression of the nuclear gene methionine adenosyltransferase I, α (MAT1A), which is required for SAM production thus regulating methylation patterns in the nuclear genome [60]. Therefore genetic variations in mtDNA are capable of influencing epigenetic modifications in both the mitochondrial and nuclear genomes. As such, it is possible that mitochondrial dysfunction in AD could lead to alterations in mtDNA methylation, affecting nuclear gene expression.
The mitochondria comprises approximately 1500 proteins, however of these, only 13 are encoded by the mitochondrial genome; the remainder are encoded by the nuclear genome and imported into the mitochondria. A recent study found that greater than 600 of these genes have tissue-specific differentially methylated regions, ultimately leading to changes in mitochondrial function dependent upon tissue type [61]. This suggests that there is an additional level of complexity to consider in the study of mitochondrial epigenetics, whereby epigenetic changes in one genome may affect transcriptional control in another in a tissue-specific manner.
Interrogating the mitochondrial epigenome: technical caveats
Despite the potential importance of mitochondrial DNA modifications in AD, there are a number of technical challenges specific to interrogating the mitochondrial epigenome that have hampered widespread studies to date. These issues can be broadly summarized as encompassing genetic issues and specificity issues, which are outlined briefly with potential solutions in Table 2.
Table 2.
A summary of the major issues and potential solutions in the field of mitochondrial epigenetics.
| Caveat | Potential issues | Potential solutions | Ref. |
|---|---|---|---|
| Genetic issues | Incorrect determination of pseudogenes as mtDNA affects the validity of results Genetic mutations in mtDNA may have specific associated methylation signatures |
Isolate mitochondria before mtDNA extraction to avoid nuclear contamination Specific primers designed with the consideration of NUMT amplification BLAST search to identify known NUMTs Haplogroup and heteroplasmy studies should consider mtDNA methylation as a potential variable |
[62] |
| Cell specificity and technical issues | Different brain regions have differential methylation patterns and different cell population compositions Reduced methylation levels in mitochondria and variation in tDNA copy number may increase noise and dilute signals Bisulfite-based methodologies cannot distinguish between 5-mC and 5-hmC |
Larger samples sizes in specific brain subregions will improve statistical significance FACS or LCM to separate cell types such as glia and neurons prior to analysis Comparative analysis of techniques for their suitability to mitochondrial methylation studies should be considered Using oxidative bisulfite-sequencing allows for the distinction of 5-mC and 5-hmC at single base resolution |
[63] |
5-hmC: 5-hydroxymethylcytosine; 5-mC: 5-methylcytosine; FACS: Fluorescence-activated cell sorting; LCM: Laser capture microdissection; NUMT: Nuclear mitochondrial pseudogene.
Genetic issues Nuclear pseudogenes
By far the greatest concern when analyzing mtDNA methylation arises from regions of homology between the mitochondrial genome and nuclear mitochondrial pseudogenes (NUMTs). These genes are nuclear paralogs of mtDNA which have been translocated and inserted into the nuclear genome during evolution of both genomes [64]. This phenomena has been shown to be evolutionarily conserved across many species including cats [65], mice, chimpanzees, rhesus macaques [66] and hominins [67]. These insertions were thought to typically occur in noncoding regions; however, more evolutionary recent translocations have actually been shown to prefer integration into coding regions, thus leading to potential alterations in gene function with implications for disease [68]. NUMTs are generally small and typically comprise approximately 0.1% of the nuclear genome [69]. However in humans, it has been shown that some NUMTs can be as large as 14.7 kb, representing a significant portion of the approximately 16.6 kb human mitochondrial genome [70]. As such, the presence of NUMTs can cause major issues in genomic analyses using presequencing enrichment methods such as custom capture or long-range PCR as the likelihood of NUMT co-amplification, or even preferential amplification, increases due to the strong sequence similarity between the two segments of genome [71]. As such, this sequence similarity can lead to the misclassification of NUMTs as mtDNA during analysis, and has led to a number of publications wrongly describing NUMTS as mtDNA [72,73]. NUMT misclassification has also been observed in AD genetic studies whereby amplification of the NUMT sequence has led to false heteroplasmies (see below) being reported [74,75]. One potential solution is to separate mitochondria prior to DNA extraction in an attempt to reduce the risk of contaminating the mitochondrial and nuclear genomes. However, despite extensive research being dedicated to mtDNA analysis, existing methods for mitochondrial isolation and mtDNA extraction via the use of fractional precipitation or gradient ultracentrifugation remain time consuming and labor intensive [76] and often leave residual nuclear DNA contamination following mitochondrial isolation [77].
