Abstract
Since estrogen is thought to protect pre-menopausal women from age-related hearing loss, we investigated whether variation in estrogen-signalling genes is linked to hearing status in the 1958 British Birth Cohort. This analysis implicated the estrogen-related receptor gamma (ESRRG) gene in determining adult hearing function and was investigated further in a total of 6134 individuals in 3 independent cohorts: (i) the 1958 British Birth Cohort; (ii) a London ARHL case-control cohort; and (iii) a cohort from isolated populations of Italy and Silk Road countries. Evidence of an association between the minor allele of single nucleotide polymorphism (SNP) rs2818964 and hearing status was found in females, but not in males in 2 of these cohorts: p = 0.0058 (London ARHL) and p = 0.0065 (Carlantino, Italy). Furthermore, assessment of hearing in Esrrg knock-out mice revealed a mild 25-dB hearing loss at 5 weeks of age. At 12 weeks, average hearing thresholds in female mice(-/-) were 15 dB worse than in males(-/-). Together these data indicate ESRRG plays a role in maintenance of hearing in both humans and mice.
Keywords: Age-related hearing loss, Estrogen, Gene, ESRRG
1. Introduction
The progressive loss of auditory function with advancing years, age-related hearing loss (ARHL), is the most common sensory ailment exhibited by the elderly population. Recent estimates suggest that 438 million individuals worldwide experience moderate or severe forms of hearing loss, a large proportion of which is adult onset (Stevens et al., 2011). The etiology of ARHL is complex; the heritability is estimated to be between 35% and 55% (Christensen et al., 2001; Gates et al., 1999; Raynor et al., 2009), and it is exacerbated by environmental factors, particularly noise (Van Eyken et al., 2007a). Histological studies in both humans (Nelson and Hinojosa, 2006) and animals (Fetoni et al., 2011) show that when the cochlea is examined, the predominant pathological feature is loss of the sensory hair cells, with defects in the stria vascularis and spiral ganglion neurons also evident. Relatively few genetic associations with ARHL have been replicated in independent populations. Associations that have been replicated include GRHL2 (Van Laer et al., 2008), KCNQ4 (Van Eyken et al., 2006), and NAT2*6A (Unal et al., 2005; Van Eyken et al., 2007b) in candidate gene studies, and 2 different metabotropic glutamate receptors, GRM7 (Friedman et al., 2009; Van Laer et al., 2010) and GRM8 (Girotto et al., 2011a) have been identified in genome wide association studies (GWAS). The GWAS so far reported for adult hearing status exhibit the phenomenon of “missing heritability” also observed in other common, complex diseases (Manolio et al., 2009). Given that more than 100 genes are known to be involved in congenital deafness then it is likely that a similar number are involved in susceptibility to ARHL. The future challenge in delineating the etiology of ARHL is to discriminate the valid associations that fall below the genome-wide significance threshold, using replication studies and functional genomics.
ARHL is more common (Cruickshanks et al., 1998; Helzner et al., 2005) and more severe (Pearson et al., 1995), with earlier onset (Davis et al., 1995), in men than in women. Historically, this has been attributed to greater occupational noise exposure in men compared to women, but it is clear that sex differences in hearing loss exist in cohorts without a significant history of noise exposure (Girotto et al., 2011b; Pearson et al., 1995). It has therefore been suggested that estrogen may act as an auditory protectant, and there is now considerable evidence linking estrogen signaling, the estrogen receptors (ER), and estrogen-related receptors (ESRR) with auditory protection (Hultcrantz et al., 2006; McCullar and Oesterle, 2009). Hence, mice carrying a targeted deletion of Erβ display an age-related hearing loss at 12 months, concurrent with a basal to apical degeneration of the organ of Corti in the cochlea (Simonoska et al., 2009). Additional studies with mice deficient for both ERβ and CYP19A1, which encodes the aromatase enzyme responsible for the aromatization of androgens into estrogens, show that these mice exhibit an impaired response of the auditory system to acoustic trauma (Meltser et al., 2008). Furthermore, mutations in the estrogen-related receptor, ESRRB, underlie autosomal recessive, non-syndromic hearing loss in humans (DFNB35) (Collin et al., 2008), and Esrrb knockout (KO) mice are deaf by 3 months of age (Chen and Nathans, 2007). A decline in hearing sensitivity has been linked to menopause in both humans (Hederstierna et al., 2010) and mice (Guimaraes et al., 2004). In addition, women with Turner's syndrome who are estrogen deficient undergo an early sensorineural hearing loss characteristic of ARHL (Beckman et al., 2004).
Estrogen-related receptor γ (ESRRG; NR3B3; ERR3) is an additional member of the ESRR family, which, together with ESRRB and a third isoform ESRRA, form the NR3B subgroup of the well-characterized, nuclear receptor superfamily. All 3 paralogues are orphan nuclear receptors and share a high structural homology with the classical ERs (Tremblay and Giguere, 2007). Esrrg mRNA has been shown to be present in the mouse embryonic inner ear in the cochlear and vestibular ganglion (Hermans-Borgmeyer et al., 2000), which suggests a role in the inner ear. Here, we investigate the relationship between ESRRG and adult hearing status in 3 independent cohorts, 2 population-based hearing cohorts and a case-control association study in a London-based ARHL cohort. In addition, we report for the first time that Esrrg knock-out mice are hearing impaired, and we characterize the expression of ESRRG in the adult mouse inner ear.
2. Methods
2.1. Ethics considerations
In regard to human participants, all studies had appropriate ethical consent, and consent forms for clinical and genetic studies were signed by each participant in the study. Ethical approval for the London ARHL cohort was granted from the Royal Free Local Research Ethics Committee (ref 6202). For the Isolated Populations Cohort, approval was granted by the relevant local ethical committee. Details of the ethical permission and consent for the 1958 British Birth Cohort (B58C) can be found at http://www.b58cgene.sgul.ac.uk/consent.php. In regard to animal use and care, Sprague-Dawley rats and C57BL/6J mice used in this study were sacrificed according to the UK Scientific Procedures Act, 1986. Generation and care of the animals and experimental procedures were in accordance with institutional guidelines and national laws for protection of experimental animals, and were approved by the local animal ethics committee (Hamburg 69/01).
2.2. Subjects
2.2.1. B58C cohort
The B58C and the collection of hearing data have been described previously (http://www.b58cgene.sgul.ac.uk/; Ecob et al., 2008; Strachan et al., 2007). In brief, participants were drawn up from 17,638 individuals born in England, Scotland, and Wales in 1 week of March 1958. Of the original cohort, 9377 members were revisited by a research nurse for a biomedical follow-up in 2002–2004. Hearing measure consisted of pure tone audiometry at 1 kHz and 4 kHz at age 44–45 years and were adjusted for sex, nuisance variables (noise at test, nurse performing test, audiometer used in test), conductive loss, and hearing loss in childhood. DNA was collected from 3900 of these individuals and genotyped for 555,164 single nucleotide polymorphisms (SNPs) on the Illumina Infinium Human Hap550 array (data deposited by Dr Panos Deloukas, Wellcome Trust, Sanger Institute, Cambridge, UK). These genetic data have been used extensively as part of the Wellcome Trust Case Control Consortium (WTCCC), (https://www.wtccc.org.uk/) (Barrett et al., 2009; WTCCC, 2007). No associations from the analysis of B58C genetic data and hearing thresholds at age 44-45 reached genome-wide significance (a version of this analysis can be accessed at: http://www.b58cgene.sgul.ac.uk/).
