Abstract
Neurotrophins play a crucial role in mediating neuronal survival and synaptic plasticity. A lack of trophic factor support in the peripheral nervous system (PNS) is associated with a transcription-dependent programmed cell death process in developing sympathetic neurons. While most of the attention has been upon events culminating in cell death in the PNS, the earliest events that occur after trophic factor withdrawal in the central nervous system (CNS) have not been investigated. In the CNS, brain-derived neurotrophic factor (BDNF) is widely expressed and is released in an activity-dependent manner to shape the structure and function of neuronal populations. Reduced neurotrophic factor support has been proposed as a mechanism to account for changes in synaptic plasticity during neurodevelopment to aging and neurodegenerative disorders. To this end, we performed transcriptional profiling in cultured rat hippocampal neurons. We used a TrkB ligand scavenger (TrkB-FC) to sequester endogenous neurotrophic factor activity from hippocampal neurons in culture. Using a high-density microarray platform, we identified a significant decrease in genes that are associated with vesicular trafficking and synaptic function, as well as selective increases in MAP kinase phosphatases. A comparison of these changes with recent studies of Alzheimer’s disease and cognitive impairment in post mortem brain tissue revealed striking similarities in gene expression changes for genes involved in synaptic function. These changes are relevant to a wide number of conditions in which levels of BDNF are compromised.
Keywords: BDNF deprivation, Microarray, Transcription, Synaptic function, Hippocampus, Neurodegeneration
INTRODUCTION
Mechanisms leading to neuronal apoptosis have been extensively studied following deprivation of nerve growth factor (NGF) in the peripheral nervous system (PNS) (Levi-Montalcini and Booker, 1960; Gorin and Johnson, 1979; Oppenheim, 1991; Deckwerth and Johnson, 1993). A lack of trophic factor support in sympathetic neurons and PC12 cells results in a transcription-dependent programmed cell death process that could be prevented by inhibitors of gene transcription (Martin et al, 1988; Batistaou and Greene, 1991). Although PNS neurons have been extensively studied in the context of cell death mechanisms, the consequences of neurotrophic factor deprivation in the CNS have not been fully studied. An underlying hypothesis has been that the lack of neurotrophin expression and/or activity may underlie many neurodegenerative disorders (Appel, 1981; Chao et al, 2003; Longo et al, 2007).
BDNF is reduced in several neurodegenerative diseases, including Alzheimer’s disease (AD) and Huntington’s diseases (HD) (Zuccato and Cattaneo, 2009). In particular, exogenous delivery of BDNF can rescue degenerating neurons in animal models of AD, HD and Parkinson’s disease (Nagahara et al, 2009; Murer et al, 2001; Zuccato and Cattaneo, 2009). For instance, loss of cortical BDNF in animal models results in age-dependent degeneration of the striatum that closely resembles HD (Baquet et al, 2004; Strand et al, 2007). Despite the overwhelming evidence that BDNF levels are reduced in neurodegeneration, it remains unclear whether low levels of BDNF are a cause, or an effect, of the progressive neuronal loss in vulnerable cell types. It is also likely that BDNF levels change during the early phases of disease onset, which then increases vulnerability of neuronal populations to degeneration.
In this study, we sought to determine whether transcriptional changes occurred as a result of depriving BDNF from primary hippocampal neurons. We were interested in investigating early transcriptional events that occur within 12 hrs following withdrawal of BDNF before the induction of proapoptotic genes. We found that apoptotic death results from BDNF withdrawal in hippocampal neurons, as assayed by caspase-3 activity (Supplementary Figure), in a similar time frame as NGF withdrawal in cultured sympathetic neurons (Deshmukh and Johnson, 1997). We anticipate that events prior to initiation of cell death could shed light on the cellular processes that are compromised before cells commit to a death program. This is an early time period that has not been examined before for a loss of trophic support. Therefore, we employed a high density microarray platform to enable extensive coverage of known transcriptional activity. In this paper, we report the results of a gene expression profiling experiment and discuss the significance of these findings in synaptic function.
METHODS
Animals
Timed pregnant Sprague Dawley rats (Charles River Laboratories) were used in all experiments. Animal handling was in compliance with the New York University Langone Medical Center guidelines for the care and use of laboratory animals.
Hippocampal Neuronal Cultures
Hippocampal neuron cultures were prepared from embryonic day 18 (E18) embryos from timed-pregnant Sprague Dawley rats. Hippocampal tissue was dissected in Hanks Balanced Salt Solution and dissociated via trypsin treatment. Following dissociation, tissue was neutralized in DMEM/10% fetal bovine serum, then triturated in neurobasal medium using fire polished glass micropipettes. Cells were plated at a 1×106 cells/well in 6-well dishes pre-coated with poly-D-lysine. Neuronal cultures were then maintained in neurobasal medium supplemented with B27 for 7 days before BDNF deprivation. 5-fluorouracil was added to the medium to prevent glial proliferation. After 7 days, cultures were treated with a recombinant human TrkB fusion protein (TrkB-FC; 688-TK; R&D Systems, Minneapolis, MN; 100 ng/ml) to sequester endogenous BDNF, as described previously (Jeanneteau et al, 2010); no prior treatment of neuronal cultures with exogenous BDNF had been performed. TrkB-FC was added to each well and incubated for the following timepoints: 1.5 hours (hr), 3 hr, 6 hr, and 12 hr.
RNA extraction
For each timepoint, culture medium was removed and cells were washed with phosphate buffered saline (PBS). Following washing, Trizol reagent (Invitrogen) was added and cells were scraped, RNA extracted and precipitated with phenol and chloroform and stored at −80°C until use. Untreated wells of hippocampal neurons served as controls. RNA quality was assessed via bioanalysis (2100, Agilent Technologies, Santa Clara, CA). cRNA probes were synthesized and labeled using the GeneChip WT cDNA Synthesis and Amplification Kit (Affymetrix, Santa Clara, CA)
Microarray Hybridization and data analysis
Microarray analysis was carried out with cRNA probes synthesized and labeled using the GeneChip WT cDNA Synthesis and Amplification assay (Affymetrix), and subjected to hybridization with GeneChip® Rat Exon 1.0 ST array (Affymetrix) according to the manufacturer’s instructions. Microarrays were hybridized with cRNA derived from experimental duplicates (n=2) of each time point along with duplicate untreated control samples.
Analysis of microarray data was performed using GeneSpring v11 (Agilent Technologies). The expression value of each probe set was determined after standard normalization of the CEL files by Robust Multichip Average (RMA), which includes quantile normalization step for probe intensity level (Bolstad et al, 2003). Baseline normalization of every gene to the average of the control samples was performed. Analysis of variance (ANOVA), (P < 0.05, alpha setting, no corrections) was used to identify reproducible modulation of transcript abundance across all conditions for the entire timecourse. ANOVA compared all conditions against each other and assigned a p-value for any significant differences based on reproducible replicate measurements. A threshold of 20% fold-change at any given condition different from baseline (untreated control) was applied to further strengthen the lists identified by ANOVA. Probesets were considered for functional analysis if the probe set intensity in one or more of the timepoints was greater than the 20% threshold in the two biological replicates. Hierarchical cluster analysis was used to cluster gene groups defined by the ANOVA statistical filtering. Functional annotation was performed with the Gene Ontology (GO) classification system using the web based DAVID software (Huang et al, 2009). Genes were grouped into classes using the gene enrichment clustering tool in DAVID. Significant association of a gene with a specified functional class was determined by the EASE Score (cut off p-value≤0.1); a modified Fisher Exact statistical test used to measure gene enrichment based on functional annotation in the DAVID system (Huang et al, 2009). The EASE score for each class is shown as the gene enrichment p-value on the functional enrichment tables.
qPCR validation
Genes relevant to synaptic function were validated via qPCR. Samples were assayed on a real-time qPCR cycler (7900HT, Applied Biosystems) using Taqman probes for these genes: Vesicle-Associated Membrane Protein (VAMP4) assay ID:Rn01490252_m1, Dual Specificity Phosphatase 5 (DUSP5) assay ID:Rn00592122_m1, Golgin5 (Golga5) assay ID:Rn01517894_m1, Spry2 assay ID: (Rn02534289_s1), Acan assay ID:Rn00573424_m1 Rab8b assay ID:Rn00596360_m1. qPCR assays were performed in triplicate per sample on a 96 well platform. The ddCT method was employed to determine relative gene level differences with glyceraldehyde-3 phosphate dehydrogenase (GAPDH) assay ID:Rn01775763_g1, Beta Actin (Actb) assay ID: Rn00667869_m1 or Ribophorin assay ID:Rn00565052_m1 as endogenous controls as described previously (Alldred et al, 2008; 2009).
Statistical analysis for quantitative PCR
Statistical analysis for the qPCR data was performed with Graph Pad Prism® (version 6.0a). Data was analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test (p-value *p<0.05, **<0.01, and ***p<0.001; 95% Confidence Interval of difference.
RESULTS
Transcriptional profiling and gene clustering
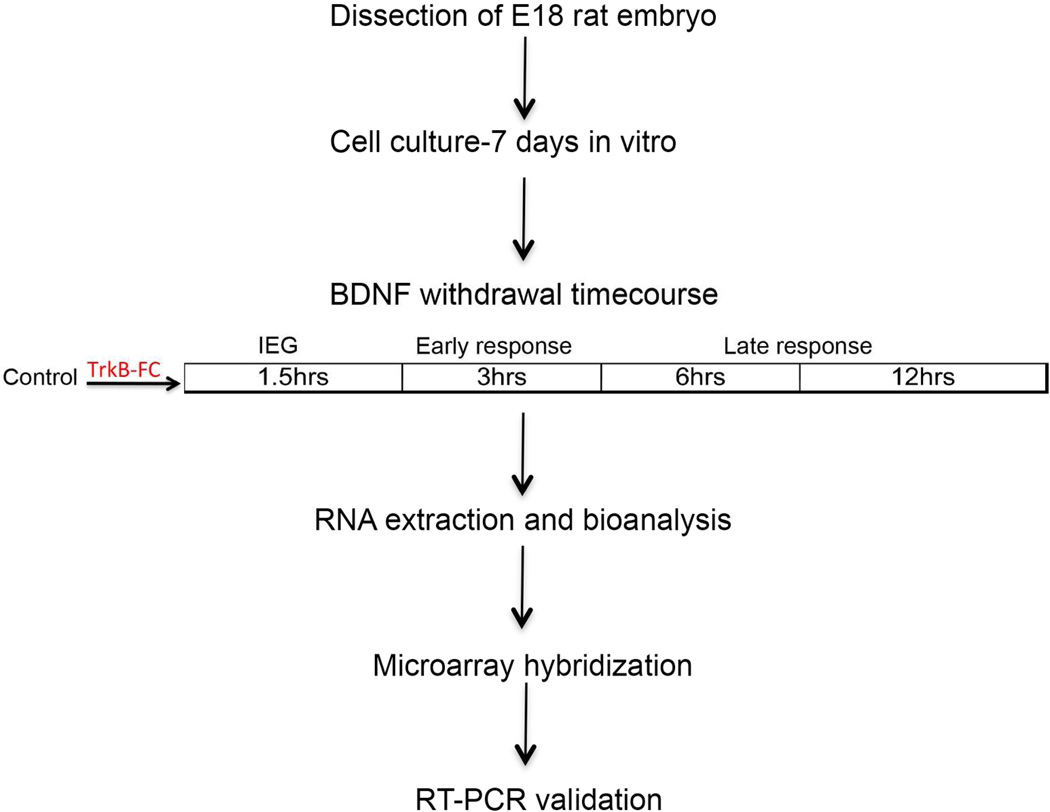
To address early events following deprivation of BDNF, we performed transcriptional profiling in primary hippocampal cultures after BDNF deprivation at four early timepoints (1.5 hr, 3 hr, 6 hr and 12 hr) to capture different phases of transcriptional activity. A schematic diagram of the experimental approach is presented in [Fig. 1].
Figure 1.
Experimental design. Outline of the BDNF withdrawal assay and high-density microarray profiling experiments. Hippocampal neurons were generated from E18 rat embryos and cultured for 7 days. On DIV 7, endogenous BDNF activity was sequestered by adding TrkB-Fc (100ng/mL) to the culture medium at different timepoints. Each timepoint was hypothesized to capture distinct transcriptional changes. The 1.5hr time point was anticipated to capture immediate early gene activity. This would activate a subsequent wave of early response genes spanning the 3–6hr timeframe. Late response gene expression would be captured from 6hrs up to the 12hr timepoint. After BDNF withdrawal, RNA was extracted from the samples followed by high density transcriptional profiling.
