Abstract
We have determined the three-dimensional structures of both native and expanded forms of turnip crinkle virus (TCV), using cryo-electron microscopy, which allows direct visualization of the encapsidated single-stranded RNA and coat protein (CP) N-terminal regions not seen in the high-resolution X-ray structure of the virion. The expanded form, which is a putative disassembly intermediate during infection, arises from a separation of the capsid-forming domains of the CP subunits. Capsid expansion leads to the formation of pores that could allow exit of the viral RNA. A subset of the CP N-terminal regions becomes proteolytically accessible in the expanded form, although the RNA remains inaccessible to nuclease. Sedimentation velocity assays suggest that the expanded state is metastable and that expansion is not fully reversible. Proteolytically cleaved CP subunits dissociate from the capsid, presumably leading to increased electrostatic repulsion within the viral RNA. Consistent with this idea, electron microscopy images show that proteolysis introduces asymmetry into the TCV capsid and allows initial extrusion of the genome from a defined site. The apparent formation of polysomes in wheat germ extracts suggests that subsequent uncoating is linked to translation. The implication is that the viral RNA and its capsid play multiple roles during primary infections, consistent with ribosome-mediated genome uncoating to avoid host antiviral activity.
Abbreviations: CP, coat protein; TCV, turnip crinkle virus; cryo-EM, cryo-electron microscopy; ssRNA, single-stranded RNA; TBSV, tomato bushy stunt virus; EDTA, ethylenediaminetetraacetic acid; 3-D, three-dimensional; VLP, virus-like particle; PDB, Protein Data Bank; CCC, cross-correlation coefficient
Keywords: cryo-electron microscopy, TCV, ssRNA virus, genomic RNA structure, genome uncoating
Graphical Abstract
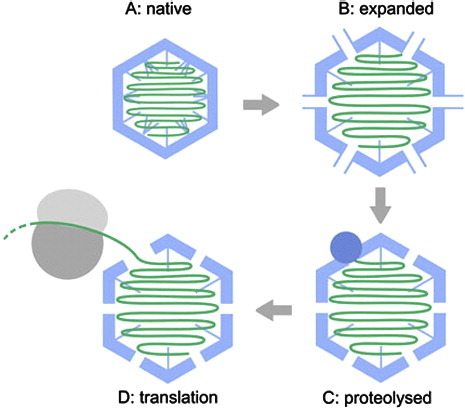
Highlights
► Native and expanded TCV cryo-electron microscopy structures reveal RNA dynamics. ► Expansion leads to the formation of pores large enough to allow exit of viral RNA. ► RNA remains resistant to nuclease, but capsid proteins become protease sensitive. ► Proteolysis causes initial extrusion of viral RNA, creating asymmetry in capsids. ► Subsequent ribosome binding prevents silencing and promotes genome extrusion.
Introduction
Single-stranded RNA (ssRNA) viruses cause very significant material and financial losses in essential crop plants every year.1 Recently, it has been recognized that infection by, and systemic spread of, many of these viruses are controlled by a series of host defense mechanisms including RNA silencing.2 The host's defenses have consequences for the mechanisms used by these viruses to introduce their genomic RNAs into cells. For turnip crinkle virus (TCV3), a member of the Tombusviridae family that includes the close structural homologue tomato bushy stunt virus (TBSV4) (Fig. 1), the coat protein (CP) subunit has been shown to have multiple functions that include overcoming some of these barriers.6 To probe the roles played by the capsid in facilitating infection, we have determined cryo-electron microscopy (cryo-EM) reconstructions of both a native form and an expanded form of TCV, the latter being a putative intermediate in viral disassembly during infection.
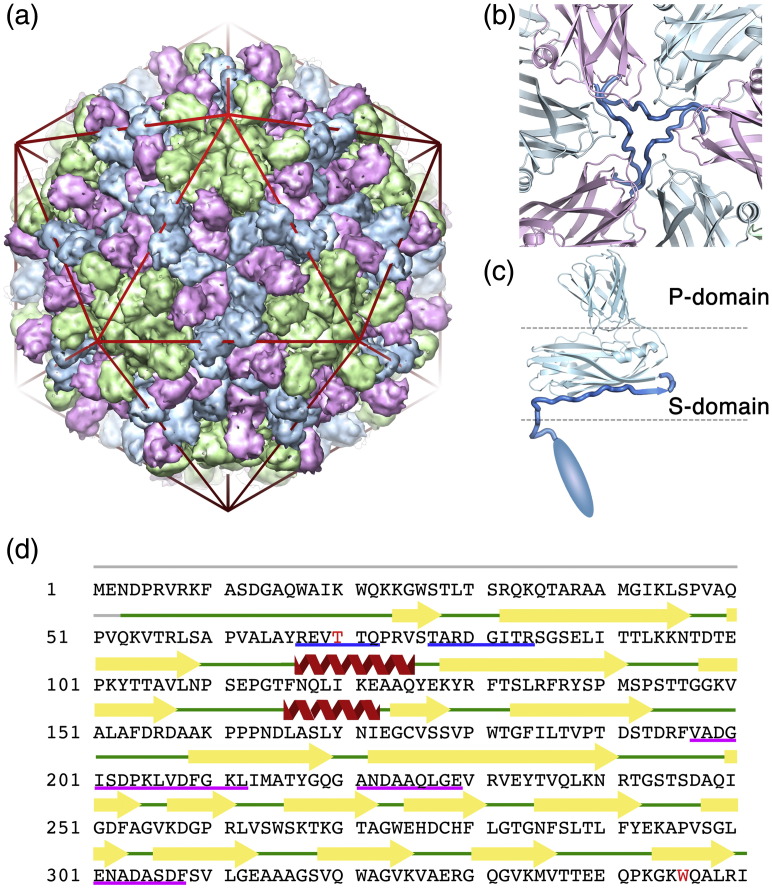
Fig. 1.
Architecture of the TCV virion. (a) The T = 3 capsid of TCV contains 180 copies of the CP subunit in three quasi-equivalent conformations. The A subunits pack around the 5-fold symmetry axes (shown in green), and subunits in the B (pink) and C (blue) conformations interdigitate around the particle 3-fold axes. The CP subunits are rendered as a surface to simplify the representation. An icosahedron is shown in red to highlight the locations of the symmetry axes. (b) At the particle 3-fold axes, the N-terminal arms of the C subunits are partially ordered and intertwine to form the β-annulus structure. Protein subunits are shown as a cartoon representation using the same color scheme as in (a). The β-annulus structure is shown as a thicker segment and highlighted in a stronger blue. (c) The domain structure of the TCV CP subunit. The P domain forms the surface spikes of the protein, while the S domain forms the continuous protein shell. The N-terminal regions are largely unstructured and have been proposed to bind genomic RNA via their basic amino acid side chains. (d) The amino acid sequence of TCV CP. The secondary structure elements determined by crystallography are shown above the sequence. Amino acids colored red (T70 and W346) indicate sites of variation in our sequence from the original publication.5 Underlined amino acids have been directly sequenced by tandem mass spectrometry (red line) or by Edman degradation (blue line).
Both TBSV and TCV are examples of T = 3 capsids3,4,7 and were among the first virus structures determined by X-ray crystallography. Each is ∼ 330 Å in diameter and is composed of 180 copies of a single polypeptide chain whose conformations differ in a symmetry-related fashion, creating three distinct quasi-equivalent conformers (denoted A, B and C). These CPs form non-covalent dimers via interactions between their C-terminal projecting (P) domains, which connect via short hinge regions to globular domains that form the shell of the capsid (S domain, Fig. 1c). In the capsid, the 90 CP dimers exist as either asymmetric A/B dimers (60 in total) or symmetric C/C dimers (30 in total). These differ in the hinge angles between S and P domains but are principally differentiated by their N-terminal regions. Here the protein forms an extended arm that is partially disordered in the X-ray structures. This region is more ordered in C/C dimers, forming an additional β-strand at the edge of the S domain that extends to make contact with the symmetry-related arms to form an additional structure termed the β-annulus at the particle 3-fold axis (Fig. 1b). The equivalent regions in the A and B subunits are disordered and not seen in the X-ray electron density map. The polypeptide chains N-terminal to the β-annulus (in C subunits) or the S domain (in A and B subunits) disappear toward the center of the virus particle where they presumably interact with genomic RNA, possibly via positively charged N-terminal regions.