2.2.2. London ARHL cohort
A total of 260 patients with sensorineural hearing loss (SNHL) consistent with an age-related decline were recruited from the adult hearing aid clinic at the Royal National Throat Nose and Ear Hospital, London; this formed our initial patient group (ARHL_1). All were interviewed by an audiological physician and underwent an audiometric examination. Air conduction and bone conduction thresholds at 0.25, 0.5, 1, 2, 4, and 8 kHz and 0.5, 1, 2, and 4 kHz, respectively were measured with masking according to BSA Recommended Procedures (http://www.thebsa.org.uk/docs/RecPro/PTA.pdf). At interview, a questionnaire was completed that recorded relevant medical history, family history of hearing loss, and history of noise exposure. This questionnaire was then amended based on answers to stage 1 questions to become self-directional, and patient recruitment was extended to the Royal Free Hospital, London. An additional 323 patients were recruited across both hospitals, forming our replication group (ARHL_2) and bringing the total number of patients to 583 (ARHL_COM). Patients were subsequently categorized for family history and for noise exposure. Noise exposure was graded using the occupations listed by Lynch and Kil, 2005 and Tak and Calvert, 2008 as a guide: Grade 0 = no noise exposure documented; Grade 1 = low to medium noise exposure; and Grade 2 = medium-to-high noise exposure. (For questionnaires, see Supplementary information S1). Patients were not excluded from the study if there was an asymmetric hearing loss as long as the better hearing ear was consistent with the criteria for late-onset SNHL. (Full details of the exclusion criteria are available upon request). With regard to controls, the control sample group comprised ECACC Human Random Control (HRC) DNA panels. In association analysis, all samples were of white European origin.
2.2.3. Isolated populations cohort
Isolated populations were recruited from Italy and Silk Road countries (for an overall number of 1651 subjects) belong to the International Consortium G-EAR, described previously (Girotto et al., 2011a). In brief, several quantitative measures of hearing function were undertaken: air conduction thresholds were determined at 0.25 kHz, 0.5 kHz, 1 kHz, 2 kHz, 4 kHz, and 8 kHz and pure tone averages (PTAs) of air conduction thresholds were determined for: PTAlow (0.25, 0.5 and 1 kHz), PTAmedium (0.5, 1 and 2 kHz) and PTAhigh (4 and 8 kHz).
2.3. Genotyping
2.3.1. London ARHL cohort
Genomic DNA was extracted from blood, using a standard phenol-chloroform extraction procedure, or from saliva, using Oragene DNA extraction kits (DNAgenoTek, Kanata, Ontario, Canada). The ESRRG rs2818964 SNP genotyping was performed using ABI TaqMan SNP genotyping assay (C_222941_10) on a SDS7500 Real Time PCR System (Life Technologies, Paisley, UK). Each plate contained a control sample of each genotype. Genotyping rates for rs2818964 were: 99.7%, and 99.4% in ARHL_COM and control populations, respectively. Genotype frequencies for rs2818964 did not deviate from Hardy–Weinburg equilibrium (HWE) in ARHL_1 (p = 0.4000), ARHL_2 (p = 0.7651), ARHL_COM (p = 0.4310), or the control population (p = 0.7199). Genotyping for the B58C and International Consortium, G-Ear has been described previously (http://www.b58cgene.sgul.ac.uk/; Barrett et al., 2009; WTCCC, 2007; Girotto et al., 2011a).
2.4. Reverse transcription–polymerase chain reaction
Cochlea, kidney, and spleen tissues were dissected from P2-P30 female rats and transferred to RNAlater (Qiagen, Manchester, UK), and total RNA was extracted using a RNeasy kit (Qiagen, Manchester, UK). Samples were treated with RQ1 RNase-Free DNase (Promega, Southampton, UK), and 1 μg RNA was reverse transcribed in a 20-μL reaction with Omniscript reverse transcriptase (Qiagen, Manchester, UK) and random primers (Promega, Southampton, UK). Esrrg forward (5′-agagttggtggttatcattggatg-3′) and reverse (5′-agaaggctcatctgatccgc-3′) primers and Gapdh forward (5′-aacgggaagcccatcacc-3′) and reverse (5′-cagccttggcagcaccag-3′) primers were used to amplify an 81 bp Esrrg and 442 bp Gapdh cDNA fragment, respectively using 2 μL cDNA and 0.5 U GoTaq DNA polymerase (Promega, Southampton, UK) in a total volume of 20 μL. Cycling conditions were as follows. Esrrg: an initial denaturation step at 95 °C for 2 minutes, followed by 40 cycles at 95 °C for 15 seconds; 60 °C for 60 seconds; and 72 °C for 30 seconds. Gapdh: an initial denaturation step at 95 °C for 2 minutes followed by 25 cycles at 94 °C for 30 seconds; 60 °C for 30 seconds; and 72 °C for 30 seconds. Polymerase chain reaction (PCR) products were separated by standard agarose gel electrophoresis.
2.5. Antibodies
The rabbit polyclonal anti-ESRRG antibody has been described previously and was a gift from Dr Ronald Evans, Salk Institute for Biological Studies, San Diego, CA, USA (Dufour et al., 2007) and used at 1:200, whereas goat anti-rabbit AlexaFluor488 secondary antibody was used at 1:1000 dilution (Life Technologies, Paisley, UK).
2.6. Vibratome sectioning and immunofluorescence
Auditory bullae were dissected from female mice, fixed in 4% paraformaldehyde for 1 hour at room temperature, washed in phosphate-buffered saline solution (PBS) and decalcified in 4.13% ethylenediaminetetraacetic acid (EDTA), pH7.4, in PBS for 72 hours at 4 °C. Bullae were mounted in 4% low melting point agarose (Sigma-Aldrich, Gillingham, UK) and sectioned at 300 μm using a 1000 Plus Vibratome (Intracel, Royston, UK). Vibratome slices were permeabilized and blocked in 0.5% Triton-X 100 with 10% goat serum for 2 hours at room temperature and incubated with primary antibodies at 4 °C overnight. After PBS washes, slices were incubated with secondary antibodies for 2 hours at room temperature in the dark. Hair cell stereocilia were stained with Phalloidin-Atto 647N to f-actin (Sigma-Aldrich, Gillingham, UK), which was added to the secondary antibody incubations at 1:200; nuclei were visualized with 1 μmol/L DAPI. Imaging was performed with a laser scanning confocal microscope (LSM Meta 510; Zeiss, Oberkochen, Germany) using ×10 (0.3 NA), ×20 (0.75 NA), and ×63 (1.2 NA) objectives.
2.7. Generation of Esrrg KO mice
A bacterial artificial chromosome (BAC) clone containing the Esrrg gene was identified from mouse strain 129/SvJ BAC library pools (Genome Systems, St. Louis, MO, USA) by PCR-based screening. A 1665-bp PCR fragment ending with a synthetic NruI site in the second coding exon was amplified from the BAC clone and used as the 5′ homology region. It was placed in front of a lacZ marker gene of pTLZN such that the start methionines of Esrrg and that of the β-galactosidase (β-gal) reporter coincide. In addition, the parental vector harbors a PGK-neomycin-bpA (neo) cassette oriented in the opposite direction. A 4356-bp BamHI fragment starting further downstream at a BamHI site in the same exon and ending in the neighboring intron was used as 3′ homology region. Both the combination of 5′ homology region with the lacZ neomycin cassette and the 3′ homology region were inserted in pKO with a flanking diphtheria toxin subunit cassette (dta) as a negative selection marker. The construct was linearized with NotI and purified by a standard phenol-chloroform extraction. The NotI-linearized Esrrg targeting construct was introduced by electroporation into R1 ES cells (129 X1 × 129 S1) (Nagy et al., 1993), a kind gift from Prof. A. Nagy (Samuel Lunenfeld Research Institute, Toronto, ON, Canada), which were then selected with 200 μg G418 per milliliter of medium. Southern blot analysis was performed to identify ES clones that had undergone homologous recombination. EcoRI-digested genomic DNA prepared from resistant ES cell clones was probed with a 509-bp fragment derived from genomic sequences located 3′ of the targeting vector. The radiolabeled probe recognized a 20-kb fragment of the wild-type allele and an 11-kb fragment of the targeted allele. Appropriate integration at the 5′ end of the targeting construct was verified by rehybridization of the blot with a 912-bp fragment located 5′ of the targeting vector. This probe hybridized to a 14.5-kb fragment in the targeted allele, as opposed to the 20-kb fragment in the wild-type allele. Cells from 3 correctly targeted lines were injected into C57BL/6J blastocysts to generate chimeras, and chimeric males mated to C57BL6/J females transmitted the targeted allele to their offspring. Homozygous mutant mice were generated by heterozygote intercrosses. Genotyping was routinely performed by Southern blot analysis. DNA was isolated with a genomic DNA kit (Applichem, Darmstadt, Germany) from tail tips. No mRNA transcripts or protein for the Esrrg gene were detected in the Esrrg KO mice (data not shown). Studies were performed with littermates of the F2 and F3 generation of Esrrg KO mice.
