Abstract
Coagulase-negative staphylococci (CoNS) are reported to be the leading cause of nosocomial bloodstream infections. Staphylococcus pettenkoferi is a novel member of CoNS that was first isolated from the human blood and bursitis wound in 2002. We have reported cases of 6 S. pettenkoferi strains isolated from blood specimens, including one pathogen and 5 contaminants and catheter colonizers. Brucker Biotyper (Brucker Daltonics, Bremen, Germany) and molecular typing with 16S rRNA gene sequencing confirmed the 6 isolates as S. pettenkoferi. The conventional phenotypic identification of these isolates is not reliable owing to their inconsistent biochemical characteristics. Five of the 6 isolates were found to be resistant to oxacillin, and all isolates showed susceptibility to vancomycin and linezolid. For accurate identification of this novel species, advanced methods by using Brucker Biotyper or molecular methods such as 16S rRNA gene sequencing are required.
Keywords: Staphylococcus pettenkoferi, MALDI-TOF MS, 16S ribosomal RNA
INTRODUCTION
Staphylococcus pettenkoferi is a novel member of coagulase-negative staphylococci (CoNS) first isolated from a blood specimen and bursitis wound in Germany [1]. Three more S. pettenkoferi isolates from blood cultures were reported later [2]. Cases of osteomyelitis [3] and bloodstream infection [4, 5] were thought to be caused by S. pettenkoferi. Isolates resistant to methicillin and linezolid were also reported recently [6, 7].
Although the characteristics of S. pettenkoferi were described in detail previously [1, 2], their identification by conventional methods was difficult in a routine microbiology laboratory. We identified six S. pettenkoferi isolates from human blood specimens and confirmed their identity through 16S rRNA gene sequencing. Patients' clinical data and microbiological characteristics were retrospectively reviewed.
CASE REPORTS
The clinical data of 6 patients with S. pettenkoferi strains are summarized in Table 1.
Table 1.
The clinical characteristics of six patients with S. pettenkoferi isolates
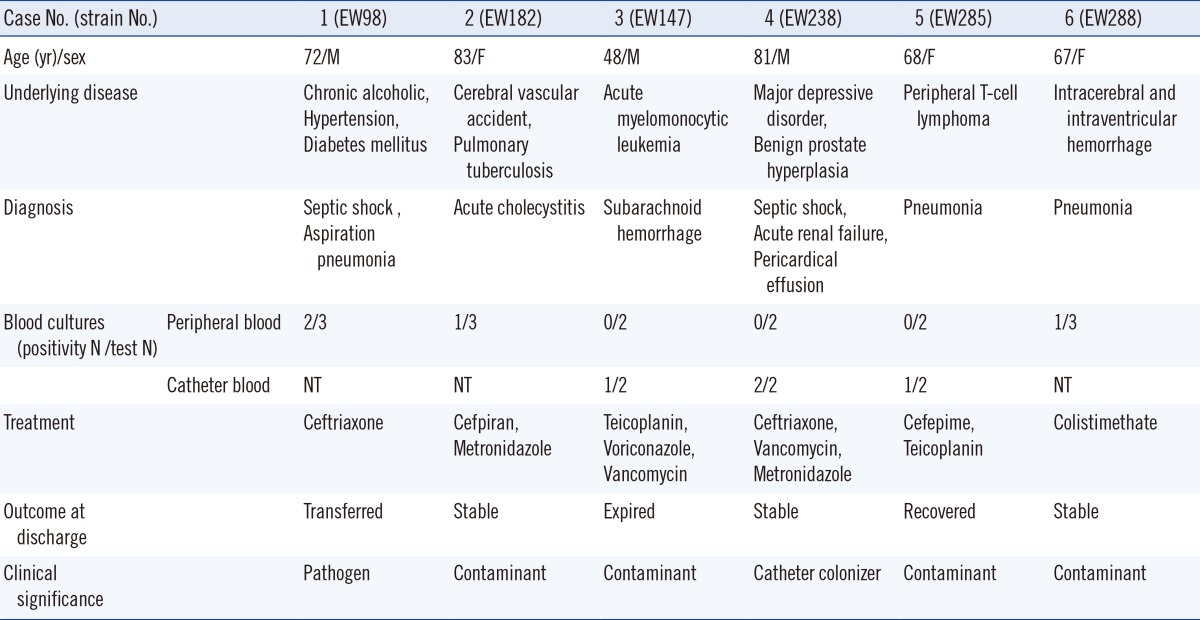
Abbreviations: M, male; F, female; NT, not tested; PTBD, percutaneous transbiliary drainage.
