Abstract
Background:
Staphylococcus aurous is a major pathogen, causing variety of diseases and death in Iran and in the world. Despite the use of a spectrum of new antibiotics, this organism has caused severe infections in burns as well as in different parts of the body, due to acquired drug resistance. Widespread inappropriate use of antibiotics in treating bacterial infections has led to the selection and circulation of resistant strains and the growing risk of transferring resistant genes to sensitive bacteria. One of the causes of antibiotic resistance in S. aurous strains is the gain of resistance genes including integrase and qac/sul1.
Objectives:
The purpose of this study was to investigate the presence of class 1 integron in S. aurous strains isolated from clinical samples for the first time in Iran.
Materials and Methods:
This descriptive study was performed on 200 strains of S. aurous isolated from patients admitted to Baqiyatallah Hospital in Tehran in 2013. These strains were confirmed using biochemical and serological tests and the presence of class 1 integron was determined by polymerase chain reaction (PCR).
Results:
Among the 200 samples, 1% of the strains (two isolates) contained the class 1 integron gene. The results of this study showed that the highest frequency of the obtained samples belonged to males and the isolates occurred mostly in individuals aged 51-60 years old. The highest number of strains was found in wound samples. The strains were most frequently isolated from the emergency ward and the intensive care unit (ICU).
Conclusions:
Findings of this study showed that integron can have a limited frequency in S. aurous isolated from clinical sample in Tehran.
Keywords: Staphylococcus aurous, Drug Resistance, Polymerase Chain Reaction
1. Background
The role of Staphylococcus aurous as a pathogen causing serious infections in humans has long been confirmed (1). S. aurous is considered as an opportunistic pathogen, especially in people who are immunosuppressed and in wounds caused by severe burns (2). S. aurous is the second most common cause of nosocomial infections after Escherichia coli, constituting about 20% of all nosocomial infections (3). In recent years, in despite of introducing a variety of antibiotics, this organism has created severe infections in burns as well as in different parts of the body due to acquired drug resistance (4). Misuse of antibiotics in treating bacterial infections has led to the selection of resistant strains, and unfortunately, the risk of transfer of the resistance gene to sensitive bacteria is growing (5). Up until recently, it has been thought that transfer of antibiotic resistance in bacteria takes place mainly through conjugation and transduction by plasmids, phages and transposons carrying the resistance genes. Hall and Collis identified another mechanism of antibiotic resistance gene transfer in 1995, which was delivery by some elements called integrons (6). Integrons are genetic sets capable of integrating mobile genetic elements called gene cassettes and displacing them. Since integrons contain a promoter, they can express genes existing in a gene cassette. Therefore, integrons act both as gene expression vectors and as a natural cloning system (7). There are two conserved nucleotide sequences in the 5' and 3' regions of integrons. Essential components of the 5' region of all integrons are: 1) integrase gene (int I), coded for recombining the enzyme dedicated to the site; 2) attI sequence, a specific recombinant site located adjacent to the int I gene; 3) the promoter sequence which is necessary for the expression of genes in the gene cassette integrated in the integron. There are three components as follows in the 3' regions of class 1 integron: 1) the qac gene, which is responsible for resistance to quaternary ammonium compounds and disinfectants; 2) the sul1 gene, which codes for resistance to sulfonamides. 3) ORF5, which codes for a protein with unknown function (7).
Currently, five classes of integrons have been identified based on the heterogeneity of Integrase proteins. These integrons can be located in integrated sequences (IS), plasmids capable of conjugation, transposons or chromosomes, and all can be used as a means of genetic material transfer between species (7). Since the discovery of antibiotics, their inappropriate prescription in bacterial infections has led to selection and spread of resistant strains of bacteria, turning them to a global problem (8-10). The main outcome of this wrong treatment strategy has been the replacement and development of some colonies from resistant strains of bacteria rather than sensitive strains (10). These strains can transfer the resistance factors to other strains and even sensitive species of bacteria and create an exponentially growing population of resistant bacteria. The presence of integron, found in bacterial species, has created a two-fold capacity to obtain the resistance factors. Strains containing integron cannot be determined by routine microbiological methods and their presence is therefore missed. The presence of integron in bacteria, whether lacking or possessing the gene cassette, is just the tip of the iceberg that potentially may create a catastrophic and uncontrollable spread of such resistant bacteria (11).