2.8. Auditory brainstem responses
To evaluate hearing thresholds ABR to clicks were recorded in anesthetized animals (xylazin hydrochloride 16 mg/kg body weight, and S-ketamin hydrochloride 60 mg/kg body weight) in a sound-proof chamber. Alternating acoustic stimuli were delivered monaurally at a rate of 21/s using a Beyer DT-48 earphone and monitored with a probe microphone (MK301, Microtech Gefell, Gefell, Germany) integrated into the earpiece. Bioelectric potentials were recorded by subcutaneous silver electrodes at both mastoids and responses averaged (400-2000). Stimulus intensities were varied starting at 117 peak equivalent dB SPL [pe dB SPL] in increments of 20 dB except near threshold where 5 dB steps were used. The hearing threshold was defined as the lowest intensity to generate a reproducible ABR waveform.
2.9. Statistical analysis
For B58C, hearing thresholds at 1 kHz and 4 kHz were log transformed and adjusted for sex, nuisance variables, and conductive hearing loss in childhood; they were analyzed by performing a 1-df “per allele” significance test for association between mean hearing threshold and number of minor alleles (0, 1, or 2), and a 2-df significance test for association between mean hearing threshold and genotype. This analysis has been described previously (http://www.b58cgene.sgul.ac.uk/; Ecob et al., 2008; Strachan et al., 2007). Results were ranked according to p value and the 500 SNPs with lowest p value analyzed as a candidate gene set (see Supplementary information S2, available online). For the London ARHL cohort, deviation from HWE and differences in genotype and allele frequencies were evaluated by χ2 test with a significance threshold of p < 0.05. Unless otherwise stated, MedCalc (version 11.2) was used to calculate the odds ratios (OR) and 95% confidence intervals (CI). Backward stepwise logistic regression was performed using the likelihood ratio procedure. Patient diagnosis was declared the dependent variable (1 = affected, 0 = unaffected), and sex and SNP genotype the independent variables. Sex was set as a categorical variable (1 = female, 0 = male). For the recessive and dominant genetic models, genotypes were set as a categorical variable: recessive (GG = 1, GA+AA = 0); dominant (GA+GG = 1, AA = 0). For the additive genetic model, genotypes were treated as a continuous covariate (0 = AA, 1 = GA and 2 = GG). For the co-dominant model, genotypes were coded as for the additive model, but treated as independent categorical variables and the AA genotype set as the reference. All statistical tests were conducted using SPSS :Statistics (version 17.0) unless otherwise stated. For the candidate gene association analysis in the London ARHL cohort, we used a p value of 0.05 as a test of significance, based on the primary analysis being that of a single SNP in a single candidate gene. Subsequent stratification analysis for effects of sex, family history, and noise exposure is a secondary analysis of the effect of the same SNP and a Bonferroni correction for multiple testing was therefore considered overly conservative. Isolated Populations Cohort: association analysis was adjusted for age by regressing it out and the residuals were then normalized using rank normal transformation. Association analysis was conducted using a mixed model linear regression, where the kinship matrix was the random effect and the SNP the fixed effect as implemented in GenABEL (Aulchenko et al., 2007).
3. Results
3.1. B58C
Hearing thresholds at age 44 to 45 years collected as part of the B58C (Ecob et al., 2008) were compared to genetic data obtained on the same individuals as part of the Wellcome Trust Case Control Consortium (Barrett et al., 2009; WTCCC, 2007) (these data are available at http://www.b58cgene.sgul.ac.uk/phenosearch.php?pheno=6). To investigate the role of estrogen in adult hearing, the 500 SNPs most strongly associated with hearing thresholds in the B58C GWAS data were screened for presence of estrogen signaling genes. The rs2818964 SNP located in the last intron of the long isoform of the ESRRG gene (transcript ENST00000361525) was the only SNP within an estrogen candidate gene locus identified in the top 500 SNPs. Rs2818964 was associated with hearing status at 4 kHz, allelic effect p = 0.0008 (Table 1). Because of the different roles of estrogen in males and females, the data were also dichotomized according to sex; evidence of association was found to be stronger in males than in females for both allelic (p = 0.0157 males; p = 0.0212 females) and genotype (p = 0.0092 males; p = 0.0624 females) effects. Detection of stronger associations with hearing loss in males can be a surrogate for the effect of greater noise exposure. However, stratification of data according to number of years of noise exposure (>5 years or <5 years) identified an association only in subjects reporting less than 5 years of noise exposure (allelic effect p = 0.0004). In each of these associations, the trend direction was consistent with the major allele, A, being associated with poorer hearing. No association was identified between hearing thresholds at 1 kHz and SNP rs2818964.
Table 1.
Evidence of association for ESRRG rs2818964 with hearing status in the 1958 British Birth Cohort
| Pure tone frequency | Trend directiona | (n) | Allele (p value)b | Genotype (p value)c |
|---|---|---|---|---|
| 4 kHz | A | 3614 | 0.0008 | 0.0008 |
| 4 kHz (male) | A | 1776 | 0.0157 | 0.0092 |
| 4 kHz (female) | A | 1838 | 0.0212 | 0.0624 |
| 4 kHz −N | A | 3230 | 0.0004 | 0.0010 |
| 4 kHz +N | A | 384 | 0.8232 | 0.2127 |
| 1 kHz | A | 3632 | 0.5372 | 0.2713 |
| 1 kHz −N | A | 3246 | 0.6959 | 0.4243 |
| 1 kHz +N | A | 386 | 0.4600 | 0.4528 |
Allele associated with poorer hearing thresholds.
A 1-df significance test for association between mean hearing threshold and number of minor alleles (0, 1, or 2); “per allele model”.
A 2-df significance test for association between mean hearing threshold and genotype; “genotype model”; p values less than 0.05 are shown in bold. −N: less than 5 years of noise exposure, +N: more than 5 years of noise exposure.
3.1.1. London ARHL cohort
To determine whether the ESRRG rs2818964 SNP may be associated with risk of ARHL, we performed a case-control association study in a London ARHL cohort (Table 2). The minor allele, G, of SNP rs2818964 was more common in our initial patient group, ARHL_1, compared to controls, but only in women (p = 0.0385) (Table 2). This sex-specific association was consistent in our replication group, ARHL_2, and in the combined cohort, ARHL_COM, for both allele (p = 0.0139 and p = 0.0058, respectively) and genotype effects (p = 0.0384 and p = 0.0238, respectively). Backward logistic regression analysis with rs2818964 genotypes re-coded based on dominant, co-dominant, recessive, and additive inheritance models showed that the additive model alone was significant and identified a sex–genotype interaction (p = 0.0073, OR = 1.40, 95% CI = 1.10–1.80). In contrast to the trend direction observed in the B58C, these data suggested at least 1 copy of the minor allele, G, of SNP rs2818964 is associated with risk of ARHL in women. Secondary analysis to examine the effect of noise exposure and family history of ARHL on this association identified the strongest evidence of association in non–noise-exposed women who report a family history of ARHL (allelic effect p = 0.0023, ORG/A = 1.61, 95% CI = 1.84–2.20; see Supplementary information S3, available online). In contrast, no evidence of an association was identified in males for any of the subgroups examined.