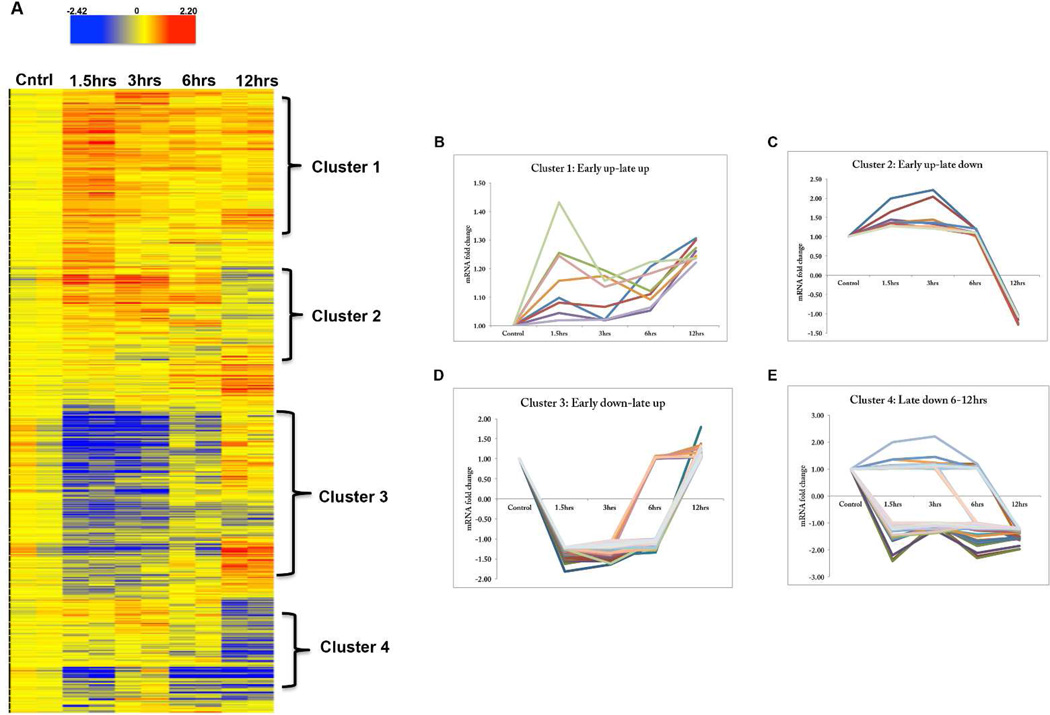
Microarray analysis of cultured hippocampal neurons following BDNF withdrawal identified 1467 genes with significant reproducibility between experimental replicates. Gene clustering (GeneSpring supervised clustering tool; Agilent Technologies) revealed several patterns or mosaics of altered gene expression [Fig. 2A]. Cluster 1 (Early up, late up) is comprised of genes that increased upon BDNF withdrawal and remained upregulated throughout the duration of the time course [Fig. 2(A), (B)]. However, a majority of these genes had Affymetrix probe IDs but no gene symbol, suggesting that they had not been functionally characterized. Only a few functionally annotated genes from this cluster had fold change expression levels above the +1.2 fold change cut off mentioned in the methods [Supplemental Table 1.]. Two striking transcriptional profiles displayed immediate upregulation cluster 2; [Fig. 2(A), (C), Table 1] and down-regulation, cluster 3; [Fig. 2(A), (D), Table 2] at early timepoints (1.5 hrs, 3 hrs), which returned to baseline by 6–12 hrs. Genes were also uncovered that showed a marked decrease (cluster 4) [Fig. 2(A), (E), Table 2 Supplemental] in the late phases of the time course. Among these were genes that have been implicated in ribosomal and golgi function as well as protein transport. These genes showed at least a 50% decrease in expression, which may point to a significant compromise in function. Other clusters that were also distinctly represented were genes that decreased throughout the entire timecourse [cluster 5, Table 3. Supplemental] and genes that increased at the end of the timecourse [cluster 6, Table 4. Supplemental]. Some of the genes in these clusters were overlapping with genes identified in clusters 2 and 3. Taken together, these results suggest that withdrawal of BDNF elicits distinct transcriptional events for different genes during the deprivation process.
Figure 2.
Hierarchical clustering post BDNF deprivation. (A) Heat map illustrating profiles of change for normalized probe sets after the ANOVA statistical filtering. Red (upregulation), Blue (downregulation), Yellow (no change). Genes showing similar profiles of changes with time were grouped to identify profile subgroups in a semi-supervised manner in GeneSpring GX. (B–E) Representative gene expression profiles (∼50 genes per cluster) were used to generate line graph plots for trends of change over time for each cluster. The graphs show relative fold change (y-axis) with time (x-axis). Four clusters are shown; Early up-late up (B), Early up-late down (C), Early down-late up (D), Late down (E)
Table 1. Genes that increased early in the time course then decreased at the end of the time course.
Gene list from cluster 2 showing genes that exhibit the early up-late down profile. The 1.5hr timepoint is highlighted in pink to show the fold increase of each gene early in the time course. The fold change cut off is +1.2. A majority of the genes within this cluster also reflect an increase in expression at the 3hr time point before plateauing to baseline by 6–12hrs. The p-value associated with each gene is based on reproducible replicate measurements and reflects a significant change (p≤ 0.05) in probe intensity for one or more timepoints relative to the untreated control.
| CLUSTER 2: EARLY UP-LATE DOWN | FOLD CHANGE RELATIVE TO CONTROL | ||||||
|---|---|---|---|---|---|---|---|
| Affymetrix ID | p-value | Gene Symbol | Gene Name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| 10716080 | 0.0078 | Dusp5 | dual specificity phosphatase 5 | 2.00 | 2.21 | 1.19 | −1.25 |
| 10708021 | 0.0495 | Acan | aggrecan | 1.65 | 2.04 | 1.20 | −1.16 |
| 10861303 | 0.0092 | Rnf148 | ring finger protein 148 | 1.45 | 1.14 | 1.20 | 1.08 |
| 10785773 | 0.0230 | Spry2 | sprouty homolog 2 (Drosophila) | 1.45 | 1.34 | 1.11 | −1.09 |
| 10889919 | 0.0305 | Gpr33 | G protein-coupled receptor 33 | 1.43 | 1.16 | 1.22 | 1.24 |
| 10769672 | 0.0203 | Rgs4 | regulator of G-protein signaling 4 | 1.36 | 1.45 | 1.01 | −1.28 |
| 10767565 | 0.0107 | Mfsd4 | major facilitator superfamily domain containing 4 | 1.36 | 1.37 | 1.20 | −1.03 |
| 10702412 | 0.0199 | Rspo3 | R-spondin 3 homolog (Xenopus laevis) | 1.35 | 1.23 | 1.06 | −1.28 |
| 10779835 | 0.0462 | Angl | angiogenin, ribonuclease A family, member 1 | angiogenin, ribonuclease, RNase A family, 5 | 1.32 | 1.24 | 1.10 | 1.17 |
| 10857382 | 0.0312 | Fgd5 | FYVE, RhoGEF and PH domain containing 5 | 1.30 | 1.19 | 1.22 | 1.08 |
| 10837435 | 0.0024 | Olr446 | olfactory receptor 446 | 1.30 | 1.14 | 1.15 | 1.15 |
| 10883071 | 0.0184 | RGD1304963 | similar to hypothetical protein MGC38716 | 1.30 | 1.21 | 1.07 | 1.07 |
| 10899967 | 0.0271 | Olr877 | olfactory receptor 877 | 1.29 | 1.02 | 1.03 | 1.09 |
| 10724351 | 0.0437 | Olrl45 | olfactory receptor 145 | 1.29 | 1.22 | 1.05 | 1.19 |
| 10726223 | 0.0128 | LOC499276 | similar to RIKEN cDNA 1700022C21 | 1.28 | 1.15 | 1.07 | 1.14 |
| 10717278 | 0.0082 | Taar7d | trace-amine-associated receptor 7d | 1.28 | 1.09 | 1.09 | 1.08 |
| 10879343 | 0.0328 | Olr869 | olfactory receptor 869 | 1.28 | 1.14 | 1.21 | 1.15 |
| 10824388 | 0.0137 | Fdps | farnesyl diphosphate synthase | 1.28 | 1.28 | 1.10 | −1.06 |
| 10768642 | 0.0378 | Lamc2 | laminin, gamma 2 | 1.27 | 1.07 | 1.08 | 1.00 |
| 10715531 | 0.0376 | Cyp2c23 | cytochrome P450, family 2, subfamily c, polypeptide 23 | 1.27 | 1.19 | 1.18 | 1.18 |
| 10860499 | 0.0285 | Sema3d | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D | 1.27 | 1.21 | 1.10 | −1.08 |
| 10729057 | 0.0322 | Olr379 | olfactory receptor 379 | 1.27 | 1.16 | 1.08 | 1.10 |
| 10718351 | 0.0398 | Fprl | formyl peptide receptor 1 | 1.27 | 1.11 | 1.07 | 1.11 |
| 10724307 | 0.0335 | Olr127 | olfactory receptor 127 | 1.27 | −1.04 | 1.10 | 1.08 |
| 10773146 | 0.0316 | Fgfbpl | fibroblast growth factor binding protein 1 | 1.26 | 1.17 | 1.10 | 1.06 |
| 10722858 | 0.0113 | Agbl1 | ATP/GTP binding protein-like 1 | 1.26 | 1.13 | −1.01 | 1.24 |
| 10855084 | 0.0389 | Olr806 | olfactory receptor 806 | 1.25 | 1.19 | 1.12 | 1.27 |
| 10722237 | 0.0439 | Mrgprb2 | MAS-related GPR, member B2 | 1.25 | 1.16 | 1.13 | 1.11 |
| 10905602 | 0.0076 | Dnajb7 | DnaJ (Hsp40) homolog, subfamily B, member 7 | 1.25 | 1.08 | 1.11 | 1.07 |
| 10774361 | 0.0286 | Rab1 | RAB1, member RAS oncogene family | 1.25 | 1.20 | 1.09 | −1.06 |
| 10709544 | 0.0496 | Olr190 | olfactory receptor 190 | 1.25 | 1.03 | −1.06 | 1.06 |
| 10932612 | 0.0103 | RGD1565862 | similar to Spindlin-like protein 2 (SPIN-2) | 1.25 | 1.14 | 1.08 | 1.09 |
| 10704565 | 0.0203 | Ptgir | prostaglandin 12 (prostacyclin) receptor (IP) | 1.24 | 1.17 | 1.14 | 1.13 |
| 10855079 | 0.0185 | Tas2r126 | taste receptor, type 2, member 126 | 1.24 | 1.14 | 1.18 | 1.24 |
| 10735765 | 0.0073 | Olr1515 | olfactory receptor 1515 | 1.24 | 1.18 | 1.04 | 1.16 |
| 10724270 | 0.0344 | Olr104 | olfactory receptor 104 | 1.24 | 1.07 | 1.09 | 1.15 |
| 10728977 | 0.0072 | Olr318 | olfactory receptor 318 | 1.24 | 1.13 | 1.09 | 1.18 |
| 10795077 | 0.0420 | Nrsn1 | neurensin 1 | 1.24 | 1.37 | 1.08 | 1.05 |
| 10769765 | 0.0435 | Fcrla | Fc receptor-like A | 1.24 | 1.08 | 1.03 | 1.13 |
| 10935245 | 0.0371 | Actrt1 | actin-related protein T1 | 1.23 | 1.09 | 1.19 | 1.10 |
| 10725841 | 0.0498 | RGD1563217 | similar to RIKEN cDMA 4930451l11 | 1.23 | 1.14 | 1.08 | 1.13 |
| 10835873 | 0.0405 | Olr419 | olfactory receptor 419 | 1.23 | 1.17 | 1.07 | 1.09 |
| 10724678 | 0.0235 | Olr259 | olfactory receptor 259 | 1.23 | 1.10 | 1.10 | 1.08 |
| 10817845 | 0.0429 | Vtcn1 | V-set domain containing T cell activation inhibitor 1 | 1.23 | 1.08 | 1.00 | 1.07 |
| 10904769 | 0.0061 | Nrbp2 | nuclear receptor binding protein 2 | 1.23 | 1.31 | 1.25 | −1.13 |
| 10749816 | 0.0215 | Olr1355 | Olr1525 | olfactory receptor 1355 | olfactory receptor 1525 | 1.23 | 1.14 | 1.09 | 1.12 |
| 10936841 | 0.0246 | RGD1559951 | similar to 60S ribosomal protein L37a | 1.23 | 1.17 | 1.14 | 1.11 |
| 10817074 | 0.0357 | Pglyrp4 | peptidoglycan recognition protein 4 | 1.23 | 1.13 | 1.09 | 1.15 |
| 10701802 | 0.0018 | Plagl1 | pleiomorphic adenoma gene-like 1 | 1.23 | 1.19 | 1.08 | −1.15 |
| 10811832 | 0.0422 | Nupl33 | nucleoporin 133 | 1.23 | 1.16 | 1.10 | 1.03 |
| 10823363 | 0.0122 | P2ry13 | purinergic receptor P2Y, G-protein coupled, 13 | 1.23 | 1.10 | 1.07 | 1.03 |
| 10734740 | 0.0296 | Pik3r6 | phosphoinositide-3-kinase, regulatory subunit 6 | 1.22 | 1.06 | 1.03 | 1.12 |
| 10724609 | 0.0462 | Olr221 | olfactory receptor 221 | 1.22 | 1.10 | 1.06 | 1.12 |
| 10798475 | 0.0268 | Hist1h2bc | histone cluster 1, H2bc | 1.22 | 1.18 | −1.11 | 1.19 |
| 10907904 | 0.0234 | Mmp10 | matrix metallopeptidase 10 | 1.22 | 1.10 | 1.03 | 1.06 |
| 10851628 | 0.0238 | Spinlw1 | serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) | 1.22 | 1.15 | 1.18 | 1.16 |
| 10782387 | 0.0083 | Nid2 | nidogen 2 | 1.22 | 1.29 | 1.00 | 1.03 |
| 10819052 | 0.0238 | Lef1 | lymphoid enhancer binding factor 1 | 1.21 | 1.35 | 1.05 | −1.08 |
| 10845607 | 0.0361 | Rbms1 | RNA binding motif, single stranded interacting protein 1 | 1.21 | 1.23 | 1.06 | −1.18 |
| 10876765 | 0.0053 | Olr851 | olfactory receptor 851 | 1.21 | 1.06 | 1.11 | 1.17 |
| 10916734 | 0.0400 | C2cd2l | C2 calcium-dependent domain containing 2-like | 1.20 | 1.13 | 1.08 | −1.13 |
| 10795460 | 0.0196 | Psme1-ps1 | proteasome [prosome, macropain) activator subunit 1 (PA28 alpha), pseudogene 1 | 1.20 | 1.12 | 1.12 | 1.07 |
| 10719824 | 0.0166 | Cnfn | cornifelin | 1.20 | 1.11 | 1.09 | 1.08 |
Table 2. Genes that decreased early in the time course then increased.