Unlike TBSV, TCV is easily dissociated in vitro by transfer into mildly alkaline buffers containing high salt and ethylenediaminetetraacetic acid (EDTA), yielding a CP–RNA complex and RNA-free CP dimers,8 which permits in vitro assembly studies. Reassembly of the T = 3 shell occurs following mixing of these components and dialysis against neutral buffers containing calcium ions.9 From estimates of the stoichiometry, the CP–RNA complex consists of up to three CP dimers and p80, a minor protein that appears to be present as a single copy per virion. p80 has been shown to consist of two covalently linked CP subunits.10 A molecular mechanism for capsid assembly was proposed based on this CP–RNA complex, which is assumed to contain a β-annulus and act as an initiation complex.9 This model is consistent with the subsequent identification of a short RNA fragment that acts as a minimal packaging signal in vivo.11
The Tombusviridae are similar to a much larger class of plant virus genera, including those with multipartite genomes,12 all of which have extended CP arms. Many human and animal viruses, such as papilloma virus,13 also have extended arms. A further similarity across plant and animal viruses is that many of these, including human immunodeficiency virus and brome mosaic virus, have arginine-rich RNA-binding domains within their extended arms.12 The N-terminal regions in TBSV/TCV are also positively charged but do not contain such a motif, although it is assumed that they play similar roles in binding viral RNA.9
For simple plant viruses, genome uncoating commonly requires the conformational rearrangement of the capsid. This is driven by the expulsion of divalent metal ions, which play structural roles within the native virion. In most cases, the result is formation of an expanded form of these virus particles, and these have been postulated to be disassembly/uncoating intermediates. A number of these states have been analyzed structurally, starting with the expanded form of TBSV determined by X-ray crystallography at 8 Å,14 and this remains the highest-resolution view of an expanded virus. A number of other structures have been determined by cryo-EM reconstruction at medium to low resolution.15–17
To further understand the functional relevance of capsid expansion, we have determined the three-dimensional (3-D) structures of the native and expanded forms of TCV, using cryo-EM. We have also assayed the protease and nuclease sensitivity of the various virion components in the native and expanded particles and used sedimentation velocity experiments to interrogate their hydrodynamic radii and stability. Our results help to rationalize a number of disparate observations for a range of different viruses, by elucidating a single experimentally tractable system. We show that expanded capsids are metastable and that their greater volume allows the packaged RNA to be more dynamic. They are also porous, in principle permitting the RNA to access the translational apparatus of the cell while still providing protection from host nucleases. However, proteolysis of expanded virions in the absence of other factors causes loss of cleaved CP subunits and efficient initial extrusion of viral RNA from a single site in the capsid. Taken together, these results provide the first clear structural rationale for the capsid expansion by plant virus particles during infection. Expanded capsids are readily proteolyzed, exposing the viral RNA to the host's translational machinery in a controlled way to avoid host antiviral responses. This coupling of biological and structural mechanisms may be a common feature among this class of viruses, although the precise molecular details may vary in each.
Results and Discussion
The structure of the native TCV virion
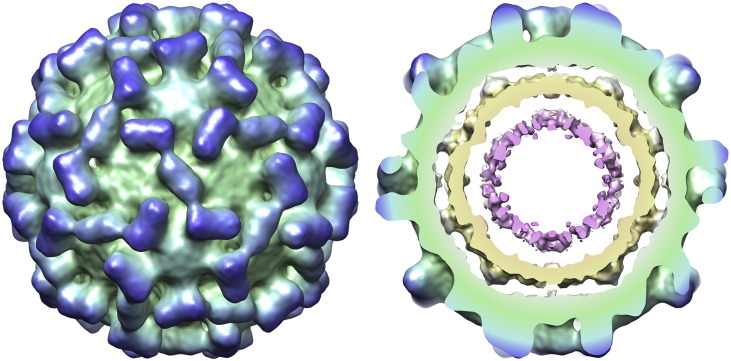
The 3-D structure of the native TCV virion was determined using cryo-EM, single-particle image processing and icosahedral symmetry averaging, at ∼ 11 Å resolution (Fig. 2). The EM map has the characteristic “spiked” appearance of Tombusviridae. To enhance our understanding of the electron density, we fitted the crystal structure of the TCV S and P domains (coordinates courtesy of Prof. S. C. Harrison, Harvard Medical School, Harvard, Boston, MA, USA3) into the map.
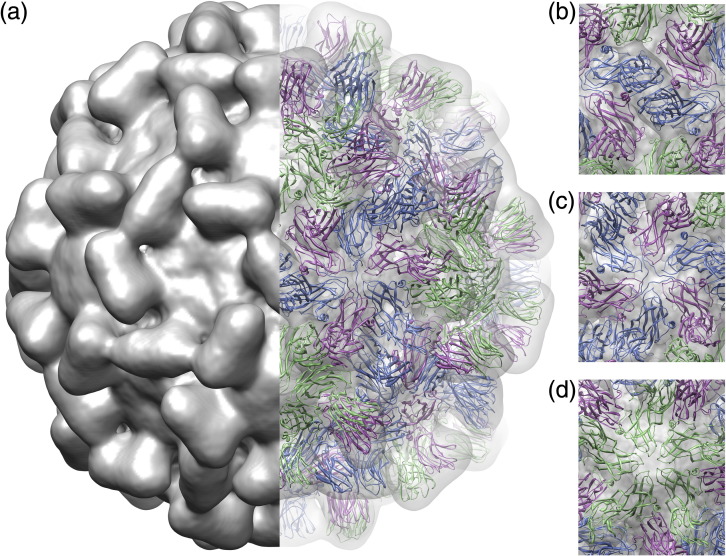
Fig. 2.
The structure of the TCV virion. (a) Surface view (left-hand side) and atomic structure fitting (right-hand side) of the native virion. On the right, the density has been made transparent, revealing the fitted coordinates for the TCV crystal structure shown as a ribbon, colored as in Fig. 1. (b–d) Close-up views of the fit of the atomic structure, along the 2-fold (b), 3-fold (c) and 5-fold (d) symmetry axes.
In cross-section, the electron density map reveals that the virion is composed of three distinct, but connected, shells of electron density. The atomic coordinates for the S and P domains fit the outermost of these shells (lying between radii of 110–180 Å) extremely well. We ascribe both shells of electron density at lower radii to a mixture of packaged viral RNA and the N-terminal regions of the CPs. The middle shell of electron density, between radii of 83–109 Å, is connected to the protein capsid at the 2-fold, 5-fold and 3-fold symmetry axes (Fig. 3a). At higher contour levels, where only the strongest electron density is visible, this shell has a distinct cage-like structure that is best described as a truncated icosahedron [consisting of pentagonal rings surrounded by “flattened” hexagons that are centered on the 3-fold symmetry axes (Fig. 3b)]. At similar contour levels, the innermost shell, located between radii of 50–73 Å, is almost completely absent. This suggests that the innermost shell has a lower occupancy or that the molecular components that contribute to it are less ordered or both. When weaker electron density is included, the middle and inner shells are connected by columns of density with a radius of ∼ 7 Å (at the contour level shown), along the 2-fold symmetry axes.
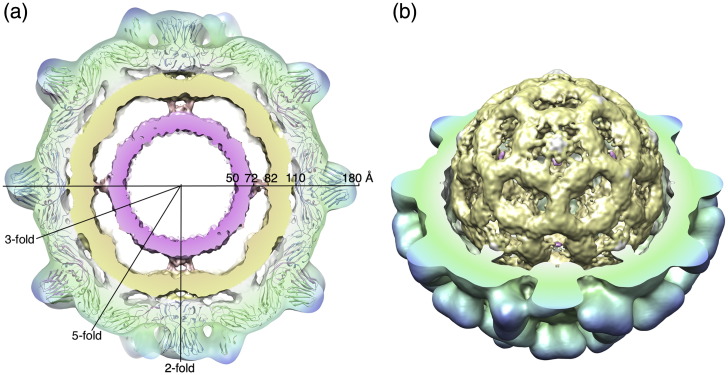
Fig. 3.
Density within the native TCV virion. (a) A central 25-Å-thick section through the cryo-EM structure of the virion. The EM density is transparent in the capsid region, revealing the fitted atomic coordinates. At lower radii, the cut surface of the density is opaque for visual clarity and is colored using a radial scheme (from pink at a radius of 50 Å to blue at 180 Å). Icosahedral symmetry axes are also indicated. (b) Cutaway view of the virus showing the cage-like structure of the internal density.