Subculturing of all isolates onto blood agar plates leads to the growth of 1-2-mm-sized, circular, opaque, glistening colonies with no hemolysis. The organisms were gram-positive cocci, catalase positive, and coagulase negative. For phenotypic identification, the Vitek 2 System (bioMérieux, Marcy l'Etoile, France) gram-positive identification card was used (Table 2). The isolates were further identified by using the Vitek MS IVD Database Version 2 (bioMérieux) and the Microflex LT RUO with Biotyper 3.0 Software (Brucker Daltonics, Bremen, Germany). We confirmed the identification of these isolates by 16S rRNA gene sequencing as per the methods described by a previous study [8]. DNA was extracted by boiling with the Chelex 100 Resin (Bio-Rad Laboratories, Hercules, CA, USA). The primer pair used for amplification was 27F (5'-AGA GTT TGA TC[A/C] TGG CTC AG-3') and 1492R (5'-G[C/T]T ACC TTG TTA CGA CTT-3'). This primer pair can amplify approximately ~1,500-bp fragment of the 16S rRNA gene between positions 8 and 1,509 of the Escherichia coli 16S rRNA gene, and it is considered to be universal for the domain bacteria [9]. Direct sequencing was performed by using an automatic DNA sequencer (ABI 3730XL; Life Technology Co., Grand Island, NY, USA). The 16S rRNA sequences were analyzed with the NCBI Genbank and EzTaxon (www.eztaxon.org, version 2.1) databases.
Table 2.
The biochemical characteristics and microbiological identification of the six S. pettenkoferi isolates
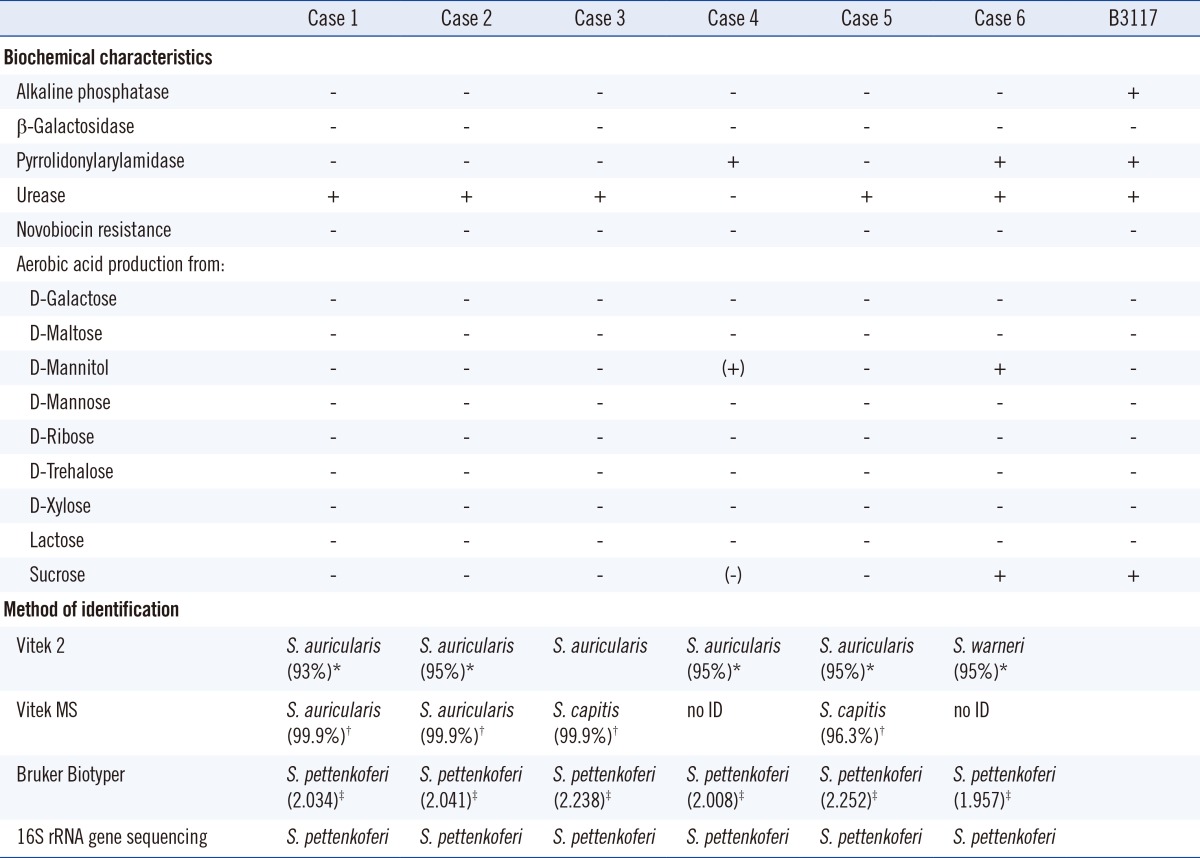
*% probability by Vitek 2; †% probability by Vitek MS; ‡the score for Brucker Biotyper.
