Abstract
Background
Depressive disorder (DD), including recurrent DD (rDD), is a severe psychological disease, which affects a large percentage of the world population. Although pathogenesis of the disease is not known, a growing body of evidence shows that inflammation together with oxidative stress may contribute to development of DD. Since reactive oxygen species produced during stress may damage DNA, we wanted to evaluate the extent of DNA damage and efficiency of DNA repair in patients with depression.
Material/Methods
We measured and compared the extent of endogenous DNA damage – single- and double-strand breaks, alkali-labile sites, and oxidative damage of the pyrimidines and purines – in peripheral blood mononuclear cells isolated from rDD patients (n=40) and healthy controls (n=46) using comet assay. We also measured DNA damage evoked by hydrogen peroxide and monitored changes in DNA damage during repair incubation.
Results
We found an increased number DNA breaks, alkali-labile sites, and oxidative modification of DNA bases in the patients compared to the controls. Exposure to hydrogen peroxide evoked the same increased damage in both groups. Examination of the repair kinetics of both groups revealed that the lesions were more efficiently repaired in the controls than in the patients.
Conclusions
For the first time we showed that patients with depression, compared with non-depresses individuals, had more DNA breaks, alkali-labile sites, and oxidative DNA damage, and that those lesions may be accumulated by impairments of the DNA repair systems. More studies must be conducted to elucidate the role of DNA damage and repair in depression.
MeSH Keywords: Depression, DNA Damage, DNA Repair, Oxidative Stress, Reactive Oxygen Species
Background
Depressive disorder (DD, including recurrent depressive disorder [rDD]) is a severe psychiatric illness, which at its worst can lead to suicide. It is characterized by persistent low mood, loss of interest or pleasure, feeling of tiredness, loss of appetite, and disturbed sleeping. According to the World Health Organization, 350 million people are affected by the disease worldwide [1]. DD is a growing problem in Western countries, where it may affect larger percentages of people than in the worldwide population – up to 10% of the people in some Western countries. Some estimates show that by 2020 the disease will be the second-most serious illness in terms of social and economic burden, exceeded only by ischemic heart disease [2,3]. Moreover, there are some reports suggesting that depression may coexist with other diseases, like cardiovascular disease [4]. To make matters worse, approximately one-third of DD patients do not respond to traditional pharmacological medications such as monoamine reuptake inhibitors [5].
Despite the common occurrence and extensive research of depression, its pathophysiology is still not fully understood. Currently, studies on the disease have focussed on its molecular aspects. People with DD have changes in the expression of neurotransmitters, alterations to the hypothalamus-adrenal-pituitary (HPA) axis, genes, and structural changes within the brain [6]. Increasing number of reports indicated that inflammation may play a pivotal in depression [7]. Patients with DD had elevated levels of interleukin-1β (IL-1β), interleukin-6 (IL-6), interleukin-8, and TNFα, which are inflammatory cytokines activate in pro-inflammatory pathways [8–10]. Moreover, it was shown that anti-inflammatory drugs act as antidepressants, and medication used in DD therapy has anti-inflammatory effects [11–14]. IL-1β and IL-8 are known to be activated by the inflammasome complex. This protein complex contains an intracellular sensor, most often one of the Nod-like receptors (NLRs), which is a precursor of procaspase-1 and apoptosis-associated speck-like protein containing a CARD (ASC) [15]. Increased gene expression of NLRP3 was found in peripheral blood mononuclear cells (PBMCs) of DD patients, and elevated levels of IL-1β and IL-8 in the patients’ serum [10]. NLRP3 is one of the most versatile NLRs, activated by a large spectrum of stressors. It was shown that alternations of mitochondria and reactive oxygen species (ROS) generated in mitochondria are the main activators of NLRP3 [16]. In agreement with this, activation of NLRP3 in PBMCs of the patients was accompanied by increased lipid peroxidation, which can be attributed to increased oxidative stress and elevated mitochondrial ROS (mtROS) production [10]. The NLRP3 inflammasome was also found to be involved in DNA damage response (DDR). After oxidative and genotoxic stress, the wild-type murine dendritic cells (DCs) had lower expression of genes involved in double-strand and base-excision repair (BER), and increased apoptosis rate, when compared to NLRP3 knock-out DCs [17]. It appears that the NLRP3 inflammasome suppresses DNA repair and enhances p53-mediated apoptosis. Currently, the extent of DNA damage in depression has only been estimated only by measurement of the level of 8-oxoguanine (8-oxoG) – a marker of oxidative DNA damage – and the results are contradictory. Increased concentration of serum and urinary 8-oxoG was found in patients with clinical depression, depressed patients with gastric adenocarcinoma, chronic heart failure, myalgic encephalomyelitis, and chronic fatigue syndrome [18–22]. Moreover, higher levels of 8-oxoG was detected in peripheral blood lymphocytes in depressed Japanese office workers when compared to controls [23]. However, there was no association between occurrence of depressive symptoms in Japanese office workers and urinary levels of 8-oxoG [24].