The organization of the interior of the virion
The overall pattern of density observed in our structure of native TCV is similar to that seen previously inside other small plant viruses, including TBSV and cucumber necrosis virus.15,18 All show an ordered protein capsid with two additional shells of density. In detail, the picture is subtly different for each virus. The way in which the middle shell contacts the capsid protein and the contacts between middle and inner shells is not consistent. In cucumber necrosis virus, the main contacts between the capsid and the middle shell are at the 5-fold axes, while the contacts between the middle and inner shells are along the 2-fold axes.18 In TBSV, the main contacts between protein and the middle shell are at the 2-fold axes, while the interior shells connect along the 5-fold axes.15 TCV shows a unique pattern of contacts (Fig. 3b). Here the capsid and the middle shell are connected at the 3-fold axes, where the β-annulus is located, and at the 5-fold axes. The interior shells connect along the 2-fold axes. These differences argue against a single arrangement for packaged RNA in the Tombusviridae and suggest that the relationship between the symmetrical CP layer and virion contents may also vary. Similar multi-shelled structures have been seen for the RNA phages, whose CPs lack extended arms, so that in those cases the inner shells are composed solely of viral RNA.19–23
Structure of the expanded particle
Both TCV and TBSV capsids are stabilized by Ca2+ ions, which are coordinated by negatively charged amino acid side chains at the interfaces between subunits (Fig. 4). The initial entry of TCV into the host cell follows mechanical damage caused by its insect vector. Conditions within the cell result in loss of these structural calcium ions.24 The resulting electrostatic repulsion between acid side chains leads to expansion. From the X-ray structure of expanded TBSV, and presumably for TCV as well, it appears that the expanded structure is stabilized by an increased ordering of the N-terminal regions of the A and B subunits adjacent to the S domain.14 For TCV, unlike TBSV, addition of high salt concentrations to the expanded form leads to dissociation of the capsid.
Fig. 4.
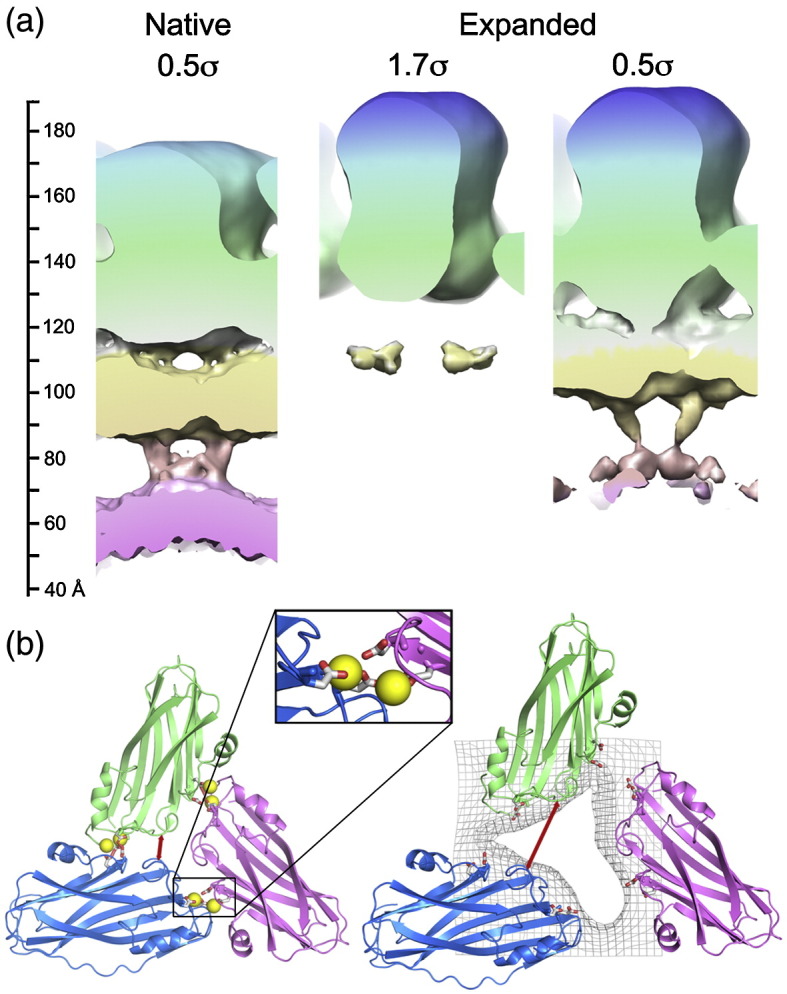
The structural consequences of expansion. (a) On the left, the radial distribution of density is shown for the native virion. The middle image shows the expanded virion contoured to show the same volume for the CP, as in Fig. 3. On the right, the expanded virion is contoured to show the weaker inner shells of density. The vertical scale bar on the left indicates the radial distance from the center of the particle in angstroms. (b) Result of viral expansion. Only the asymmetric trimer of TCV is shown for the native virus on the left and the expanded particle on the right (colored as in Fig. 1). The calcium ions (yellow spheres) are shown in the locations they occupy in the homologous TBSV structure (PDB code 2TBV). In the native virus, these ions are coordinated by acidic side chains (D155 and D157 opposite E127 and D199) from adjacent monomers (inset). For the expanded virus, the edges of the pores observed in the cryo-EM reconstruction are shown as gray mesh. The effect of expansion on the relative separation of two amino acids that remain at the same radial level (D203 and S202) is shown by the red arrows. This distance is 6.5 Å in the native particle and 17 Å in the expanded virus.
To examine expansion in TCV, we incubated native virions in low-ionic-strength buffer containing EDTA at pH 8.5. This resulted in particles that were visibly expanded in the raw cryo-electron micrographs, and these were used to determine an icosahedrally averaged, 3-D reconstruction for the expanded TCV virion at ∼ 17 Å resolution. Although the expanded form is ∼ 5% larger than the native virion (outer radius of ∼ 190 Å compared to ∼ 180 Å), the two structures are superficially similar. The expanded capsid again has three shells of density, albeit each lying at a slightly higher radius. However, significant differences exist between the inner shells of the native and expanded forms. The middle shell of electron density becomes weaker and less cage like in the expanded state. However, as in the native virus, at lower contour levels, electron density at the 2-fold axes connects the innermost and middle shells of density (Fig. 5). The density forming the interior shells is much weaker in the expanded state (cf. Fig. 5, in which the electron density in each state is contoured such that the CP layer encloses the same molecular volume). The loss of density could be the result of the larger volume of the expanded state (a 5% increase in radius leads to a 17% increase in volume) or because the RNA and/or N-terminal regions become more dynamic. For TBSV, NMR measurements are consistent with the RNA, at least, becoming more dynamic upon expansion.25
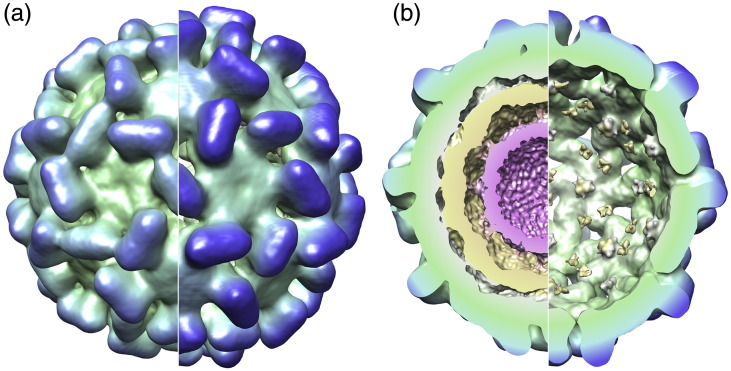
Fig. 5.
Comparison of native and expanded forms of TCV. (a) Surface representation of the exterior of the two capsids, with the native virion on the left and expanded form on the right. The capsids are colored using an identical radial scheme, so the larger radius of the expanded particle is highlighted by the deeper blue of the tips of the surface spikes. Both models are contoured to incorporate approximately the same volume of density for the CP region. (b) Back half of the capsid showing the reduction in internal density in the expanded form.
There is also a major difference between the capsid structures of expanded and native forms (Fig. 5). In the native virus, the capsid layer is contiguous, resulting in a sealed container. However, the expanded form is porous, with distinct holes (with a diameter of ∼ 15–20 Å) in the CP layer. To understand better the structural changes that form these pores, we fitted the atomic coordinates of the CP domains into the expanded density. No significant alteration in the relative positions of the S and P domains of the individual CP subunits (i.e., hinge angles) was observed at this resolution, and the pentamers and hexamers making up the T = 3 capsid remain intact. The pores arise because intact pentamers and hexamers have moved apart by ∼ 10 Å, movements that should allow the β-annuli, which are crucial to capsid stability, to remain intact. However, at this resolution, we cannot confirm this. The conformational rearrangement seen for TCV, with respect to the nature and size of the shifts, and the approximate size of the pores that result are entirely consistent with the 6-Å structure of TBSV seen by X-ray diffraction, in which the β-annuli are intact.14 Another expanded plant virus shows similar pores, although their locations within their icosahedral shells and their biochemical details differ,26 implying that pores may be a generic functional feature.