Abbreviation: ID, identification.
Using the Vitek 2 System, all the isolates were susceptible to novobiocin and negative for alkaline phosphatase and β-galactosidase. Except for the isolate 4, all organisms were positive for urease. The isolates 4 and 6 differed from others by being pyrrolidonylarylamidase positive and being positive for acid production from D-mannitol. Five isolates were identified as Staphylococcus auricularis and one isolate as Staphylococcus warneri.
Isolates 1 and 2 showed the same results on both Vitek 2 and Vitek MS. Four isolates showed different results between Vitek 2 and the Vitek MS. We tested the isolates by Brucker Biotyper (Brucker Daltonics) to compare the accuracy of the Vitek MS systems and identified all 6 isolates as S. pettenkoferi.
Since Vitek 2, Vitek MS, and Biotyper showed contradicting results, 16S rRNA gene sequencing of these isolates was conducted. All 6 strains were identified as S. pettenkoferi (100%) on the basis of the NCBI Genbank databases. Second possible identification was Staphylococcus cohnii (98%). According to the EzTaxon databases, the sequences of all 6 isolates were identical to those of S. pettenkoferi strain B3117 (accession no. AF322002) (100%), as reported by Trülzsch et al. [1] The second possible identification was of S. cohnii subsp. cohnii (97.8-98.1%). According to the CLSI molecular method 18-A [10], we concluded that all 6 isolates were S. pettenkoferi.
With the VITEK 2 antimicrobial susceptibility testing cards (Table 3), five of the six organisms showed susceptability to gentamicin, vancomycin, teicoplanin, and linezolid and resistance to oxacillin, penicillin, ciprofloxacin, and erythromycin.
Table 3.
Antimicrobial susceptibility profiles of the six S. pettenkoferi isolates
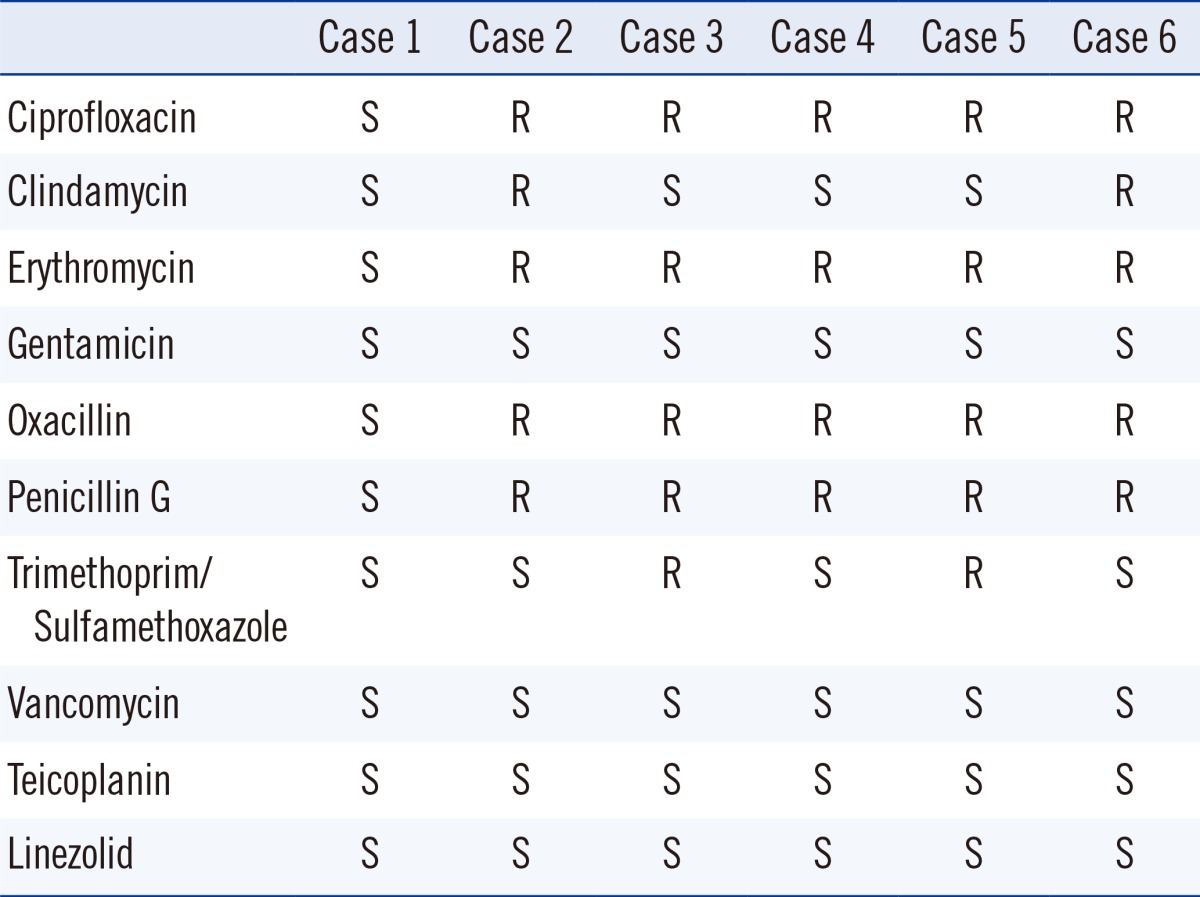
Abbreviations: S, susceptible; R, resistant.
DISCUSSION