Gene list from cluster 3 showing genes that exhibit the early down-late up trend. The 1.5hr timepoint is highlighted in green to show the fold decrease of each gene early the time course. The fold change cut off is −1.2. A majority of the genes within this cluster also reflect a decrease in expression at the 3hr time point before plateauing to baseline by 6–12hrs. The p-value associated with each gene is based on reproducible replicate measurements and reflects a significant change (p≤ 0.05) in probe intensity for one or more timepoints relative to the untreated control.
| CLUSTER 3: EARLY DOWN-LATE UP | FOLD CHANGE RELATIVE TO CONTROL | ||||||
|---|---|---|---|---|---|---|---|
| Affymetrix ID | p-value | Gene Symbol | Gene Name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| 10902409 | 0.0161 | LOC688019 | similar to THAP domain containing, apoptosis associated protein 2 | −1.82 | −1.65 | −1.13 | 1.17 |
| 10912412 | 0.0081 | Tfdp2 | transcription factor Dp-2 (E2F dimerization partner 2) | −1.71 | −1.55 | −1.04 | −1.07 |
| 10705431 | 0.0175 | LOC499110 | similar to Zinc finger protein 354A (Transcription factor 17) (Renal transcription factor Kid-1) | −1.62 | −1.44 | −1.18 | 1.20 |
| 10876567 | 0.0164 | Xpa | Ncbp1 | xeroderma pigmentosum, complementation group A | nuclear cap binding protein subunit 1 | −1.55 | −1.56 | −1.17 | 1.01 |
| 10760760 | 0.0469 | LOC288521 | similar to Leukosialin precursor (Leucocyte sialoglycoprotein) (Sialophorin) (CD43) (W3/13 antigen) | −1.53 | −1.41 | −1.34 | 1.79 |
| 10849813 | 0.0495 | Zc3h8 | zinc finger CCCH type containing 8 | −1.53 | −1.48 | −1.19 | 1.06 |
| 10717779 | 0.0307 | Fbxo5 | F-box protein 5 | −1.51 | −1.38 | −1.21 | 1.37 |
| 10752007 | 0.0191 | Dgkg | diacylglycerol kinase, gamma | −1.49 | −1.52 | −1.13 | 1.00 |
| 10833668 | 0.0368 | Gpbp1l1 | GC-rich promoter binding protein 1-like 1 | −1.49 | −1.45 | −1.12 | 1.14 |
| 10836633 | 0.0195 | Phospho2 | phosphatase, orphan 2 | −1.48 | −1.36 | −1.19 | 1.18 |
| 10804371 | 0.0328 | Ccdc112 | coiled-coil domain containing 112 | −1.47 | −1.28 | −1.15 | −1.17 |
| 10917087 | 0.0105 | Fam55b | family with sequence similarity 55, member B | −1.47 | −1.58 | −1.18 | −1.04 |
| 10871250 | 0.0361 | Gpbp1l1 | GC-rich promoter binding protein 1-like 1 | −1.47 | −1.42 | −1.10 | 1.16 |
| 10819130 | 0.0343 | Gpbp1l1 | GC-rich promoter binding protein 1-like 1 | −1.47 | −1.45 | −1.09 | 1.15 |
| 10908990 | 0.0348 | Pus3 | pseudouridylate synthase 3 | −1.47 | −1.27 | −1.16 | 1.02 |
| 10798376 | 0.0279 | Ttrap | Traf and Tnf receptor associated protein | −1.46 | −1.30 | −1.19 | −1.00 |
| 10811216 | 0.0380 | Adat1 | adenosine deaminase, tRNA-specific 1 | −1.46 | −1.37 | −1.13 | 1.09 |
| 10774435 | 0.0449 | Gpbp1l1 | GC-rich promoter binding protein 1-like 1 | −1.46 | −1.45 | −1.09 | 1.14 |
| 10822961 | 0.0378 | Mfsd8 | major facilitator superfamily domain containing 8 | −1.46 | −1.51 | −1.18 | 1.10 |
| 10867688 | 0.0444 | Rbml2b | RNA binding motif protein 12B | −1.46 | −1.53 | −1.03 | 1.01 |
| 10876896 | 0.0180 | Ctnnal1 | catenin (cadherin associated protein), alpha-like 1 | −1.45 | −1.34 | −1.10 | −1.22 |
| 10750812 | 0.0433 | Cep97 | centrosomal protein 97kDa | −1.45 | −1.39 | −1.10 | −1.08 |
| 10931638 | 0.0251 | Cbx8 | chromobox homolog 8 (Pc class homolog, Drosophila) | −1.45 | −1.10 | −1.11 | −1.15 |
| 10719829 | 0.0214 | Lipe | lipase, hormone sensitive | −1.45 | −1.17 | −1.08 | 1.16 |
| 10885931 | 0.0269 | RGD1560978 | similar to hypothetical protein | −1.44 | −1.39 | −1.17 | −1.07 |
| 10742104 | 0.0464 | Gpbp1l1 | GC-rich promoter binding protein 1-like 1 | −1.44 | −1.44 | −1.08 | 1.15 |
| 10759241 | 0.0217 | Hscb | HscB iron-sulfur cluster co-chaperone homolog (E. coli) | −1.43 | −1.23 | −1.11 | 1.00 |
| 10704752 | 0.0300 | Fbxo46 | F-box protein 46 | −1.43 | −1.16 | −1.18 | 1.05 |
| 10826158 | 0.0494 | Prmt6 | protein arginine methyltransferase 6 | −1.42 | −1.33 | −1.15 | 1.16 |
| 10811208 | 0.0033 | Tmem231 | transmembrane protein 231 | −1.41 | −1.37 | −1.15 | 1.08 |
| 10705772 | 0.0063 | Zfp84 | zinc finger protein 84 | −1.41 | −1.39 | −1.19 | 1.08 |
| 10704234 | 0.0417 | Zfp128 | zinc finger protein 128 | −1.39 | −1.20 | −1.19 | 1.19 |
| 10846173 | 0.0489 | Cir1 | corepressor interacting with RBPJ, 1 | −1.39 | −1.27 | −1.13 | 1.11 |
| 10731787 | 0.0467 | Tcfap4 | transcription factor AP4 | −1.38 | −1.29 | −1.08 | −1.04 |
| 10918374 | 0.0460 | Rab8b | RAB8B, member RAS oncogene family | −1.38 | −1.35 | −1.06 | 1.07 |
| 10885464 | 0.0160 | Fut8 | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | −1.38 | −1.42 | −1.01 | 1.07 |
| 10807464 | 0.0057 | Pla2g15 | phospholipase A2, group XV | −1.37 | −1.18 | −1.06 | 1.02 |
| 10794099 | 0.0040 | Uimc1 | ubiquitin interaction motif containing 1 | −1.37 | −1.27 | −1.09 | −1.07 |
| 10704918 | 0.0285 | RGD1564214 | similar to Zfp93 protein | −1.37 | −1.20 | −1.14 | 1.08 |
| 10778579 | 0.0116 | Etaa1 | Ewing tumor-associated antigen 1 | −1.37 | −1.29 | −1.09 | 1.04 |
| 10883606 | 0.0338 | Rdh14 | retinol dehydrogenase 14 (all-trans/9-cis/11-cis) | −1.37 | −1.27 | −1.10 | −1.05 |
| 10829703 | 0.0436 | Tfam | transcription factor A, mitochondrial | −1.37 | −1.26 | −1.15 | 1.02 |
| 10905148 | 0.0136 | Znf250 | zinc finger protein 250 | −1.36 | −1.12 | −1.12 | 1.16 |
| 10891818 | 0.0331 | Atxn3 | ataxin 3 | −1.36 | −1.30 | −1.13 | −1.13 |
| 10708235 | 0.0008 | Zfp29 | zinc finger protein 29 | −1.36 | −1.26 | −1.14 | 1.15 |
| 10757140 | 0.0417 | Zfp68 | zinc finger protein 68 | −1.35 | −1.43 | −1.15 | 1.08 |
| 10807504 | 0.0022 | Zfp90 | zinc finger protein 90 | −1.35 | −1.34 | −1.10 | 1.03 |
| 10933494 | 0.0060 | LOC302680 | similar to CXORF15 | −1.35 | −1.32 | −1.15 | 1.06 |
| 10832460 | 0.0156 | Zfp280b | zinc finger protein 280b | −1.35 | −1.15 | −1.09 | 1.35 |
| 10908248 | 0.0174 | Zfp42612 | zinc finger protein 426-like 2 | −1.35 | −1.26 | −1.07 | −1.12 |
| 10803953 | 0.0006 | Sra1 | steroid receptor RNA activator 1 | −1.35 | −1.05 | −1.19 | −1.17 |
| 10894221 | 0.0262 | Zfp472 | zinc finger protein 472 | −1.34 | −1.22 | −1.11 | −1.00 |
| 10743608 | 0.0131 | Znf286a | zinc finger protein 286A | −1.33 | −1.18 | −1.09 | 1.10 |
| 10782695 | 0.0158 | Abhd6 | abhydrolase domain containing 6 | −1.33 | −1.35 | −1.07 | 1.21 |
| 10865630 | 0.0433 | Ncapd2 | non-SMC condensin 1 complex, subunit D2 | −1.32 | −1.25 | −1.11 | 1.05 |
| 10864979 | 0.0389 | Ankrd26 | ankyrin repeat domain 26 | −1.32 | −1.32 | 1.00 | 1.04 |
| 10924199 | 0.0148 | Smarcal1 | Swi/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 | −1.32 | −1.19 | −1.02 | 1.14 |
| 10878912 | 0.0240 | Toe1 | target of EGR1, member 1 (nuclear) | −1.32 | −1.12 | 1.01 | 1.34 |
| 10860226 | 0.0281 | Lrrcl7 | Fbxl13 | leucine rich repeat containing 17 | F-box and leucine-rich repeat protein 13 | −1.31 | 1.05 | −1.13 | −1.02 |
| 10733726 | 0.0081 | Mfap3 | microfibrillar-associtaed protein 3 | −1.31 | −1.36 | −1.09 | 1.10 |
| 10742431 | 0.0217 | Rufy1 | RUN and FYVE domain containing 1 | −1.31 | −1.12 | −1.09 | −1.12 |
| 10726655 | 0.0300 | Bet1l | blocked early in transport 1 homolog (S. cerevisiae) like | −1.31 | −1.10 | −1.14 | −1.08 |
| 10739099 | 0.0106 | Map3k3 | mitogen activated protein kinase kinase kinase 3 | −1.31 | −1,16 | −1.14 | −1.02 |
| 10826764 | 0.0237 | Rrh | retinal pigment epithelium derived rhodopsin homolog | −1.31 | −1.34 | −1.08 | 1.18 |
| 10758094 | 0.0067 | Slc15a4 | solute carrier family 15, member 4 | −1.30 | −1.35 | −1.19 | −1.00 |
| 10779668 | 0.0092 | Socs4 | suppressor of cytokine signaling 4 | −1.30 | 1.01 | −1.13 | −1.07 |
| 10883321 | 0.0361 | Asxl2 | additional sex combs like 2 (Drosophila) | −1.30 | −1.19 | −1.20 | 1.04 |
| 10889660 | 0.0385 | Ahr | aryl hydrocarbon receptor | −1.30 | −1.47 | −1.08 | −1.04 |
| 10904242 | 0.0163 | RGD1308133 | similar to RIKEN cDNA 1700010C24 | −1.30 | −1.16 | −1.02 | 1.01 |
| 10830989 | 0.0322 | Ppp1r10 | protein phosphatase 1, regulatory subunit 10 | −1.29 | −1.19 | −1.09 | 1.36 |
| 10891364 | 0.0239 | Alkbh | alkB, alkylation repair homolog (E. coli) | −1.29 | −1.21 | −1.08 | 1.23 |
| 10838197 | 0.0239 | Cstf3 | cleavage stimulation factor, 3’ pre-RNA, subunit 3 | −1.29 | −1.29 | −1.19 | 1.03 |
| 10736509 | 0.0413 | Gosr1 | golgi SNAP receptor complex member 1 | −1 79 | −1 18 | −1.07 | 1.07 |
| 10800603 | 0.0245 | Pik3c3 | phosphoinositide-3-kinase, class 3 | −1.29 | −1.19 | −1.09 | 1.03 |
| 10924392 | 0.0147 | Ttll4 | tubulin tyrosine ligase-like family, member 4 | −1.29 | −1.34 | −1.14 | 1.08 |
| 10753231 | 0.0498 | Setd4 | SET domain containing 4 | −1.29 | −1.24 | −1.07 | 1.04 |
| 10852034 | 0.0350 | Zfp64 | Zfp93 | zinc finger protein 64 | zinc finger protein 93 | −1.29 | −1.15 | −1.12 | −1.01 |
| 10751362 | 0.0365 | Stxbp5l | syntaxin binding protein 5-like | −1.29 | −1.40 | −1.08 | 1.04 |
| 10807430 | 0.0191 | Pskh1 | protein serine kinase H1 | −1.28 | −1.10 | −1.03 | 1.07 |
| 10801209 | 0.0459 | Pcdhb21 | protocadherin beta 21 | −1.28 | −1.26 | −1.07 | 1.08 |
| 10891271 | 0.0481 | Nek9 | NIMA (never in mitosis gene a)- related kinase 9 | −1.28 | −1.12 | −1.01 | 1.04 |
| 10904233 | 0.0163 | RGD1308133 | similar to RIKEN cDNA 1700010C24 | −1.28 | −1.20 | 1.08 | 1.16 |
| 10799893 | 0.0396 | LOC686314 | similar to dachshund b | −1.28 | −1.28 | −1.15 | 1.14 |
| 10910222 | 0.0449 | Hmg20a | high mobility group 20A | −1.28 | −1.25 | −1.12 | 1.03 |
| 10782689 | 0.0114 | Rpp14 | ribonuclease P 14 subunit (human) | −1.28 | −1.28 | −1.28 | 1.02 |
| 10817396 | 0.0288 | Arnt | aryl hydrocarbon receptor nuclear translocator | −1.27 | −1.23 | −1.04 | 1.08 |
| 10768874 | 0.0441 | Cep350 | centrosomal protein 350kDa | −1.27 | −1.25 | −1.19 | −1.17 |
| 10876717 | 0.0410 | Mrpl50 | mitochondrial ribosomal protein L50 | −1.27 | −1.20 | −1.02 | −1.19 |
| 10706619 | 0.0465 | Vrk3 | vaccinia related kinases 3 | −1.27 | −1.18 | −1.05 | 1.09 |