The role(s) of the expanded state in genome uncoating
At the resolution of our map, the size of the pores cannot be precisely determined; however, an obvious possibility is that their creation allows the RNA genome or other molecular components to exit the capsid. To test the idea that genomic RNA and/or sequences in the N-terminal region of the CP become externalized upon expansion, we assayed the protease and nuclease susceptibilities of the expanded particles (Fig. 6).
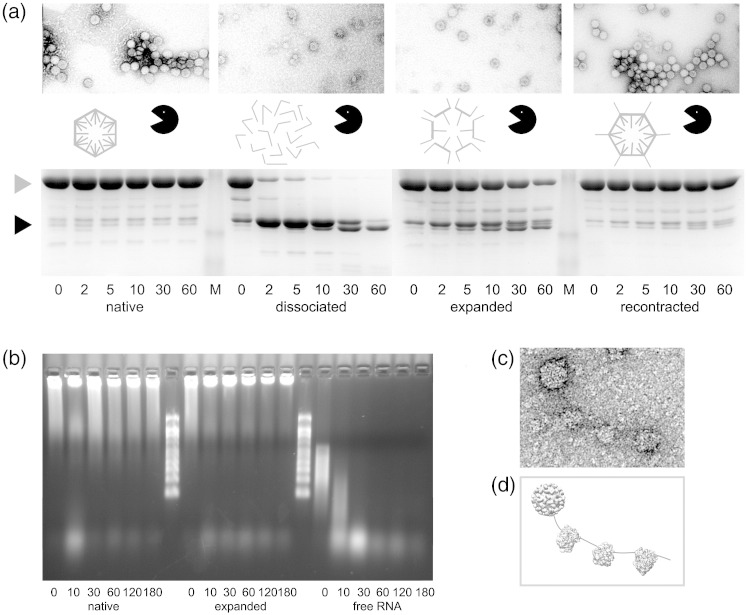
Fig. 6.
Assaying the consequences of expansion. (a) Results of chymotrypsin digestion of the virus in various states. The native virus appears unaffected by the protease, while in dissociated virus, all CPs are rapidly cleaved to the ∼ 30-kDa products that roughly correspond to the S and P domains. In expanded virus, about two-thirds of the CP subunits cleave within 1 h. In the recontracted virus few, if any, CP subunits are cleaved, implying that expansion is fully reversible (see Supplementary Fig. 3). Gray and black arrows indicate the positions of full-length CP and the ∼ 30-kDa cleavage products, respectively. Negative stain transmission electron microscopy images of the samples prior to addition of chymotrypsin are shown above each gel. The stain is largely excluded from the centers of native and recontracted virus but is clearly able to penetrate to the center of the expanded form. (b) Denaturing agarose gel showing the results of ribonuclease A incubation of native and expanded TCVs, as well as protein-free TCV genomic RNA. Time points are indicated under each lane in minutes. The protein-free genomic RNA was incubated in expansion buffer to allow direct comparison. (c) Negative stain transmission electron microscopy of a virus complexed with three ribosome-like particles, formed when expanded TCV was incubated with wheat germ extract. (d) Schematic representation based on known 3-D structures of the presumed components (c).
Preparations of TCV and other Tombusviridae contain two major protein species visible by SDS-PAGE, corresponding to the full-length CP subunit (∼ 38 kDa) and the p80 protein. Minor bands at ∼ 30 kDa are also observed. This characteristic pattern is unaltered when native virus is challenged with chymotrypsin, reflecting the great stability of native TCV virions. In contrast, incubation of dissociated capsids with protease results in rapid disappearance of the full-length CP and the appearance of metastable products at ∼ 30 kDa, as has previously been observed.8 These co-migrate with the bands described above for the virion. Mass spectrometry of these digestion products and N-terminal amino acid sequencing (Fig. 1, and Supplementary Figs. 1 and 2) confirm earlier suggestions that they are proteolysis products of the CP consisting of the S and P domains only.8,10 In this analysis, we observed small differences from the masses expected from the originally published TCV CP sequence.5 We therefore resequenced the CP gene using cDNA derived from the TCV genomic RNA used here (see Materials and Methods). This identified two amino acid changes (S70T and L346W). Use of this sequence resulted in precise agreement with the masses observed (Fig. 1 and Supplementary Figs. 1 and 2).
Supplementary Figure 1.
Alignment of TCV protein sequences. The sequences shown are from (a) the RNA sequence determined here; (b) the original sequence report5 and (c) the published crystal structure.3 Highlighted in red are sequences changes due to differences between the two nucleotide sequences. Highlighted in blue are residues where the density could not be interpreted in the crystal structure which was built prior to the availability of the nucleotide sequence.
Supplementary Figure 2.
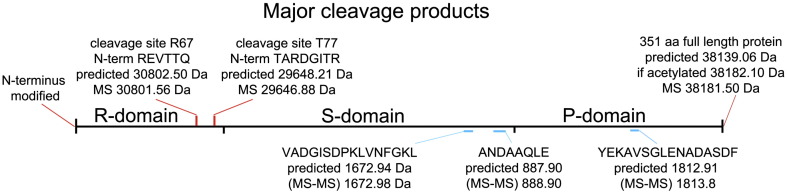
Schematic of TCV coat protein proteolysis. The red cleavage sites at residues R67 and T77 were characterised by N-terminal sequencing (sequences shown) and mass spectrometry (calculated and observed masses shown). N-terminal sequencing showed that the N-terminus of the CP was covalently modified, and mass spectrometry was consistent with an N-terminal acetylation. Blue cleavage sites were characterised by mass spectrometry and sequenced by MS-MS (calculated and observed masses shown). The positions of the structural domains are shown.
Treatment of expanded virus capsids with protease leads to the same proteolytic cleavages as those seen in the dissociated virion. However, the kinetics of these cleavages are remarkably different. A significant fraction (∼ 1/3) of the full-length CP is still present after 60 min, suggesting that only some of the subunits are proteolytically sensitive. The approximate ratio of the gel bands is consistent with cleavage of only the A- and B-type subunits. Since the pores in the expanded capsid are too small to allow chymotrypsin to access the interior directly, the cleavage sites in C-type subunits involved in β-annular contacts are likely to be inaccessible.
When the expanded virus is recontracted by the addition of excess calcium ions before the addition of chymotrypsin, minimal proteolysis is observed, implying that expansion is largely reversible. This was confirmed by carrying out a low-resolution icosahedral reconstruction of the recontracted particle (Supplementary Fig. 3). This shows electron density for all three shells in the same positions as they are in native virus. Note that the p80 protein shows an identical susceptibility to protease as the CP subunit under these conditions, consistent with it being a cross-linked dimer of the CP subunit.10
Supplementary Figure 3.
Reconstruction of recontracted TCV virion. Virus was incubated in expansion buffer for 10 min, before being recontracted by lowering the pH and adding a surplus of calcium. 3,885 particles were used, with defocus from 2.1-4.3 μm. Data was collected as for native and expanded forms of TCV.
As expected, the RNA in the native virus is protected from nuclease (Fig. 6). Interestingly, given the sensitivity of CPs to protease treatment, the expanded virus still protects its RNA. This is not simply a consequence of the buffer conditions needed to expand the capsid inhibiting the nuclease, as protein-free TCV genomic RNA under the same buffer conditions as the expanded virus is rapidly degraded (Fig. 6b). Thus, it appears that expansion exposes proteolytic cleavage sites in the N-terminal region of the CP but does not expose the genome to nucleases. Despite these results, when expanded TCV was added to wheat germ translational extract and examined by negative stain EM, particles were seen extruding a thin tether, presumably genomic RNA, bound by structures that appear to be ribosomes (Fig. 6c). The images are consistent with a transfer of the TCV RNA from the expanded capsid to the host's translational machinery. Other plant viruses also form such striposomes,24 although the requirement for expansion and/or proteolysis in each case is not known.