S. pettenkoferi was newly introduced as a member of CoNS in 2002 [1]. Its clinical importance and epidemiology are therefore not completely elucidated. We identified six strains of S. pettenkoferi isolated from six hospitalized patients in different wards. All strains were isolated from the human blood. In case 1, S. pettenkoferi was determined to be the pathogen of septic shock on the basis of the clinical signs and culture positivity (2/3 peripheral blood cultures). In the remaining patients, the isolates were considered to be skin contaminants or catheter colonizers.
We confirmed the presence of these novel strains in the patients, although they were not identified by the conventional methods (Vitek 2 System) and Vitek MS. Since not all laboratories used the same microbiological identification system, these novel strains are probably underestimated in clinical microbiology. One of the first known strains of S. pettenkoferi, B3117, has distinct biochemical characteristics such as susceptibility to novobiocin, and positivity for alkaline phosphatase, urease activity, nitrate reduction, and pyrrolidonylarylamidase. This strain is also positive for aerobic production of acid from β-D-fructose, glucose, and sucrose [1, 2]. In this study, the phenotypic characteristics were variable and not consistent for all six isolates. For more accurate identification of S. pettenkoferi, an approach other than the conventional methods is needed. Between the two MALDI-TOF MS systems, Biotyper was superior to Vitek MS with regards to identification of S. pettenkoferi.
16S rRNA gene sequencing is a useful tool to identify bacterial isolates [11]. For analysis of the sequence data of these isolates, we found that both the NCBI Genbank and EzTaxon are useful for identification of S. pettenkoferi. Although 16S rRNA gene sequences are highly similar between closely related species [12], 16S rRNA gene sequencing allows for identification of S. pettenkoferi.