Table 2.
Genotype and allele frequency distribution of ESRRG rs2818964 in the London Age Related Hearing Loss cohort
| Control | ARHL_1 | p value | ARHL_2 | p value | ARHL_COM | p value | ORG/A (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| All | ||||||||
| N | 477 | 260 | 323 | 583 | ||||
| Genotype frequency (n) | ||||||||
| AA | 0.38 (180) | 0.35 (91) | 0.4783 | 0.33 (105) | 0.1237 | 0.34 (196) | 0.2079 | |
| AG | 0.47 (223) | 0.46 (120) | 0.48 (156) | 0.47 (276) | ||||
| GG | 0.15 (74) | 0.19 (49) | 0.19 (62) | 0.19 (111) | ||||
| Allele frequency (n) | ||||||||
| A | 0.61 (583) | 0.58 (302) | 0.2559 | 0.57 (366) | 0.0751 | 0.57 (668) | 0.0751 | |
| G | 0.39 (371) | 0.42 (218) | 0.43 (280) | 0.43 (498) | ||||
| Females | ||||||||
| N | 243 | 111 | 162 | 273 | ||||
| Genotype frequency (n) | ||||||||
| AA | 0.41 (99) | 0.31 (34) | 0.1209 | 0.33 (53) | 0.0384 | 0.32 (87) | 0.0238 | |
| AG | 0.46 (111) | 0.49 (55) | 0.44 (72) | 0.46 (127) | ||||
| GG | 0.13 (33) | 0.20 (22) | 0.23 (37) | 0.22 (59) | ||||
| Allele frequency (n) | ||||||||
| A | 0.64 (309) | 0.55 (123) | 0.0385 | 0.55 (178) | 0.0139 | 0.55 (301) | 0.0058 | 1.42 (1.11–1.82) |
| G | 0.36 (177) | 0.45 (99) | 0.45 (146) | 0.45 (245) | ||||
| Males | ||||||||
| N | 233 | 148 | 161 | 309 | ||||
| Genotype frequency (n) | ||||||||
| AA | 0.35 (81) | 0.38 (56) | 0.7671 | 0.32 (52) | 0.6669 | 0.35 (108) | 0.9723 | |
| AG | 0.48 (111) | 0.44 (65) | 0.52 (84) | 0.48 (149) | ||||
| GG | 0.17 (41) | 0.18 (27) | 0.16 (25) | 0.17 (52) | ||||
| Allele frequency (n) | ||||||||
| A | 0.59 (273) | 0.60 (177) | 0.7401 | 0.58 (188) | 0.9563 | 0.59 (365) | 0.8743 | |
| G | 0.41 (193) | 0.40 (119) | 0.42 (134) | 0.41 (253) |
Differences in genotype and allele frequencies between patients and controls were assessed by χ2-test; p values that are significant at the 5% level are shown in bold. ORs and 95% CIs were calculated only for significant p values relative to the major allele as reference (G/A).
3.1.2. Isolated populations cohort
The ESRRG SNP rs2818964 was genotyped in cohorts collected as part of the G-EAR International Consortium (Girotto et al., 2011a) incorporating cohorts from several distinct European isolated populations including Carlantino, Friuli Venezia Giulia Genetic Park, and different countries located along the Silk Road (Georgia, Azerbaijan, Uzbekistan, Kazakhstan, and Tajikistan). The relationship between the ESRRG rs2818964 SNP and quantitative measures of hearing were analyzed for each population and for the combined cohort. Given the evidence of a sex-specific association in the London ARHL cohort and the role of estrogen, males and females were examined separately (Table 3). Analysis of each population showed evidence of a significant association only in women from the Carlantino population for thresholds averaged across Pure Tone Average (PTA) at low and medium frequencies and at each of the individual low and medium pure tone thresholds (0.125, 0.25, and 0.5 kHz, 1 kHz, and 2 kHz), with the strongest association occurring at 2 kHz (p = 0.0065). No evidence for association with hearing thresholds was found for women in the Friuli Venezia Giulia Genetic Park and Silk Road populations or for males in any of the 3 populations. When data from the 3 cohorts were combined, evidence of an association was found with PTA at medium frequencies (allelic effect p = 0.0385) and at 1 kHz (p = 0.0157) in women only. For each of these associations, the minor allele, G, was associated with poorer hearing, which is consistent with the trend direction observed in the London ARHL cohort but in contrast to the B58C cohort.
Table 3.
Evidence of association for ESRRG rs2818964 with hearing status in Isolated Populations Cohort
| Quantitative trait | Combined populations |
Carlantino project |
Silk Road |
Friuli Venezia Giulia project |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n) | p value | Trend directiona | (n) | p value | Trend directiona | (n) | p value | Trend directiona | (n) | p value | Trend directiona | |
| Female 0.125 kHz | 804 | 0.0638 | G | 171 | 0.0795 | G | No data | No data | No data | 633 | 0.2390 | G |
| Female 0.25 kHz | 977 | 0.4509 | G | 172 | 0.0172 | G | 171 | 0.5018 | G | 634 | 0.9847 | A |
| Female 0.50 kHz | 977 | 0.2563 | G | 172 | 0.0262 | G | 171 | 0.1020 | G | 634 | 0.3235 | A |
| Female 1 kHz | 977 | 0.0157 | G | 172 | 0.0286 | G | 171 | 0.7825 | G | 634 | 0.0577 | A |
| Female 2 kHz | 976 | 0.0640 | G | 172 | 0.0065 | G | 170 | 0.2899 | G | 634 | 0.1875 | A |
| Female 4 kHz | 962 | 0.4220 | G | 172 | 0.2290 | G | 171 | 0.5997 | G | 619 | 0.5483 | A |
| Female 8 kHz | 947 | 0.3370 | G | 172 | 0.6325 | G | 171 | 0.7240 | G | 604 | 0.2844 | A |
| Female PTAlow | 977 | 0.1454 | G | 172 | 0.0156 | G | 171 | 0.2886 | G | 634 | 0.3157 | A |
| Female PTAmedium | 977 | 0.0385 | G | 172 | 0.0101 | G | 171 | 0.2401 | G | 634 | 0.0889 | A |
| Female PTAhigh | 962 | 0.3582 | G | 172 | 0.4407 | G | 171 | 0.6116 | G | 619 | 0.3471 | A |
| Male 0.125 kHz | 547 | 0.5105 | G | 120 | 0.6431 | G | No data | No data | No data | 427 | 0.6330 | G |
| Male 0.25 kHz | 674 | 0.4498 | G | 121 | 0.2777 | A | 126 | 0.7735 | G | 427 | 0.1943 | G |
| Male 0.50 kHz | 674 | 0.0847 | G | 121 | 0.6631 | G | 126 | 0.3915 | G | 427 | 0.1558 | A |
| Male 1 kHz | 674 | 0.1362 | G | 121 | 0.4953 | G | 126 | 0.9596 | G | 427 | 0.1556 | G |
| Male 2 kHz | 674 | 0.8231 | A | 121 | 0.8827 | A | 126 | 0.2542 | G | 427 | 0.7008 | A |
| Male 4 kHz | 667 | 0.5413 | G | 121 | 0.5997 | A | 126 | 0.0850 | G | 420 | 0.0568 | A |
| Male 8 kHz | 659 | 0.7290 | A | 121 | 0.4821 | A | 126 | 0.4501 | G | 412 | 0.7326 | A |
| Male PTAlow | 674 | 0.1692 | G | 121 | 0.9900 | A | 126 | 0.6938 | G | 427 | 0.1460 | G |
| Male PTAmedium | 674 | 0.4879 | G | 121 | 0.7997 | G | 126 | 0.6869 | G | 427 | 0.3584 | A |
| Male PTAhigh | 667 | 0.7590 | G | 121 | 0.4522 | A | 126 | 0.3115 | G | 420 | 0.1991 | A |
Allele associated with poorer hearing thresholds. Association analysis was conducted using mixed model linear regression; p values that are significant at the 5% level are shown in bold.