Since the findings described above are inconsistent, we wanted to determine if the oxidative modification of purines, like 8-oxoG, and pyrimidines are present in a higher degree in patients with depression than in controls. Moreover, we also wanted to know if the patients have elevated levels of other kinds of DNA damage, such as strand breaks. Finally, we wanted to determine if the increased DNA damage is caused, at least partly, by impairments in DNA damage repair. Elucidation of those molecular factors and mechanisms may be helpful in development of new methods for diagnosing depression in the future. To achieve these objectives, we measured and compared the extent of endogenous DNA damage – single- and double-strand breaks, alkali-labile sites, and oxidative damage of the pyrimidines and purines – in PBMCs isolated from DD patients and healthy controls. Moreover, we induced oxidative DNA damage in those PBMCs by incubating them with hydrogen peroxide, measured the kinetics of removing of such damage, and compared the results between the patients and the controls.
Material and Methods
Patients
The study was carried out in a group of 86 subjects aged 22–65 (M=40.75 years, SD=15.02). The participants were divided into 2 groups: patients with rDD (n=40) and healthy subjects (a control group, CG, n=46). All the patients were native Poles, inhabitants of central Poland and unrelated. Assignment of individuals to the test group was random, without replacement sampling. Respondents, before deciding to participate in the study, had been informed of its purpose, ensured that the participation was voluntary, and were guaranteed that personal data and test results would not be distributed, but only used in the overall statistics.
Patients were selected for the study according to the inclusion criteria of ICD-10 (F32.0–7.32.2, F33.0–F33.8) [25]. All the subjects were examined during the course of their hospitalization. The presence of axis I and II disorders other than depressive episode, and the diagnosis of somatic diseases and injuries of the central nervous system (CNS) were regarded as exclusion criteria. Other exclusion criteria were inflammatory or autoimmune disorders and unwilling to give informed consent.
In all the included subjects, case history was obtained prior to main study procedure, using the standardized Composite International Diagnostic Interview (CIDI) [26]. During hospitalization all the patients received antidepressant pharmacotherapy.
Ethics
Informed, written consent for participation in the study was obtained from each subject, according to the protocol approved by the Bioethics Committee of the Medical University of Lodz (No. RNN/70/14/KE).
Peripheral blood mononuclear cells isolation
PBMCs were isolated from 3 ml of fresh blood diluted with 3 ml of phosphate-buffered saline (PBS) by isopycnic centrifugation (30 min, 400 × g, 4°C) in Histopaque-1077 (Sigma-Aldrich, St. Louis, MO, USA). Then, the cells were washed twice with PBS and counted using a Bürcker chamber.
Induction of DNA damage and repair incubation
DNA damage was induced by exposing PBMCs to 20 μM H2O2 (POCH S.A., Gliwice, Poland) for 10 min at 4°C in PBS. DNA damage induced by H2O2 was measured in the cells, which were immediately subjected to comet assay after the exposure. To estimate the efficiency of DNA damage repair, the cells were washed with PBS, suspended in fresh RPMI-1640 medium, and kept at 37°C for 15, 30, 60, and 120 min. After each time period the cells were subjected to comet assay procedure.
Comet assay
In this study we used the alkaline version (pH >13) of the comet assay according to the procedure described by Singh et al. with later modifications [27–29]. Although this technique recognizes both double- and single-strand breaks along with alkali-labile sites, it does not detect oxidative DNA damage. Thus, the level of this kind of DNA damage was assessed by the use of 2 DNA glycosylases: human 8-oxoguanine DNA glycosylase or Nth (hOOG1 and endonuclease III, respectively; New England Biolabs, Ipswich, MA, USA) according to the procedure described earlier [30]. Both enzymes are bifunctional glycosylases and they possess AP-lyase activity. Therefore, after the recognition and removal of the damage bases, they introduce single-strand breaks, which can be detected by the alkaline version of the comet assay. hOOG1 recognizes and removes 8-oxoadenine when paired with cytosine, 7,8-dihydro-8-oxoguanine (8-oxoguanine) when paired with cytosine, methyfapy-guanine and foramidopyrimidine (fapy)-guanine [31,32]. Nth recognizes and removes urea, thymine glycol, 5,6-dihydroxythymine, uracil glycol, 5-hydroxy-5-methylhydanton, 6-hydroxy-5,6-dihdrothimine, and methyltartronylurea [33,34].