There is an increase in the incidences of methicillin-resistant staphylococcal infections due to acquisition of resistance to several antimicrobial agents [13], and the number of methicillin-resistant CoNS have also dramatically increased to 60-70% [14, 15]. In this study, all the isolates, except for isolate 1, were oxacillin-resistant and multidrug resistant. Recently, linezolid-resistant S. pettenkoferi isolates were also reported [7]. In our cases, all isolates were susceptible to linezolid.
In conclusion, S. pettenkoferi isolated from blood was found to be the causative agent, catheter colonizer, and skin contaminants. For accurate identification of this novel species, advanced approaches such as Biotyper or molecular methods such as 16S rRNA gene sequencing are needed.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Trülzsch K, Rinder H, Trček J, Bader L, Wilhelm U, Heesemann J. "Staphylococcus pettenkoferi," a novel staphylococcal species isolated from clinical specimens. Diagn Microbiol Infect Dis. 2002;43:175–182. doi: 10.1016/s0732-8893(02)00399-1. [DOI] [PubMed] [Google Scholar]
- 2.Trülzsch K, Grabein B, Schumann P, Mellmann A, Antonenka U, Heesemann J, et al. Staphylococcus pettenkoferi sp. nov., a novel coagulase-negative staphylococcal species isolated from human clinical specimens. Int J Syst Evol Microbiol. 2007;57:1543–1548. doi: 10.1099/ijs.0.64381-0. [DOI] [PubMed] [Google Scholar]
- 3.Loïez C, Wallet F, Pischedda P, Renaux E, Senneville E, Mehdi N, et al. First case of osteomyelitis caused by "Staphylococcus pettenkoferi". J Clin Microbiol. 2007;45:1069–1071. doi: 10.1128/JCM.02328-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song SH, Park JS, Kwon HR, Kim SH, Kim HB, Chang HE, et al. Human bloodstream infection caused by Staphylococcus pettenkoferi. J Med Microbiol. 2009;58:270–272. doi: 10.1099/jmm.0.004697-0. [DOI] [PubMed] [Google Scholar]
- 5.d'Azevedo PA, Comin G, Cantarelli V. Characterization of a new coagulase-negative Staphylococcus species (Staphylococcus pettenkoferi) isolated from blood cultures from a hospitalized patient in Porto Alegre, Brazil. Rev Soc Bras Med Trop. 2010;43:331–332. doi: 10.1590/s0037-86822010000300023. [DOI] [PubMed] [Google Scholar]
- 6.Garza-González E, López D, Pezina C, Muruet W, Bocanegra-García V, Muñoz I, et al. Diversity of staphylococcal cassette chromosome mec structures in coagulase-negative staphylococci and relationship to drug resistance. J Med Microbiol. 2010;59:323–329. doi: 10.1099/jmm.0.015800-0. [DOI] [PubMed] [Google Scholar]
- 7.Mihaila L, Defrance G, Levesque E, Ichai P, Garnier F, Derouin V, et al. A dual outbreak of bloodstream infections with linezolid-resistant Staphylococcus epidermidis and Staphylococcus pettenkoferi in a liver Intensive Care Unit. Int J Antimicrob Agents. 2012;40:472–474. doi: 10.1016/j.ijantimicag.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Moon HW, Lee SH, Chung HS, Lee M, Lee K. Performance of the Vitek MS matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for identification of Gram-positive cocci routinely isolated in clinical microbiology laboratories. J Med Microbiol. 2013;62:1301–1306. doi: 10.1099/jmm.0.062950-0. [DOI] [PubMed] [Google Scholar]
- 9.Schuurman T, de Boer RF, Kooistra-Smid AM, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004;42:734–740. doi: 10.1128/JCM.42.2.734-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline, MM18-A. Wayne: PA: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 11.Clarridg JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 13.Drozenová J, Petrás P. Characteristics of coagulase-negative staphylococci isolated from hemocultures. Epidemiol Mikrobiol Imunol. 2000;49:51–58. [PubMed] [Google Scholar]
- 14.Koksal F, Yasar H, Samasti M. Antibiotic resistance patterns of coagulase-negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiol Res. 2009;164:404–410. doi: 10.1016/j.micres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Carbon C. MRSA and MRSE: is there an answer? Clin Microbiol Infect. 2000;6:17–22. doi: 10.1046/j.1469-0691.2000.00005.x. [DOI] [PubMed] [Google Scholar]