Variation in mtDNA: haplogroups & genetic & epigenetic heteroplasmy
Each mitochondrion contains between two and ten copies of mtDNA. However, not all mtDNA in each mitochondrion share the same DNA sequence. Indeed, mutations in some copies of mtDNA mean that the cell itself may be made up of a mixture of different sequences. This phenomenon is known as mitochondrial heteroplasmy and has been linked to various mitochondrial diseases [78]. It is a potential confounder in studies of mitochondrial diseases, because inter- and intra-individual heteroplasmic variation can confuse the association between a haplogroup with its corresponding phenotype. The importance of this issue, in the context of this review, is highlighted by a recent study demonstrating that mitochondrial heteroplasmy alters DNA methylation across the nuclear-encoded mitochondrial genes TFAM and POLMRT [79]. Finally, if mtDNA methylation is altered across different mtDNA in the same mitochondrion, it could create an epigenetic mosaic within the mitochondrion, the cell and across the tissue, whereby each copy of mtDNA may possess its own methylation profile. If this ‘methylomic heteroplasmy’ were to occur it could be very difficult to tease apart the effects of such a mosaic in functional studies.
On a larger scale, mutations in mtDNA can be used to help group cohorts or ‘haplogroups’. Throughout evolution, mutations in mtDNA may be conserved and passed on through maternal inheritance, thus allowing for the tracing of common ancestral lineage by comparing haplogroups. Numerous studies have identified both contributory and protective effects of different haplogroups in AD. For example, haplogroup K reduces the risk of developing sporadic AD in apolipoprotein ε4 (APOε4) carriers in an Italian population [80] but not in the Polish population [81]. This presents an additional potential caveat in mitochondrial epigenetics, as mitochondrial haplogroups have been found to affect global levels of DNA methylation [60]. As such, extra care should be taken to account for haplogroup variability in AD mitochondrial epigenetic studies.
Specificity & technical issues
The brain is a complex, heterogeneous organ with numerous functionally distinct subregions, each with their own different composition of cell types. Unsurprisingly, there are clear tissue-specific epigenetic differences across brain regions [82,83]. There is an added level of complexity with respect to the mitochondrial epigenome because each mitochondrion contains between 2–10 copies of mtDNA and each cell contains varying levels of mitochondria; therefore the amount of mtDNA copies in each cell can vary between 100 and 10,000, dependent upon cell type. In neurodegenerative diseases such as AD, the issue becomes more complicated in that the disease itself is characterized by the loss of neuronal cells and the activation of glia, a process that has been associated with changes in mitochondrial morphology and fission [84,85]. A recent study using laser capture microdissection demonstrated that alterations in mitochondrial 5-hmC are seen with age in dissected mouse cerebellar purkinje cells, which was not evident in whole cerebellar tissue [58], demonstrating the importance of cell-specific analyses in heterogeneous tissue, particularly when investigating functional impact.
Currently, the most common method of measuring DNA methylation is via the conversion of DNA with sodium bisulfite followed by subsequent sequence analysis. However, these approaches are unable to distinguish between 5-mC and 5-hmC [86], an important limitation given recent studies confirmed the presence of 5-hmC in mitochondria in brain tissue [58]. Studies have found that although both DNA modifications are present in the mitochondria, they occur at much lower levels compared with in ncDNA [37,57], and thus methods used for quantification may need to be more sensitive. Furthermore, variation in mitochondrial copy number may lead to the dilution of signals and reduce detection if the tissue is largely heterogeneous. Importantly, the mitochondrial genome is not interrogated using tools such as the Illumina Infinium 450K methylation array, the current gold standard for methylomic analyses in large numbers of samples; thus methods for detecting mtDNA modifications across the entire mitochondrial genome are largely restricted to antibody-based enrichment, such as MeDIP-Seq, which may be less sensitive for detecting low levels of modified cytosine and does not interrogate methylation levels at single base resolution [87].
