Abstract
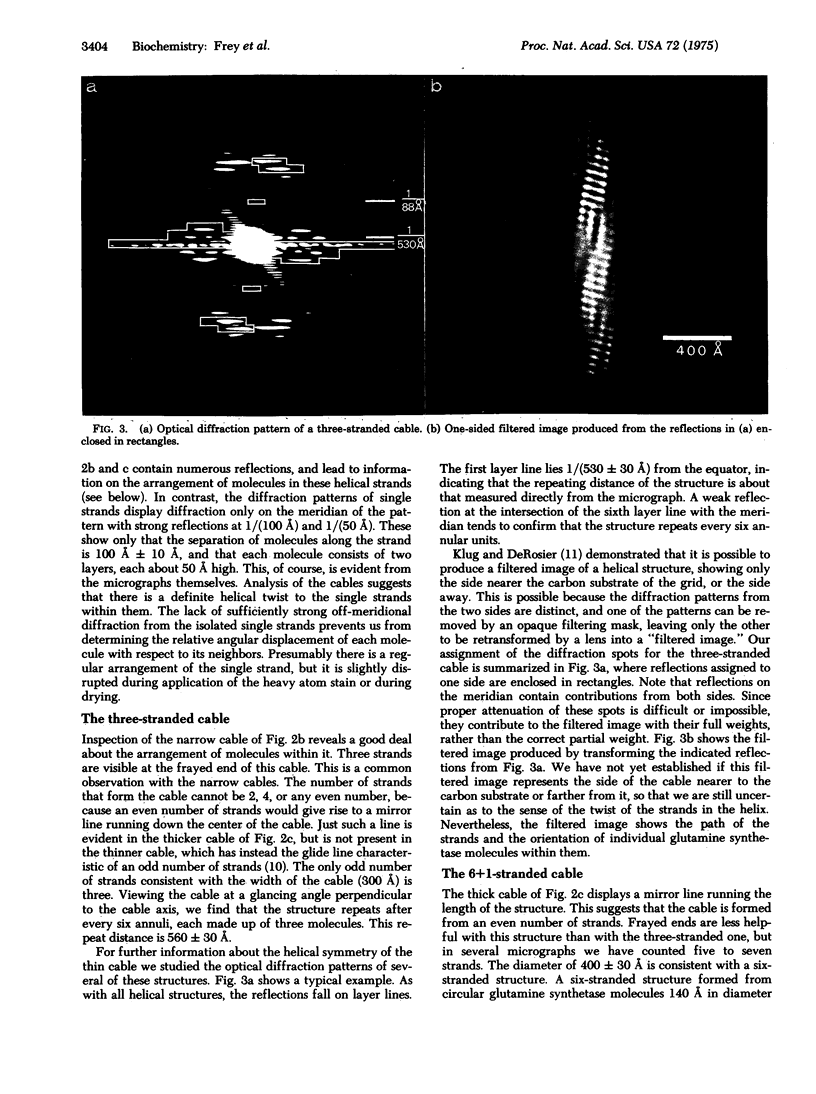
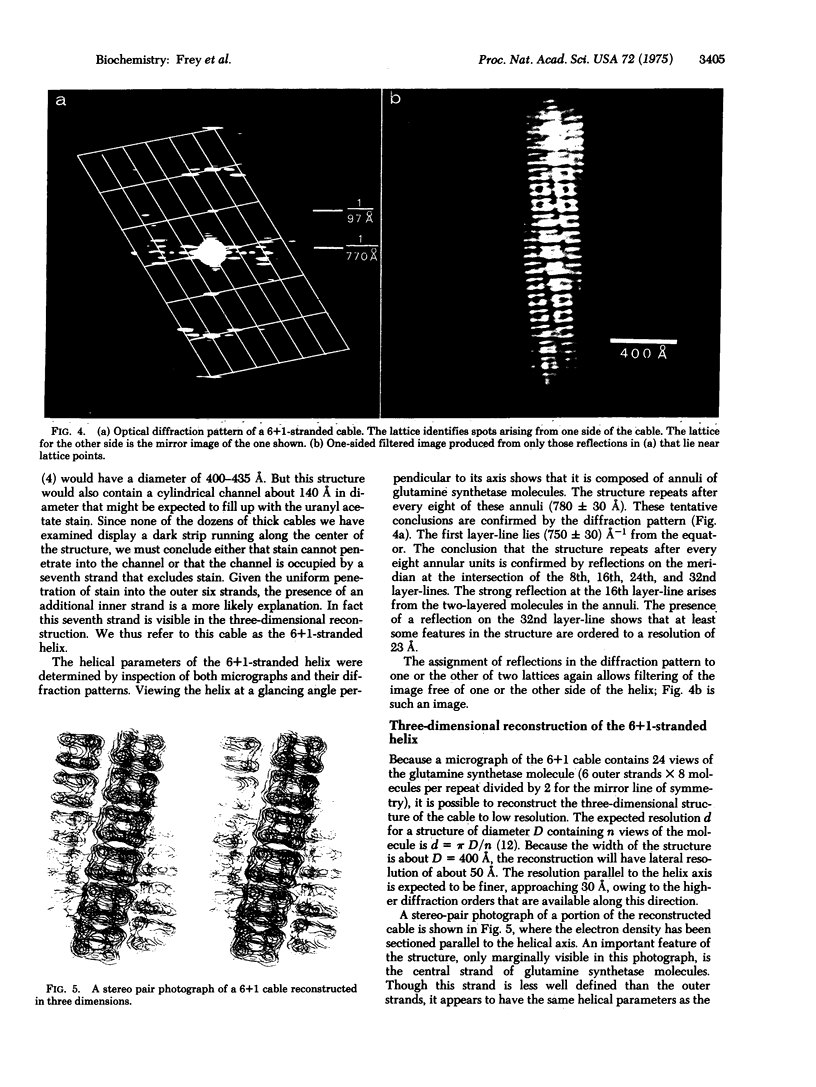
When cobaltous ion is bound to glutamine synthetase [L-glutamate:ammonia ligase (ADP-forming), EC 6.3.1.2], the two-layered hexagonal molecules polymerize face-to-face, to form long strands. The strands then wind round each other to form three- and seven-stranded cables. The structures of these cables are not immediately evident from electron micrographs because of the confusing superposition of front and back portions of the cables. But optical diffraction and filtering by the procedure of Klug and DeRosier leads to interpretable images of the cables. Because a micrograph of the seven-stranded cable contains 24 views of the glutamine synthetase molecule, it is possible to reconstruct the three-dimensional electron density of a cable and its constituent molecules at a resolution of 30--50 A. This reconstruction confirms that the symmetry of a glutamine synthetase molecule is D6. It suggests that the single subunit is an oblate ellipsoid with its minor axis (about 48 A) roughly parallel to the 6-fold axis of the molecule and its major axis (about 63 A) perpendicular to the 6-fold axis of the molecule. The subunits of the two hexagonal layers of a molecule are eclipsed. Neighboring molecules along a strand also have their hexagonal faces together, but they are rotated about the strand axis by about 7 degrees with respect to one another, rather than being eclipsed. Six outer strands are coiled about a straight central strand, and each forms identical contacts with the central strand. Moreover, these contacts between central and outer strands are apparently similar to the contacts between neighboring outer strands.