3.2. Esrrg KO mice are hearing impaired
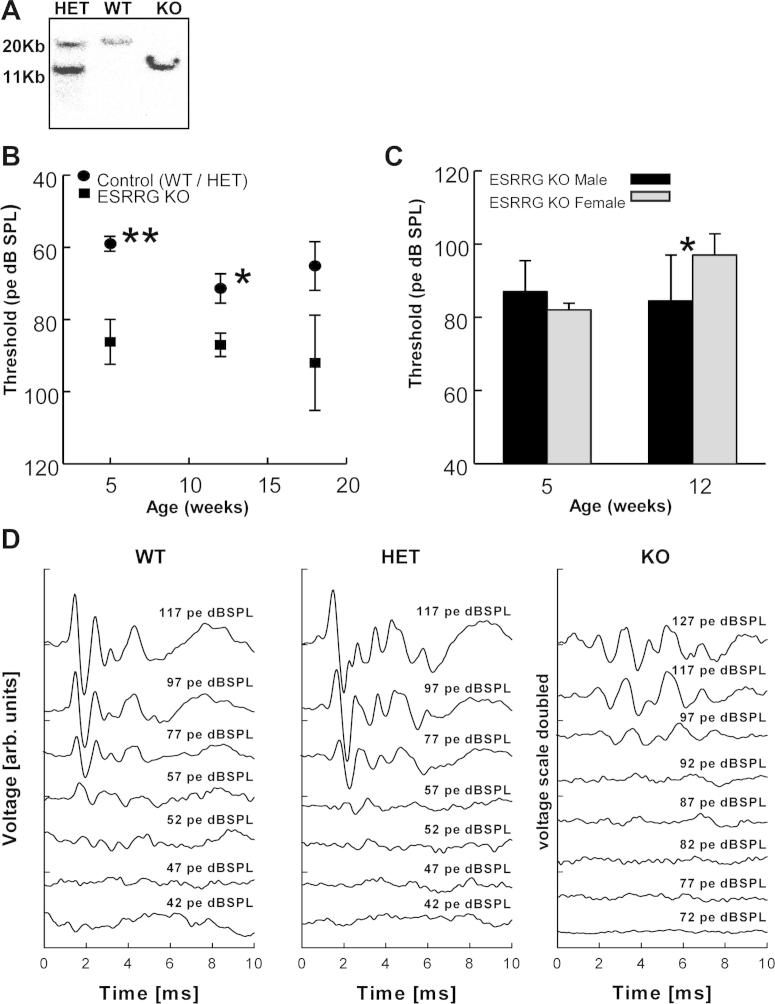
To explore the role of ESRRG in the cochlea, we generated a novel Esrrg KO mouse model by targeting the second exon of the mouse Esrrg gene with a lacZ-neomycin fusion cassette by homologous recombination in mouse embryonic stem cells. Consistent with the published data for the 2 previously reported Esrrg KO mouse models (S strain, Salk Institute, Alaynick et al., 2007; G strain, GlaxoSmithKline/Deltagen, Alaynick et al., 2010) only a fraction of our mice homozygous for the Esrrg deletion survived into adulthood; most died in the early postnatal period before the onset of hearing. This limited the number of homozygous Esrrg KO mice that could be analyzed. Auditory evoked brain stem responses (ABRs) hearing thresholds in Esrrg heterozygotes were not statistically different from those in wild-type animals, and therefore both groups were pooled for controls. Hearing thresholds in Esrrg KO mice were significantly elevated at 5 weeks and 12 weeks of age (Fig. 1B). In the oldest group, at 18 weeks, this difference was still present, but only very few KO animals survived to 18 weeks of age (n = 3) and this finding remained below statistical significance. When age groups were examined for sex effects, a statistically significant higher threshold in female compared to male Esrrg KO mice was found at 12 weeks of age (p < 0.05, Mann–Whitney rank sum test, Fig. 1C). Recorded potentials in wild-type and heterozygous mice displayed no obvious differences, whereas potentials in Esrrg KO mice were of lower amplitude and altered in waveform (Fig. 1D).
Fig. 1.
ABR hearing thresholds in Esrrg KO mice. (A) Genotyping by Southern blotting of offspring from Esrrg heterozygote hybrid crosses produces predicted fragments of 20 kb for the WT allele and 11 kb for the null allele. (B) ABR thresholds on click stimuli were recorded at 5 (n = 6 Esrrg KO vs. 11 controls), 12 (n = 9 Esrrg KO vs. 8 controls), and 18 (n = 3 Esrrg KO vs. 8 controls) weeks of age. Esrrg KO mice exhibit significant hearing loss compared to controls (MV ± SE, **p = 0.001; *p < 0.05, Mann–Whitney rank sum test). (C) Sex-specific difference in hearing thresholds for 5 (n = 2 female vs. 4 male) and 12 (n = 3 female vs. 6 male) weeks of age. Esrrg KO female mice exhibit greater hearing loss compared to males at 12 weeks (MV ± SE, *p < 0.05, Mann–Whitney rank sum test). (D) Sample recordings for Esrrg WT, Esrrg Het and Esrrg KO mice; hearing thresholds in the Esrrg KO (voltage scale doubled) are approximately 40 dB worse compared to the WT and Het controls.
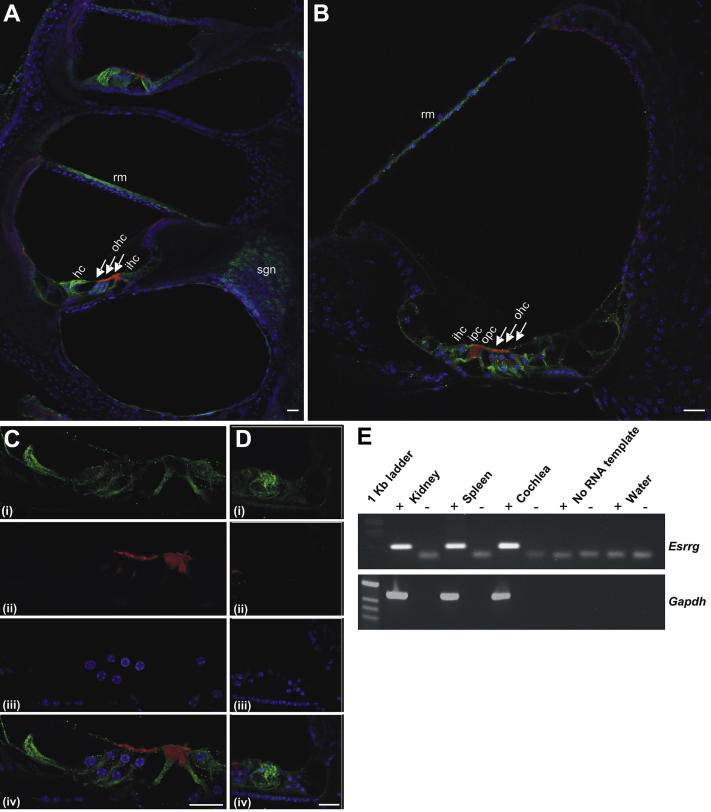
3.3. ESRRG expression in the inner ear
In the adult mouse cochlea, strong ESRRG immunoreactivity was localized to Reissner's membrane (Fig. 2A and B) and a distinct cluster of supporting cells adjacent to the third row of outer hair cells (Fig. 2A, B, and D) which is consistent with the location of Hensen's cells. Intense ESRRG immunoreactivity was also detected in the elongated cell bodies of the inner and outer pillar cells (Fig. 2A–C) that were identified by Phalloidin staining of f-actin in the apical and basal region. Some ESRRG expression was also detected in inner hair cells (Fig. 2A–C). ESRRG staining was not detected in outer hair cells, but appeared to localize in the supporting cell bodies of Deiters' cells and their phalangeal processes intervening the outer hair cells (Fig. 2B and C). Less intense immunoreactivity was detected in the spiral ganglion neurons (Fig. 2A). Abundant Esrrg mRNA was detected in P2 rat cochlea as well as positive control tissue P30 rat kidney and spleen (Fig. 2E).
