Abstract
Background
Viridans group streptococci (VGS) are both commensal microbes and potential pathogens. Increasing resistance to penicillin in VGS is an ongoing issue in the clinical environment. We investigated the difference in susceptibility and resistance to penicillin among various VGS species.
Methods
In total 1,448 VGS isolated from various clinical specimens were analyzed over a two-yr period. Identification and antimicrobial susceptibility test was performed by the automated VITEK 2 system (bioMerieux, France) or the MicroScan MICroSTREP system (Siemens, Germany).
Results
Among the 1,448 isolates, 412 were isolated from blood (28.4%). Streptococcus mitis group was the most frequently isolated (589 isolates, 40.7%), followed by the S. anginosus group (290 isolates, 20.0%), S. sanguinis group (179 isolates, 12.4%) and S. salivarius group (57 isolates, 3.9%). In total, 314 isolates could not be identified up to the species level. The overall non-susceptibility to penicillin was observed to be 40.0% (resistant, 11.2% and intermediately resistant, 28.8%) with uneven distribution among groups; 40.2% in S. sanguinis group (resistant, 5.0% and intermediately resistant, 35.2%), 60.3% in S. mitis group (resistant, 20.9% and intermediately resistant, 39.4%), 78.9% in S. salivarius group (resistant, 8.8% and intermediately resistant, 70.1%), and 6.2% in S. anginosus group (resistant, 1.7% and intermediately resistant, 4.5%).
Conclusions
Antimicrobial resistance patterns towards penicillin show differences among various VGS; this should be considered while devising an effective antimicrobial treatment against VGS.
Keywords: Viridans streptococci, Penicillin, Resistance, Microbial
INTRODUCTION
Viridans group streptococci (VGS) represent a diverse group of potentially pathogenic gram-positive cocci that form a part of the normal flora in the oropharynx, urogenital tract, and gastrointestinal tract. Doern and Burnham [1] commented that VGS are the "grab bag" that remains on exclusion of beta-hemolytic streptococci, enterococci, and pneumococci from the streptococci. Indeed, VGS includes various groups of streptococci such as Streptococcus mitis, S. salivarius, S. mutans, S. sanguinis, S. anginosus, and S. bovis [2].