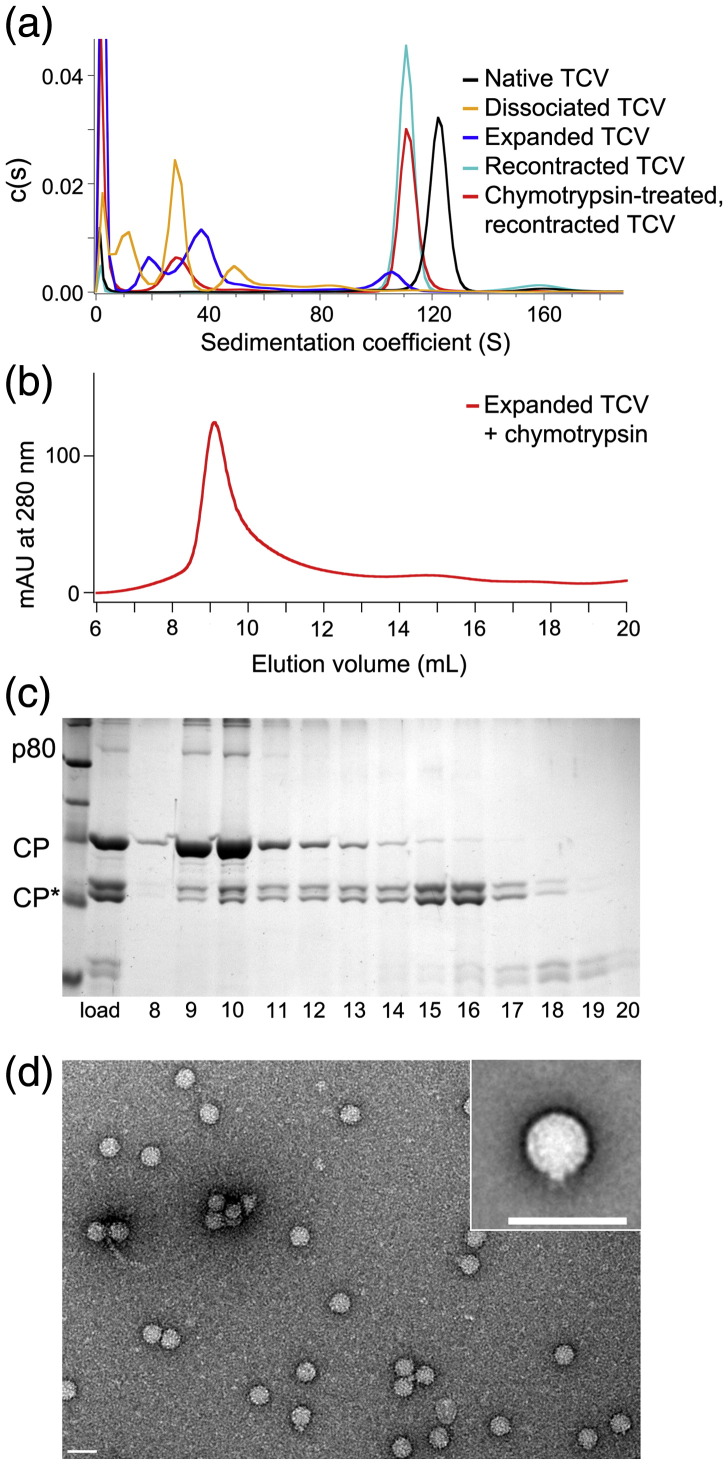
In order to examine the stability of the expanded state and the changes that occur during expansion, we carried out sedimentation velocity analysis (Fig. 7a). When spun at high velocity, the expanded virions dissociated into smaller components, implying that they are metastable. In contrast, expanded virions that were recontracted by addition of excess calcium ions sediment as virus-like particles (VLPs), albeit with a slightly lower sedimentation coefficient than native virions. This is apparently inconsistent with the low-resolution EM reconstruction of such particles, which suggests that they are identical with the starting material. The reconstruction was, however, determined at relatively low resolution and employed icosahedral averaging. The resulting structure would not, therefore, reveal the presence of small nonsymmetric features. Expanded, proteolyzed, recontracted virus sediments similarly, although it has clearly lost some lower-molecular-weight material (Fig. 7a).
Fig. 7.
Expansion and proteolysis lead to partial extrusion of the viral RNA. (a) Sedimentation velocity analysis of the different states of TCV studied. (b) Size-exclusion chromatogram of expanded and proteolyzed TCV VLPs. (c) SDS-PAGE of fractions from the size-exclusion chromatogram shown in (b). Densitometry measurements indicate that the percentage of full-length CP in the starting sample (“load”) is ∼ 50% but increases to ∼ 80% in fractions 9–11. This is consistent with the dissociation of proteolytically cleaved CP dimers from VLPs. (d) Negative stain electron micrograph of the expanded and proteolyzed TCV VLPs, showing the increased heterogeneity and disruption to the structure of the VLP. Inset is an aligned average of this type of particle, calculated using multivariate statistical classification. The scale bar in both main panel and inset represents 50 nm.
The results of the proteolysis experiments could arise in two ways. They might reflect the behavior of all viral particles in the sample, or they could represent the average behavior of intact and unchanged particles in the presence of a fraction of the material that has completely dissociated. To determine which of these scenarios is more likely, we prepared expanded proteolyzed TCV in which cleavage of roughly half of the subunits had occurred (Fig. 7b and c). This sample was then analyzed by gel filtration, and the protein content of various fractions was determined by SDS-PAGE. The column profile reveals a VLP peak (fractions 8–11) that contains virtually all the A254 and by implication the input viral RNA and has a protein content with significantly less cleaved CP than the material that was loaded. Lower-molecular-weight fractions contain fragments encompassing the cleaved S and P domains, and at later elution times, the cleaved N-terminal fragments are observed. This result is consistent with dissociation of cleaved CP subunits from a VLP rather than a mixture of dissociated and intact particles. Presumably, such release would require A/B CP dimers to dissociate. This may occur in stages after initial cleavage of a single subunit with an externalized N-terminal domain. Loss of CP subunits in this way would make the expanded and proteolyzed VLP even more porous, although clearly it remains stable when gently handled.
Negative stain EM of the expanded proteolyzed fraction is instructive (Fig. 7d). Although expansion results in an icosahedrally symmetric structure (Fig. 5), subsequent proteolysis leads to a mixed population of VLPs, many of which show a clear loss of symmetry. Such images are consistent with extrusion of the viral RNA from a unique site within the capsids. The degree of extrusion appears to vary between VLPs. However. the majority of VLPs appear to contain small disruptions to the capsid structure to some degree (Fig. 7d and inset).
Implications for the viral lifecycle
In native TCV virions and many other ssRNA viruses, the packing density of the nucleic acid is close to, or even exceeds that of, double-stranded DNA in tailed phages.27 The latter virions are the most successful entities in the biosphere. Their lifecycle includes formation of a prohead (immature capsid), which is filled with the DNA genome using a phage-encoded, ATP-dependent packaging motor protein attached at one vertex of the spherical capsid.28,29 This motor complex is replaced by the phage tail in mature particles. Upon location and binding to the membrane of a host bacterium, the free energy stored in electrostatic repulsion between DNA phosphates is released as the DNA exits the capsid via a protective tail structure.
Although in principle able to exploit similar physical forces, no equivalent structures exist for ssRNA viruses, and in vitro reassembly results demonstrate that genome encapsidation does not require the net input of free energy. Presumably, this is because positively charged side chains on the inward face of the S domains and the N-terminal regions (∼ 2700 side chains per particle) can significantly neutralize the repulsive forces within the RNA. If this is the case, disassembly/uncoating must make use of a distinct mechanism that destabilizes the native virion structure. TCV and many other ssRNA plant viruses exploit the chelation of divalent metal ions within their capsids to trigger a conformational change associated with cell entry.14 For the majority of these viruses, this creates an expanded capsid containing pores large enough to permit exit of ssRNA. In TCV, the expanded form is not very stable, dissociating on addition of higher salt concentrations or in response to high-speed centrifugation (Fig. 7a). Such dissociation would expose the viral RNA to host cell defense mechanisms such as RNA silencing and so would presumably be strongly selected against in vivo. As described above, the structure of expanded TBSV shows that additional protein–protein contacts stabilize the expanded state. We assume that similar but nonidentical contacts are made in expanded TCV, implying that a metastable expanded state is preprogrammed into the viral lifecycle.
It is from this expanded structure that the viral genome must be brought into contact with host factors. The reduced ordering of the internal TCV density on expansion suggests that the RNA and CP N-terminal regions are more dynamic following expansion, thereby facilitating such interactions, although in our experiments the RNA remained resistant to nuclease (Fig. 6). In expanded TBSV, it appears that the N-terminal regions of B subunits become extruded to the exterior of the particle, although there was no density for this portion of the CP in the electron density map. Since the viral RNA in the expanded TCV particle remains inaccessible to nuclease, the implication is that some CP N-terminal regions have dissociated from the RNA and become externalized. Such a process would presumably be assisted by both expansion and ingress of cellular counterions.