Fig. 2.
Expression of ESRRG in mouse inner ear. (A–D) Results of immunofluoresence with anti-ESRRG in mouse inner ear at P36: anti-ESRRG (green), DAPI (blue), and Phalloidin staining to f-actin (red). (A) Low magnification of mid-apical coil. Strong ESRRG immunoreactivity is localized to Reissner's membrane (rm), Hensen's cells (hc) can be seen in the inner (ihc) and outer (ohc) hair cell region (solid arrows). Weaker immunoreactivity is shown in the spiral ganglion neurons (sgn). (B) Mid magnification of the opposite mid-cochlear coil showing ESRRG immunoreactivity in the organ of Corti localizes to the inner (ipc) and outer (opc) pillar cells and supporting cell bodies and their processes intervening the outer hair cells. (C) High magnification of the organ of Corti. (D) Mid magnification of the opposite apical coil showing strong immunoreactivity in Hensen's cells. Scale bar is 20 μm. (E) Detection of Esrrg mRNAs in P2-P30 rat kidney, spleen, and cochlea tissue using reverse transcription–polymerase chain reaction. (RT-PCR). Samples were tested with (+) or without (−) reverse transcriptase (RT) enzyme. No bands were detected in RT (−), in the absence of RNA template or negative control for PCR (water only). Gapdh served as a positive control for mRNA quality.
4. Discussion
Despite some recent progress in the genetic analysis of adult hearing, very little is known about the genetic risk factors for ARHL, and virtually nothing about their role in the pathogenesis of this common sensory loss. In this study, based on the hypothesis that estrogen plays a role in protecting pre-menopausal women from ARHL, we investigated whether variants in estrogen signaling genes may be risk factors for adult-onset hearing loss. The rs2818964 SNP in the ESRRG gene was associated with hearing status in a London ARHL cohort, the minor allele being associated with poorer hearing but only in women. This association was replicated in the Carlantino and the combined cohort of Isolated Populations from Italy and Silk Road countries, both in the direction of the allelic effect and in the female-specific association. Although this association had also been detected in the discovery cohort B58C, the effect was in the opposite direction with the major allele associated with poorer hearing and here the evidence of association was stronger in males than in females. Interpretation of such so-called “flip-flop” replications should be cautious, as they may be spurious; alternatively they can be indicators of real effects due to genetic architecture or differences in cohort characteristics (Lin et al., 2007). Audiometric data are not trivial to collect, and therefore large, well-characterized cohorts are limited. Here, we have sought to replicate an association in 3 of the largest cohorts, in a total of 6134 individuals. However, the cohorts have different characteristics that are not ideal for genetic replication. The B58C is a cross-sectional study of the UK population conducted at age 44–45 years, younger than the subjects recruited for the London ARHL case-control study. The average age in the London cohort is 71 years, and 96% of the women are more than 50 years of age and predominantly post-menopausal. The Isolated Populations Cohort is population-based, 18–92 years of age, and incorporating women more than 50 years of age, especially in Carlantino. Data from the London ARHL and Carlantino cohorts indicate a sex-specific predisposition to a low-to-mid frequency hearing loss in women is conferred by variation in the ESRRG gene inherited as an additive trait. It is possible that the menopausal state of the women is a key determinant of the association with hearing status and ESRRG genotype; with the minor allele of the rs2818964 SNP being a risk factor for ARHL after estrogen declines in women after menopause and therefore not detected in the younger women in the B58C. This SNP has previously been linked to risk of osteoporosis in women, an effect that also shows an interaction with the menopause status of women (Elfassihi et al., 2010).
Evidence of replication in our genetic cohorts are suggestive of a role for ESRRG in maintenance of hearing; but, given the difference in trend direction observed in the B58C, we sought functional evidence to establish the role of ESRRG in hearing. To explore the function of ESRRG in the auditory system, we generated a novel Esrrg KO mouse model, and ABRs to click stimuli were measured. Our data showed that absence of the Esrrg gene in mice leads to a mild hearing impairment at 5 weeks of age; this hearing loss, although present, had not progressed by 12 and 18 weeks of age. Strikingly, auditory thresholds were significantly worse in female mice compared to males at 12 weeks of age, resembling the sex difference observed with the rs2818964 SNP in the London ARHL and Carlantino cohorts. Relatively little is known regarding the function of ESRRG in the inner ear. Previous studies with Esrrg null mice show that these mice die in the first few weeks after birth because of abnormal heart function (Alaynick et al., 2007). Gene expression profiling and chromatin immunoprecipitation (ChIP)-on-chip analysis shows that Esrrg regulates a network of genes that control oxidative metabolic function in embryonic and adult heart (Alaynick et al., 2007; Dufour et al., 2007), as well as genes involved in cellular ion homeostasis in tissues subject to high metabolic demand (Alaynick et al., 2010). A number of these genes are involved in potassium transport, some of which are implicated in inner ear homeostasis (Wangemann, 2002).
In this study, we have shown for the first time that ESRRG is expressed in the adult mouse inner ear. Similar to the expression pattern of ESRRB in the inner ear (Meltser et al., 2008), we find that expression of ESRRG is widespread but predominantly confined to discrete supporting cell populations of the organ of Corti, Reissner's membrane, and the inner hair cells. Further work is required to establish the role of ESRRG in the inner ear; however, the known role of ESRRGs in regulation of genes involved in ion homeostasis in other tissues with high metabolic demand is one that is also critical for cochlear function. One such gene involved in potassium ion transport that is reported to be down-regulated in a Essrg KO mouse model is Kcne1 (Alaynick et al., 2010). Interestingly, mice with targeted deletion of the Kcne1 gene exhibit a profound hearing loss (Warth and Barhanin, 2002). In humans, mutations in KCNE1 account for approximatelly 10% of cases of Jervell and Lange-Nielsen syndrome, a syndromic form of deafness that presents with cardiac defects (Bitner-Glindzicz and Tranebjaerg, 2000). Therefore, 1 putative mechanism by which ESRRG might underlie ARHL is through down-regulation of the KCNE1 gene, leading to impaired potassium ion homeostasis.
In summary, our genetic association study, characterization of the expression of ESRRG in the inner ear, and impaired hearing in our Esrrg KO mouse model support an important role for ESRRG in hearing, particularly for maintenance of hearing in women after menopause. It also provides further evidence of GWAS as a discovery tool in complex disease to provide a candidate gene set for further investigation by the use of replication and functional genomics.
Disclosure statement
The authors declare no conflicts of interest.