Future perspective: the potential for biomarkers in AD
Two important goals of research into the etiology of AD are a fast, noninvasive, inexpensive and reliable biomarker and an effective treatment that targets the underlying neuropathology. A potential utility for DNA methylation biomarkers has been proposed for diseases in which traditional biomarkers are either too expensive, invasive, unspecific or insensitive for clinical purposes [88]. Epigenetic modifications have been widely studied in a variety of different cancers and other conditions such as pre-eclampsia to check for their suitability as prognostic and/or diagnostic bio-markers [89,90,91] Differential methylation of mtDNA has yet to be examined with respect to its potential utility as an AD biomarker, but certainly warrants further investigation.
Conclusion
With mitochondrial epigenetics only recently emerging as a focus for biomedical research, the role of the mitochondrial epigenome in AD has yet to receive much attention. However, it is possible that deregulation of the mitochondrial methylome may lead to aberrant changes in many of the intricately controlled processes that it helps to govern, such as apoptosis, which may play a key role in pathogenesis. Furthermore, as mitochondrial dysfunction occurs early in AD pathogenesis, it is plausible that alterations in the mitochondrial methylome may play a major role in the onset and development of the disease. Despite the field presenting numerous challenges, the links between mitochondrial epigenetics and AD provide good bounds for future research directions.
Executive summary.
Mitochondrial dysfunction: a prominent feature of AD
The mitochondrial genome plays a vital role in a variety of key biological functions, including apoptosis via caspase dependent and independent mechanisms, regulating calcium homeostasis and production of ROS.
Mitochondrial dysfunction is reported to occur in both the brain and blood of Alzheimer's disease (AD) patients.
EWAS & AD
Studies focusing on global levels of 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) have found a reduction in levels of both marks in AD brain.
Three recent epigenome-wide association studies have found differential methylation at specific loci in AD brain.
Epigenetic regulation of the mitochondria genome
Despite early controversial results, both 5-methylcytosine and 5-hmC have been recently reported in mitochondria.
MtDNA is not tightly wrapped by histones and is instead condensed by nucleoids, suggesting methylation could play an important role in gene regulation.
DNMT1 can bind to the D-Loop of the mitochondrial genome and can influence gene expression.
MtDNA methylation occurs at both CpG sites and non-CpG site in the mitochondrial genome. MtDNA methylation: a key player in AD?
Very few empirical studies have examined the role of mtDNA methylation in brain.
Decreased mtDNA 5-hmC levels and increased expression of some mitochondrial-encoded genes has been seen in the prefrontal cortex of aged mice.
Technical caveats
Nuclear mitochondrial pseudogenes (NUMT) misclassification has been observed in AD genetic studies whereby amplification of the NUMT sequence has led to false heteroplasmies being reported.
MtDNA methylation could be altered in different mitochondria, creating a methylomic heteroplasmy.
MtDNA methylation patterns could be cell specific and are an important consideration when investigating heterogeneous tissues such as brain.
Acknowledgments
This work was supported by an Alzheimer's Research UK pilot grant to KL and a NIH grant AG036039 to JM.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
•• of considerable interest
- 1.Prince M, Albanese E, Guerchet M, Prina M. World Alzheimer Report 2014. Alzheimer's Disease International. World Alzheimer Report 2014. Alzheimer's Disease International. www.alz.co.uk/research/WorldAlzheimerReport2014.pdf.