In contrast to the apparent resistance of the viral RNA in the expanded virion to nuclease, the formation of striposomes in the presence of wheat germ extract is suggestive of the fact that one end of the viral RNA can exit the expanded particle and is able to contact a ribosome. Since the striposomes seen resemble polysomes with up to five ribosomes on an RNA, the implication is that the 5′ end exits the capsid first, although we have been unable to detect translation products using biotinylated amino acids. A high-affinity ribosome binding site at the 3′ end of TCV RNA has also been described,30 but if ribosomes bind functionally, the free energy of translation could be used to further extrude the RNA from its container, coating it with ribosomes to prevent silencing. The protein capsid would thus retain its protective role right up until the genome is fully extruded and bound to the translational apparatus of the host cell.
The asymmetric VLPs seen following proteolysis of the expanded form of TCV are obvious potential intermediates in such a reaction sequence and provide the first structural glimpse of how genome uncoating proceeds for this class of virus (Fig. 8). The partial extrusion of the genomic RNA from these particles suggests that charge repulsion is able to promote the earliest stage of uncoating. The loss of positively charged N-terminal regions via proteolysis would increase this effect. This, combined with the exquisite proteolytic sensitivity of the CP, strongly suggests that proteolysis is a prerequisite for this step. The putative capsid protein(s) attached to the end of the extruded RNA, visible in a number of the VLPs, may mediate initial ribosomal contacts. As the negative stain EM suggests that RNA is extruded from a unique site on the capsid, it is plausible that this protein moiety may be p80.
Fig. 8.
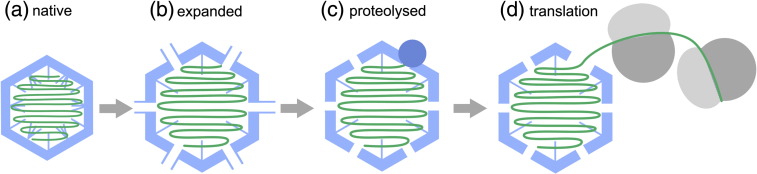
A model for uncoating the TCV genome. The native TCV virion (a) undergoes an expansion linked to release of Ca2+ and electrostatic repulsion (b). The expanded VLP is porous and exposes sites in the N-terminal region that are sensitive to proteolysis. Proteolytic cleavage at these sites leads to a loss of CP subunits (c), introducing asymmetry into the VLP and externalization of genomic RNA. This then allows the translational machinery of the plant cell to access the genomic RNA, resulting in the formation of virus-linked polysomes or “striposomes.”
The structural similarity of many plant viral CPs and the common occurrence of expanded forms suggest that many viruses use similar uncoating mechanisms. RNA extrusion has also been postulated to be the uncoating mechanism for picornaviridae but in those cases occurs at the external cellular receptor or within the endosome.31,32 Uncoating in the RNA phage MS2 also occurs adjacent to a pore at a 5-fold vertex identified as the location of the single copy of maturation protein per capsid.20,21 In MS2, there is extensive evidence for the roles played by the viral RNA in assembling an uncoating-competent capsid,33,34 starting with a sequence-specific RNA–protein assembly initiation complex, similar to the one proposed for TCV.9,35,36
Our results provide novel insights into the functioning of a complex RNA–protein machine whose dynamic behavior is tailored to release its precious genomic cargo only at the appropriate point of the infection cycle.
Materials and Methods
EM data collection and image preprocessing of native TCV
TCV was purified from Nicotina benthamiana leaves, infected with 35S-TCV (derived from TCV-M cDNA) in an Agrobacterium tumifaciens vector, as described previously.3,37,38 Three microliters of 2 mg mL− 1 native TCV in 10 mM sodium phosphate, pH 7.4, and 10 mM magnesium sulfate was applied to 400 mesh copper grids overlaid with holey carbon film (Quantifoil R1.2/1.3; Quantifoil Microtools GmbH), blotted for 1.6 s and flash-frozen in liquid ethane using a pneumatic plunging device.39 Grids were stored in liquid nitrogen. Samples were imaged using an FEI Tecnai-F20 electron microscope at 200 kV, with a nominal magnification of 50,000× (calibrated at 52,911× against an external standard). The dose used for each exposure was between 10 and 20 electrons/Å2. Micrograph defocus ranged from − 1.5 to − 3 μm. Images were recorded on a Kodak SO-163 photographic film. Film was digitized using a Zeiss SCAI microdensitometer at 7 μm/pixel, resulting in a final object sampling of 1.323 Å/pixel.
The contrast transfer function was calculated for each micrograph using the program CTFFIND3.40 Particles were interactively selected using the program BOXER.41 A total number of 18,681 molecular views of native TCV were selected. Correction for the contrast transfer function was applied in SPIDER by computational flipping of image phases.42 A band-pass filter was applied with a lower limit of 550 Å and upper limit of 6 Å. All subsequent image-processing steps were performed in SPIDER. All images were normalized to a constant mean and standard deviation and then centered by iterative translational alignment.
For negative stain EM, the sample was applied to a carbon-coated grid (Agar Scientific), which had been treated with UV light for 15 min, and removed by blotting after 30 s, and 1% (w/v) uranyl acetate was applied and removed by blotting after 10 s.
Single particle reconstruction of native TCV
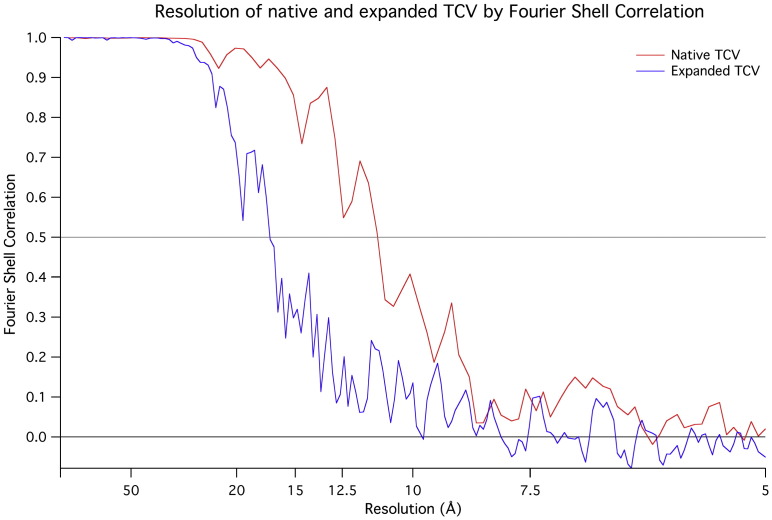
All images were rotationally and translationally aligned to a single reference image (the rotationally averaged total sum of the data set) and subsequently divided into 100 classes on the basis of multivariate statistical analysis using IMAGIC.43 A starting model was obtained by selecting a class representing a 5-fold view and using back-projection with icosahedral symmetry imposed. For each iteration, the reconstruction was reprojected over a range of angles (53–331 angles, representing an angular spacing of 3–1.5°) to obtain reference images that evenly covered the icosahedral asymmetric unit. All image data were then aligned to these references. The images corresponding to each view were ranked according to their cross-correlation coefficient (CCC), and the best-aligning members for each class were averaged, such that the most populated class never contained more than three times the number of images of the least populated class. The class averages were back-projected to obtain a new reconstruction, which was then used to generate a new series of reference images. The resolution of the final reconstruction was determined using the criterion of 0.5 Fourier shell correlation44 (Supplementary Fig. 4).
Expanded and recontracted TCV capsids
Expanded TCV capsids were obtained by incubating native TCV in a 0.1-M Tris buffer, pH 8.0, containing 5 mM EDTA and, except where stated, a protease inhibitor cocktail (Roche Complete). Sample preparation and microscopy were performed as for the native virus. Micrographs recorded (at a calibrated magnification of 52,911×) were scanned on an Imacon FlexTight 848 scanner at 10 μm/pixel, corresponding to an object sampling of 1.89 Å/pixel. Image preprocessing and single-particle reconstruction were done as for the native virus. Micrograph defocus ranged from − 1.7 to − 4 μm. The total data set was 5121 particles, of which 2610 were used in the final reconstruction. For the recontracted TCV particle reconstruction, images were collected at a calibrated magnification of 69,020 on a Gatan US4000 SP CCD camera, giving a final object sampling of 2.17 Å/pixel. We used 3885 particles.