Screening for integrons in bacterial strains and its relationship with the severity and extent of antibiotic resistance can be helpful in management of infection. In addition, this study provided national and international information about the extent and frequency of integron sequences among strains of S. aurous in Iran. Simultaneous occurrence of multiple drug resistance in bacteria has been associated with integron class 1. No report has been provided on these important genetic elements in S. aurous in Iran
2. Objectives
This study was performed to detect and identify class 1 integron in S. aurous isolates.
3. Materials and Methods
This descriptive study was performed on 200 S. aurous isolates from patients admitted to Baqiyatallah Hospital in Tehran in 2013. These strains were confirmed using biochemical and serological tests and the presence of class 1 integron was determined by polymerase chain reaction (PCR). Gram-positive isolates with positive DNase, catalase, mannitol and coagulase tests were considered as S. aurous (12).
3.1. DNA Extraction
The bacteria were cultured in Luria-Bertani (LB) medium (CinnaGen, Iran) and incubated at 37°C overnight. The inoculated medium was centrifuged at 5000 rpm for six minutes. The supernatant was removed and the pellet was washed with 200 µL distilled water and the previous step was repeated once more; 100 µL Tris-EDTA (TE) buffer, 10 µL sodium acetate and 10 µL lysozyme 100 were added and the final mixture was incubated for half an hour at room temperature and then for 30 minutes at 43°C. Afterwards, 20 µL of proteinase K was added and the mixture was maintained at 60°C for one hour. Thereafter, 20 µL 20% SDS was added and the final volume was incubated for 10 minutes at 37°C, followed by adding 160 µL chloroform and centrifuging at 10000 rpm for 10 minutes. Afterwards, 70 µL cold isopropanol and 5 µL NaCl 5N were added to the supernatant and it was incubated for two hours at -20°C and also one hour at -70°C and then centrifuged at 13000 rpm for 20 minutes. The supernatant was discarded and ethanol, 70%, 750 µL was added to the pellet and it was centrifuged with the previous conditions. The supernatant was removed and the pellet was incubated for one hour at 37°C to dry out and then 50 µL TE buffer was added to the dried pellet.
3.2. Polymerase Chain Reaction to Detect integrase and qac/sul1 Genes
This technique was used to screen for the integrase and qac/sul1 genes in the extracted DNA from samples of S. aurous (Table 1).
Table 1. Nucleotide Sequences of Primers and the Polymerase Chain Reaction Product Size (13, 14).
| Primer | Target Gene | Primer Sequence | PCR Product, bp |
|---|---|---|---|
| Forward- int1 | integrate | GTTCGGTCAAGGTTCTG | 490 |
| Revers- int1 | integrate | GCCAACTTTCAGCACATG | 490 |
| Forward- qac3 | conserved segment | ATCGCAATAGTTGGCGAAGT | 570 |
| Revers- sul1-3 | conserved segment | TCCATCGATAATATACCTAA | 570 |
The PCR reaction mixtures included 2 μL 10 x buffer, 2.5 μL dNTP 10 Mm, 2 μL and 1.5 µL MgCl2 50 mM for the integrase and qac/sul1 genes, respectively, 1 μL of each primer (10 mM), 2 μL Taq DNA polymerase 0.5 U, 1 μL template DNA, and 14 and 14.5 µL deionized distilled water for the integrase and qac/sul1 genes, respectively. The final volume was 25 µL (all of the materials were purchased from CinnaGen Company, Iran).