Although VGS comprises diverse groups of the genus Streptococcus, they are grouped together owing to their shared status as potential pathogens. This concept, while underrated in the past has been gaining attention recently. VGS are known to be the most frequently isolated in infective endocarditis [3], and VGS bacteremia is being viewed as a serious cause of sepsis in neutropenic patients [4]. In addition, VGS could function as a reservoir of resistance genes, transferring resistance traits to S. pneumoniae or S. pyogenes [5].
Another VGS-related concern is the possibility of increased antimicrobial resistance to β-lactams and other such antimicrobial agents [4, 6]. We conducted a two-years study on the antimicrobial resistance of VGS isolated from various clinical specimens and compared the differences in the levels of antimicrobial resistance according to species. This study elucidates the antimicrobial resistance pattern of VGS isolated from various specimens.
METHODS
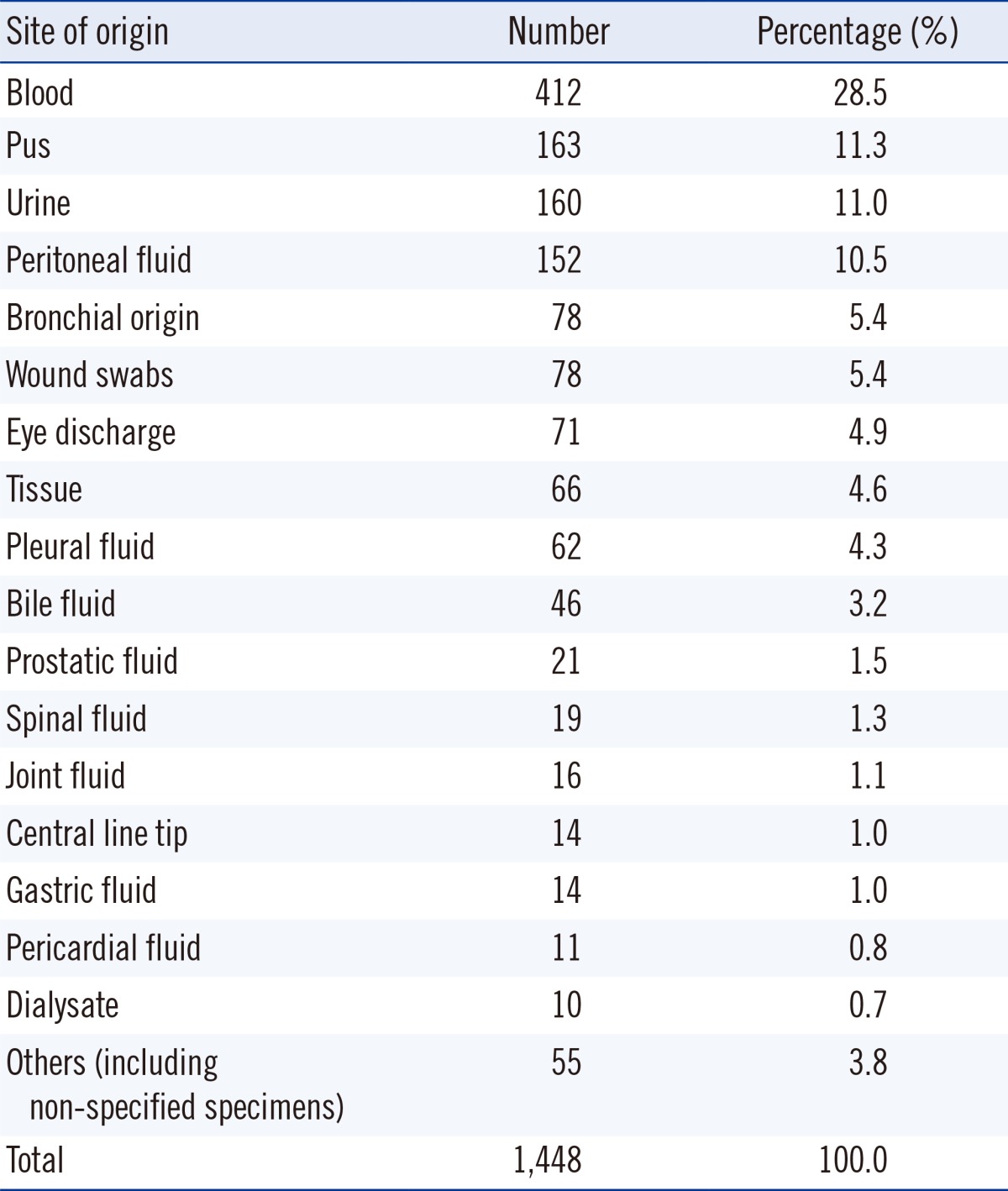
From May 2011 to April 2013, all cases showing VGS growth from any kind of specimen that was identified and reported for antimicrobial susceptibility at the Samsung Medical Center, Seoul, Korea were included in this study. The criteria for identification and antimicrobial resistance testing were determined as per the specimen. Growth of VGS in sources considered sterile was also included. Isolates from voided urine and indwelling urine catheter cultures were identified when the number of colonies exceeded 100,000 colony- forming unit (CFU)/mL. Isolates from straight or suprapubic urinary catheters were reported when the number of colonies exceeded 1,000 CFU/mL. Isolates from sites generally considered to harbor commensal flora were included when VGS were observed as being the dominantly growing group when cultured on a blood agar plate. VGS retrieved from bronchoalveolar lavage fluid or transtracheal aspiration, wound swabs, or bile fluid were identified and tested for antibiotic susceptibility. Isolates from specimens where