For each sample, aliquots of 5×105 PBMCs were centrifuged (250 × g, 15 min 4°C), the supernatant was removed and the pellet was re-suspended in 40 μl of 1.13% low gelling temperature agarose type XI (Sigma-Aldrich, St. Louis, MO, USA) in PBS cooled to 37°C. Then, the suspension was cast on a microscope slide, which was earlier coated with 0.5% low electroendosmosis agarose type I (Sigma-Aldrich, St. Louis, MO, USA) in distillated water, covered with a cover glass, and gelled on a cold plate for 10 min. After this time, the cover glass was removed, the slide was submerged in a chilled lysis solution (100 mM EDTA, 2.5 mM NaCl, 10 mM TRIS, 1% Triton X-100, pH 10) and stored at 4°C overnight. On the next day, the enzyme-treated samples were washed with enzyme buffer (0.5 mM EDTA, 40 mM HEPES, 0.1 M KCl, 0.2 mg/ml bovine serum albumin; pH 8). Then, 100 μl of the buffer containing 1 U of either hOGG1 or Nth enzymes, or, as a control, the buffer alone, were placed onto gels. Those samples were covered with cover glasses and incubated at 37°C for 60 min in a moist chamber. After the incubation, cover slips were removed, the slides were submerged in a electrophoresis buffer (1 mM EDTA and 300 mM NaOH, pH >13), and left for 20 min for unwinding of DNA. DNA of undigested samples was unwound directly after the lysis step. Then, electrophoresis was conducted at an electric field strength of 0.73 V/cm (300 mA) for 20 min.
After electrophoresis, samples were washed with distillated water and drained. Approximately 60 min before analysis, gels were stained with DAPI (1 μg/ml). The analysis was done using an Eclipse fluorescence microscope (Nikon, Tokyo, Japan) at 200× magnification. Fifty images of the comets were randomly selected from each sample, captured using a COHU 4910 video camera (Cohu, San Diego, CA, USA) and their parameters were measured with the Lucia-Comet image analysis system (Laboratory Imaging, Praha, Czech Republic). The level of DNA damage of the samples was assessed by calculating the mean of the percentage of DNA in the tail of the comets. Results for samples digested with the DNA repair enzymes were normalized by subtracting the level of DNA damage evoked by enzyme buffer only.
Statistical analysis
Data shown in this paper are presented as a mean ±SEM from 2 separate experiments. We applied the Shapiro-Wilk test to assess normality of distribution of the studied group. If it was normally distributed, we used the t test to determine differences between means, if not, we used the Mann-Whitney test. Analysis of the data was done using STATISTICA (StatSoft, Tulsa, OK).
Results
Basal endogenous DNA damage
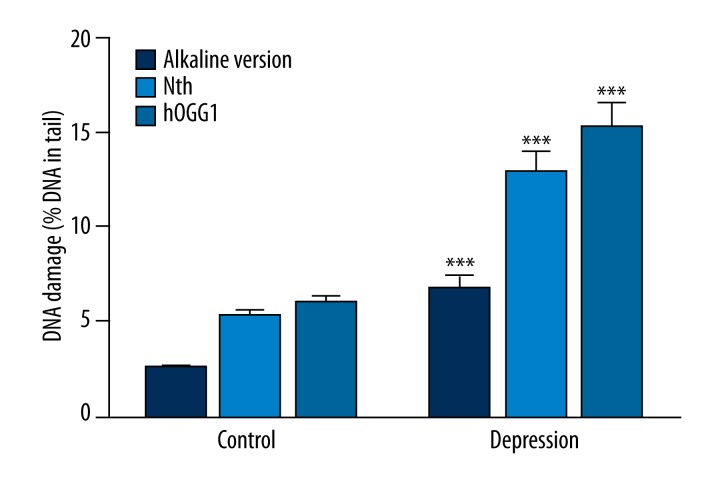
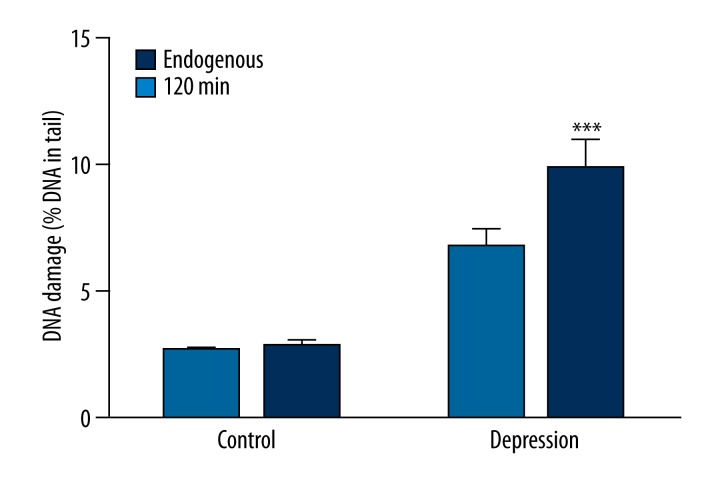
We evaluated the level of basal endogenous DNA damage by subjecting PBMCs to comet assay procedure immediately after isolation from blood. We used an alkaline version of comet assay to measure the amount of DNA alkali label sites and strand breaks, and the results are presented in the Figure 1. We found that this kind of DNA damage was significantly higher in the rDD patients than in the controls (p<0.001). Moreover, we estimated the extent of oxidative DNA damage by employing modified comet assay with 2 glycosylases: Nth removing oxidized pyrimidines and hOGG1 excising oxidized purines. The results obtained using Nth and hOGG1 are presented in Figure 1. In both cases the damage was significantly higher in the patients than in the controls (p<0.001).