Acknowledgements
The authors would like to thank Uwe Borgmeyer and Ute Süssens for excellent technical expertise in generation and characterisation of the Esrrg knockout mice and Dr Jonathan Gale and Dr Dan Jagger for help and expertise with confocal microscopy and its interpretation. We acknowledge use of phenotype and genotype data from the British 1958 Birth Cohort DNA collection (http://www.b58cgene.sgul.ac.uk/). The authors would also like to thank all the participants of the study.
This work was funded under the following: the Haigh Fellowship in age related deafness, Deafness Research UK (444:UEI:SD); the London ARHL cohort collection was initiated by funding from Research into Ageing (Ref. 223, SD), Teresa Rosenbaum Golden Charitable Trust, Telethon Foundation (GGP09037); Italian Ministry of Health (RC16/06) (to P.G.). We acknowledge use of phenotype and genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. (http://www.b58cgene.sgul.ac.uk/). Genotyping for the B58C-WTCCC subset was funded by the Wellcome Trust grant 076113/B/04/Z. The B58C-T1DGC genotyping used resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. B58C-T1DGC GWAS data were deposited by the Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research (CIMR), University of Cambridge, which is funded by Juvenile Diabetes Research Foundation International, the Wellcome Trust, and the National Institute for Health Research Cambridge Biomedical Research Centre; the CIMR is in receipt of a Wellcome Trust Strategic Award (079895). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.neurobiolaging.2013.02.009.
Appendix A. Supplementary data
References
- Alaynick W.A., Kondo R.P., Xie W., He W., Dufour C.R., Downes M., Jonker J.W., Giles W., Naviaux R.K., Giguere V., Evans R.M. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell. Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Alaynick W.A., Way J.M., Wilson S.A., Benson W.G., Pei L., Downes M., Yu R., Jonker J.W., Holt J.A., Rajpal D.K., Li H., Stuart J., McPherson R., Remlinger K.S., Chang C.Y., McDonnell D.P., Evans R.M., Billin A.N. ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 2010;24:299–309. doi: 10.1210/me.2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y.S., de Koning D.J., Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., Plagnol V., Pociot F., Schuilenburg H., Smyth D.J., Stevens H., Todd J.A., Walker N.M., Rich S.S. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman A., Conway G.S., Cadge B. Audiological features of Turner's syndrome in adults. Int. J. Audiol. 2004;43:533–544. doi: 10.1080/14992020400050068. [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M., Tranebjaerg L. The Jervell and Lange-Nielsen syndrome. Adv. Otorhinolaryngol. 2000;56:45–52. doi: 10.1159/000059080. [DOI] [PubMed] [Google Scholar]
- Chen J., Nathans J. Estrogen-related receptor beta/NR3B2 controls epithelial cell fate and endolymph production by the stria vascularis. Dev. Cell. 2007;13:325–337. doi: 10.1016/j.devcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Christensen K., Frederiksen H., Hoffman H.J. Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J. Am. Geriatr Soc. 2001;49:1512–1517. doi: 10.1046/j.1532-5415.2001.4911245.x. [DOI] [PubMed] [Google Scholar]
- Collin R.W., Kalay E., Tariq M., Peters T., van der Zwaag B., Venselaar H., Oostrik J., Lee K., Ahmed Z.M., Caylan R., Li Y., Spierenburg H.A., Eyupoglu E., Heister A., Riazuddin S., Bahat E., Ansar M., Arslan S., Wollnik B., Brunner H.G., Cremers C.W., Karaguzel A., Ahmad W., Cremers F.P., Vriend G., Friedman T.B., Leal S.M., Kremer H. Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am. J. Hum. Genet. 2008;82:125–138. doi: 10.1016/j.ajhg.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks K.J., Wiley T.L., Tweed T.S., Klein B.E., Klein R., Mares-Perlman J.A., Nondahl D.M. Prevalence of hearing loss in older adults in Beaver Dam, Wisconsin. The Epidemiology of Hearing Loss Study. Am. J. Epidemiol. 1998;148:879–886. doi: 10.1093/oxfordjournals.aje.a009713. [DOI] [PubMed] [Google Scholar]
- Davis A., Wood S., Healy R., Webb H., Rowe S. Risk factors for hearing disorders: epidemiologic evidence of change over time in the UK. J. Am. Acad. Audiol. 1995;6:365–370. [PubMed] [Google Scholar]
- Dufour C.R., Wilson B.J., Huss J.M., Kelly D.P., Alaynick W.A., Downes M., Evans R.M., Blanchette M., Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ecob R., Sutton G., Rudnicka A., Smith P., Power C., Strachan D., Davis A. Is the relation of social class to change in hearing threshold levels from childhood to middle age explained by noise, smoking, and drinking behaviour? Int. J. Audiol. 2008;47:100–108. doi: 10.1080/14992020701647942. [DOI] [PubMed] [Google Scholar]
- Elfassihi L., Giroux S., Bureau A., Laflamme N., Cole D.E., Rousseau F. Association with replication between estrogen-related receptor gamma (ESRRgamma) polymorphisms and bone phenotypes in women of European ancestry. J. Bone Miner. Res. 2010;25:901–911. doi: 10.1359/jbmr.091014. [DOI] [PubMed] [Google Scholar]
- Fetoni A.R., Picciotti P.M., Paludetti G., Troiani D. Pathogenesis of presbycusis in animal models: a review. Exp. Gerontol. 2011;46:413–425. doi: 10.1016/j.exger.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Friedman R.A., Van Laer L., Huentelman M.J., Sheth S.S., Van Eyken E., Corneveaux J.J., Tembe W.D., Halperin R.F., Thorburn A.Q., Thys S., Bonneux S., Fransen E., Huyghe J., Pyykko I., Cremers C.W., Kremer H., Dhooge I., Stephens D., Orzan E., Pfister M., Bille M., Parving A., Sorri M., Van de Heyning P.H., Makmura L., Ohmen J.D., Linthicum F.H., Jr., Fayad J.N., Pearson J.V., Craig D.W., Stephan D.A., Van Camp G. GRM7 variants confer susceptibility to age-related hearing impairment. Hum. Mol. Genet. 2009;18:785–796. doi: 10.1093/hmg/ddn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates G.A., Couropmitree N.N., Myers R.H. Genetic associations in age-related hearing thresholds. Arch. Otolaryngol. Head Neck Surg. 1999;125:654–659. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- Girotto G., Pirastu N., Gasparini A., D'Adamo P., Gasparini P. Frequency of hearing loss in a series of rural communities of five developing countries located along the Silk Road. Audiol. Med. 2011;9:135–140. [Google Scholar]
- Girotto G., Pirastu N., Sorice R., Biino G., Campbell H., d'Adamo A.P., Hastie N.D., Nutile T., Polasek O., Portas L., Rudan I., Ulivi S., Zemunik T., Wright A.F., Ciullo M., Hayward C., Pirastu M., Gasparini P. Hearing function and thresholds: a genome-wide association study in European isolated populations identifies new loci and pathways. J. Med. Genet. 2011;48:369–374. doi: 10.1136/jmg.2010.088310. [DOI] [PubMed] [Google Scholar]