VGS are considered the dominant flora, such as oral swabs, tissue samples from small intestinal tract, and specimens from female genitalia, were excluded from this study. The origin of all isolates is reported in Table 1. On the basis of the 1,448 isolates evaluated, blood was the most frequent source of VGS (412 isolates, 28.5%).
Table 1.
Site of origin of the isolates
The isolates were identified by routine bacteriological methods and by using the automated VITEK 2 system (bioMérieux, Marcy l'Étoile, France). After pure cultures of the isolates were obtained, genus-, group-, or species-level identification was performed by using the VITEK2 GP identification system.
Antimicrobial resistance testing was performed by using two automated devices: the MicroScan MICroSTREP Plus Antimicrobial Panel (Siemens Healthcare Diagnostics, Erlangen, Germany) was utilized for testing specimens obtained between May 1, 2011 and April 13, 2012 and the VITEK 2 system was used for testing specimens obtained between April 14, 2012 and April 30, 2013.
Antimicrobial susceptibility was compared among the VGS groups by using the chi-square test. All statistical analyses were performed by using SPSS 21 (IBM Inc., Chicago, IL., USA). All statistical testing was conducted at the 0.05 level.
RESULTS
1. Identification
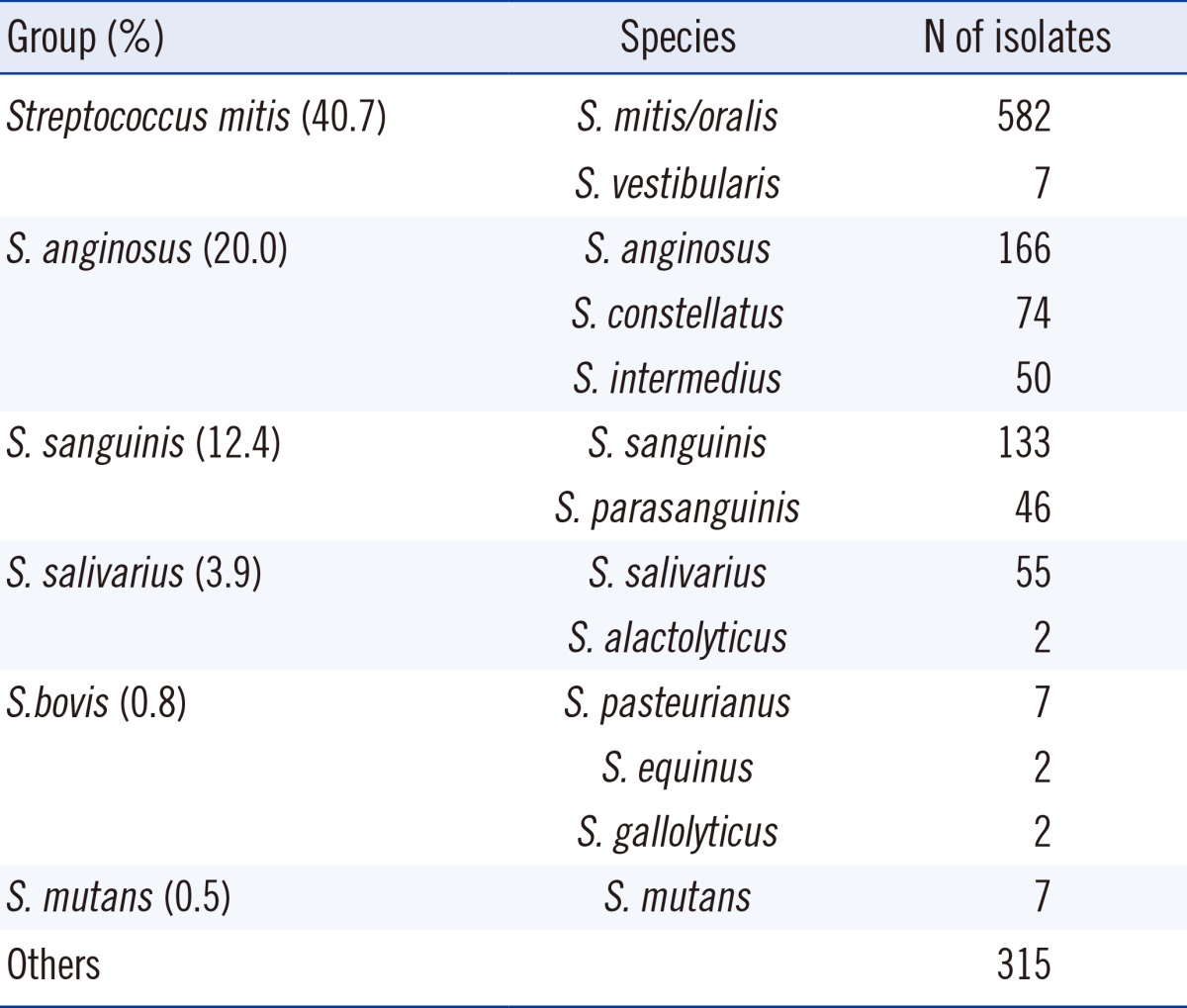
Among 1,448 unique isolates, species-level identification was achieved for 1,133 isolates (78.2%), with the S. mitis group being the most frequently identified species (40.7%). A comprehensive overview of the identified VGS bacterial groups is provided in Table 2.
Table 2.
Identification of isolates up to the species level
2. Antimicrobial susceptibility testing
1) Overall results
In total, 869 isolates (60.0%) were susceptible to penicillin, 416 isolates (28.7%) showed intermediate resistance, and 163 isolates (11.3%) were resistant. Similar results were observed when ampicillin susceptibility was tested; 924 isolates (63.8%) were susceptible to ampicillin, 334 isolates (23.1%) showed intermediate resistance, and 190 isolates (13.1%) were resistant. Resistance patterns to the two third-generation