- 2.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE. Beta-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J. Neurosci. 1992;12(2):376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacor PN, Buniel MC, Furlow PW, et al. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer's disease. J. Neurosci. 2007;27(4):796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadowaki H, Nishitoh H, Urano F, et al. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 2005;12(1):19–24. doi: 10.1038/sj.cdd.4401528. [DOI] [PubMed] [Google Scholar]
- 7••.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer's disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromol. Med. 2004;5(2):147–162. doi: 10.1385/NMM:5:2:147. [Investigated expression levels of mitochondrial-encoded electron transport chain genes in Alzheimer's disease, reporting decreased expression of complex I and increased expression of complex III and IV in early- and late-stage disease.] [DOI] [PubMed] [Google Scholar]
- 8.Ankarcrona M, Mangialasche F, Winblad B. Rethinking Alzheimer's disease therapy: are mitochondria the key? J. Alzheimers Dis. 2010;20(Suppl. 2):S579–S590. doi: 10.3233/JAD-2010-100327. [DOI] [PubMed] [Google Scholar]
- 9.Lunnon K, Ibrahim Z, Proitsi P, et al. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer's disease blood. J. Alzheimers Dis. 2012;30(3):685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
- 10.Lunnon K, Sattlecker M, Furney S, et al. A blood gene expression marker of early Alzheimer's disease. J. Alzheimers Dis. 2013;33(3):737–753. doi: 10.3233/JAD-2012-121363. [DOI] [PubMed] [Google Scholar]
- 11.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 12.Pradelli LA, Beneteau M, Ricci JE. Mitochondrial control of caspase-dependent and -independent cell death. Cell. Mol. Life Sci. 2010;67(10):1589–1597. doi: 10.1007/s00018-010-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SL, Liu D, Kyriazis GA, Bagsiyao P, Ouyang X, Mattson MP. Mitochondrial uncoupling protein-4 regulates calcium homeostasis and sensitivity to store depletion-induced apoptosis in neural cells. J. Biol. Chem. 2006;281(49):37391–37403. doi: 10.1074/jbc.M605552200. [DOI] [PubMed] [Google Scholar]
- 14.Fu W, Ruangkittisakul A, MacTavish D, Baker GB, Ballanyi K, Jhamandas JH. Activity and metabolism-related Ca2+ and mitochondrial dynamics in co-cultured human fetal cortical neurons and astrocytes. Neuroscience. 2013;250:520–535. doi: 10.1016/j.neuroscience.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Zhao B. Oxidative stress and the pathogenesis of Alzheimer's disease. Oxid. Med. Cell. Longev. 2013;2013:316523. doi: 10.1155/2013/316523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devi L, Ohno M. Mitochondrial dysfunction and accumulation of the beta-secretase-cleaved C-terminal fragment of APP in Alzheimer's disease transgenic mice. Neurobiol. Dis. 2012;45(1):417–424. doi: 10.1016/j.nbd.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto M, Pickrell AM, Fukui H, Moraes CT. Mitochondrial DNA damage in a mouse model of Alzheimer's disease decreases amyloid beta plaque formation. Neurobiol. Aging. 2013;34(10):2399–2407. doi: 10.1016/j.neurobiolaging.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swerdlow RH, Burns JM, Khan SM. The Alzheimer's disease mitochondrial cascade hypothesis. J. Alzheimers Dis. 2010;20(Suppl. 2):S265–S279. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinnery PF, Elliott HR, Hudson G, Samuels DC, Relton CL. Epigenetics, epidemiology and mitochondrial DNA diseases. Int. J. Epidemiol. 2012;41(1):177–187. doi: 10.1093/ije/dyr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manczak M, Jung Y, Park BS, Partovi D, Reddy PH. Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J. Neurochem. 2005;92(3):494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- 21.Reddy PH, McWeeney S, Park BS, et al. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer's disease. Hum. Mol. Genet. 2004;13(12):1225–1240. doi: 10.1093/hmg/ddh140. [DOI] [PubMed] [Google Scholar]
