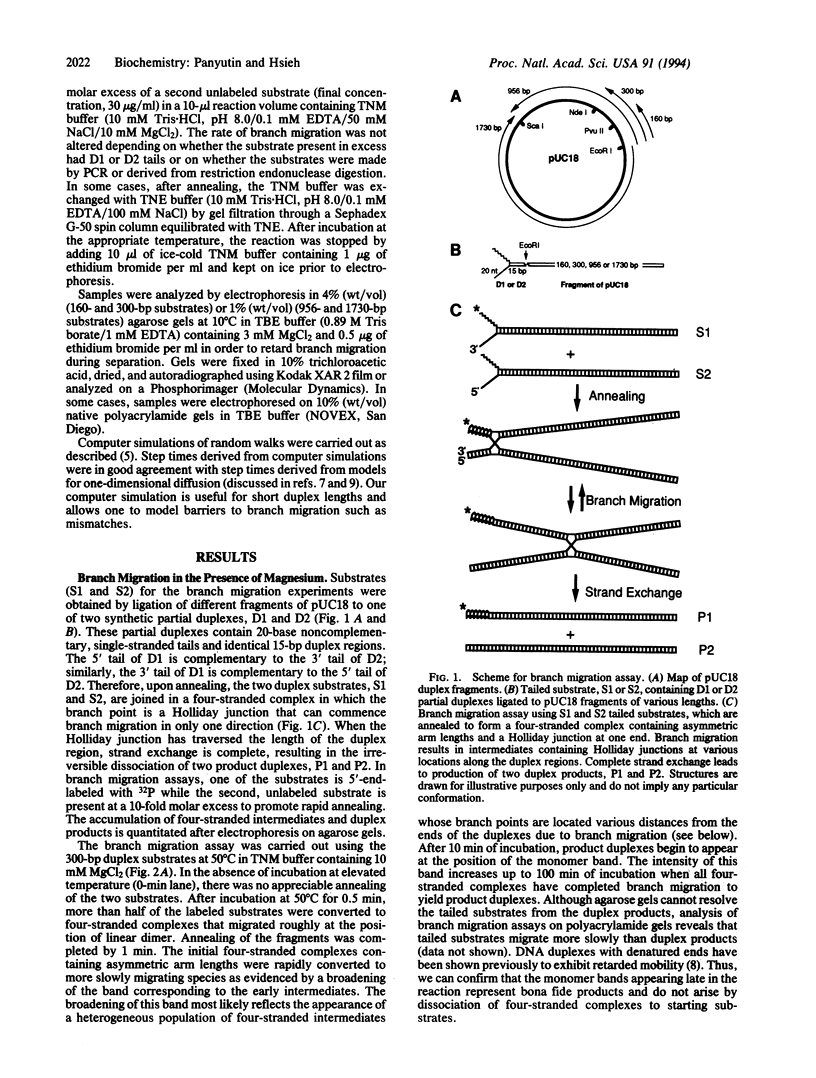
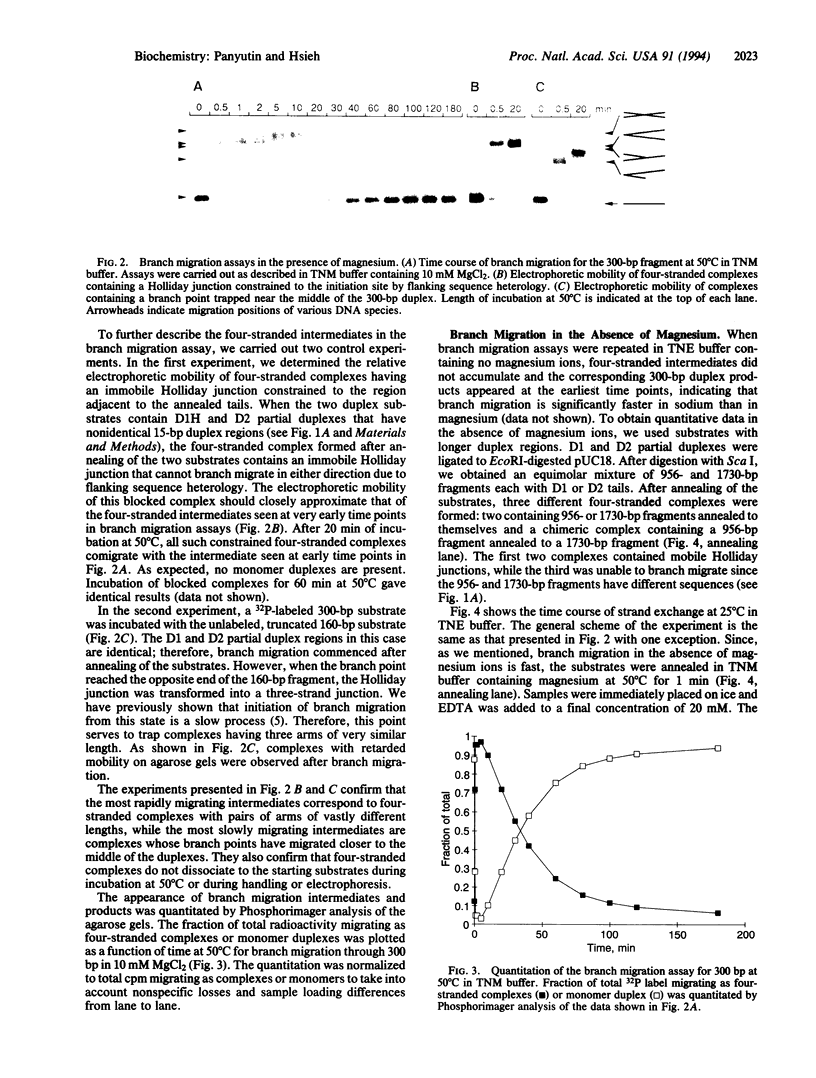
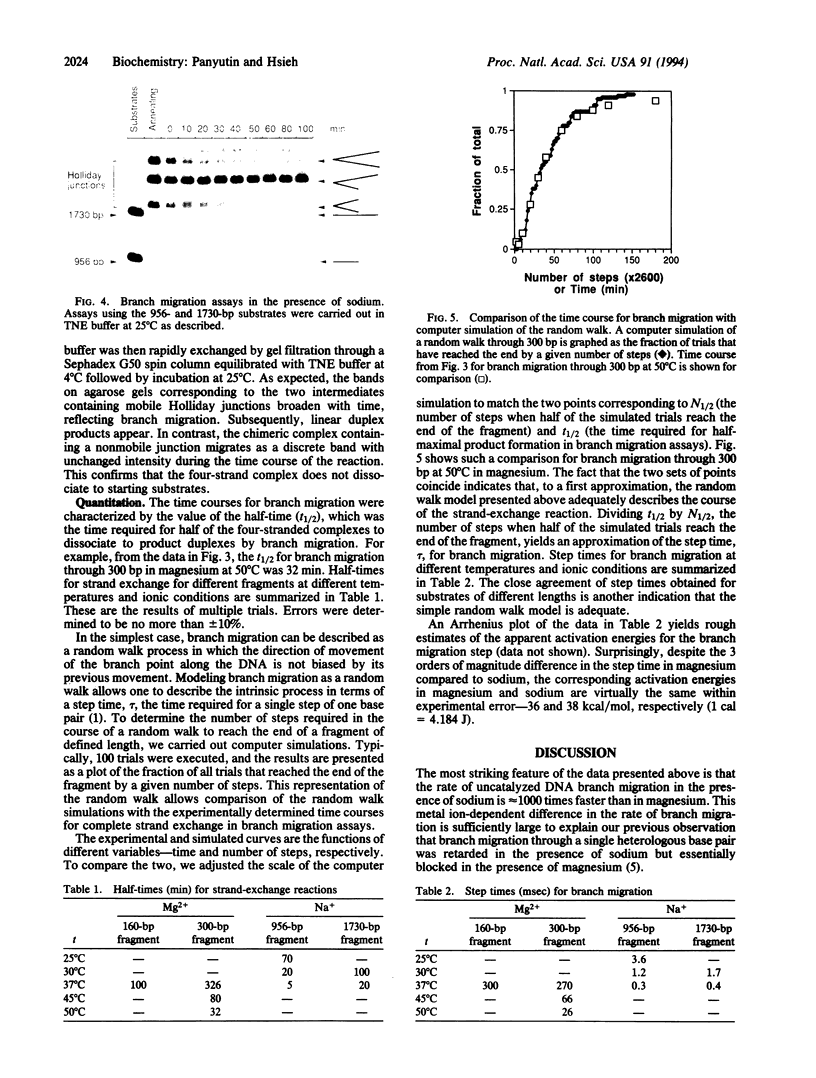
Abstract
An important step in genetic recombination is DNA branch migration, the movement of the Holliday junction or exchange point between two homologous duplex DNAs. We have determined kinetic parameters of spontaneous branch migration as a function of temperature and ionic conditions. The branch migration substrates consist of two homologous duplex DNAs each having two single-strand tails at one end that are complementary to the corresponding single-strand tails of the other duplex. Upon rapid annealing of the two duplex DNAs, a four-stranded intermediate is formed that has a Holliday junction at one end of the duplexes. Branch migration to the opposite end of the duplexes results in complete strand exchange and formation of two duplex products. The rate of branch migration is exceedingly sensitive to the type of metal ions present. In magnesium, branch migration is quite slow with a step time, tau, equal to 300 msec at 37 degrees C. Surprisingly, branch migration in the absence of magnesium was 1000 times faster. Despite this difference in