cephalosporins were similar, with 11.2% of the isolates exhibiting resistance in each case. The least active antimicrobial agent was tetracycline, with 616 isolates (43.3%) exhibiting resistance and 765 isolates (53.7%) exhibiting susceptibility, while vancomycin showed universal susceptibility (Table 3). While isolates classified into the S. salivarius, S. bovis, and S. mutans groups were too few to be subjected to proper statistical analysis, the remaining samples showed that antimicrobial resistance differed between species. An overall chi-square test revealed a statistically significant difference in resistance to penicillin and erythromycin between groups (P<0.001). All possible pairwise comparisons were reported by using the chi-square test and were indicated as being statistically significant (P<0.001).
Table 3.
Antimicrobial resistance patterns exhibited by the viridans group streptococci group
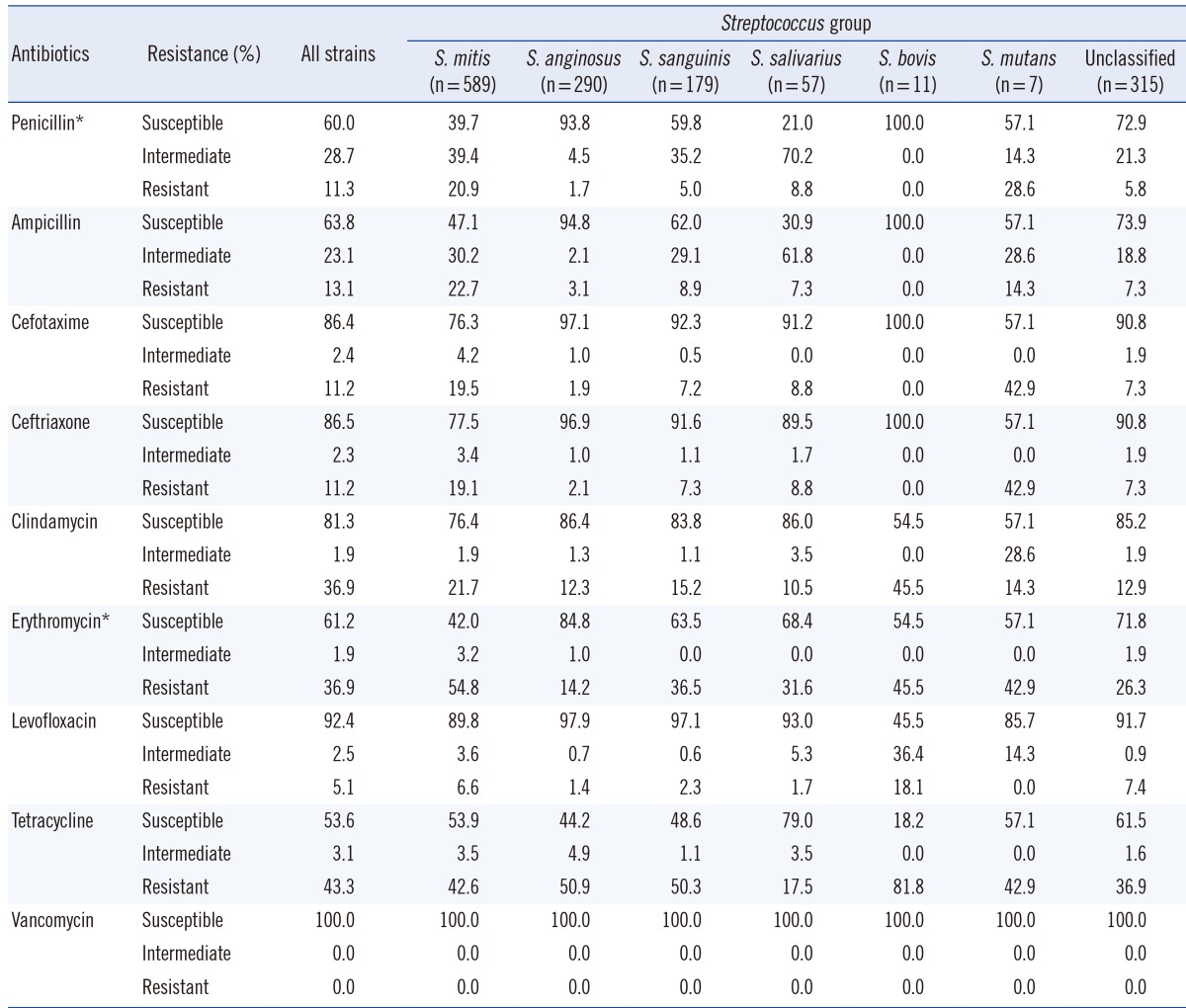
*Chi-square test was used to compare the difference in susceptibility pattern in S. mitis, S. anginosus, and S. sanguinis groups; P value was <0.001.
2) S. mitis group
Overall resistance shown by isolates identified as being members of the S. mitis group was higher than that shown by members of other groups. The number of isolates susceptible to penicillin was 234 (39.7%), while that of isolates displaying intermediate and complete resistance were 232 (39.4%) and 123 (20.9%), respectively. Ampicillin resistance patterns were similar to penicillin resistance patterns. Resistance to third-generation cephalosporins exhibited by S. mitis showed a nearly two-fold increase compared to that exhibited by the resistant strains of the entire VGS group analyzed.
Of the 589 isolates, 582 were identified as S. oralis or S. mitis, and seven were identified as S. vestibularis. The resistance pattern of S. vestibularis seen in this study may not be reflective of the species' true resistance patterns owing to its low numbers. Despite this, it was noted that one isolate was resistant to penicillin and ampicillin, and two isolates showed intermediate resistance to the two agents. The isolate that was resistant to penicillin and ampicillin also showed resistance to cefotaxime and ceftriaxone.