Figure 1.
Basal endogenous DNA damage as mean percent of DNA in comets’ tails in peripheral blood mononuclear cells isolated from the patients with depression and the controls without psychiatric diseases. Dark blue bars denote strand breaks and alkali-labile sites measured by an alkaline version of comet assay. Light blue and blue bars represent oxidative DNA damage recognized by either (Nth) or human 8-oxoguanine DNA glycosylase (hOGG1), respectively, and measured by the alkaline version of comet assay. Forty patients and 46 controls were analyzed. The number of cells scored for each individual was 100 and the analysis was repeated twice. Error bars denote SEM, p<0.001 as compared to the controls.
Hydrogen peroxide-induces DNA damage
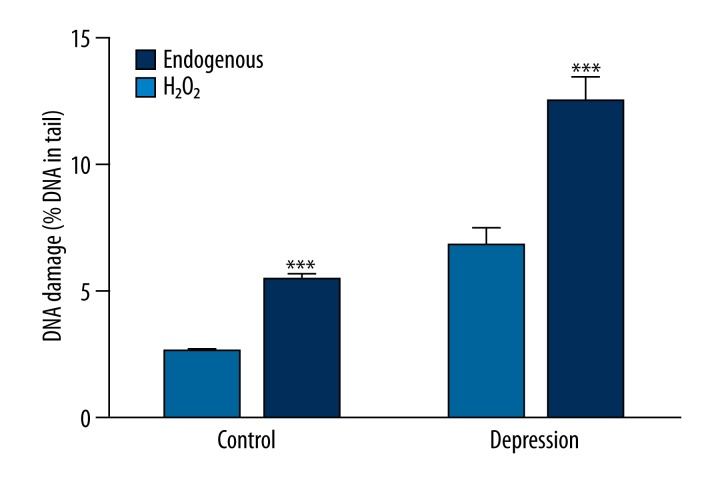
Figure 2 shows basal endogenous DNA damage and the damage induced after 10-min incubation with 20 μM H2O2 in PBMCs isolated from the patients and controls without psychiatric disturbances. For both groups the incubation caused a significant increase of the damage (p<0.001). There was no difference between increase of the damage in the patients and the controls (p=0.090), which equaled 197.29% and 228.46%, respectively. Additionally, DNA damage after treatment with H2O2 was significantly higher in PBMCs of the patients than in those of the controls (p<0.001).
Figure 2.
Basal endogenous DNA damage and DNA damage induced by hydrogen peroxide measured by an alkaline version of comet assay in peripheral blood mononuclear cells isolated from the patients with depression and the controls without psychiatric disturbances. Forty patients and 46 controls were analyzed. The number of cells scored for each individual was 100 and the analysis was repeated twice. Error bars denote SEM, p<0.001 as compared to endogenous DNA damage.