- Guimaraes P., Zhu X., Cannon T., Kim S., Frisina R.D. Sex differences in distortion product otoacoustic emissions as a function of age in CBA mice. Hear. Res. 2004;192:83–89. doi: 10.1016/j.heares.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Hederstierna C., Hultcrantz M., Collins A., Rosenhall U. The menopause triggers hearing decline in healthy women. Hear. Res. 2010;259:31–35. doi: 10.1016/j.heares.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Helzner E.P., Cauley J.A., Pratt S.R., Wisniewski S.R., Zmuda J.M., Talbott E.O., de Rekeneire N., Harris T.B., Rubin S.M., Simonsick E.M., Tylavsky F.A., Newman A.B. Race and sex differences in age-related hearing loss: the Health, Aging and Body Composition Study. J. Am. Geriatr. Soc. 2005;53:2119–2127. doi: 10.1111/j.1532-5415.2005.00525.x. [DOI] [PubMed] [Google Scholar]
- Hermans-Borgmeyer I., Susens U., Borgmeyer U. Developmental expression of the estrogen receptor-related receptor gamma in the nervous system during mouse embryogenesis. Mech. Dev. 2000;97:197–199. doi: 10.1016/s0925-4773(00)00422-6. [DOI] [PubMed] [Google Scholar]
- Hultcrantz M., Simonoska R., Stenberg A.E. Estrogen and hearing: a summary of recent investigations. Acta Otolaryngol. 2006;126:10–14. doi: 10.1080/00016480510038617. [DOI] [PubMed] [Google Scholar]
- Lin P.I., Vance J.M., Pericak-Vance M.A., Martin E.R. No gene is an island: the flip-flop phenomenon. Am. J. Hum. Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch E.D., Kil J. Compounds for the prevention and treatment of noise-induced hearing loss. Drug Discov. Today. 2005;19:1291–1298. doi: 10.1016/S1359-6446(05)03561-0. [DOI] [PubMed] [Google Scholar]
- Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., Cho J.H., Guttmacher A.E., Kong A., Kruglyak L., Mardis E., Rotimi C.N., Slatkin M., Valle D., Whittemore A.S., Boehnke M., Clark A.G., Eichler E.E., Gibson G., Haines J.L., Mackay T.F., McCarroll S.A., Visscher P.M. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullar J.S., Oesterle E.C. Cellular targets of estrogen signaling in regeneration of inner ear sensory epithelia. Hear. Res. 2009;252:61–70. doi: 10.1016/j.heares.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltser I., Tahera Y., Simpson E., Hultcrantz M., Charitidi K., Gustafsson J.A., Canlon B. Estrogen receptor beta protects against acoustic trauma in mice. J. Clin. Invest. 2008;118:1563–1570. doi: 10.1172/JCI32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E.G., Hinojosa R. Presbycusis: a human temporal bone study of individuals with downward sloping audiometric patterns of hearing loss and review of the literature. Laryngoscope. 2006;116:1–12. doi: 10.1097/01.mlg.0000236089.44566.62. [DOI] [PubMed] [Google Scholar]
- Pearson J.D., Morrell C.H., Gordon-Salant S., Brant L.J., Metter E.J., Klein L.L., Fozard J.L. Gender differences in a longitudinal study of age-associated hearing loss. J. Acoust. Soc. Am. 1995;97:1196–1205. doi: 10.1121/1.412231. [DOI] [PubMed] [Google Scholar]
- Raynor L.A., Pankow J.S., Miller M.B., Huang G.H., Dalton D., Klein R., Klein B.E., Cruickshanks K.J. Familial aggregation of age-related hearing loss in an epidemiological study of older adults. Am. J. Audiol. 2009;18:114–118. doi: 10.1044/1059-0889(2009/08-0035). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonoska R., Stenberg A.E., Duan M., Yakimchuk K., Fridberger A., Sahlin L., Gustafsson J.A., Hultcrantz M. Inner ear pathology and loss of hearing in estrogen receptor-beta deficient mice. J. Endocrinol. 2009;201:397–406. doi: 10.1677/JOE-09-0060. [DOI] [PubMed] [Google Scholar]
- Stevens G., Flaxman S., Brunskill E., Mascarenhas M., Mathers C.D., Finucane M. Global and regional hearing impairment prevalence: an analysis of 42 studies in 29 countries. Eur. J. Public Health. 2013 doi: 10.1093/eurpub/ckr176. 23, 146–152. [DOI] [PubMed] [Google Scholar]
- Strachan D.P., Rudnicka A.R., Power C., Shepherd P., Fuller E., Davis A., Gibb I., Kumari M., Rumley A., Macfarlane G.J., Rahi J., Rodgers B., Stansfeld S. Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int. J. Epidemiol. 2007;36:522–531. doi: 10.1093/ije/dyl309. [DOI] [PubMed] [Google Scholar]
- Tak S.W., Calvert G.M. Hearing difficulty attributable to employment by industry and occupation: an analysis of the National Health Interview Survey- United States, 1997 to 2003. J. Occup. Environ. Med. 2008;50:46–56. doi: 10.1097/JOM.0b013e3181579316. [DOI] [PubMed] [Google Scholar]
- Tremblay A.M., Giguere V. The NR3B subgroup: an ovERRview. Nucl. Recept. Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal M., Tamer L., Dogruer Z.N., Yildirim H., Vayisoglu Y., Camdeviren H. N-acetyltransferase 2 gene polymorphism and presbycusis. Laryngoscope. 2005;115:2238–2241. doi: 10.1097/01.mlg.0000183694.10583.12. [DOI] [PubMed] [Google Scholar]
- Van Eyken E., Van Camp G., Fransen E., Topsakal V., Hendrickx J.J., Demeester K., Van de Heyning P., Maki-Torkko E., Hannula S., Sorri M., Jensen M., Parving A., Bille M., Baur M., Pfister M., Bonaconsa A., Mazzoli M., Orzan E., Espeso A., Stephens D., Verbruggen K., Huyghe J., Dhooge I., Huygen P., Kremer H., Cremers C.W., Kunst S., Manninen M., Pyykko I., Lacava A., Steffens M., Wienker T.F., Van Laer L. Contribution of the N-acetyltransferase 2 polymorphism NAT2*6A to age-related hearing impairment. J. Med. Genet. 2007;44:570–578. doi: 10.1136/jmg.2007.049205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken E., Van Camp G., Van Laer L. The complexity of age-related hearing impairment: contributing environmental and genetic factors. Audiol. Neurootol. 2007;12:345–358. doi: 10.1159/000106478. [DOI] [PubMed] [Google Scholar]
- Van Eyken E., Van Laer L., Fransen E., Topsakal V., Lemkens N., Laureys W., Nelissen N., Vandevelde A., Wienker T., Van De Heyning P., Van Camp G. KCNQ4: a gene for age-related hearing impairment? Hum. Mutat. 2006;27:1007–1016. doi: 10.1002/humu.20375. [DOI] [PubMed] [Google Scholar]
- Van Laer L., Huyghe J.R., Hannula S., Van Eyken E., Stephan D.A., Maki-Torkko E., Aikio P., Fransen E., Lysholm-Bernacchi A., Sorri M., Huentelman M.J., Van Camp G. A genome-wide association study for age-related hearing impairment in the Saami. Eur. J. Hum. Genet. 2010;18:685–693. doi: 10.1038/ejhg.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L., Van Eyken E., Fransen E., Huyghe J.R., Topsakal V., Hendrickx J.J., Hannula S., Maki-Torkko E., Jensen M., Demeester K., Baur M., Bonaconsa A., Mazzoli M., Espeso A., Verbruggen K., Huyghe J., Huygen P., Kunst S., Manninen M., Konings A., Diaz-Lacava A.N., Steffens M., Wienker T.F., Pyykko I., Cremers C.W., Kremer H., Dhooge I., Stephens D., Orzan E., Pfister M., Bille M., Parving A., Sorri M., Van de Heyning P.H., Van Camp G. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- Wangemann P. K(+) cycling and its regulation in the cochlea and the vestibular labyrinth. Audiol. Neurootol. 2002;7:199–205. doi: 10.1159/000063736. [DOI] [PubMed] [Google Scholar]
- Warth R., Barhanin J. The multifaceted phenotype of the knockout mouse for the KCNE1 potassium channel gene. Am. J. Physiol. Regul. Integr Comp. Physiol. 2002;282:R639–R648. doi: 10.1152/ajpregu.00649.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.