3) S. anginosus group
S. anginosus group, formerly known as S. milleri [2], comprises three species: S. anginosus, S. intermedius, and S. constellatus.
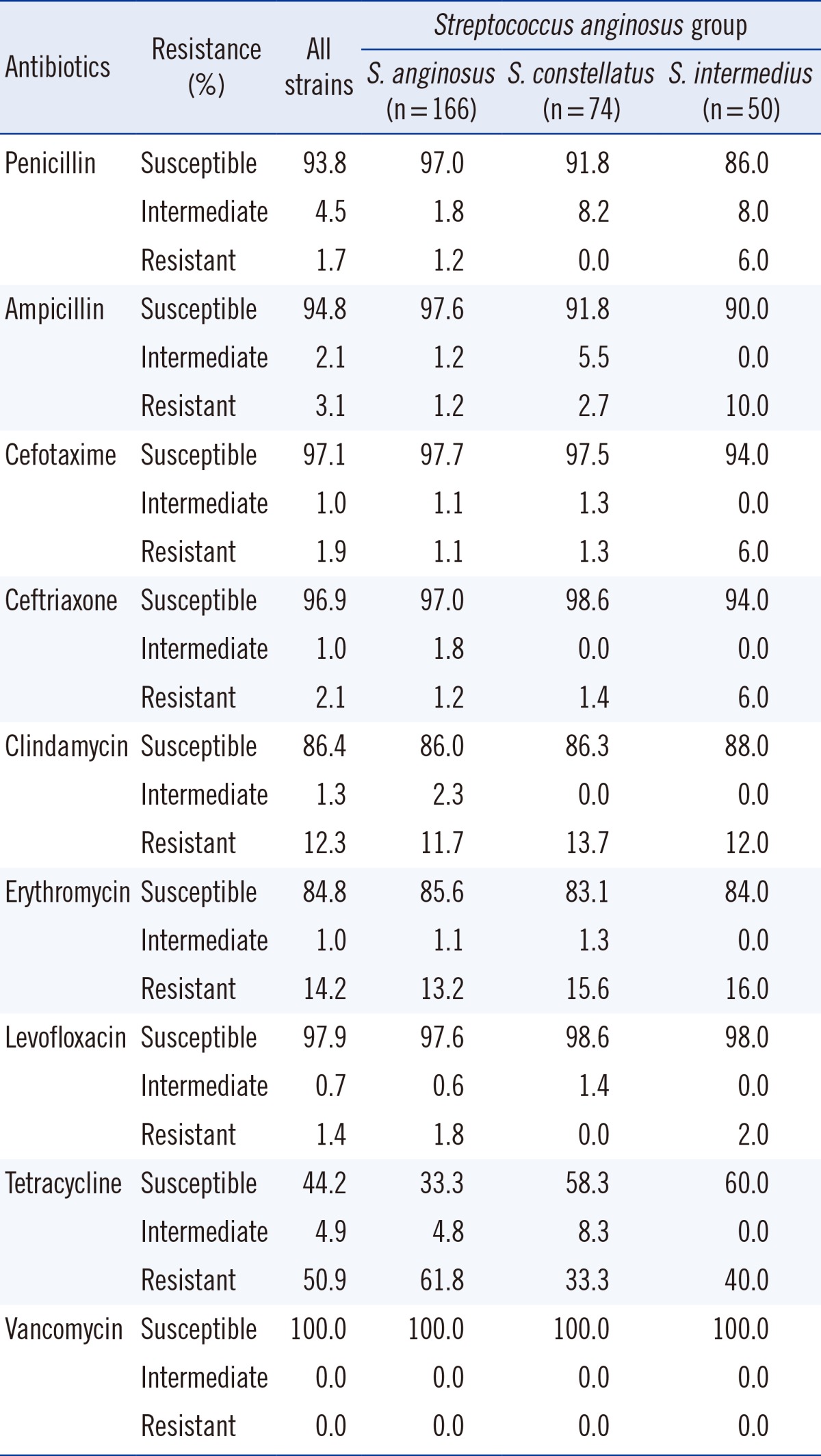
Penicillin proved to be an effective antimicrobial agent, with only five isolates (1.7%) showing penicillin resistance and 13 isolates (4.5%) showing intermediate resistance out of a total of 290 isolates (Table 4). Resistance to third-generation cephalosporins was at approximately 2%, and approximately 1% of isolates showed intermediate resistance. However, a large portion of isolates of this group (163 isolates, 55.8%) showed resistance or intermediate resistance to tetracycline. All isolates assigned to this group were identified up to the species level. 166 isolates were identified as S. anginosus, of which 161 were susceptible to penicillin, 3 showed intermediate resistance, and two were resistant. Resistance to tetracycline was high and seen in 103 isolates (61.8%). 74 isolates were identified as S. constellatus, of which 68 (91.9%) were susceptible to penicillin and the remaining six (8.1%) were intermediately resistant. 50 isolates were identified as S. intermedius, of which 43 were susceptible to penicillin (86.0%), four isolates (8.0%) exhibited intermediate resistance, and three isolates (6.0%) were resistant (Table 4).
Table 4.
Antimicrobial resistance pattern in the Streptococcus anginosus group
4) S. sanguinis group
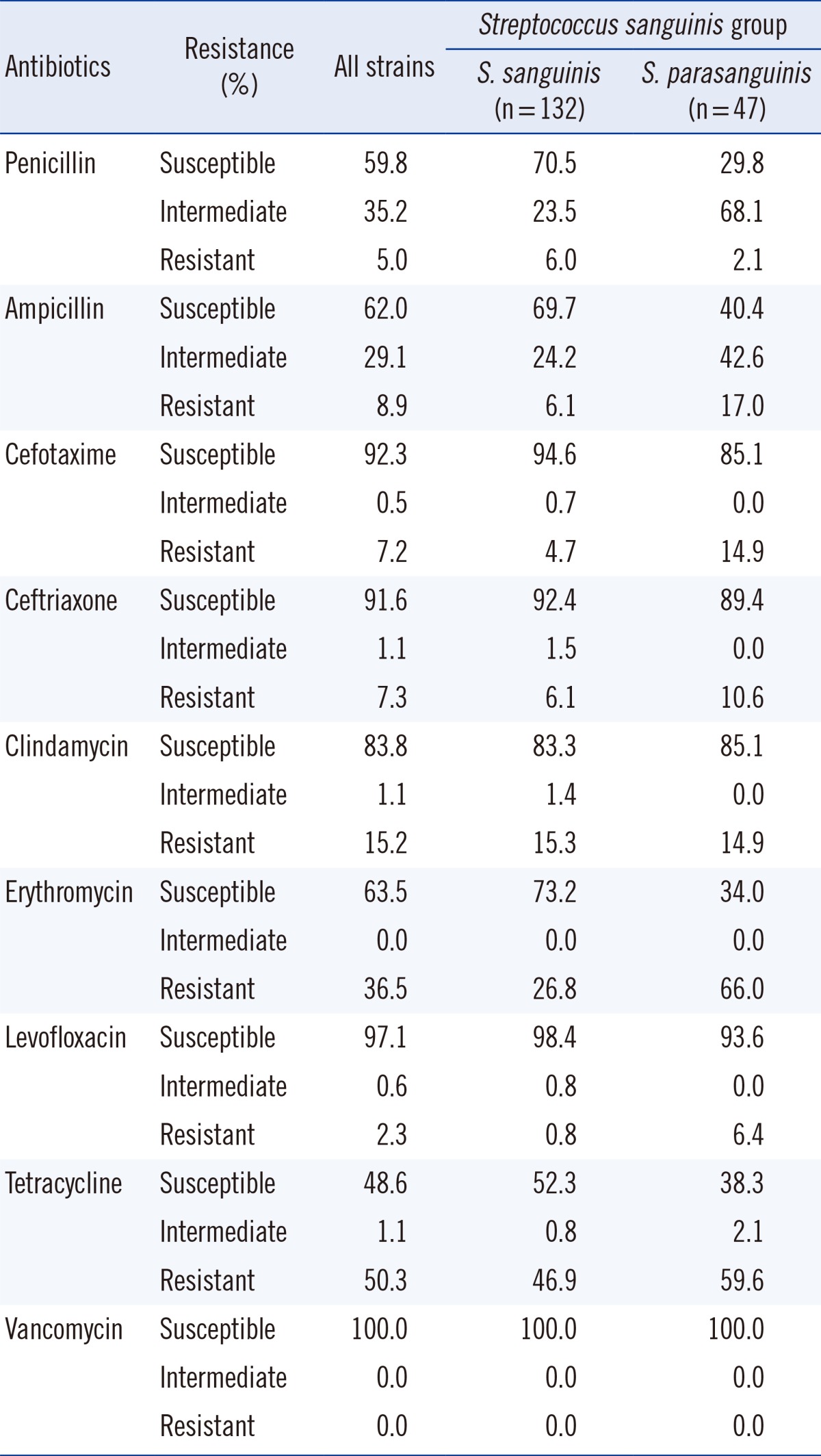
Isolates that were identified as being members of the S. sanguinis group were classified as S. sanguinis (132 isolates) or S. parasanguinis (47 isolates). The overall susceptibility to penicillin was seen in 107 isolates (59.8%) with 93 isolates being S. sanguinis and 14 being S. parasanguinis (Table 5). It should be noted that while 70.4% of S. sanguinis isolates were susceptible to penicillin, only 29.7% of S. parasanguinis isolates were susceptible to penicillin (chi-square test, P<0.001). Such significant differences were also observed in susceptibility to erythromycin (P<0.001).