The PCR reactions had 35 cycles, including 45 seconds at 94°C for denaturation, 50 seconds at 45°C for annealing, and one minute at 72°C for amplification of the target gene (extension). After the PCR reaction, the PCR product was investigated on a 1% agarose gel and the bands were visualized in a transilluminator under the UV light.
4. Results
The results of this study showed that the highest frequency of the obtained samples belonged to males (69.3%). The mode of age frequency was in the 51-60 age groups (20.3%). The samples were most frequently isolated from wounds (59.9%), sputum (25%) and bronchoalveolar lavage (15.1%). The emergency ward and the intensive care unit (ICU) were the most infected sites of the hospital (18.2% and 19.8%, respectively). PCR detection revealed that only 1% (two isolates) of 200 S. aurous isolates had the integrase and qac/sul1 genes, which are responsible for the presence of class 1 integron (Figures 1 and 2).
Figure 1. Frequency of the Staphylococcus aurous Isolates in Different Age Groups.
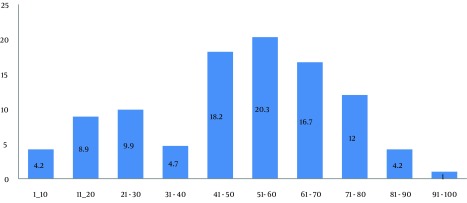
S. aurous was mostly isolated from the individuals 51-60 years old.
Figure 2. Frequency of Samples in Different Wards.
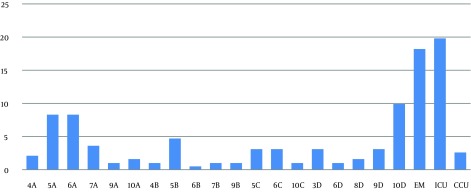
The S. aurous samples were mostly isolated from the emergency ward and ICU (18.2% and 19.8%, respectively).
4.1. Polymerase Chain Reaction Detection of the integrase Gene
PCR was performed to detect the integrase gene. The PCR products were run on 1% agarose gel. The integrase gene of S. aurous s was visualized at the 490-bp region (Figure 3).
Figure 3. The Polymerase Chain Reaction Products Separated by Electrophoresis on 1% Agarose Gel.
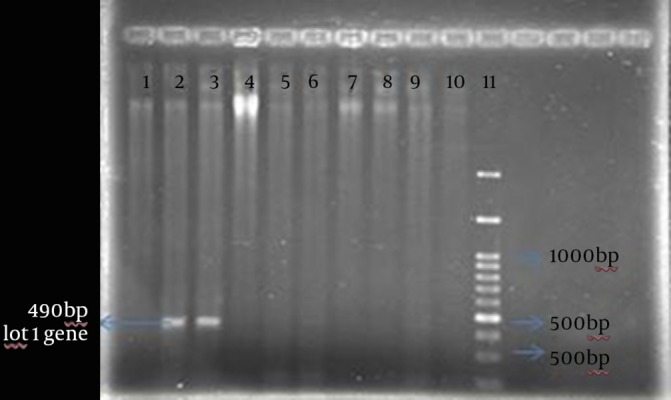
Lanes 1, 4, 5, 6, 7, 8 and 9: clinical samples showed the absence of the integrase gene; lanes 2 and 3: int1 gene; lane 10: negative control; lane 11: low-bp ladder.
4.2. Polymerase Chain Reaction Detection of the qac/sul1 Gene
PCR was performed to detect the qac/sul1 gene. The PCR product was visualized on 1% agarose gel at the 570-bp region (Figure 4).
Figure 4. The Polymerase Chain Reaction Products Separated by Electrophoresis on 1% Agarose Gel.

Lanes 1, 4, 5, 6, 7, 8, 9: clinical samples lacking the integrase gene; lanes 2 and 3: qac/sul1 gene; lane 10: negative control; lane 11: low-bp ladder.