- 22.Manczak M, Reddy PH. Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer's disease. Hum. Mol. Genet. 2012;21(23):5131–5146. doi: 10.1093/hmg/dds360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du H, Guo L, Fang F, et al. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat. Med. 2008;14(10):1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 25.Lunnon K, Mill J. Epigenetic studies in Alzheimer's disease: current findings, caveats, and considerations for future studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013;162B(8):789–799. doi: 10.1002/ajmg.b.32201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS ONE. 2009;4(8):e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31(12):2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chouliaras L, Mastroeni D, Delvaux E, et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging. 2013;34(9):2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Condliffe D, Wong A, Troakes C, et al. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer's disease brain. Neurobiol. Aging. 2014;35(8):1850–1854. doi: 10.1016/j.neurobiolaging.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakulski KM, Dolinoy DC, Sartor MA, et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 2012;29(3):571–588. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Lunnon K, Smith R, Hannon EJ, et al. Cross-tissue methylomic profiling in Alzheimer's disease implicates a role for cortex-specific deregulation of ANK1 in neuropathology. Nat. Neurosci. 2014;17(9):1164–1170. doi: 10.1038/nn.3782. [Used Illumina Infinium 450K methylation bead array to demonstrate DNA methylation changes in Alzheimer's disease cortex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeJager PL, Srivastava G, Lunnon, et al. Alzheimer'sdisease pathology is associated with early alterations in brain DNA methylationat ANK1, BIN1 and other loci. Nat. Neurosci. 2014;17(9):1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nass MM. Differential methylation of mitochondrial and nuclear DNA in cultured mouse, hamster and virus-transformed hamster cells. In vivo and in vitro methylation. J. Mol. Biol. 1973;80(1):155–175. doi: 10.1016/0022-2836(73)90239-8. [DOI] [PubMed] [Google Scholar]
- 34.Dawid IB. 5-methylcytidylic acid: absence from mitochondrial DNA of frogs and HeLa cells. Science. 1974;184(4132):80–81. doi: 10.1126/science.184.4132.80. [DOI] [PubMed] [Google Scholar]
- 35.Cummings DJ, Tait A, Goddard JM. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta. 1974;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- 36.Groot GS, Kroon AM. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in -CCGG- sequences. Biochim. Biophys. Acta. 1979;564(2):355–357. doi: 10.1016/0005-2787(79)90233-8. [DOI] [PubMed] [Google Scholar]
- 37••.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 2011;108(9):3630–3635. doi: 10.1073/pnas.1012311108. [Reported 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) in mtDNA, leading to a resurgence of interest in the field of mitochondrial epigenetics. This paper also identified an isoform of DNMT1, mtDNMT1, in the mitochondria.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alan L, Zelenka J, Jezek J, Dlaskova A, Jezek P. Fluorescent in situ hybridization of mitochondrial DNA and RNA. Acta Biochim. Pol. 2010;57(4):403–408. [PubMed] [Google Scholar]
- 39.Tauber J, Dlaskova A, Santorova J, et al. Distribution of mitochondrial nucleoids upon mitochondrial network fragmentation and network reintegration in HEPG2 cells. Int. J. Biochem. Cell Biol. 2013;45(3):593–603. doi: 10.1016/j.biocel.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Kaufman BA, Durisic N, Mativetsky JM, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18(9):3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manev H, Dzitoyeva S, Chen H. Mitochondrial DNA: a blind spot in neuroepigenetics. Biomol. Concepts. 2012;3(2):107–115. doi: 10.1515/bmc-2011-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 2009;37(20):6701–6715. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31(46):16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DNMT3a was localized in the mitochondria of motor neurons, potentially indicating tissue-specific localization of this methyltransferase. This study also found 5-mC in mitochondria in vivo, suggesting that mitochondrial methylation may play a role in motor neurons.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Bellizzi D, D'Aquila P, Scafone T, et al. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013;20(6):537–547. doi: 10.1093/dnares/dst029. [Reported both 5-mC and 5-hmC in mtDNA at both CpG and non-CpG sites. The study also found that inactivation of DNMT1, DNMT3a and DNMT3b reduced CpG methylation levels markedly, but failed to impact non-CpG methylation to the same extent. As such, this study poses the question as to whether DNMT activity is important for mitochondrial methylation, or whether other factors may also be important.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133(7):1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, Terragni J, Borgaro JG, et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 2013;3(2):567–576. doi: 10.1016/j.celrep.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol. Med. Rep. 2012;6(1):125–130. doi: 10.3892/mmr.2012.870. [DOI] [PubMed] [Google Scholar]
- 49.Phillips AC, Sleigh A, McAllister CJ, et al. Defective mitochondrial function in vivo in skeletal muscle in adults with Down's syndrome: a 31P-MRS study. PLoS ONE. 2013;8(12):e84031. doi: 10.1371/journal.pone.0084031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coyle JT, Oster-Granite ML, Gearhart JD. The neurobiologic consequences of Down syndrome. Brain Res. Bull. 1986;16(6):773–787. doi: 10.1016/0361-9230(86)90074-2. [DOI] [PubMed] [Google Scholar]
- 51.Wisniewski KE, Wisniewski HM, Wen GY. Occurrence of neuropathological changes and dementia of Alzheimer's disease in Down's syndrome. Ann. Neurol. 1985;17(3):278–282. doi: 10.1002/ana.410170310. [DOI] [PubMed] [Google Scholar]
- 52.Byun HM, Panni T, Motta V, et al. Effects of airborne pollutants on mitochondrial DNA methylation. Part. Fibre Toxicol. 2013;10:18. doi: 10.1186/1743-8977-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pirola CJ, Gianotti TF, Burgueno AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 54.Infantino V, Castegna A, Iacobazzi F, et al. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down's syndrome. Mol. Genet. Metab. 2011;102(3):378–382. doi: 10.1016/j.ymgme.2010.11.166. [DOI] [PubMed] [Google Scholar]
- 55.Wong M, Gertz B, Chestnut BA, Martin LJ. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front. Cell. Neurosci. 2013;7:279. doi: 10.3389/fncel.2013.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen SL, Zhang F, Feng S. Decreased copy number of mitochondrial DNA: a potential diagnostic criterion for gastric cancer. Oncol. Lett. 2013;6(4):1098–1102. doi: 10.3892/ol.2013.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer's disease. Mech. Ageing Dev. 2013;134(10):486–495. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging. 2012;33(12):2881–2891. doi: 10.1016/j.neurobiolaging.2012.02.006. [Found that the global levels of 5-hmC in mtDNA show an age-associated decrease in murine frontal cortex and that this was inversely correlated with the expression of some mitochondrial genes, suggesting a potential role of mtDNA methylation in aging.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 2008;7(8):1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellizzi D, D'Aquila P, Giordano M, Montesanto A, Passarino G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics. 2012;4(1):17–27. doi: 10.2217/epi.11.109. [DOI] [PubMed] [Google Scholar]
- 61.Takasugi M, Yagi S, Hirabayashi K, Shiota K. DNA methylation status of nuclear-encoded mitochondrial genes underlies the tissue-dependent mitochondrial functions. BMC Genomics. 2010;11:481. doi: 10.1186/1471-2164-11-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl Acad. Sci. USA. 2008;105(36):13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Booth MJ, Ost TW, Beraldi D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013;8(10):1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 65.Antunes A, Pontius J, Ramos MJ, O'Brien SJ, Johnson WE. Mitochondrial introgressions into the nuclear genome of the domestic cat. J. Hered. 2007;98(5):414–420. doi: 10.1093/jhered/esm062. [DOI] [PubMed] [Google Scholar]
- 66.Calabrese FM, Simone D, Attimonelli M. Primates and mouse NumtS in the UCSC Genome Browser. BMC Bioinform. 2012;13(Suppl. 4):S15. doi: 10.1186/1471-2105-13-S4-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ovchinnikov IV. Hominin evolution and gene flow in the Pleistocene Africa. Anthropol. Anz. 2013;70(2):221–227. doi: 10.1127/0003-5548/2013/0313. [DOI] [PubMed] [Google Scholar]