DNA repair
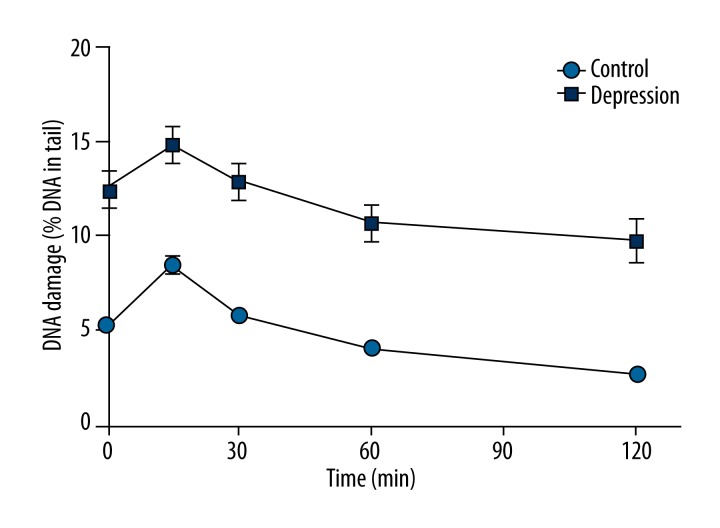
After the induction of DNA damage, H2O2 was removed and the cells were suspended in fresh medium and left for 120 min of repair incubation. The extent of DNA damage was assessed using the alkaline version of comet assay at the beginning of the incubation, and after 15, 30, 60, and 120 min. Figure 3 shows mean DNA damage changes in PBMCs of the patients with depression and the controls without psychiatric disturbances during the repair incubation. At each time, DNA damage was higher in the patients than in the controls (p<0.001). The observed initial elevation of the DNA damage in the repair kinetics can be explained by the actions of enzymes involved in DNA repair. H2O2 introduces mainly oxidative modifications of the DNA bases and single-strand breaks. The latter ones, mainly repaired by base excision repair (BER), are not recognized by the alkaline version of comet assay. One of the earliest steps of this repair pathway is excision of the damaged base, which creates structure recognized by the alkaline version of comet assay, thus causing increased DNA damage.
Figure 3.
Kinetics of DNA damage repair in peripheral blood mononuclear cells isolated from the patients with depression and controls without psychiatric disturbances measured by an alkaline version of comet assay as a percent DNA in comets’ tails. Forty patients and 46 controls were analyzed. The number of cells scored for each individual was 100 and the analysis was repeated twice. Error bars denote SEM, p<0.001 at each time point of the kinetics when comparing patients to controls.
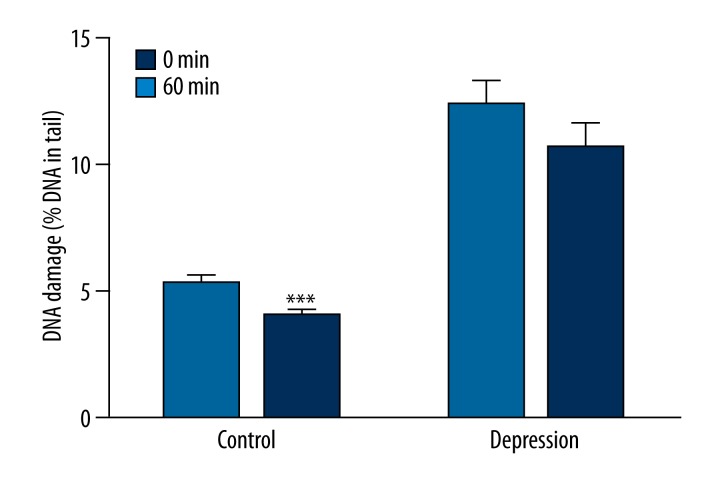
Figure 4 presents comparison of DNA damage at the beginning (0 min) and after 60 min of the repair incubation in PBMCs of the patients and the controls measured by the alkaline version of comet assay. In the controls, DNA damage was lower after 60 min than at the beginning of the incubation (p<0.001), and in the patients it was at the same level (p=0.231). Figure 5 compares basal endogenous DNA damage and the level of this parameter at the end of the repair incubation in PBMCs of the patients and the controls measured by the alkaline version of comet assay. In the controls, DNA damage induced by H2O2 was fully repaired after 120 min of incubation, since the DNA damage at this point was at the same level as an endogenous DNA damage (p=0.812). In contrast, in patients, DNA damage at the end of the repair was higher than the endogenous levels of this parameter (p<0.001).
Figure 4.
Efficiency of DNA damage repair by comparing the DNA damage evoked by hydrogen peroxide (0 min) and DNA damage after 60 min of repair incubation measured by the alkaline version of comet assay as a percent DNA in comets’ tails. Forty patients and 46 controls were analyzed. The number of cells scored for each individual was 100 and the analysis was repeated twice. Error bars denote SEM, p<0.001 when compared to the 0 min.
Figure 5.
Efficiency of DNA damage repair by comparing the basal endogenous DNA damage and DNA damage after 120 min of repair incubation measured by an alkaline version of comet assay as percent DNA in comets’ tails. Forty patients and 46 controls were analyzed. The number of cells scored for each individual was 100 and the analysis was repeated twice. Error bars denote SEM, p<0.001 when compared to the 0 min.