rates, apparent activation energies for the branch migration step in the presence and absence of magnesium are similar. Since metal ions have a profound effect on the structure of the Holliday junction, it appears that the structure of the branch point plays a key role in determining the rate of spontaneous DNA branch migration. We discuss the role of proteins in promoting the branch migration step during homologous recombination.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Cowart M., Benkovic S. J., Nash H. A. Behavior of a cross-linked attachment site: testing the role of branch migration in site-specific recombination. J Mol Biol. 1991 Aug 5;220(3):621–629. doi: 10.1016/0022-2836(91)90105-f. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Nakata A., Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992 Nov;6(11):2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Johnson R. D., Symington L. S. Crossed-stranded DNA structures for investigating the molecular dynamics of the Holliday junction. J Mol Biol. 1993 Feb 20;229(4):812–820. doi: 10.1006/jmbi.1993.1087. [DOI] [PubMed] [Google Scholar]
- Kodadek T., Alberts B. M. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987 Mar 19;326(6110):312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993 Apr 25;21(8):1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Lyubchenko YuL Gel electrophoresis of partially denatured DNA. Retardation effect: its analysis and application. Nucleic Acids Res. 1982 Aug 11;10(15):4813–4826. doi: 10.1093/nar/10.15.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P., Hejna J. A., Ehrlich S. D., Cassuto E. Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res. 1993 Jul 11;21(14):3205–3209. doi: 10.1093/nar/21.14.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Burdett I., West S. C. Unusual stability of recombination intermediates made by Escherichia coli RecA protein. EMBO J. 1992 Jul;11(7):2685–2693. doi: 10.1002/j.1460-2075.1992.tb05334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Hsieh P. Formation of a single base mismatch impedes spontaneous DNA branch migration. J Mol Biol. 1993 Mar 20;230(2):413–424. doi: 10.1006/jmbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Cruciform transitions in DNA. J Biol Chem. 1984 May 25;259(10):6593–6600. [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]