Table 5.
Antimicrobial resistance pattern in the Streptococcus sanguinis group
5) Isolates from blood
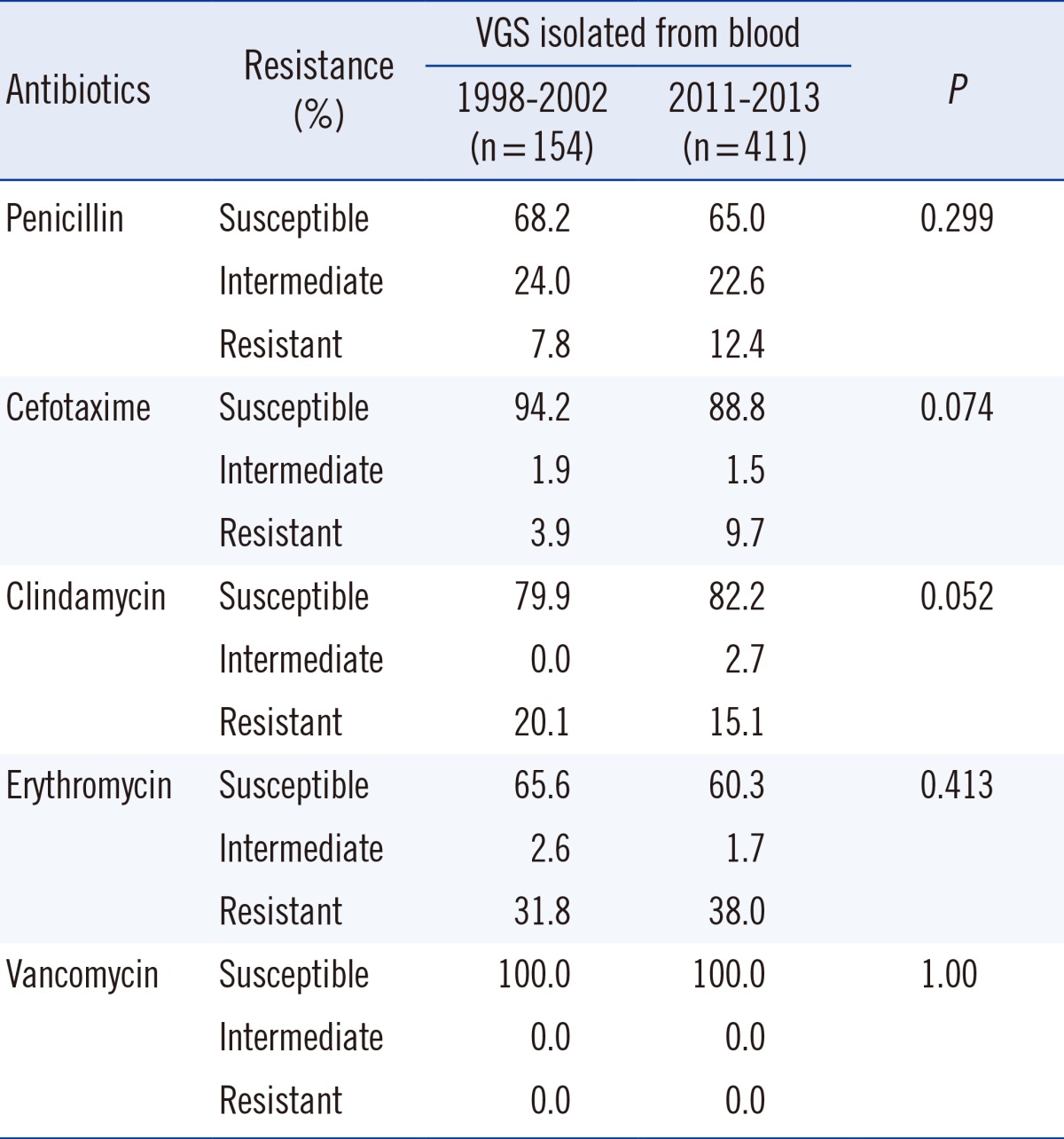
Isolates from blood were analyzed separately. Data from the previous study conducted at our institute [6] was compared with the present data (Table 6). No significant change was observed in the antimicrobial resistance pattern.