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi U., Smith P. R., Dubochet J., Henry C., Kellenberger E. A study of the structure of the T-layer of Bacillus brevis. J Supramol Struct. 1973;1(6):498–522. doi: 10.1002/jss.400010606. [DOI] [PubMed] [Google Scholar]
- DeRosier D. J., Moore P. B. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J Mol Biol. 1970 Sep 14;52(2):355–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Three-dimensional reconstruction of the stacked-disk aggregate of tobacco mosaic virus protein from electron micrographs. Philos Trans R Soc Lond B Biol Sci. 1971 May 27;261(837):211–219. doi: 10.1098/rstb.1971.0053. [DOI] [PubMed] [Google Scholar]
- Klug A., De Rosier D. J. Optical filtering of electron micrographs: reconstruction of one-sided images. Nature. 1966 Oct 1;212(5057):29–32. doi: 10.1038/212029a0. [DOI] [PubMed] [Google Scholar]
- Miller R. E., Shelton E., Stadtman E. R. Zinc-induced paracrystalline aggregation of glutamine synthetase. Arch Biochem Biophys. 1974 Jul;163(1):155–171. doi: 10.1016/0003-9861(74)90465-2. [DOI] [PubMed] [Google Scholar]
- Moody M. F. Structure of the sheath of bacteriophage T4. II. Rearrangement of the sheath subunits during contraction. J Mol Biol. 1967 Apr 28;25(2):201–208. doi: 10.1016/0022-2836(67)90137-4. [DOI] [PubMed] [Google Scholar]
- PAULING L., COREY R. B. Compound helical configurations of polypeptide chains: structure of proteins of the alpha-keratin type. Nature. 1953 Jan 10;171(4341):59–61. doi: 10.1038/171059a0. [DOI] [PubMed] [Google Scholar]
- Valentine R. C., Shapiro B. M., Stadtman E. R. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968 Jun;7(6):2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]