| 10757555 | 0.0049 | Prkrip1 | Prkr interacting protein 1 (IL11 inducible) | −1.27 | −1.07 | −1.02 | 1.11 |
| 10905362 | 0.0318 | Ankrd54 | ankyrin repeat domain 54 | −1.27 | −1.23 | −1.19 | 1.09 |
| 10704902 | 0.0026 | Zfp112 | zinc finger protein 112 | −1.27 | −1.34 | −1.15 | 1.13 |
| 10789470 | 0.0310 | Dcun1d2 | DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) | −1.27 | −1.35 | −1.17 | 1.10 |
| 10876169 | 0.0351 | RGD1306576 | similar to hypothetical protein | −1.27 | −1.06 | −1.13 | 1.08 |
| 10711127 | 0.0409 | Phkg2 | phosphorylase kinase, gamma 2 (testis) | −1.26 | −1.11 | −1.12 | 1.15 |
| 10737389 | 0.0178 | Tubd1 | LOC100363 | tubulin, delta 1 | tubulin, delta 1-like | −1.26 | −1.19 | −1.05 | −1.01 |
| 10833806 | 0.0358 | Armc2 | armadillo repeat containing 2 | −1.26 | −1.25 | −1.09 | 1.13 |
| 10862376 | 0.0157 | Zfp786 | zinc finger protein 786 | −1.26 | −1.25 | −1.05 | −1.00 |
| 10912959 | 0.0398 | Tusc4 | tumor suppressor candidate 4 | −1.26 | −1.12 | −1.10 | 1.10 |
| 10743600 | 0.0353 | Znf287 | zinc finger protein 287 | −1.26 | −1.33 | −1.09 | 1.23 |
| 10742236 | 0.0151 | Thg1l | tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) | −1.26 | −1.15 | −1.11 | 1.04 |
| 10757239 | 0.0327 | Cnpy4 | canopy 4 homolog (zebrafish) | −1.26 | −1.12 | −1.06 | 1.10 |
| 10875425 | 0.0370 | RGD1309085 | similar to F23N19.9 | −1.25 | −1.21 | −1.14 | 1.03 |
| 10907268 | 0.0129 | Tcfcp2 | transcription factor CP2 | −1.25 | −1.05 | −1.05 | −1.02 |
| 10767175 | 0.0068 | Insig2 | insulin induced gene 2 | −1.25 | −1.26 | −1.18 | −1.17 |
| 10725286 | 0.0251 | Gpr139 | G protein-coupled receptor 139 | −1.25 | −1.40 | 1.04 | 1.26 |
| 10908827 | 0.0359 | Prdm10 | PR domain containing 10 | −1.25 | −1.62 | −1.04 | 1.02 |
| 10914940 | 0.0478 | Kdm4d | lysine (K)-specific demethylase 4D | −1.25 | −1.20 | −1.14 | 1.25 |
| 10912099 | 0.0374 | RGD1561074|LOC | similar to tripartite motif-containing 43 | hypothetical protein LOC100233177 | −1.25 | −1.26 | −1.15 | −1.02 |
| 10812438 | 0.0139 | Acot12 | acyl-CoA thioesterase 12 | −1.25 | −1.35 | −1.24 | 1.18 |
| 10875771 | 0.0030 | Klhl32 | kelch-like 32 (Drosophila) | −1.25 | −1.31 | −1.08 | 1.15 |
| 10710028 | 0.0445 | Arntl | aryl hydrocarbon receptor nuclear translocator-like | −1.25 | −1.08 | −1.0S | 1.14 |
| 10836570 | 0.0462 | Nostrin | nitric oxide synthase trafficker | −1.25 | −1.14 | −1.12 | 1.24 |
| 10824300 | 0.0354 | Slc25a44 | solute carrier family 25, member 44 | −1.25 | −1.15 | −1.19 | 1.01 |
| 10839423 | 0.0416 | Galk2 | galactokinase 2 | −1.25 | −1.18 | −1.01 | 1.13 |
| 10772107 | 0.0407 | Cenpc1 | centromere protein C 1 | −1.24 | −1.36 | −1.10 | 1.11 |
| 10891445 | 0.0248 | Ston2 | stonin 2 | −1.24 | −1.24 | −1.04 | −1.11 |
| 10754506 | 0.0402 | Ptplb | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b | −1.24 | −1.21 | −1.06 | 1.01 |
| 10840038 | 0.0033 | RGD1561852 | similar to Protein C20orf29 | −1.24 | −1.14 | −1.03 | 1.01 |
| 10767102 | 0.0336 | Epb4.1l5 | erythrocyte protein band 4.1-like 5 | −1.24 | −1.14 | −1.01 | −1.04 |
| 10870929 | 0.0393 | Ttc39a | tetratricopeptide repeat domain 39A | −1.24 | −1.22 | −1.22 | 1.25 |
| 10843634 | 0.0416 | Ubac1 | UBA domain containing 1 | −1.24 | −1.17 | −1.15 | −1.09 |
| 10863187 | 0.0343 | Rnf181 | ring finger protein 181 | −1.24 | −1.03 | −1.06 | 1.12 |
| 10774825 | 0.0332 | RGD1562229 | similar to hypothetical protein FLJ40298 | −1.24 | −1.22 | −1.18 | −1.00 |
| 10885951 | 0.0316 | Vsx2 | visual system homeobox 2 | −1.24 | −1.20 | −1.15 | −1.00 |
| 10839803 | 0.0184 | Rpl22l1 | Rpl22l2 | ribosomal protein L22 like 1 | ribosomal protein L22-like 2 | −1.24 | −1.22 | −1.20 | −1.00 |
| 10797344 | 0.0013 | Zfp346 | zinc finger protein 346 | −1.24 | −1.12 | −1.05 | 1.04 |
| 10901563 | 0.0170 | Chpt1 | choline phosphotransferase 1 | −1.24 | −1.09 | 1.03 | 1.05 |
| 10921141 | 0.0281 | Fyco1 | FYVE and coiled-coil domain containing 1 | −1.23 | −1.21 | −1.11 | 1.05 |
| 10833013 | 0.0172 | Unc5b | unc-5 homolog B (C. elegans) | −1.23 | −1.15 | −1.00 | −1.19 |
| 10845322 | 0.0015 | Stam2 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | −1.23 | −1.02 | −1.18 | −1.20 |
| 10806698 | 0.0334 | Mri1 | methylthioribose-1-phosphate isomerase homolog (S. cerevisiae) | −1.23 | −1.05 | −1.00 | 1.26 |
| 10901253 | 0.0109 | Pwp1 | PWP1 homolog (S. cerevisiae) | −1.23 | −1.10 | 1.04 | −1.06 |
| 10786422 | 0.0036 | Actr8 | ARP8 actin-related protein 8 homolog (yeast) | −1.23 | −1.17 | −1.11 | 1.03 |
| 10806709 | 0.0052 | Zswim4 | zinc finger, SWIM-type containing 4 | −1.23 | −1.13 | −1.09 | −1.10 |
| 10809856 | 0.0016 | Orc6l | origin recognition complex, subunit 6 like (yeast) | −1.23 | −1.19 | −1.03 | 1.05 |
| 10786204 | 0.0004 | Fam116a | family with sequence similarity 116, member A | −1.23 | −1.19 | −1.00 | −1.01 |
| 10731385 | 0.0090 | Parn | poly(A)-specific ribonuclease (deadenylation nuclease) | −1.23 | −1.13 | −1.02 | −1.00 |
| 10770566 | 0.0379 | Rpl7a | RGD156295 | ribosomal protein L7a | similar to Rpl7a protein | similar to 60S ribosomal protein L7a | −1.23 | −1.11 | −1.07 | −1.04 |
| 10701717 | 0.0021 | Katna1 | katanin p60 (ATPase-containing) subunit A1 | −1.23 | −1.09 | −1.02 | 1.01 |
| 10916016 | 0.0394 | St3gal4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | −1.23 | −1.09 | −1.09 | −1.03 |
| 10748361 | 0.0445 | Smurf2 | SMAD specific E3 ubiquitin protein ligase 2 | −1.23 | −1.17 | −1.15 | −1.15 |
| 10891026 | 0.0283 | Zfyve1 | zinc finger, FYVE domain containing 1 | −1.23 | −1.11 | −1.04 | 1.05 |
| 10908960 | 0.0285 | Rpusd4 | RNA pseudouridylate synthase domain containing 4 | −1.22 | −1.07 | −1.03 | −1.03 |
| 10927233 | 0.0318 | Zfp451 | zinc finger protein 451 | −1.22 | −1.10 | −1.06 | 1.00 |
| 10846340 | 0.0346 | Fkbp7 | FK506 binding protein 7 | −1.22 | −1.21 | 1.05 | −1.05 |
| 10777337 | 0.0094 | Stx18 | syntaxin 18 | −1.22 | 1.04 | −1.15 | −1.12 |
| 10814398 | 0.0441 | Mtfr1 | mitochondrial fission regulator 1 | −1.22 | −1.27 | −1.09 | −1.37 |
| 10859864 | 0.0493 | Rbm33 | RNA binding motif protein 33 | −1.22 | −1.19 | −1.07 | −1.00 |
| 10730287 | 0.0175 | RGD1307934 | similar to DNA segment, Chr 19, ERATO Doi 386, expressed | −1.22 | −1.13 | −1.03 | 1.33 |
| 10911016 | 0.0056 | Csnk1g1 | casein kinase 1, gamma 1 | −1.22 | −1.19 | −1.04 | −1.04 |
| 10892381 | 0.0348 | Nudt14 | nudix (nucleoside diphosphate linked moiety X)-type motif 14 | −1.22 | 1.18 | −1.28 | −1.23 |
| 10910966 | 0.0165 | Mtfmt | mitochondrial methionyl-tRNA formyltransferase | −1.22 | −1.08 | −1.02 | 1.20 |
| 10710627 | 0.0369 | Plk1 | polo-like kinase 1 (Drosophila) | −1.22 | −1.22 | −1.08 | 1.11 |
| 10736000 | 0.0302 | Myo1c | myosin IC | −1.22 | −1.03 | 1.03 | −1.07 |
| 10805029 | 0.0183 | Ptpn2 | protein tyrosine phosphatase, non-receptor type 2 | −1.21 | −1.17 | −1.09 | 1.05 |
| 10891436 | 0.0393 | Gtf2a1 | general transcription factor IIA, 1 | −1.21 | −1.12 | −1.02 | −1.17 |
| 10905465 | 0.0306 | Unc84b | unc-84 homolog B (C.elegans) | −1.21 | −1.19 | −1.04 | −1.03 |
| 10750373 | 0.0160 | Morc3 | MORC family CW-type zinc finger 3 | −1.21 | −1.20 | −1.10 | −1.06 |
| 10886322 | 0.0123 | Ttc8 | tetratricopeptide repeat domain 8 | −1.21 | −1.12 | 1.00 | 1.02 |
| 10859264 | 0.0093 | Ddx47 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 | −1.21 | 1.05 | −1.17 | −1.04 |
| 10720539 | 0.0133 | LOC499124 | mouse zinc finger protein 14-like | −1.21 | −1.21 | −1.03 | 1.04 |
| 10787117 | 0.0126 | Med26 | mediator complex subunit 26 | −1.21 | −1.10 | −1.14 | 1.22 |
| 10865077 | 0.0172 | Lrtm2 | leucine-rich repeats and transmembrane domains 2 | −1.21 | −1.13 | −1.00 | −1.16 |
| 10905270 | 0.0494 | Rabl4 | RAB, member of RAS oncogene family-like 4 | −1.21 | −1.18 | 1.01 | 1.01 |
| 10771119 | 0.0115 | Znf644 | zinc finger protein 644 | −1.21 | −1.14 | −1.05 | 1.06 |
| 10710806 | 0.0082 | LOC361646 | similar to K04F10.2 | −1.21 | −1.09 | −1.12 | −1.17 |
| 10850793 | 0.0074 | Zcchc3 | zinc finger, CCHC domain containing 3 | −1.21 | −1.13 | −1.07 | 1.07 |
| 10796455 | 0.0226 | Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | −1.20 | −1.14 | −1.10 | −1.12 |
| 10748021 | 0.0158 | Map3k14 | mitogen-activated protein kinase kinase kinase 14 | −1.20 | −1.03 | −1.03 | 1.03 |
| 10729715 | 0.0146 | Sgms1 | sphingomyelin synthase 1 | −1.20 | −1.19 | −1.01 | 1.09 |
| 10727867 | 0.0343 | Mus81 | MUS81 endonuclease homolog (S. cereuisiae) | −1.20 | 1.01 | 1.01 | −1.00 |
| 10800173 | 0.0282 | Rbbp8 | retinoblastoma binding protein 8 | −1.20 | −1.16 | −1.03 | 1.05 |
Functional classification of the transcriptomic changes
Following microarray hybridization and gene clustering, we performed functional analysis of genes in the 1467 ANOVA dataset. Functional classification of the microarray data was conducted using a gene ontology module of the DAVID software, which maps genes to function. We identified distinct groups of genes that reflected a high degree of functional similarity using the gene enrichment EASE score (modified Fisher exact statistical test threshold p≤ 0.1) in the DAVID system. Enrichment in select classes of genes was present in cluster 2 and 3. Cluster 2 (early up, late down) had significant enrichment in genes coding for G-protein coupled receptor signaling as well as extracellular matrix components and cell adhesion [Table 3]. Cluster 3 (early down, late up) was enriched in genes coding for Golgi function, protein trafficking and localization, vesicle mediated transport and transcription regulators [Table 4]. These genes were characterized by an initial decrease in expression, which persisted even after 6hrs then gradually returned to baseline levels by 12hrs. A few genes seemed to increase by 12hrs, however the increases were small and very close to the untreated control. Genes such as syntaxin 18, Rab8b showed functional overlap for protein transport and localization.