Table 6.
Comparison of antimicrobial susceptibility of viridans group streptococci (VGS) isolated from blood with data from a previous study [6]
DISCUSSION
In the past, VGS were considered to show little resistance to penicillin. However, strains showing penicillin resistance were reported to be isolated from the gingival flora of patients receiving penicillin prophylaxis for rheumatic fever in 1962 [7]. The virulence of penicillin-resistant VGS was further reported in S. anginosus [8], S. mitis [9, 10], and S. intermedius [10]. There were also speculations that this resistance was associated with the emergence of penicillin-resistant pneumococci [11]. Streptococci develop resistance to penicillin owing to an alteration in the penicillin-binding protein; thus, it was speculated that penicillin resistance could be translated into resistance to other β-lactams, all of which depend on penicillin-binding proteins to exert their activity [12]. This mechanism was described in S. pneumoniae [13], and it was theorized that VGS might exhibit a similar phenomenon regarding acquisition of resistance, since S. pneumoniae was suggested to have acquired its resistance from S. mitis [14].
Previously, Alcaide et al. [15] investigated the difference in penicillin resistance among stains isolated from VGS bacteremia. They revealed that the most frequently isolated species among VGS were S. mitis, S. anginosus, and S. sanguinis. It was also observed that antimicrobial resistance to penicillin differed among these species; 41.5% of S. mitis, 41.7% of S. sanguinis, 28.1% of S. salivarius, and 14% of S. anginosus (P<0.01) showed intermediate or high resistance to penicillin. Their findings are in concordance with our findings, in that S. mitis, S. anginosus, and S. sanguinis were the most frequently isolated species. The penicillin resistance patterns exhibited by the various species were also similar to those observed in this study; isolates from the S. anginosus group showed the greatest susceptibility to penicillin, while a large number of isolates of both S. mitis and S. sanguinis groups were resistant to penicillin. However, our investigation shows a high portion of resistance or intermediate resistance of S. salivarius to penicillin (79% non-susceptible, n=57), which is in contrast to the 28.1% resistance reported by Alcaide et al. [15]. It must be noted that the number of isolates in our study was relatively smaller compared with that in the former study. Another different finding from the former study is that compared to S. sanguinis, S. mitis is distinctly more resistant to penicillin (60.3% vs. 40.2%, P<0.001). It was noticed that the rise of penicillin resistant strains was mostly observed in the S. mitis and S. sanguinis groups, whereas the S. anginosus group remains mostly susceptible to penicillin. It was also observed that some studies disregarded the difference in antimicrobial susceptibility among species when investigating VGS bacteremia [16, 17, 18]. We suggest that this premise can be misleading, since our data clearly shows that there is such a difference, specifically among the resistant strains of the different species.
The percentage of strains resistant to penicillin is seen to differ from region to region. Lower resistance rates were reported in Germany [19] compared with the rates observed in our study or that observed in the study from Spain [15]. Such geological variance has also been noted in a surveillance program that covered multiple hospitals from the United States, Canada, and Latin America [12]. β-Lactam-resistant VGS are particularly prevalent in regions with high-incidence of penicillin-resistant pneumococci, which, in turn, correlated with outpatient sales of β-lactams [20].
Also, our previous study that was conducted 10 yr ago showed a non-susceptibility of 32% in VGS isolated from blood cultures [6]. When comparing the rates of penicillin alone, this shows no significant difference compared to the non-susceptibility rate of 40% (35% when including isolates from blood cultures only) seen in our study. However, our data show that the rate of cefotaxime-resistant VGS has tripled in the last 10 yr. In addition, when considering non-susceptibility to antibiotic agents other than β-lactams, a comparison with our previous results showed no significant difference [6]. Considering the mechanism behind the resistance to other antimicrobial agents being different from that involved in resistance to penicillin, this finding is not surprising.
Multiple sources have demonstrated that our chosen identification method shows results that are similar to those obtained using other methods with regards to the genus Streptococcus [16, 21, 22]. However, it should also be noted that the identification of alpha-hemolytic streptococci to the species level is one of the limitations of the VITEK2 system [23]. The large number of isolates that were identified simply as VGS without mention of specific species type reflects such limitations.