Discussion
The goal of our research was to examine the susceptibility of rDD patients to DNA damage induced by oxidative stress by measuring the level of endogenous DNA damage, including oxidative DNA damage, the amount of DNA damage induced by H2O2, and efficiency of DNA damage repair in the patients as compared to the controls without psychological disturbances. The biological material for this study was PBMCs isolated from blood samples. Although we are fully aware that these cells are not affected by depression, there some reasons why we have chosen to do our experiments on these cells. Firstly, in Polish conditions it is not possible to carry out large-scale studies based on the analysis of preparations taken from the brains of patients with DD, because such samples can be obtained only post-mortem. Secondly, even though the results acquired from experiments done on PBMCs cannot be simply extrapolated to the cells of the central nervous system (CNS), PBMCs are affected by the same environmental conditions as CNS cells, thus any disturbances found in the former may reflect the condition of the latter. Finally, results obtained from PBMCs may reveal evidence of inherited defects in the process of repairing DNA damage, because the genetic background of those cells reflects the constitution of the whole body, including CNS cells.
We found that patients with depression, when compared to controls without psychiatric disturbances, had more single- and double-strand breaks and alkali labile sites as measured by the alkaline version of comet assay (Figure 1). Moreover, we showed, using modified comet assay with hOGG1 and Nth, that the patients in comparison to the controls had elevated levels of oxidative DNA damage, particularly oxidative modifications of the purines and pyrimidines (Figure 2). As mentioned above, to date, the study of DNA damage in the context of depression has been limited to measurements of 8-oxoG levels in serum, urine, or peripheral blood lymphocytes. It is worth noting that this technic measures only levels of free, not incorporated into DNA 8-oxoG, thus it does not measure oxidative DNA damage per se. Our results are consistent with most studies [18–23]. The exception is work done by Yi et al., who found no significant correlation between the amount of urinary 8-oxoG and occurrence of depressive symptoms [24]. Those discrepancies can be explained by the fact, as also emphasized by other authors, that study populations consisted of rather healthy people with milder forms of depressive syndromes and not the more severe, clinically diagnosed depression cases, as in our work. It is possible that increased oxidative DNA damage occurs only in patients with more severe forms of depression, or in later stages of the disease development.
Apart from measuring the extent of endogenous DNA damage, we also estimated the amount of DNA damage induced by the incubation of PBMCs with H2O2 and efficiency of its repair. We found that this incubation caused a significant increase of DNA damage, both in the patients and the controls (Figure 2). The increase was similar in both groups, and higher DNA damage after exposure to H2O2 in the patient when compared to the controls was rather a result of the differences in endogenous DNA damage levels, thus we can conclude that DNA of the cells isolated from the patients and the controls had similar susceptibility to H2O2. Additionally, we monitored the repair efficiency of the induced DNA damage. Kinetics of the repair were similar for both studied groups, except that the damage levels were higher in the patients than in the controls (Figure 3). We observed initial increase of DNA damage caused by the action of the repair enzymes, mainly glycosylates, which excise damaged bases and create structures recognized by the alkaline version of comet assay. The maximum of DNA damage was observed after 15 min of repair incubation in both patients and controls, and after this point the amount of DNA damage was systematically reduced. Because of the significantly higher levels of DNA damage after incubation with H2O2 in the patients than in the controls, the efficiency of DNA repair cannot be evaluated simply by comparing the extent of DNA damage between the groups. Thus, we compared DNA damage within the groups. We found that in the controls, after 60 min of repair, incubation DNA damage was reduced below the level of DNA damage induced by H2O2, when in the patients it was still at the same level (Figure 4). Moreover, at the end of the incubation, in the controls the DNA damage was repaired and matched to the extent of endogenous DNA damage, in contrast to the patients, where DNA damage after 120 min was higher than the endogenous levels of this parameter (Figure 5). These results indicate that in the patients, oxidative DNA damage is less efficiently removed than in the controls.
Our results do not show whether the increased DNA damage and less efficient DNA repair are the hallmarks of rDD patients and at some point they contribute to development of the diseases, or rather they are caused by the depression itself. On the one hand, the latter hypothesis is proved by the results obtained in the subject with milder depressive syndromes, where levels of 8-oxoG were not elevated [23]. Moreover, NLRP3 inflammasome, activation of which was detected in the patients’ PBMCs, was also found to inhibit DNA repair after induction of oxidative stress [10,17]. Additionally, depression was also found to be associated with activation of innate immune response, which may cause oxidation of lipids and DNA [35–37]. On the other hand, correlation analysis done in the patients with depression between levels of 8-iso-PGF2α – a marker of oxidative damage to lipids – and 8-oxoG showed that those disparities were independent of each other [18,38]. Apart from this, a growing body of evidence shows that disturbances of mitochondria, mainly increased ROS production, are present in depression [10]. Because most of enzymes involved in mitochondrial BER are splicing variants or are the same proteins as those used in nuclear BER [39], one can speculate that disturbances of the nuclear DNA repair pathway may reflect impairments of its mitochondrial counterpart. When at some point mitochondrial DNA damage levels cross a specific threshold, they may interfere with the electron transport chain, causing increased production of mtROS. mtROS were found to be the main activators of NLRP3, which could suggest that DNA damage triggers the inflammatory state and by this may contribute to development of depression. This could prove the former hypothesis.