Table 3. Functional classification of genes in cluster 2: early up, late down.
Functional classification of genes in cluster 2 (Table 1). The table highlights gene enrichment for G-protein coupled receptor signaling and extracellular matrix components for genes in the early up-late down dataset and their corresponding trends of change throughout the time course. Functional annotation was performed using DAVID gene ontology tool. Gene enrichment is based on the EASE Score (threshold p≤ 0.1), a modified Fisher Exact test used to measure gene enrichment in annotation terms in the DAVID system. The EASE score for each class is shown as the gene enrichment p-value.
| G-PROTEIN COUPLED RECEPTOR SIGNALING AND CELL SURFACE SIGNALING TRANSDUCTION | FOLD CHANGE RELATIVE TO CONTROL | ||||
|---|---|---|---|---|---|
| Enrichment p-value: 3.61E-06 | |||||
| Gene symbol | Gene Name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Gpr33 | G protein-coupled receptor 33 | 1.43 | 1.16 | 1.22 | 1.24 |
| Mrgprb2 | MAS-related G protein-coupled receptor, member X2-like; MAS-related GPR, member B2 | 1.25 | 1.16 | 1.13 | 1.11 |
| Rspo3 | R-spondin 3 homolog (Xenopus laevis) | 1.35 | 1.23 | 1.06 | −1.28 |
| Fpr1 | formyl peptide receptor 1 | 1.27 | 1.11 | 1.07 | 1.11 |
| Lef1 | lymphoid enhancer binding factor 1 | 1.21 | 1.35 | 1.05 | −1.08 |
| Olr104 | olfactory receptor 104 | 1.24 | 1.07 | 1.09 | 1.15 |
| Olr127 | olfactory receptor 127 | 1.27 | −1.04 | 1.10 | 1.08 |
| Olr145 | olfactory receptor 145 | 1.29 | 1.22 | 1.05 | 1.19 |
| Olr1515 | olfactory receptor 1515 | 1.24 | 1.18 | 1.04 | 1.16 |
| Olr190 | olfactory receptor 190 | 1.25 | 1.03 | −1.06 | 1.06 |
| Olr221 | olfactory receptor 221 | 1.22 | 1.10 | 1.06 | 1.12 |
| Olr259 | olfactory receptor 259 | 1.23 | 1.10 | 1.10 | 1.08 |
| Olr318 | olfactory receptor 318 | 1.24 | 1.13 | 1.09 | 1.18 |
| Olr379 | olfactory receptor 379 | 1.27 | 1.16 | 1.08 | 1.10 |
| Olr419 | olfactory receptor 419 | 1.23 | 1.17 | 1.07 | 1.09 |
| Olr446 | olfactory receptor 446 | 1.30 | 1.14 | 1.15 | 1.15 |
| Olr806 | olfactory receptor 806 | 1.25 | 1.19 | 1.12 | 1.27 |
| Olr851 | olfactory receptor 851 | 1.21 | 1.06 | 1.11 | 1.17 |
| Olr869 | olfactory receptor 869; olfactory receptor 862 | 1.28 | 1.14 | 1.21 | 1.15 |
| Olr877 | olfactory receptor 877 | 1.29 | 1.02 | 1.03 | 1.09 |
| Ptgir | prostaglandin I2 (prostacyclin) receptor | 1.24 | 1.17 | 1.14 | 1.13 |
| P2ry13 | purinergic receptor P2Y, G-protein coupled, 13 | 1.23 | 1.10 | 1.07 | 1.03 |
| Tas2r126 | taste receptor, type 2, member 126 | 1.24 | 1.14 | 1.18 | 1.24 |
| Taar7d | trace-amine-associated receptor 7d | 1.28 | 1.09 | 1.09 | 1.08 |
| EXTRACELLULAR MATRIX COMPONENTS AND CELL ADHESION | FOLD CHANGE RELATIVE TO CONTROL | ||||
| Enrichment p-value : 0.013095162 | |||||
| Gene symbol | Gene Name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Acan | aggrecan | 1.65 | 2.04 | 1.20 | −1.16 |
| Ang1 | angiogenin, ribonuclease A family, member 1 | 1.32 | 1.24 | 1.10 | 1.17 |
| Lamc2 | laminin, gamma 2 | 1.27 | 1.07 | 1.08 | 1.00 |
| Mmp10 | matrix metallopeptidase 10 | 1.22 | 1.10 | 1.03 | 1.06 |
| Nid2 | nidogen 2; similar to nidogen 2 protein | 1.22 | 1.29 | 1.00 | 1.03 |
Table 4. Functional classification of genes in cluster 3: early down late up.
Functional classification of genes in cluster 3 (Table 2). The table highlights different gene classes that were enriched in the early down-late up dataset and their corresponding trends of change throughout the time course. Functional annotation was performed using DAVID gene ontology tool. Gene enrichment reflects genes that are highly associated with the indicated functional terms (i.e SNARE interaction, protein transport, protein localization and intracellular transport). It is based on the EASE Score (threshold p-value< 0.1), a modified Fisher Exact statistical test used to measure gene enrichment in annotation terms in the DAVID system. The EASE score for each class is shown as the gene enrichment p-value. Genes are enriched for SNARE interaction, protein trafficking, vesicle mediated transport and regulation of transcription. A number of genes show overlapping functions in vesicle trafficking and protein localization
| SNARE INTERACTION IN VESICULAR TRANSPORT | FOLD CHANGE RELATIVE TO CONTROL | ||||
|---|---|---|---|---|---|
| Enrichment p-value: 0.004060282 | |||||
| Gene symbol | Gene name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Bet 1l | blocked early in transport 1 homolog (S. cerevisiae) like | −1.31 | −1.10 | −1.14 | −1.08 |
| Gosr1 | golgi SNAP receptor complex member 1 | −1.29 | −1.18 | −1.07 | 1.07 |
| Stx18 | syntaxin 18 | −1.22 | 1.04 | −1.15 | −1.12 |
| PROTEIN TRANSPORT | FOLD CHANGE RELATIVE TO CONTROL | ||||
| Enrichment p-value: 0.030887946 | |||||
| Gene symbol | Gene name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Rab8b | RAB8B, member RAS oncogene family | −1.38 | −1.35 | −1.06 | 1.07 |
| Rufy1 | RUN and FYVE domain containing 1 | −1.31 | −1.12 | −1.09 | −1.12 |
| Arntl | aryl hydrocarbon receptor nuclear translocator-like | −1.25 | −1.08 | −1.08 | 1.14 |
| Bet1l | blocked early in transport 1 homolog (S. cerevisiae) like | −1.31 | −1.10 | −1.14 | −1.08 |
| Gosr1 | golgi SNAP receptor complex member 1 | −1.29 | −1.18 | −1.07 | 1.07 |
| Myo1c | myosin IC | −1.22 | −1.03 | 1.03 | −1.07 |
| Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | −1.20 | −1.14 | −1.10 | −1.12 |
| Stam2 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | −1.23 | −1.02 | −1.18 | −1.20 |
| Zfp280b | similar to suppressor of hairy wing homolog 2; zinc finger protein 280b | −1.35 | −1.15 | −1.09 | 1.35 |
| Slc15a4 | solute carrier family 15, member 4 | −1.30 | −1.35 | −1.19 | −1.00 |
| Stx18 | syntaxin 18 | −1.22 | 1.04 | −1.15 | −1.12 |
| PROTEIN LOCALIZATION | FOLD CHANGE RELATIVE TO CONTROL | ||||
| Enrichment p-value: 0.041166313 | |||||
| Gene symbol | Gene name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Rab8b | RAB8B, member RAS oncogene family | −1.38 | −1.35 | −1.06 | 1.07 |
| Rufy1 | RUN and FYVE domain containing 1 | −1.31 | −1.12 | −1.09 | −1.12 |
| Arntl | aryl hydrocarbon receptor nuclear translocator-like | −1.25 | −1.08 | −1.08 | 1.14 |
| Bet1l | blocked early in transport 1 homolog (S. cerevisiae) like | −1.31 | −1.10 | −1.14 | −1.08 |
| Gosr1 | golgi SNAP receptor complex member 1 | −1.29 | −1.18 | −1.07 | 1.07 |
| Myo1c | myosin IC | −1.22 | −1.03 | 1.03 | −1.07 |
| Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | −1.20 | −1.14 | −1.10 | −1.12 |
| Stam2 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | −1.23 | −1.02 | −1.18 | −1.20 |
| Zfp280b | similar to suppressor of hairy wing homolog 2; zinc finger protein 280b | −1.35 | −1.15 | −1.09 | 1.35 |