Overall, our study displays antimicrobial resistance in VGS in a large collection of isolates obtained from various specimens. We show that while the percentage of penicillin-resistant isolates may have not increased substantially, our focus should shift to other β-lactams. There is also a significant difference in antimicrobial resistance pattern among species; this must be taken into consideration when isolating and identifying VGS in order to decide the first-line empirical antimicrobial therapy. We suggest that the practice of reporting isolates simply as VGS may not be sufficient in a clinical setting and recommend identification up to the species level.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Doern CD. It's not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J Clin Microbiol. 2010;48:3829–3835. doi: 10.1128/JCM.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson WR, Karchmer AW, Dajani AS, Taubert KA, Bayer A, Kaye D, et al. Antibiotic treatment of adults with infective endocarditis due to streptococci, enterococci, staphylococci, and HACEK microorganisms. American Heart Association. JAMA. 1995;274:1706–1713. [PubMed] [Google Scholar]
- 4.Tunkel AR. Infections caused by viridans streptococci in patients with neutropenia. Clin Infect Dis. 2002;34:1524–1529. doi: 10.1086/340402. [DOI] [PubMed] [Google Scholar]
- 5.Bryskier A. Viridans group streptococci: a reservoir of resistant bacteria in oral cavities. Clin Microbiol Infect. 2002;8:65–69. doi: 10.1046/j.1198-743x.2001.00398.x. [DOI] [PubMed] [Google Scholar]
- 6.Cho EH. Clinical significance of viridans streptococcal bacteremia. Korean J Lab Med. 2003;23:246–250. [Google Scholar]
- 7.Naiman RA. Pencillin-resistant bacteria in the mouths and throats of children receiving continuous prophylaxis against rheumatic fever. Ann Intern Med. 1963;58:768–772. doi: 10.7326/0003-4819-58-5-768. [DOI] [PubMed] [Google Scholar]
- 8.Gossling J. Occurrence and pathogenicity of the Streptococcus milleri group. Rev Infect Dis. 1988;10:257–285. doi: 10.1093/clinids/10.2.257. [DOI] [PubMed] [Google Scholar]
- 9.Farber BF, Eliopoulos GM, Ward JI, Ruoff KL, Syriopoulou V, Moellering RC., Jr Multiply resistant viridans streptococci: susceptibility to beta-lactam antibiotics and comparison of penicillin-binding protein patterns. Antimicrob Agents Chemother. 1983;24:702–705. doi: 10.1128/aac.24.5.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn JP, DiVincenzo CA, Lucks DA, Luskin RL, Shatzer KL, Lerner SA. Serious infections due to penicillin-resistant strains of viridans streptococci with altered penicillin-binding proteins. J Infect Dis. 1988;157:764–769. doi: 10.1093/infdis/157.4.764. [DOI] [PubMed] [Google Scholar]
- 11.Goldfarb J, Wormser GP, Glaser JH. Meningitis caused by multiply antibiotic-resistant viridans streptococci. J Pediatr. 1984;105:891–895. doi: 10.1016/s0022-3476(84)80072-4. [DOI] [PubMed] [Google Scholar]
- 12.Diekema DJ, Beach ML, Pfaller MA, Jones RN SENTRY Participants Group. Antimicrobial resistance in viridans group streptococci among patients with and without the diagnosis of cancer in the USA, Canada and Latin America. Clin Microbiol Infect. 2001;7:152–157. doi: 10.1046/j.1198-743x.2001.00230.x. [DOI] [PubMed] [Google Scholar]
- 13.Doern GV, Brueggemann AB, Huynh H, Wingert E. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997-98. Emerg Infect Dis. 1999;5:757–765. doi: 10.3201/eid0506.990603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowson CG, Coffey TJ, Kell C, Whiley RA. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 15.Alcaide F, Liñares J, Pallares R, Carratala J, Benitez MA, Gudiol F, et al. In vitro activities of 22 beta-lactam antibiotics against penicillin-resistant and penicillin-susceptible viridans group streptococci isolated from blood. Antimicrob Agents Chemother. 1995;39:2243–2247. doi: 10.1128/aac.39.10.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han SB, Bae EY, Lee JW, Lee DG, Chung NG, Jeong DC, et al. Clinical characteristics and antibiotic susceptibility of viridans streptococcal bacteremia in children with febrile neutropenia. Infection. 2013;41:917–924. doi: 10.1007/s15010-013-0470-7. [DOI] [PubMed] [Google Scholar]
- 17.Husain E, Whitehead S, Castell A, Thomas EE, Speert DP. Viridans streptococci bacteremia in children with malignancy: relevance of species identification and penicillin susceptibility. Pediatr Infect Dis J. 2005;24:563–566. doi: 10.1097/01.inf.0000164708.21464.03. [DOI] [PubMed] [Google Scholar]
- 18.Marron A, Carratalà J, González-Barca E, Fernández-Sevilla A, Alcaide F, Gudiol F. Serious complications of bacteremia caused by Viridans streptococci in neutropenic patients with cancer. Clin Infect Dis. 2000;31:1126–1130. doi: 10.1086/317460. [DOI] [PubMed] [Google Scholar]
- 19.Reinert RR, von Eiff C, Kresken M, Brauers J, Hafner D, Al-Lahham A, et al. Nationwide German multicenter study on the prevalence of antibiotic resistance in streptococcal blood isolates from neutropenic patients and comparative in vitro activities of quinupristin-dalfopristin and eight other antimicrobials. J Clin Microbiol. 2001;39:1928–1931. doi: 10.1128/JCM.39.5.1928-1931.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bronzwaer SL, Cars O, Buchholz U, Mölstad S, Goettsch W, Veldhuijzen IK, et al. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg Infect Dis. 2002;8:278–282. doi: 10.3201/eid0803.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorgensen JH, Barry AL, Traczewski MM, Sahm DF, McElmeel ML, Crawford SA. Rapid automated antimicrobial susceptibility testing of Streptococcus pneumoniae by use of the bioMerieux VITEK 2. J Clin Microbiol. 2000;38:2814–2818. doi: 10.1128/jcm.38.8.2814-2818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SJ, Uh Y, Jang IH, Lee KS, Park SD, Yoon KJ. Evaluation of the MicroScan MICroSTREP plus antimicrobial panel for testing β-hemolytic streptococci and viridans group streptococci. Korean J Lab Med. 2011;31:185–190. doi: 10.3343/kjlm.2011.31.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haanperä M, Jalava J, Huovinen P, Meurman O, Rantakokko-Jalava K. Identification of alpha-hemolytic streptococci by pyrosequencing the 16S rRNA gene and by use of VITEK 2. J Clin Microbiol. 2007;45:762–770. doi: 10.1128/JCM.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