5. Discussion
The spread of antibiotic resistance genes including integrons has become a major problem in treatment of infections caused by pathogenic bacteria (15). Over the past few decades, different genera of S. aurous have increasingly become resistant to commonly used antibiotics (16). The rise of resistance to antibiotics may be due to improper use of antimicrobial agents in animal feeds or inappropriate use of antibiotics in treating infections (17). Recently, the emergence of strains with multi-antibiotic resistance has caused some problems in treatment of infections in humans and animals (18). According to national and international reports, the issue of multidrug resistance of S. aurous has emerged in the recent few years and has become a public health problem in Iran (19). The aim of this study was to identify and investigate the prevalence of class 1 integron in S. aurous strains isolated from clinical samples. Our findings indicated that the integrase gene was observed only in two isolates (1%) among 200 samples of S. aurous.
Antunes and colleagues used the integrase gene as a target for identification of class 1 and 2 integrons and found that the frequency of integron in Salmonella enterica was 75% (20). Krauland et al. also used both regions as targets for identification of integrons and showed that clonal expansion and horizontal transfer of the integrase gene has led to the worldwide spread of resistance to antimicrobial drugs against Salmonella (15). Rodriguez et al. used variable regions as targets for identification of integrons in Salmonella samples (21). In another study by Xu et al. in 2007 in North China on 53 isolates of methicillin-resistant coagulase-negative S. aurous, 30 isolates had class 1 integron (13). In 2008, Zhenbo Xu and colleagues reported methicillin-resistant S. aurous in China for the first time, in which they showed that six strains of S. aurous had class 1 integron (14).
Zhang et al. in 2004 studied 33 isolates of Salmonella among healthy people in China and found that most of them were resistant to sulfamethoxazole, tetracycline, and streptomycin. All of the isolates were susceptible to ceftriaxone and 11 isolates had class 1 integron (22). In another study, Mirnejad et al. identified class 1 and 2 integrons in Acinetobacter baumannii strains and showed that 42% and 82% of the strains contained class 1 and class 2 integrons, respectively (23). Few studies have been performed on integrons in clinical strains of S. aurous in the world. However, there is no report of that from Iran. Further research is required in this field. On the other hand, geographical maps of antibiotic resistance can be used to guide clinicians in the choice of antibiotics, before culture results become available (24). Prudence and foresight in this area has been limited to the countries that are aware of their past and present reports in this field. Therefore, the results from other parts of the world may not be useful for policy makers of public health in Iran. Undoubtedly, further studies are required to achieve these goals, especially in the field of epidemiology and identification of dominant strains using sensitive genotypic and phenotypic typing techniques in the country.
Our findings indicated that integrons can be found limitedly among S. aurous isolated from clinical samples in Tehran. The surveillance on antibiotic resistance including integron screening as an indicator and sign of acquisition and spread of antibiotic resistance can be an important strategy in combating antibiotic resistance in S. aurous.
Footnotes
Authors’ Contributions:Abbas Ali Imani Fooladi supervised, developed the study concept, and design and critical revision of the manuscript; Emad Yahaghi researched and contributed to the development drafting of the manuscript and critical revision of the manuscript; Mohsen Amin was the main editor and contributed to the design of this research; Reza Mirnejad and Reza Nezamzade advised and contributed to the design of this research.