| Slc15a4 | solute carrier family 15, member 4 | −1.30 | −1.35 | −1.19 | −1.00 |
| Stx18 | syntaxin 18 | −1.22 | 1.04 | −1.15 | −1.12 |
| INTRACELLULAR TRANSPORT | FOLD CHANGE RELATIVE TO CONTROL | ||||
| Enrichment p-value: 0.086073852 | |||||
| Gene symbol | Gene name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Ankrd54 | ankyrin repeat domain 54 | −1.27 | −1.23 | −1.19 | 1.09 |
| Arntl | aryl hydrocarbon receptor nuclear translocator-like | −1.25 | −1.08 | −1.08 | 1.14 |
| Bet1l | blocked early in transport 1 homolog (S. cerevisiae) like | −1.31 | −1.10 | −1.14 | −1.08 |
| Gosr1 | golgi SNAP receptor complex member 1 | −1.29 | −1.18 | −1.07 | 1.07 |
| Golga 5 | golgi autoantlgen, golgin subfamily a, 5 | −1.61 | −1.21 | −1.42 | −1.35 |
| Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | −1.20 | −1.14 | −1.10 | −1.12 |
| Stam2 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | −1.23 | −1.02 | −1.18 | −1.20 |
| Zfp280b | similar to suppressor of hairy wing homolog 2; zinc finger protein 280b | −1.35 | −1.15 | −1.09 | 1.35 |
| Stx18 | syntaxin 18 | −1.22 | 1.04 | −1.15 | −1.12 |
| Stxbp5l | syntaxin binding protein 5-like | −1.29 | −1.40 | −1.08 | 1.04 |
| Nostrin | nitric oxide synthase trafficker | −1.25 | −1.14 | −1.12 | 1.24 |
| REGULATION OF TRANSCRIPTION | FOLD CHANGE RELATIVE TO CONTROL | ||||
| Enrichment p-value: 1.50E-05 | |||||
| Gene symbol | Gene name | 1.5hrs | 3hrs | 6hrs | 12hrs |
| Cir1 | CBF1 interacting corepressor | −1.39 | −1.27 | −1.13 | 1.11 |
| Gpbp1l1 | GC-rich promoter binding protein 1-like 1; similar to GC-rich promoter binding protein 1-like 1 | −1.49 | −1.45 | −1.12 | −1.14 |
| Smarcal1 | Swi/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 | −1.32 | −1.19 | −1.02 | 1.14 |
| Ahr | Aryl hydrocarbon receptor | −1.30 | −1.47 | −1.08 | −1.04 |
| Arnt | Aryl hydrocarbon receptor nuclear translocator | −1.27 | −1.23 | −1.04 | 1.08 |
| Arntl | Aryl hydrocarbon receptor nuclear translocator-like | −1.25 | −1.08 | −1.08 | 1.14 |
| Atxn3 | Ataxin 3 | −1.36 | −1.30 | −1.13 | −1.13 |
| Gtf2a1 | General transcription factor IIA, 1 | −1.21 | −1.12 | −1.02 | −1.17 |
| Med 26 | Mediator complex subunit 26 | −1.21 | −1.10 | −1.14 | 1.22 |
| LOC499124 | Mouse zinc finger protein 14-like | −1.21 | −1.21 | −1.03 | 1.04 |
| Prmt6 | Protein arginine methyltransferase 6 | −1.42 | −1.33 | −1.15 | 1.16 |
| Ppp1r10 | Protein phosphatase 1, regulatory subunit 10 | −1.29 | −1.19 | −1.09 | 1.36 |
| Sra1 | Steroid receptor RNA activator 1 | −1.35 | −1.05 | −1.19 | −1.17 |
| Tfam | Transcription factor A, mitochondrial | −1.37 | −1.26 | −1.15 | 1.02 |
| Tfdp2 | Transcription factor Dp-2 (E2F dimerization partner 2) | −1.71 | −1.55 | −1.04 | −1.07 |
| Uimc1 | Ubiquitin interaction motif containing 1 | −1.37 | −1.27 | −1.09 | −1.07 |
| Vsx2 | Visual system homeobox 2 | −1.24 | −1.20 | −1.15 | −1.00 |
| Zc3h8 | Zinc finger CCCH type containing 8 | −1.53 | −1.48 | −1.19 | 1.06 |
| Zfp112 | Zinc finger protein 112 | −1.27 | −1.34 | −1.15 | 1.13 |
| Zfp128 | Zinc finger protein 128 | −1.39 | −1.20 | −1.19 | 1.19 |
| Znf287 | Zinc finger protein 287 | −1.26 | −1.33 | −1.09 | 1.23 |
| Zfp426l2 | Zinc finger protein 426-like 2 | −1.35 | −1.26 | −1.07 | −1.12 |
| Zfp472 | Zinc finger protein 472 | −1.34 | −1.22 | −1.11 | −1.00 |
| Zfp68 | Zinc finger protein 68 | −1.35 | −1.43 | −1.15 | 1.08 |
| Zfp786 | Zinc finger protein 786 | −1.26 | −1.25 | −1.05 | −1.00 |
| Zfp84 | Zinc finger protein 84 | −1.41 | −1.39 | −1.19 | 1.08 |
| Zfp90 | Zinc finger protein 90 | −1.35 | −1.34 | −1.10 | 1.03 |
Quantitative RT-PCR validation of potential candidates genes
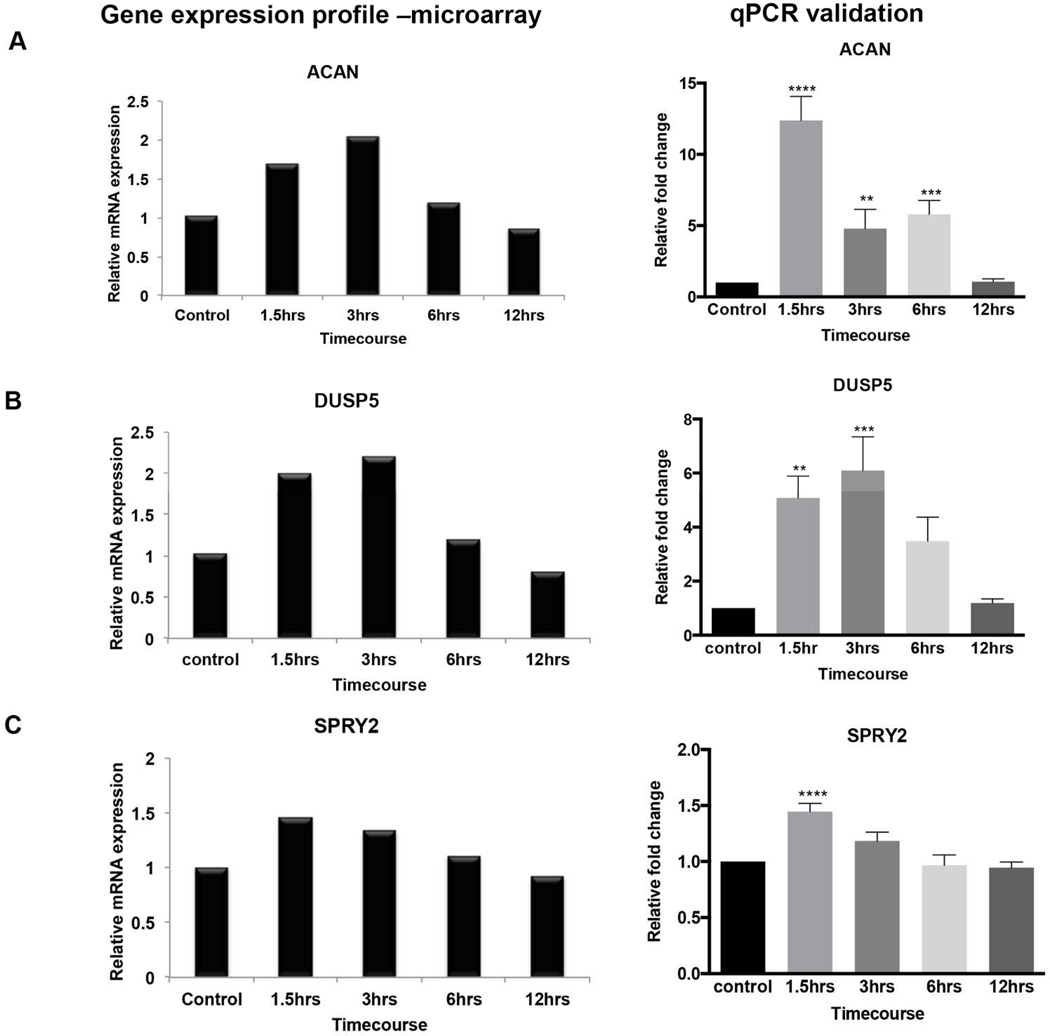
We carried out validation of select candidate genes using quantitative RT-PCR (qPCR) to assess changes in levels of gene expression. We selected genes for qPCR validation based on high fold change increase or decrease (±1.2 and above), substantial characterization in previous literature as well as gene expression changes associated with cellular processes (synaptic function and vesicular trafficking) that were enriched in the gene clusters. We hypothesized that these processes could increase vulnerability of neurons to degeneration if impaired. We validated three representative genes from clusters showing significant downregulation of expression upon BDNF withdrawal and three genes from clusters that reflected significant upregulation [Table 5]. Validated gene targets showed expression patterns that were similar to the microarray expression profiles [Figures (3) and (4) left panel]. We found significant changes in the gene coding for the extracellular matrix component Acan (Aggrecan). In the microarray profiling, Acan increased by 0.5 fold after 1.5hrs of BDNF withdrawal peaking to 2 fold by 3hrs [Figure (3A) left panel]. By 6hrs Acan expression was decreasing, returning to baseline by 12hrs. A similar profile of change was seen in the qPCR validation [Figure (3A) right panel]; however Acan increased more than 10-fold as early as 1.5hrs and by 3hrs its expression had already begun to decline although it was still more than 4 fold above control. At 6hrs, expression was still approximately 4 fold relative to control then declined to baseline by 12hrs.
Table 5. Gene list validated by qPCR.
Representative genes selected for qPCR validation. Table shows the six genes from select classes highlighting the trend of change throughout the time course. Highest changes in expression occurred between 1.5hrs and 3hrs.
| UPREGULATED GENES | FOLD CHANGE RELATIVE TO CONTROL | |||||||
|---|---|---|---|---|---|---|---|---|
| Affymetrix ID | p-value | Gene Symbol | Gene Name | Function | 1.5hrs | 3hrs | 6hrs | 12hrs |
| 10708021 | 0.0495 | Acan | Aggrecan | Extracellular matrix component | 1.65 | 2.04 | 1.20 | −1.16 |
| 10716080 | 0.0078 | Dusp5 | Dual specificity phosphatase S | Map kinase phosphatase, dephosphorvlates ERK1/2 | 2.00 | 2.21 | 1.19 | −1.25 |
| 10785773 | 0.0230 | Spry2 | Sprouty homolog 2 (Drosophila) | Negative regulation of Map kinase activity | 1.45 | 1.34 | 1.11 | −1.09 |
| DOWNREGULATED GENES | ||||||||
| Affymetrix ID | p-value | Gene Symbol | Gene Name | Function | 1.5hrs | 3hrs | 6hrs | 12hrs |
| 10886465 | 0.0278 | Golga5 | Golgi autoantigen, golgin subfamily a, 5 | Golgi structure maintainance | −1.61 | −1.21 | −1.42 | −1.35 |
| 10918374 | 0.0460 | Rab8b | RAB8B, member RAS oncogene family | Vesicle mediated transport | −1.38 | −1.35 | −1.06 | 1.07 |
| 10765115 | 0.0077 | Vamp4 | Vesicle-associated membrane protein 4 | Vesicle mediated transport; neurotransmitter release | −1.53 | 1.11 | −1.51 | −1.54 |
Figure 3.