Atomic structure fitting
Coordinates for the C subunit conformer of TCV CP [Protein Data Bank (PDB) code 3ZXA], as separate S and P domains, were obtained from Prof. S. C. Harrison (Harvard Medical School). To generate the complete capsid, we aligned individually the two separate domains of the TCV CP to the TBSV crystal structure (PDB accession code 2TBV45) to create the CP trimer that is the repeating unit of the capsid. The N-terminal fragment (residues 53–80) of the C-type subunit was then removed as this region is disordered in A and B subunits. The P domains for the A- and B-type subunits of the CP trimer were then further manually rotated and translated, as rigid bodies, to fit the native TCV cryo-EM density better. This is consistent with the observation that the hinge angle between the S and P domains is different for TBSV and TCV.3 The resulting asymmetric unit was then rigid body refined against the native TCV cryo-EM data with icosahedral symmetry applied, using CNS.46 This resulted in an excellent fit to the cryo-EM electron density, reflected in the improvement in CCC47 from 0.20 to 0.33, between the model and the data after modification of the S–P hinge angle and rigid-body refinement. The magnification of the map was confirmed by correlation against the fitted coordinates. The same approach was taken to fit the TCV atomic coordinates into the cryo-EM density for the expanded capsid. In this case, the density was best fitted by an expanded asymmetric unit trimer in which the individual subunits were moved apart. As for the native TCV structure, the CCC of the model and the data were improved (to 0.43) by rigid-body refinement using CNS. The relative orientations of the S and P domains of the individual subunits did not change.
Protease and nuclease accessibility
TCV was incubated under different conditions to obtain (a) native particles (10 mM MgSO4 and 10 mM Na phosphate, pH 7.4), (b) expanded particles (5 mM EDTA and 25 mM Tris, pH 8.5), (c) dissociated virus [5 mM EDTA, 100 mM Tris (pH 8.5) and 1 M NaCl] and (d) recontracted capsids prepared as in (b) followed by the addition of CaCl2 and Hepes at pH 7.5 to final concentrations of 10 mM and 100 mM, respectively.
For protease digestion, a reaction containing 1:100 w/w chymotrypsin:TCV was incubated at room temperature, and aliquots were taken after 0, 2, 5, 10, 30 and 60 min. The reactions for SDS-PAGE were stopped by boiling the sample in 4 volumes SDS loading buffer for 2 min and analyzed on a 10% w/v gel and stained with Coomassie blue. Proteolysis reactions for gel-filtration analysis were stopped by adding Roche Complete protease inhibitor (1/10th volume of a solution of 1 tablet/mL), 160 μg Pepstatin A in dimethyl sulfoxide and 2 mg Aprotinin per milliliter of reaction volume. Particles were separated on a 10/300 GL Superdex S200 column using an AKTA Explorer at a flow rate of 0.5 mL/min. Fractions (1 mL) were collected and concentrated using an Amicon Ultra (3 kDa molecular mass cutoff, Millipore) before analysis on SDS-PAGE.
For RNase digestion, 0.1 ng of ribonuclease A (Ambion, AM2274) was added to separate aliquots of TCV treated as for the protease digestion, as well as protein-free TCV genomic RNA in expansion buffer. The samples were phenol extracted and ethanol precipitated following standard protocols, and the RNA was recovered by centrifugation at 17,000g at 4 °C. Pellets were washed with 50 μL of 75% (v/v) ethanol and repelleted. The RNA was then resuspended in nuclease-free water (Fermentas), and aliquots were incubated at 65 °C with an equal volume of formaldehyde loading buffer (Ambion) containing 0.2 μg/μL ethidium bromide (Sigma) to denature the RNA before analysis on a denaturing 1.5% w/v agarose gel.
Mass spectrometry
Native, expanded, dissociated or recontracted TCV was prepared as above and incubated for 40 min with chymotrypsin. The mixtures were then desalted using C4 ziptips (Millipore) according to the manufacturer's protocol and eluted in 60% v/v acetonitrile and 3% v/v formic acid. Electro-spray ionization mass spectra were recorded using a Q-TOF mass spectrometer (Waters). Two major proteolysis products were observed at 30,802 Da and 29,647 Da, as well as a number of smaller peptides. Three of these smaller peptides were directly sequenced by tandem mass spectrometry (see Supplementary Fig. 4).
Supplementary Figure 4.
Fourier shell correlation (FSC) curves for the TCV reconstructions. For the resolutions quoted, the resolution where FSC = 0.5 is used. The red curve is for the native virus; the expanded virus is shown in blue.
N-terminal sequencing
Chymotrypsin-treated native, expanded, dissociated or recontracted TCV was prepared as above. Samples were fractionated on an SDS-PAGE gel before blotting onto a polyvinylidene fluoride membrane. The blot was stained with Coomassie blue, and the major proteolysis bands at ∼ 31 and 29.5 kDa were subjected to N-terminal sequencing by Edman degradation (see Supplementary Fig. 4).
Sequencing of the CP gene
A PCR product encompassing the CP gene was prepared by reverse transcription of TCV genomic RNA phenol extracted from native virus, using a Transcriptor High Fidelity cDNA Synthesis Sample Kit (Roche), and the reverse primer 5′-CCCTAACACAGGTCAAAATAAAGCG-3′. PCR was carried out using a KAPA2G Robust PCR Kit (Kapa Biosystems, Boston, USA) according to the manufacturer's instructions and primer 5′-CCTGAAATCAAACCGATTCACACATCC-3′. The PCR product was purified on a 1.5% w/v agarose gel in TBE running buffer (89 mM Tris–borate and 2 mM EDTA) and recovered using a QIAgen Gel Extraction Kit (QIAGEN, Washington, USA). PCR products were submitted to GATC Biotech for automated sequencing using the primers listed above, as well as two further internal primers, MF (5′-CGAGATGCAGCCAAACCTCC-3′) and MR (5′-GCTGATACCATCCGCCACAAAGC-3′), selected to ensure coverage of the entire gene.
Striposomes
Expanded TCV was added to a wheat germ extract (Promega). An excess of Mg2+ was added to the reaction after mixing in the EDTA-containing expanded TCV. After incubation at 25 °C for 30 min, cycloheximide (Sigma) was added to a final concentration of 1 mg/mL to stop translation. The sample was then centrifuged at 4 °C for 10 min before application to a UV-treated EM grid and staining with 1% w/v uranyl acetate. Samples were imaged in a F20 microscope at 50,000× nominal magnification.
Analytical ultracentrifugation
Samples (0.32 mL) were placed in 1.2-cm-path-length 2-sector meniscus-matching epon centerpiece cells built with sapphire windows. These were then centrifuged at 10,000 rpm and 20.0 °C in an An50Ti analytical rotor in an Optima XL-I analytical ultracentrifuge (Beckman Instruments, Inc., Palo Alto, CA 94304). Changes in solute concentration were detected by interference and absorbance scans at 270 nm. Buffer densities and viscosities were calculated using the program Sednterp version 1.09 (2006, Dr. Thomas Laue, Department of Biochemistry, University of New Hampshire, Durham, NH 03824). Radial absorbance plots were used for fitting to sedimentation profiles with the program Sedfit, version 12.1b (2010), using a continuous distribution c(s) Lamm equation model.48
Accession codes
The updated TCV nucleotide sequence has been deposited in GenBank with the accession code HQ589261. The cryo-EM density maps for native and expanded TCVs have been deposited in Electron Microscopy Data Bank with accession codes 1863 and 1864, respectively. The fitted atomic coordinates for native and expanded TCVs have been deposited in the PDB with accession codes 3ZX8 and 3ZX9, respectively.
The following are the supplementary materials related to this article
Supplementary materials related to this article can be found online at doi:10.1016/j.jmb.2012.01.017
Acknowledgements
We thank Drs. Jeff Keen, James Ault and Gabby Basnak for N-terminal peptide sequencing, mass spectrometry and assistance with TCV purification, respectively; Dr George Lomonossoff (John Innes Centre, Norwich, UK) for providing the TCV clone and both G.L. and Prof. Reidun Twarock (University of York) for their helpful comments on the manuscript; Prof. Stephen C. Harrison (Harvard Medical School) for the coordinates of the TCV crystal structure and his invaluable insights on the TBSV/TCV system; and Dr. S. Tsokov (University of Sheffield, UK) for use of the Zeiss SCAI microdensitometer. S.E.B. was supported by the Wellcome Trust. P.G.S., A.R.P. and N.A.R. thank the Research Councils UK, Biotechnology and Biological Sciences Research Council and Leverhulme Trust for support of various aspects of this research.
Contributor Information
Arwen R. Pearson, Email: a.r.pearson@leeds.ac.uk.
Neil A. Ranson, Email: n.a.ranson@leeds.ac.uk.