It must be noted that the study group was relatively small and was ethnically homologous. That is why, although our results are statistically significant, they should be considered as preliminary. If the hypothesis that amount of DNA damage in patients with depression can be associated with severity of the disease is going to be confirmed, such an association could provide a valuable diagnostic tool. We emphasize that there is a necessity for more advance research, including clinical, molecular, and epidemiological studies.
Conclusions
For the first time, we showed that patients with depression had elevated levels of DNA breaks, alkali-labile sites, and oxidative DNA damage, and that these lesions may be accumulated by impairments of DNA repair pathways. There is a need for further studies to define the role of nuclear and mitochondrial DNA damage and repair in people with depression, and their implications for clinical outcome.
Footnotes
Source of support: This study was supported with funding from the scientific research grants from the National Science Centre (no. 2011/01/D/HS6/05484 and no. 2012/05/B/NZ5/01452) and Medical University of Łódź (no. 502-03/5-062-02/502-54-105)
References
- 1.Oct, 2012. http://www.who.int/mediacentre/factsheets/fs369/en/
- 2.Greden JF. The burden of disease for treatment-resistant depression. J Clin Psychiatry. 2001;62(Suppl 16):26–31. [PubMed] [Google Scholar]
- 3.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: global burden of disease study. Lancet. 1997;349:1498–504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Z, Zeng Y, Huang H, et al. MicroRNA-132 may play a role in coexistence of depression and cardiovascular disease: A hypothesis. Med Sci Monit. 2013;19:438–43. doi: 10.12659/MSM.883935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwata M, Ota KT, Duman RS. The inflammasome: pathways linking psychological stress, depression, and systemic illnesses. Brain Behav Immun. 2013;31:105–14. doi: 10.1016/j.bbi.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel A. Review: the role of inflammation in depression. Psychiatr Danub. 2013;25(Suppl 2):S216–23. [PubMed] [Google Scholar]
- 7.Gardner A, Boles RG. Beyond the serotonin hypothesis: mitochondria, inflammation and neurodegeneration in major depression and affective spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(3):730–43. doi: 10.1016/j.pnpbp.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 8.Maes M, Bosmans E, Meltzer HY, et al. Interleukin-1 beta: a putative mediator of HPA axis hyperactivity in major depression? Am J Psychiatry. 1993;150(8):1189–93. doi: 10.1176/ajp.150.8.1189. [DOI] [PubMed] [Google Scholar]
- 9.Rawdin BJ, Mellon SH, Dhabhar FS, et al. Dysregulated relationship of inflammation and oxidative stress in major depression. Brain Behav Immun. 2013;31:143–52. doi: 10.1016/j.bbi.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcocer-Gómez E, de Miguel M, Casas-Barquero N, et al. NLRP3 inflammasome is activated in mononuclear blood cells from patients with major depressive disorder. Brain Behav Immun. 2014;36:111–17. doi: 10.1016/j.bbi.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Müller N, Schwarz MJ, Dehning S, et al. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol Psychiatry. 2006;11(7):680–84. doi: 10.1038/sj.mp.4001805. [DOI] [PubMed] [Google Scholar]
- 12.Nery FG, Monkul ES, Hatch JP, et al. Celecoxib as an adjunct in the treatment of depressive or mixed episodes of bipolar disorder: a double-blind, randomized, placebo-controlled study. Hum Psychopharmacol. 2008;23(2):87–94. doi: 10.1002/hup.912. [DOI] [PubMed] [Google Scholar]
- 13.Hannestad J, DellaGioia N, Bloch M. The effect of antidepressant medication treatment on serum levels of inflammatory cytokines: a metaanalysis. Neuropsychopharmacology. 2011;36(12):2452–59. doi: 10.1038/npp.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbasi SH1, Hosseini F, Modabbernia A, et al. Effect of celecoxib add-on treatment on symptoms and serum IL-6 concentrations in patients with major depressive disorder: randomized double-blind placebocontrolled study. J Affect Disord. 2012;141(2–3):308–14. doi: 10.1016/j.jad.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Leemans JC, Cassel SL, Sutterwala FS. Sensing damage by the NLRP3 inflammasome. Immunol Rev. 2011;243(1):152–62. doi: 10.1111/j.1600-065X.2011.01043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R, Yazdi AS, Menu P, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–25. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 17.Licandro G, Ling Khor H, Beretta O, et al. The NLRP3 inflammasome affects DNA damage responses after oxidative and genotoxic stress in dendritic cells. Eur J Immunol. 2013;43(8):2126–37. doi: 10.1002/eji.201242918. [DOI] [PubMed] [Google Scholar]