Microarray expression profiles and qPCR validation of genes from select clusters that were upregulated upon BDNF withdrawal. A-C shows a comparison of the microarray (left panel) and qPCR (right panel) expression profiles for each gene (A) Acan (B) Dusp5 (C) Spry2. Microarray graphs are from the two hybridization experiments described in the methods and are meant to be a comparison to the qPCR results in terms of the pattern of gene expression changes over time. For qPCR validation, at least three independent BDNF withdrawal experiments were done to confirm the expression profiling. Values were normalized using the ddCT method; normalization to the endogenous control relative to untreated control. Error Bars represent standard error of mean (SEM). *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
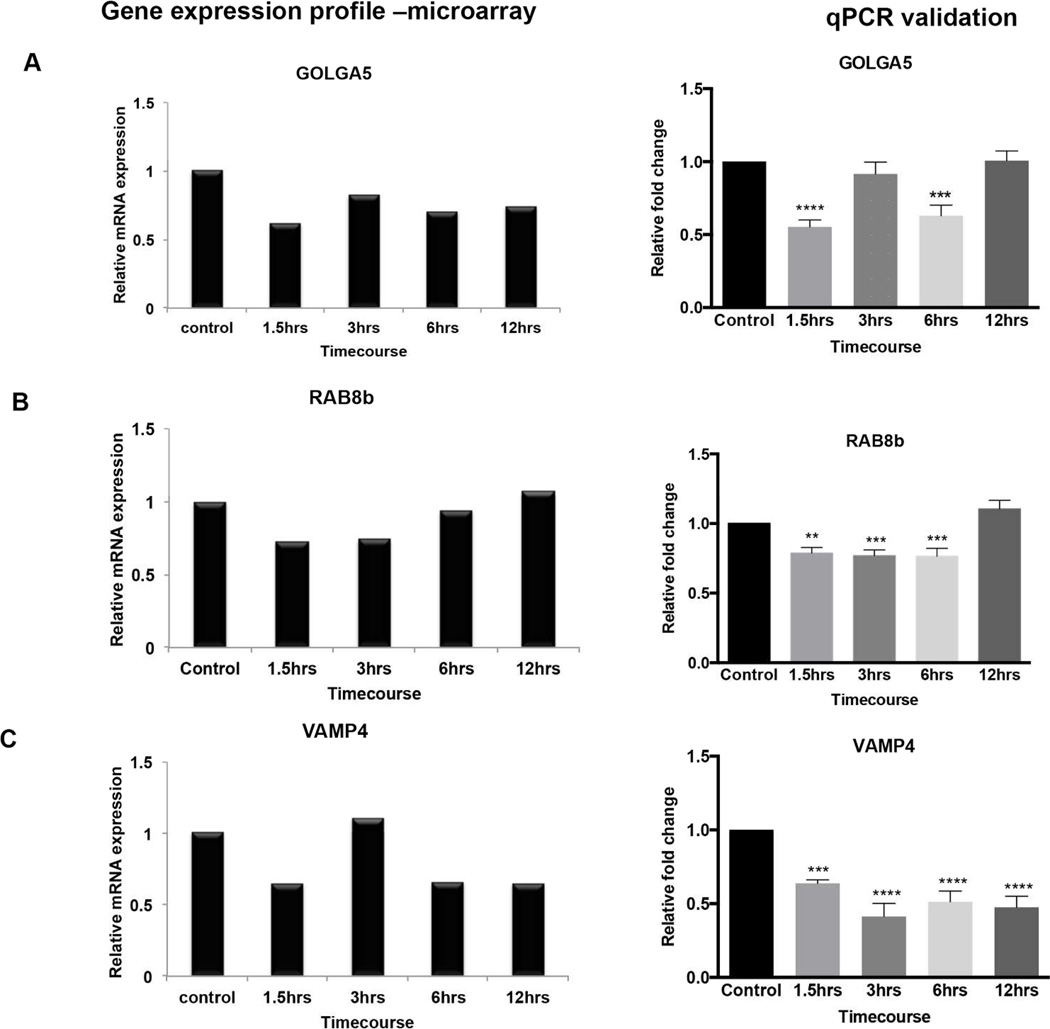
Figure 4.
Microarray expression profiles and qPCR validation of select transcripts that were downregulated following BDNF deprivation. A-C shows a comparison of the microarray (left panel) and qPCR (right panel) expression profiles for each gene (A) Golga5 (B) Rab8b (C) Vamp4. Microarray graphs are from the two hybridization experiments described in the methods and are meant to be a comparison to the qPCR results in terms of the pattern of gene expression changes over time. For qPCR validation, at least three independent BDNF withdrawal experiments were done to confirm the expression profiling. Values were normalized using the ddCT method; normalization to the endogenous control relative to untreated control. Error Bars represent standard error of mean (SEM). *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
Also from cluster 2, striking changes were observed in Dual-Specificity Phosphatases 5 (DUSP5), a MAP kinase phosphatase which dephosphorylates Erk1/2. Following BDNF withdrawal, DUSP5 increased 2-fold as early as 1.5hrs, peaked at 3hrs to a 2.3-fold increase then returned to baseline by 6–12hrs [Fig. 3(B) left panel]. This trend was reproduced by qPCR validation where DUSP5 increased 4-fold at 1.5hrs, peaked to 6-fold increase at 3hrs then returned to the same levels as control by 12hrs [Fig. 3(B) right panel].
Another gene that had consistent changes for the microarray and qPCR validation was Sprouty homolog 2 (Spry2); an inhibitor of the MAP kinase pathway. Spry2 expression increased 0.6-fold after 1.5hrs of BDNF withdrawal then gradually declined throughout the timecourse, returning to baseline by 12hrs [Fig. 3(C) left panel]. qPCR validation also showed Spry2 reproducing a similar profile of change with a 0.6 fold increase at 1.5hrs and corresponding gradual decline from 3hrs to 12hrs [Fig. 3(C) right panel ].
Downregulated genes were selected from cluster 3 (early down, late up) for qPCR validation. Golga5, a gene coding for a protein that is important for golgi structure maintenance showed a reproducible decrease in expression both by microarray and qPCR. Upon BDNF withdrawal, Golga5 expression decreased 0.4-fold then remained below baseline with another significant decrease at 6–12hrs [Fig. 4(A) left panel]. In the qCR validation, the profile of change is similar, however expression returns to baseline by 12hrs [Fig. 4(A) right panel]. Rab8b, a Rab-GTPase transport regulator, also decreased 0.3-fold at 3–6hrs following BDNF withdrawal in the microarray [Fig. 4(B) left panel] which was reliably reproducible by qPCR with a small but significant decrease of 0.3-fold at 3 and 6hrs [Fig. 4(B) right panel]. Vamp4 was also decreased in both microarray and qPCR although the 3hr timepoint had an opposite response to the treatment for qPCR compared to microarray. For the microarray, Vamp4 increased slightly above baseline by 3hrs then decreased at 6–12hrs. For qPCR Vamp4 maintained a gradual decrease; starting with a 0.4-fold decrease at 1.5hrs which was sustained up to the 12hr timepoint. Since qPCR was a validation for at least 3 independent experiments, the trend for qPCR more likely portrays an accurate and consistent Vamp4 response to BDNF withdrawal. Given the well-established functions of these genes in golgi maintenance, and vesicle trafficking, our results could be suggesting a potential disassembly of the protein trafficking and secretory machinery upon BDNF withdrawal.
DISCUSSION
The experimental goal of this study was to determine whether there are changes in transcription following neurotrophin starvation in primary hippocampal neurons. We utilized a well-established method of TrkB-FC application to sequester BDNF and NT-4, which also binds TrkB (Ninkina et al, 1997; Soppet et al, 1991; Croll et al, 1998; Jia et al, 2010). Four early timepoints (1.5 hr, 3 hr, 6 hr and 12 hr) were selected to capture different phases of transcriptional activity. These time points were specifically chosen to identify signaling pathways that are activated prior to the process of cell death initiation based on previous reports that commitment to cell death following NGF deprivation occurs approximately 16–20 hours after removal of NGF from sympathetic and sensory neurons (Deshmukh and Johnson, 1997; Nikolaev et al, 2009).
We anticipated identifying individual genes and groups of transcripts in hippocampal neurons as BDNF withdrawal proceeds. Early timepoints were predicated to identify changes in immediate early genes, among others, whereas later time points would likely activate initiation mitochondrial changes associated with programmed cell death. During NGF withdrawal, cell death occurs in sympathetic neurons with increases in c-jun, c-myb mkp-1, cyclin D1 and the pro-apoptotic Bim transcripts (Estus et al, 1994; Freeman et al, 1994; et al, 1995; Whitfield et al, 2001). However, due to the time course examined (1–12 hours), the microarray screen would not be expected to detect genes involved in cell death. Indeed, instead of proapoptotic genes, we detected significant enrichment in genes involved in synaptic function.
Hippocampal neurons were selected for microarray analysis following BDNF withdrawal, as BDNF has profound effects upon long-term potentiation, synaptic plasticity and cell morphology in the hippocampus (Park and Poo, 2013), where its receptor, TrkB is highly expressed. The functional analysis with DAVID indicated that many relevant pathways were represented although changes in expression levels were within 20–30% range. It is worth noting that in neuronal populations, relatively small changes can be significant given the nature of neuronal signaling relative to heterologous cell lines or tissue with admixed cell types (Ginsberg et al, 2012). Most importantly, BDNF withdrawal resulted in a significant decrease in genes that are associated with vesicular trafficking and synaptic function as well as selective increases in phosphatases and extracellular matrix genes.
DUSP5, a stress inducible MAP kinase phosphatase that deactivates Erk1/2 in the MAP kinase pathway, (Keyse, 2008) was significantly upregulated upon BDNF withdrawal. Recently, the role of MAP kinase phosphatases in the development of CNS primary neurons was described in which expression of DUSPs is regulated by neurotrophins to modulate structural plasticity. The induction of MKP-1/DUSP1 by BDNF is influenced by activity-dependent events that culminate in the regulation of JNK to promote axonal branching (Jeanneteau et al, 2010). MKP1/DUSP1 has also been implicated in depressive disorders (Duric et al, 2010), which are downstream of BDNF. Hence, changes in DUSP5 may reflect downstream effects of BDNF on structural plasticity, which could be relevant in disease.
Genes coding for extracellular matrix components, such as aggrecan (Acan), increased significantly (2-fold), 1.5–3 hrs after withdrawal of BDNF. Aggrecan is highly expressed and regulated by neuronal activity in hippocampal parvalbumin interneurons (McRae et al, 2007; Morawski et al, 2012) and is a major component of extracellular perineuronal nets where it is involved in the onset of critical periods. It is highly enriched on presynaptic contacts where it enwraps synaptic compartments on postsynaptic dendrites and dendritic spines in human hippocampus (Lendivai et al, 2013). Elevated levels of Aggrecan have been reported in severe cases of Alzheimer disease (Lendivai et al, 2013); Aggrecan is enriched in the vicinity of plaques around healthy neurons suggesting a role in preserving the structural integrity of the synapse. It is also known to be neuroprotective against oxidative stress in primary neuronal cultures (Suttkas et al, 2014). Thus, Aggrecan may function downstream of BDNF to preserve the integrity of synaptic contacts.
Spry2, a member of the Sprouty family of proteins that negatively regulate receptor tyrosine kinase signaling, was also increased shortly after BDNF withdrawal. In recent studies, BDNF has been shown to regulate Spry2 expression in immature primary neuronal cultures (Gross et al, 2007). Overexpression of Spry2 inhibited neurite outgrowth and increased neuronal apoptosis (Gross et al, 2007). Therefore, low BDNF may compromise structural plasticity and neuronal survival through increasing levels of Spry2.
Among the genes that changed with BDNF deprivation are small GTPases of the Rab family (Rab1A and Rab8B), intracellular membrane trafficking proteins that direct the identification and routing of vesicles and organelles, as wells as receptors and ion channels (Pfeffer, 2013). These changes are not isolated events, as other proteins, such as Vesicle Associated Membrane Protein 4 (VAMP4) and syntaxins were also identified. VAMP4 showed a significant 50% decrease in expression post BDNF withdrawal. It is also intimately associated with Rab proteins (Simonson et al, 1999); endosomal and Golgi membrane trafficking of proteins depend upon Rab regulation of SNARE (Soluble N-ethylmaleimide-sensitive-factor Attachment protein Receptor) proteins such as the VAMP4 interacting partner, syntaxin 6. Also, neurotransmission is influenced by VAMP4, which is required with syntaxin proteins for neurotransmitter release (Raingo et al, 2012). VAMP4 is significant since BDNF is known to regulate pre-synaptic functions through enhanced neurotransmitter release (Lohof et al, 1993; Yano et al, 2006). VAMP4 is also required for maintenance of the ribbon structure of the Golgi apparatus (Shitara et al, 2013). In addition to VAMP4, another gene coding for Golga5-Golgin84; a protein involved in maintaining the Golgi membrane structure was also dramatically reduced by more than 60% after 3 hrs of BDNF deprivation. The decrease in expression of vesicular trafficking and Golgi maintenance genes suggests that components of the secretory machinery are changing following BDNF deprivation.
In the present study, we found that many transcriptional changes occur in hippocampal neurons at early time points after BDNF withdrawal. Our results indicate several distinct groups of genes that are markedly and simultaneously affected by BDNF deprivation. They include molecules involved with synaptic vesicle trafficking and connectivity and enzymes that are directly involved with major signal transduction pathways, such as MAP kinase phosphatases.
What is the relevance of these alterations in BDNF-regulated transcription? Neurotrophins, such as BDNF are critical in modulating synaptic plasticity, in addition to their well-established roles in neuronal cell survival. Application of neurotrophins to peripheral and central neurons results in rapid increases in the frequency of spontaneous action potentials and excitatory synaptic activity (Park and Poo, 2013). The work we have presented suggest there are events that occur early following neurotrophin withdrawal that may have an impact upon later events leading to neurodegeneration. This is supported by previous studies that demonstrated that NGF withdrawal is linked to changes in APP metabolism (Matrone et al 2008; Nikolaev et al 2009). Synaptic defects are thought to represent early markers of aging and dementia. Our results suggest that loss of trophic factors may play a role in this process. A lack of BDNF will likely affect pre- and post-synaptic functions and may lead to morphological changes and synaptic failure. In fact, many studies have documented a decrease in BDNF levels in neurodegenerative diseases, most notably in Alzheimer’s disease (Narisawa-Saito et al, 1996; Connor et al, 1997; Nagahara et al, 2009), Huntington’s disease (Zuccato et al, 2008) and Spinocerebellar ataxia (Takahashi et al, 2012). Administration of BDNF has been shown to be neuroprotective against age-related hippocampal synaptic loss (Nagahara et al, 2009; 2013).