Peter G. Stockley, Email: p.g.stockley@leeds.ac.uk.
References
- 1.Lomonossoff G.P. Pathogen-derived resistance to plant viruses. Annu. Rev. Phytopathol. 1995;33:323–343. doi: 10.1146/annurev.py.33.090195.001543. [DOI] [PubMed] [Google Scholar]
- 2.Harvey J.J., Lewsey M.G., Patel K., Westwood J., Heimstädt S., Carr J.P., Baulcombe D.C. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogle J.M., Maeda A., Harrison S.C. Structure and assembly of turnip crinkle virus. 1. X-ray crystallographic structure-analysis at 3.2 Å resolution. J. Mol. Biol. 1986;191:625–638. doi: 10.1016/0022-2836(86)90450-x. [DOI] [PubMed] [Google Scholar]
- 4.Harrison S.C., Olson A.J., Schutt C.E., Winkler F.K., Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 5.Carrington J.C., Morris T.J., Stockley P.G., Harrison S.C. Structure and assembly of turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J. Mol. Biol. 1987;194:265–276. doi: 10.1016/0022-2836(87)90374-3. [DOI] [PubMed] [Google Scholar]
- 6.Cao M., Ye X., Willie K., Lin J., Zhang X., Redinbaugh M.G. The capsid protein of turnip crinkle virus overcomes two separate defense barriers to facilitate systemic movement of the virus in Arabidopsis. J. Virol. 2010;84:7793–7802. doi: 10.1128/JVI.02643-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspar D.L.D., Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 8.Golden J.S., Harrison S.C. Proteolytic dissection of turnip crinkle virus subunit in solution. Biochemistry. 1982;21:3862–3866. doi: 10.1021/bi00259a022. [DOI] [PubMed] [Google Scholar]
- 9.Sorger P.K., Stockley P.G., Harrison S.C. Structure and assembly of turnip crinkle virus: II. Mechanism of reassembly in vitro. J. Mol. Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 10.Stockley P.G., Kirsch A.L., Chow E.P., Smart J.E., Harrison S.C. Structure of turnip crinkle virus: III. Identification of a unique coat protein dimer. J. Mol. Biol. 1986;191:721–725. doi: 10.1016/0022-2836(86)90456-0. [DOI] [PubMed] [Google Scholar]
- 11.Qu F., Morris T.J. Encapsidation of turnip crinkle virus is defined by a specfic packaging signal and RNA size. J. Virol. 1997;71:1428–1435. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao A.L. Genome packaging for spherical plant RNA viruses. Annu. Rev. Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 13.Modis Y., Trus B.L., Harrison S.C. Atomic model of the papillomavirus. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson I.K., Harrison S.C. Structure of the expanded state of tomato bushy stunt virus. Nature. 1982;297:563–568. [Google Scholar]
- 15.Aramayo R., Merigoux C., Larquet E., Bron P., Perez J., Dumas C. Divalent ion-dependent swelling of tomato bushy stunt virus: a multi-approach study. Biochim. Biophys. Acta. 2005;1724:345–354. doi: 10.1016/j.bbagen.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Speir J.A., Munshi S., Wang G., Baker T.S., Johnson J.E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opalka N., Tihova M., Brugidou C., Kumar A., Beachy R.N., Fauquet C.M., Yeager M. Structure of native and expanded sobemoviruses by electron cryo-microscopy and image reconstruction. J. Mol. Biol. 2000;303:197–211. doi: 10.1006/jmbi.2000.4043. [DOI] [PubMed] [Google Scholar]
- 18.Katpally U., Kakani K., Reade R., Dryden K., Rochon D., Smith T.J. Structures of T = 1 and T = 3 particles of cucumber necrosis virus: evidence of internal scaffolding. J. Mol. Biol. 2007;365:502–512. doi: 10.1016/j.jmb.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 19.Toropova K., Basnak G., Twarock R., Stockley P.G., Ranson N.A. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 20.Toropova K., Stockley P.G., Ranson N.A. Visualising a viral RNA genome poised for release from its receptor complex. J. Mol. Biol. 2011;408:408–419. doi: 10.1016/j.jmb.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 21.Dykeman E.C., Grayson N.E., Toropova K., Ranson N.A., Stockley P.G., Twarock R. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J. Mol. Biol. 2011;408:399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 22.van den Worm S.H., Koning R.I., Warmenhoven H.J., Koerten H.K., van Duin J. Cryo electron microscopy reconstructions of the Leviviridae unveil the densest icosahedral RNA packing possible. J. Mol. Biol. 2006;363:858–865. doi: 10.1016/j.jmb.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 23.Koning R., van den Worm S., Plaisier J.R., van Duin J., Pieter Abrahams J., Koerten H. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J. Mol. Biol. 2003;332:415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- 24.Brisco M., Hull R., Wilson T.M.A. Swelling of isometric and of bacilliform plant-virus nucleocapsids is required for virus-specific protein-synthesis in vitro. Virology. 1986;148:210–217. doi: 10.1016/0042-6822(86)90416-2. [DOI] [PubMed] [Google Scholar]
- 25.Munowitz M.G., Dobson C.M., Griffin R.G., Harrison S.C. On the rigidity of RNA in tomato bushy stunt virus. J. Mol. Biol. 1980;141:327–333. doi: 10.1016/0022-2836(80)90185-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu H., Qu C., Johnson J.E., Case D.A. Pseudo-atomic models of swollen CCMV from cryo-electron microscopy data. J. Struct. Biol. 2003;142:356–363. doi: 10.1016/s1047-8477(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J., Rueckert R. Packaging and release of the viral genome. In: Burnett R.M., Garcea R., Chiu W., editors. Structural Biology of Viruses. Oxford University Press; Oxford, UK: 1997. pp. 269–287. [Google Scholar]
- 28.Smith D.E., Tans S.J., Smith S.B., Grimes S., Anderson D.L., Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 29.Sun S., Konabagil K., Draper B., Alam T.I., Bowman V.D., Zhang Z. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Yuan X., Shi K., Young M.Y., Simon A.E. The terminal loop of a 3′ proximal hairpin plays a critical role in replication and the structure of the 3′ region of turnip crinkle virus. Virology. 2010;402:271–280. doi: 10.1016/j.virol.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bostina M., Levy H., Filman D.J., Hogle J.M. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol. 2011;85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuthill T.J., Bubeck D., Rowlands D.J., Hogle J.M. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 2006;80:172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolfsson O., Toropova K., Ranson N.A., Stockley P.G. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J. Mol. Biol. 2010;401:309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morton V.L., Dykeman E.C., Stonehouse N.J., Ashcroft A.E., Twarock R., Stockley P.G. The impact of viral RNA on assembly pathway selection. J. Mol. Biol. 2010;401:298–308. doi: 10.1016/j.jmb.2010.05.059. [DOI] [PubMed] [Google Scholar]
- 35.Stockley P.G., Rolfsson O., Thompson G.S., Basnak G., Francese S., Stonehouse N.J. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basnak G., Morton V.L., Rolfsson O., Stonehouse N.J., Ashcroft A.E., Stockley P.G. Viral genomic single-stranded RNA directs the pathway toward a T = 3 capsid. J. Mol. Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas C.L., Leh V., Lederer C., Maule A.J. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotina benthamiana. Virology. 2003;1:33–41. doi: 10.1016/s0042-6822(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 38.Oh J.W., Kong Q., Song C., Carpenter C.D., Simon A.E. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant. Interact. 1995;8:979–987. doi: 10.1094/mpmi-8-0979. [DOI] [PubMed] [Google Scholar]
- 39.Walker M., Trinick J., White H. Millisecond time resolution electron cryo-microscopy of the M-ATP transient kinetic state of the acto-myosin ATPase. Biophys. J. 1995;68:875–915. [PMC free article] [PubMed] [Google Scholar]
- 40.Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 42.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Van Heel M., Harauz G., Orlova E.V., Schmidt R., Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 44.van Heel M., Schatz M. Fourier shell correlation threshold criteria. J. Struct. Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Hopper P., Harrison S.C., Sauer R.T. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J. Mol. Biol. 1984;177:701–713. doi: 10.1016/0022-2836(84)90045-7. [DOI] [PubMed] [Google Scholar]
- 46.Brünger A.T., Adams P.D., Clore G.M., Gros P., Grosse-Kunstleve R.W., Jiang J.S. Crystallography and NMR system (CNS), a new software suite for macromolecular software determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 47.Frank . Three Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press; Oxford, UK: 2006. pp. 95–96. [Google Scholar]
- 48.Schuck Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modelling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]