References
- 1.Rasigade JP, Vandenesch F. Staphylococcus aureus: a pathogen with still unresolved issues. Infect Genet Evol. 2014;21:510–4. doi: 10.1016/j.meegid.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues MV, Fortaleza CM, Riboli DF, Rocha RS, Rocha C, da Cunha Mde L. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a burn unit from Brazil. Burns. 2013;39(6):1242–9. doi: 10.1016/j.burns.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Biber A, Abuelaish I, Rahav G, Raz M, Cohen L, Valinsky L, et al. A typical hospital-acquired methicillin-resistant Staphylococcus aureus clone is widespread in the community in the Gaza strip. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira V, Lopes C, Castro A, Silva J, Gibbs P, Teixeira P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009;26(3):278–82. doi: 10.1016/j.fm.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z, Li L, Shi L, Shirtliff ME. Class 1 integron in staphylococci. Mol Biol Rep. 2011;38(8):5261–79. doi: 10.1007/s11033-011-0676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15(4):593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 7.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4(8):608–20. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 8.Adegoke AA, Okoh AI. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol (Praha). 2014;59(2):133–40. doi: 10.1007/s12223-013-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyzik E, Marek A. Plasmid profile analysis and evaluation of antibiotic susceptibility of Staphylococcus aureus strains isolated from table chicken eggs. Pol J Vet Sci. 2013;16(2):307–12. doi: 10.2478/pjvs-2013-0042. [DOI] [PubMed] [Google Scholar]
- 10.Kaneti G, Sarig H, Marjieh I, Fadia Z, Mor A. Simultaneous breakdown of multiple antibiotic resistance mechanisms in S. aureus. FASEB J. 2013;27(12):4834–43. doi: 10.1096/fj.13-237610. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Wang H, Huang M, Mei Y, Gu B, Wu R, et al. Analysis of antimicrobial resistance and class 1 integrons among strains from upper respiratory tract of healthy adults. J Thorac Dis. 2013;5(2):149–55. doi: 10.3978/j.issn.2072-1439.2013.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDowell B, Papasian CJ. Staphylococcus aureus identification: thermonuclease agar for direct testing of blood isolates and a new slide agglutination test. Clin Lab Sci. 1991;4(5):299–302. [PubMed] [Google Scholar]
- 13.Xu Z, Shi L, Alam MJ, Li L, Yamasaki S. Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004. FEMS Microbiol Lett. 2008;278(2):223–30. doi: 10.1111/j.1574-6968.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Li L, Alam MJ, Zhang L, Yamasaki S, Shi L. First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2008;57(3):264–8. doi: 10.1007/s00284-008-9187-8. [DOI] [PubMed] [Google Scholar]
- 15.Krauland MG, Marsh JW, Paterson DL, Harrison LH. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis. 2009;15(3):388–96. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merlino J, Watson J, Rose B, Beard-Pegler M, Gottlieb T, Bradbury R, et al. Detection and expression of methicillin/oxacillin resistance in multidrug-resistant and non-multidrug-resistant Staphylococcus aureus in Central Sydney, Australia. J Antimicrob Chemother. 2002;49(5):793–801. doi: 10.1093/jac/dkf021. [DOI] [PubMed] [Google Scholar]
- 17.van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents. 2000;14(4):327–35. doi: 10.1016/s0924-8579(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 18.da Costa PM, Loureiro L, Matos AJ. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. 2013;10(1):278–94. doi: 10.3390/ijerph10010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347(22):1770–82. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 20.Antunes P, Machado J, Peixe L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J Antimicrob Chemother. 2006;58(2):297–304. doi: 10.1093/jac/dkl242. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez I, Rodicio MR, Mendoza MC, Cruz Martin M. Large conjugative plasmids from clinical strains of Salmonella enterica serovar virchow contain a class 2 integron in addition to class 1 integrons and several non-integron-associated drug resistance determinants. Antimicrob Agents Chemother. 2006;50(4):1603–7. doi: 10.1128/AAC.50.4.1603-1607.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Shi L, Li L, Guo S, Zhang X, Yamasaki S, et al. Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from healthy humans in China. Microbiol Immunol. 2004;48(9):639–45. doi: 10.1111/j.1348-0421.2004.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 23.Mirnejad R, Mostofi S, Masjedian F. Antibiotic resistance and carriage class 1 and 2 integrons in clinical isolates of Acinetobacter baumannii from Tehran, Iran. Asian Pac J Trop Biomed. 2013;3(2):140–5. doi: 10.1016/S2221-1691(13)60038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leseva M, Arguirova M, Nashev D, Zamfirova E, Hadzhyiski O. Nosocomial infections in burn patients: etiology, antimicrobial resistance, means to control. Ann Burns Fire Disasters. 2013;26(1):5–11. [PMC free article] [PubMed] [Google Scholar]