- 18.Forlenza MJ, Miller GE. Increased serum levels of 8-hydroxy-2′-deoxyguanosine in clinical depression. Psychosom Med. 2006;68(1):1–7. doi: 10.1097/01.psy.0000195780.37277.2a. [DOI] [PubMed] [Google Scholar]
- 19.Wei YC, Zhou FL, He DL, et al. Oxidative stress in depressive patients with gastric adenocarcinoma. Int J Neuropsychopharmacol. 2009;12(8):1089–96. doi: 10.1017/S1461145709000091. [DOI] [PubMed] [Google Scholar]
- 20.Kupper N, Gidron Y, Winter J, et al. Association between type D personality, depression, and oxidative stress in patients with chronic heart failure. Psychosom Med. 2009;71(9):973–80. doi: 10.1097/PSY.0b013e3181bee6dc. [DOI] [PubMed] [Google Scholar]
- 21.Maes M1, Mihaylova I, Kubera M, et al. Increased 8-hydroxy-deoxyguanosine, a marker of oxidative damage to DNA, in major depression and myalgic encephalomyelitis/chronic fatigue syndrome. Neuro Endocrinol Lett. 2009;30(6):715–22. [PubMed] [Google Scholar]
- 22.Irie M, Asami S, Ikeda M, et al. Depressive state relates to female oxidative DNA damage via neutrophil activation. Biochem Biophys Res Commun. 2003;311(4):1014–18. doi: 10.1016/j.bbrc.2003.10.105. [DOI] [PubMed] [Google Scholar]
- 23.Irie M, Asami S, Nagata S, et al. Psychosocial factors as a potential trigger of oxidative DNA damage in human leukocytes. Jpn J Cancer Res. 2001;92(3):367–76. doi: 10.1111/j.1349-7006.2001.tb01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi S, Nanri A, Matsushita Y, et al. Depressive symptoms and oxidative DNA damage in Japanese municipal employees. Psychiatry Res. 2012;200(2–3):318–22. doi: 10.1016/j.psychres.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 25.ICD-10 Classification of Mental&Behavioural Disorders. Geneva: World Health Organization; 1993. [Google Scholar]
- 26.Patten S. Performance of the Composite International Diagnostic Interview Short Form for major depression in community and clinical samples. Chronic Dis Can. 1997;3:18–24. [PubMed] [Google Scholar]
- 27.Singh NP, McCoy MT, Tice RR, et al. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–91. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 28.Klaude M, Eriksson S, Nygren J, et al. The comet assay: mechanisms and technical considerations. Mutat Res. 1996;12:89–96. doi: 10.1016/0921-8777(95)00063-1. [DOI] [PubMed] [Google Scholar]
- 29.Blasiak J, Gloc E, Drzewoski J, et al. Free radicals scavengers can differentially modulate the genotoxicity of amsacrine in normal and cancer cells. Mutat Res. 2003;535:25–34. doi: 10.1016/s1383-5718(02)00289-9. [DOI] [PubMed] [Google Scholar]
- 30.Blasiak J, Synowiec E, Tarnawska J, et al. Dental methacrylates may exert genotoxic effects via the oxidative induction of DNA double strand breaks and the inhibition of their repair. Mol Biol Rep. 2012;39:7487–96. doi: 10.1007/s11033-012-1582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjorâs M, Luna L, Johnsen B, et al. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7, 8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–22. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boiteux S, Radicella J. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie. 1999;81:59–67. doi: 10.1016/s0300-9084(99)80039-x. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M, Laval J, Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993;32:12105–11. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 34.Hatahet Z, Kow YW, Purmal AA, et al. New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–20. [PubMed] [Google Scholar]
- 35.Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends Immunol. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Memon RA, Staprans I, Noor M, et al. Infection and inflammation induce LDL oxidation in vivo. Arterioscler Thromb Vasc Biol. 2000;20:1536–42. doi: 10.1161/01.atv.20.6.1536. [DOI] [PubMed] [Google Scholar]
- 37.Aust AE, Eveleigh JF. Mechanisms of DNA oxidation. Proc Soc Exp Biol Med. 1999;222:246–52. doi: 10.1046/j.1525-1373.1999.d01-141.x. [DOI] [PubMed] [Google Scholar]
- 38.Yager S, Forlenza MJ, Miller GE. Depression and oxidative damage to lipids. Psychoneuroendocrinology. 2010;35:1356–62. doi: 10.1016/j.psyneuen.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Boesch P, Weber-Lotfi F, Ibrahim N, et al. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813(1):186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]