Our findings of dysregulation of select synaptic and vesicle trafficking genes, as well as MAP kinase phosphatases, are consistent with significant decreases in expression of genes encoding synaptic proteins that have been documented in microarray studies of post mortem Alzheimer’s disease cases (Callahan et al, 1999, Ginsberg et al, 2010, Gutala et al, 2010, Berchtold et al, 2013). Moreover, the changes were more pronounced in the hippocampus (Berchtold et al, 2013), which matched our findings in rat hippocampal cultures. More importantly, the changes we have observed in gene transcription are consistent with the hypothesis that early events in neurodegeneration may reflect changes in synaptic function (Selkoe, 2002; Arancio and Chao, 2007). Deficits in synaptic transmission have been observed well before the detected of plaques and tangles. Therefore, a decrease in BDNF may manifest in changes that have also been seen in age-related neurodegenerative diseases. Our studies establish an in vitro system model for understanding the interplay between low trophic factor support and early synaptic loss associated with neurodegeneration and aging.
Supplementary Material
REFERENCES
- Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int J Mol Sci. 2008;9:2091–2104. doi: 10.3390/ijms9112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods. 2009;177:381–385. doi: 10.1016/j.jneumeth.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel S. A unifying hypothesis for the cause of amyotrophic lateral sclerosis, parkinsonism, and Alzheimer disease. Ann Neurol. 1981;10:499–505. doi: 10.1002/ana.410100602. [DOI] [PubMed] [Google Scholar]
- Arancio O, Chao MV. Neurotrophins, synaptic plasticity, and dementia. Current Opin in Neurobiol. 2007;17:325–330. doi: 10.1016/j.conb.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neuroscience. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batisatou A, Greene LA. Aurintricarboxylic acid rescues PC12 cells and sympathetic neurons from death caused by nerve growth factor deprivation: correlation with suppression of endonuclease activity. J Cell Biol. 1991;15:461–471. doi: 10.1083/jcb.115.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender R, Lange S. Adjusting for multiple testing-when and how? Journal of Clinical Epidemiology. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, Cotman CW. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol of Aging. 2013;34:1653–1661. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Cagnol S, Rivard N. Oncogenic KRAS and BRAF activation of the MEK/ERK signaling pathway promotes expression of dual-specificity phosphatase 4 (DUSP4/MKP2) results in nuclear ERK1/2 inhibition. Oncogene. 2013;32:564–576. doi: 10.1038/onc.2012.88. [DOI] [PubMed] [Google Scholar]
- Callahan LM, Vaules WA, Coleman PD. Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J Neuropathol Exp Neurol. 1999;58:275–287. doi: 10.1097/00005072-199903000-00007. [DOI] [PubMed] [Google Scholar]
- Caunt CJ, Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs), Shaping the outcome of MAP kinase signaling. FEBS J. 2013;280:489–504. doi: 10.1111/j.1742-4658.2012.08716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: A convergence point for many signaling pathways. Nature Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Connor B, Young D, Yan Q, Faull RL, Synek B, Dragunow M. Brain-derived neurotrophic factor is reduced in Alzheimer’s disease. Brain Res Mol Brain Res. 1997;49:71–81. doi: 10.1016/s0169-328x(97)00125-3. [DOI] [PubMed] [Google Scholar]
- Croll SD, Chesnutt CR, Rudge JS, Acheson A, Ryan TE, Siuciak JA, DiStefano PS, Wiegand SJ, Lindsay RM. Co-infusion with a TrkB-FC receptor body carrier enhances BDNF distribution in the adult brain. Experimental Neurobiology. 1998;152:20–33. doi: 10.1006/exnr.1998.6836. [DOI] [PubMed] [Google Scholar]
- Deckwerth TL, Johnson EM. Temporal analysis of events associated with programmed cell death (apoptosis) of sympathetic neurons deprived of nerve growth factor. J Cell Biol. 1993;123:1207–1222. doi: 10.1083/jcb.123.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M, Johnson EM. Programmed cell death in neurons: Focus on the pathway of nerve growth factor deprivation-induced death of sympathetic neurons. Mol Pharmacology. 1997;51:897–906. doi: 10.1124/mol.51.6.897. [DOI] [PubMed] [Google Scholar]
- Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS. A negative regulator of MAP kinase cuases depressive behavior. Nature Med. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr Altered gene expression in neurons during programmed cell death: identifcation of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman RS, Estus S, Johnson EM. Analysis of cell cycle-related gene expression in postmitotic neurons: selective induction of cyclin D1 during programmed cell death. Neuron. 1994;12:343–355. doi: 10.1016/0896-6273(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Counts SE, Wuu J, Alldred MJ, Nixon RA, Che S. Regional selectivity of rab5 and rab7 protein upregulation in mild cognitive impairment and Alzheimer's disease. J Alzheimer’s Dis. 2010;22:631–639. doi: 10.3233/JAD-2010-101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Che S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer's disease. Neurobiology Dis. 2012;45:99–107. doi: 10.1016/j.nbd.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin PD, Johnson EM. Experimental autoimmune model of nerve growth factor deprivation: effect on developing peripheral sympathetic and sensory neurons. Proc Natl. Acad Sci. 1979;76:5382–5386. doi: 10.1073/pnas.76.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Armant O, Benosman S, de Aguilar JL, Freund JN, Kedinger M, Licht JD, Gaiddon C, Loeffler JP. Sprouty2 inhibits BDNF-induced signaling and modulates neuronal differentiation and survival. Cell Death and Differentiation. 2007;14:1802–1812. doi: 10.1038/sj.cdd.4402188. [DOI] [PubMed] [Google Scholar]
- Gutala RV, Reddy PH. The use of real-time PCR analysis in a gene expression study of Alzheimer's disease post-mortem brains. J Neurosci Methods. 2004;132:101–107. doi: 10.1016/j.jneumeth.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau F, Deinhardt K, Miyoshi G, Bennett AM, Chao MV. The MAP kinase phosphatase MKP-1 regulates BDNF-induced axon branching. Nature Neurosci. 2010;13:1373–1379. doi: 10.1038/nn.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Gall CM, Lynch G. Presynaptic BDNF promotes postsynaptic long-term potentiation in the dorsal striatum. J Neurosci. 2010;30:14440–14445. doi: 10.1523/JNEUROSCI.3310-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- Lawan A, Shi H, Gatzke F, Bennett AM. Diversity and specificity of the mitogen-activated protein kinase phosphatase-1 functions. Cell Mol Life Sci. 2013;70:223–237. doi: 10.1007/s00018-012-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvai D, Morawski M, Négyessy L, Gáti G, Jäger C, Baksa G, Glasz T, Attems J, Tanila H, Arendt T, Harkany T, Alpár A. Neurochemical mapping of the human hippocampus reveals perisynaptic matrix around functional synapses in Alzheimer's disease. Acta Neuropathol. 2013;125:215–229. doi: 10.1007/s00401-012-1042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Montalcini R, Booker B. Destruction of the sympathetic ganglia in mammalians with an antiserum against nerve growth protein. Proc. Natl. Acad Sci. 1960;46:384–391. doi: 10.1073/pnas.46.3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff AM, Poo MM. Potentiation of developing neuromuscular synapses by the neurotrophins NT-3 and BDNF. Nature. 1993;363:350–353. doi: 10.1038/363350a0. [DOI] [PubMed] [Google Scholar]
- Longo FM, Yang T, Knowles JK, Xie Y, Moore LA, Massa SM. Small molecule neurotrophin receptor ligands: novel strategies for targeting Alzheimer's disease mechanisms. Curr Alzheimer Res. 2007;4:503–506. doi: 10.2174/156720507783018316. [DOI] [PubMed] [Google Scholar]
- Martin DP, Schmidt RE, DiStefano PS, Lowry OH, Carter JG, Johnson EM. Inhibitors of protein synthesis and RNA synthesis prevent neuronal death cause by nerve growth factor deprivation. J Cell Biol. 1988;106:829–844. doi: 10.1083/jcb.106.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone C, Ciotti MT, Marolda R, Mercanti D, Calissano P. NGF and BDNF control amyloidogenic route and Ab production in hyppocampal neurons. Proc Natl Acad Sci. 2008;105:13139–13144. doi: 10.1073/pnas.0806133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone C, Barbagallo APM, LaRosa LR, Florenzano F, Ciotti MT, Mercanti D, Chao MV, Calissano P, D’Adamio L. APP is phosphorylated by TrkA and regulates NGF/TrkA signaling. J Neurosci. 2011;31:11756–11761. doi: 10.1523/JNEUROSCI.1960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae PA, Rocco MM, Kelly G, Brumberg JC, Matthews RT. Sensory deprivation alters aggrecan and perineuronal net expression in the mouse barrel cortex. J Neurosci. 2007;27:5405–5413. doi: 10.1523/JNEUROSCI.5425-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski M, Bruckner G, Arendt T, Matthews RT. Aggrecan: beyond cartilage and into the brain. Int J Biochem Cell Biol. 2012;44:690–693. doi: 10.1016/j.biocel.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Murer MG, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol. 2001;63:71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Nagahara AH, Mateling M, Kovacs I, Wang L, Eggert S, Rockenstein E, Koo EH, Masliah E, Tuszynski MH. Early BDNF treatment ameliorates cell loss in the entorhinal cortex of APP transgenic mice. J. Neurosci. 2013;39:15596–15602. doi: 10.1523/JNEUROSCI.5195-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM, Wang L, Blesch A, Kim A, Conner JM, Rockenstein E, Chao MV, Koo EH, Geschwind D, Masliah E, Chiba AA, Tuszynski MH. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer’s disease. Nature Med. 2009;15:331–337. doi: 10.1038/nm.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa-Saito M, Wakabayashi K, Tsuji S, Takahashi H, Nawa H. Regional specificity of alterations in NGF, BDNF and NT-3 levels in Alzheimer’s disease. Neuroreport. 1996;7:2925–2928. doi: 10.1097/00001756-199611250-00024. [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ninkina N, Grashchuck M, Buchman VL, Davies AM. TrkB variants with deletions in the leucine-rich motifs of the extracellular domain. J Biol Chem. 1997;272:13019–13025. doi: 10.1074/jbc.272.20.13019. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW. Cell death during development of the nervous system. Ann Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nature Rev Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingo J, Khvotchev M, Liu P, Darios F, Li YC, Ramirez DM, Adachi M, Lemieux P, Toth K, Davletov B, Kavalali ET. VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission. Nature Neurosci. 2012;15:738–745. doi: 10.1038/nn.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Simonson S, Gaullier JM, D’Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J Biol Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- Shitara A, Shibui T, Okayama M, Arakawa T, Mizoguchi I, Sakakura Y, Takuma T. Vamp4 is required to maintain the ribbon structure of the Golgi Apparatus. Mol Cell Biochem. 2013;380:11–21. doi: 10.1007/s11010-013-1652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Marmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppet D, Escandon E, Maragos J, Middlemas DS, Reid SW, Blair J, Burton LE, Stanton BR, Kaplan DR, Hunter T, Nikolics K, Parada LF. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the TrkB tyrosine kinase receptor. Cell. 1991;65:895–903. doi: 10.1016/0092-8674(91)90396-g. [DOI] [PubMed] [Google Scholar]
- Strand A, Baquet Z, Aragaki A, Holmans P, Yang L, Cleren C, Beal MF, Jones L, Kooperberg C, Olson J, Jones K. Expression profiling of Huntington’s disease models suggests BDNF depletion plays a major role in striatal degeneration. J Neuroscience. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttkus A, Rohn S, Weigel S, Glöckner P, Arendt T, Morawski M. Aggrecan, link protein and tenascin-R are essential components of the perineuronal net to protect neurons against iron-induced oxidative stress. Cell Death and Disease. 2014;5:e1119. doi: 10.1038/cddis.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Ishikawa K, Sato N, Obayashi M, Niimi Y, Ishiguro T, Yamada M, Toyoshima Y, Takahashi H, Kato T, Takao M, Murayama S, Mori O, Eishi Y, Mizusawa H. Reduced brain-derived neurotrophic factor (BDNF) mRNA expression and presence of BDNF-immunoreactive granules in the spinocerebellar ataxia type 6 (SCA6) cerebellum. Neuropathology. 2012;32:595–603. doi: 10.1111/j.1440-1789.2012.01302.x. [DOI] [PubMed] [Google Scholar]
- Ueda K, Arakawa H, Nakamura Y. Dual-specificity phosphatase 5 (DUSP5) as a direct scriptional target of tumor suppressor p53. Oncogene. 2003;22:5586–5591. doi: 10.1038/sj.onc.1206845. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Selkoe D. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron. 2004;44:181–183. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Wolf L, Gao CS, Gueta K, Xie Q, Chevallier T, Podduturi NR, Sun J, Conte I, Zelenka PS, Ashery-Padan R, Zavadil J, Cvekl A. Identification and characterization of FGF2-dependent mRNA: microRNA networks during lens fiber cell differentiation. G3. 2013;3:2239–2255. doi: 10.1534/g3.113.008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Ninan I, Zhang H, Milner TA, Arancio O, Chao MV. BDNF-mediated neurotransmission relies upon a myosin VI motor complex. Nature Neuroscience. 2006;9:1009–1018. doi: 10.1038/nn1730. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices in Huntington’s disease. Brain Pathol. 2008;18:225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nature Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.