- 68.Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2(9):E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Triant DA, DeWoody JA. Molecular analyses of mitochondrial pseudogenes within the nuclear genome of arvicoline rodents. Genetica. 2008;132(1):21–33. doi: 10.1007/s10709-007-9145-6. [DOI] [PubMed] [Google Scholar]
- 70.Mourier T, Hansen AJ, Willerslev E, Arctander P. The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol. Biol. Evol. 2001;18(9):1833–1837. doi: 10.1093/oxfordjournals.molbev.a003971. [DOI] [PubMed] [Google Scholar]
- 71.Ho SY, Gilbert MT. Ancient mitogenomics. Mitochondrion. 2010;10(1):1–11. doi: 10.1016/j.mito.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J. Androl. 2003;24(3):388–392. doi: 10.1002/j.1939-4640.2003.tb02687.x. [DOI] [PubMed] [Google Scholar]
- 73.Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J. Med. Genet. 2008;45(12):769–772. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
- 74.Hirano M, Shtilbans A, Mayeux R, et al. Apparent mtDNA heteroplasmy in Alzheimer's disease patients and in normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc. Natl Acad. Sci. USA. 1997;94(26):14894–14899. doi: 10.1073/pnas.94.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis RE, Miller S, Herrnstadt C, et al. Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc. Natl Acad. Sci. USA. 1997;94(9):4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol. Genet. Metab. 2013;110(1–2):25–34. doi: 10.1016/j.ymgme.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 77.Zhou J, Liu L, Chen J. Method to purify mitochondrial DNA directly from yeast total DNA. Plasmid. 2010;64(3):196–199. doi: 10.1016/j.plasmid.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 78.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hua S, Lu C, Song Y, et al. High levels of mitochondrial heteroplasmy modify the development of ovine-bovine interspecies nuclear transferred embryos. Reprod. Fertil. Dev. 2012;24(3):501–509. doi: 10.1071/RD11091. [DOI] [PubMed] [Google Scholar]
- 80.Carrieri G, Bonafe M, De Luca M, et al. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Hum. Genet. 2001;108(3):194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 81.Maruszak A, Canter JA, Styczynska M, Zekanowski C, Barcikowska M. Mitochondrial haplogroup H and Alzheimer's disease: is there a connection? Neurobiol. Aging. 2009;30(11):1749–1755. doi: 10.1016/j.neurobiolaging.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 82.Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanchez-Mut JV, Aso E, Panayotis N, et al. DNA methylation map of mouse and human brain identifies target genes in Alzheimer's disease. Brain. 2013;136(Pt 10):3018–3027. doi: 10.1093/brain/awt237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banati RB, Egensperger R, Maassen A, Hager G, Kreutzberg GW, Graeber MB. Mitochondria in activated microglia in vitro. J. Neurocytol. 2004;33(5):535–541. doi: 10.1007/s11068-004-0515-7. [DOI] [PubMed] [Google Scholar]
- 85.Park J, Choi H, Min JS, et al. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 2013;127(2):221–232. doi: 10.1111/jnc.12361. [DOI] [PubMed] [Google Scholar]
- 86.Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48(4):317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- 87.Clark C, Palta P, Joyce CJ, et al. A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation 450 Bead Chip ((R)) for methylome profiling. PLoS ONE. 2012;7(11):e50233. doi: 10.1371/journal.pone.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications. Biochimie. 2012;94(11):2314–2337. doi: 10.1016/j.biochi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 89.Liu C, Liu L, Chen X, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS ONE. 2013;8(5):e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sandoval J, Mendez-Gonzalez J, Nadal E, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J. Clin. Oncol. 2013;31(32):4140–4147. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 91.Anderson CM, Ralph JL, Wright ML, Linggi B, Ohm JE. DNA methylation as a biomarker for preeclampsia. Biol. Res. Nurs. 2013;16(4):409–420. doi: 10.1177/1099800413508645. [DOI] [PubMed] [Google Scholar]