Abstract
The RpoS/σS sigma subunit of RNA polymerase (RNAP) activates transcription of stationary phase genes in many Gram-negative bacteria and controls adaptive functions, including stress resistance, biofilm formation and virulence. In this study, we address an important but poorly understood aspect of σS-dependent control, that of a repressor. Negative regulation by σS has been proposed to result largely from competition between σS and other σ factors for binding to a limited amount of core RNAP (E). To assess whether σS binding to E alone results in significant downregulation of gene expression by other σ factors, we characterized an rpoS mutant of Salmonella enterica serovar Typhimurium producing a σS protein proficient for EσS complex formation but deficient in promoter DNA binding. Genome expression profiling and physiological assays revealed that this mutant was defective for negative regulation, indicating that gene repression by σS requires its binding to DNA. Although the mechanisms of repression by σS are likely specific to individual genes and environmental conditions, the study of transcription downregulation of the succinate dehydrogenase operon suggests that σ competition at the promoter DNA level plays an important role in gene repression by EσS.
INTRODUCTION
In eubacteria, the dissociable σ subunit of RNA polymerase (RNAP) enables specific binding of RNAP to gene promoters and is required for transcription initiation. Besides a primary housekeeping σ, which promotes transcription of genes required for essential functions, one or more alternative σ factors direct transcription of specific subsets of genes (1–3). The alternative sigma σS/38 (RpoS) is a central regulator allowing many Gram-negative bacteria to adapt to stress conditions and specialized environments (4–7). In the wide host-range pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium), σS is not only required for general stress resistance but also for virulence, biofilm formation and development of the red dry and rough (rdar) morphotype, a colony morphology caused by production of amyloid fibers (curli) and cellulose (5,8,9). In contrast to the housekeeping sigma, σ70 (RpoD), σS is almost undetectable in early exponential phase and is induced in stationary phase or in response to various stresses (4–7). S. Typhimurium contains four other alternative σ, σE/24 (rpoE), σH/32 (rpoH), σN/54 (rpoN) and σ28 (fliA) which associate with the core RNAP (E) to form the holoenzyme Eσ (1–3). The cellular concentration of σ70 molecules exceeds that of E, suggesting that σ factors compete for binding to a limiting number of E (2–5). Levels and affinity for E are thus major determinants of σ competitiveness and its ability to form Eσ. However, in vivo, the efficiency of formation of the housekeeping and alternative Eσ is also modulated by regulatory factors that bind E and/or σ (2–5).
In this study, we have addressed an important aspect of σS-dependent control, that of a repressor. σS has a negative effect on the expression of a large number of genes, including genes involved in metabolism and membrane permeability (5,6,10–13). Consequently, the acquisition of stress resistance mediated by σS comes at the expense of growth capabilities, causing a trade-off between self-preservation and nutritional competence, the so-called SPANC balance (11,14). This trade-off favors the appearance of non-functional rpoS alleles in environments with no stress, where reduced σS activity confers a growth advantage (5,11,15–17). The negative control of bacterial growth by σS and its contribution to population polymorphisms is a subject of growing interest with relevance in molecular ecology and evolution. However, gene repression by σS is not mechanistically well documented and is thought to result primarily from σS competing with other σ for E binding (18,19).
Previous studies, using artificial manipulation of σ factor levels and the use of σ mutants with reduced affinity for E, have shown that σ competition for binding to E modulates the activity of promoters controlled by σS, σ32 and σ54 (20,21) and may explain how a σ negatively regulates transcription by other σ (2,18–21). For example, mutations in rpoD reducing the σ70 affinity for E cause an upregulation of genes controlled by alternative σ, presumably increasing the pool of available E (2,20,21). Conversely, overexpression of σS lowers the expression of genes controlled by σ70, presumably reducing the Eσ70 amounts, and affecting promoters that are sensitive to the cellular concentration of Eσ70 (18,19). Although these findings are compatible with a model in which σS competition with other σ for E binding alone accounts for negative regulation of gene transcription by σS (Figure 1A), they could be explained by alternative regulatory mechanisms (Figure 1B and C). Negative regulation by σS might be mediated by negative effectors whose expression is activated by σS (Figure 1B) (5,6,12,13). Indeed, σS controls the expression of sRNAs (12,22–24) and several regulatory proteins and metabolic/signaling enzymes (5–7,10,12,25,26) that might endow σS with repressor function. RNAP itself is also a DNA binding protein and EσS might theoretically function as a transcriptional repressor through promoter occlusion or transcriptional interference (Figure 1C) (27–35). Stable but transcriptionally inactive RNAP bound to DNA might inhibit transcription by another holoenzyme (27,28,32). Tandem or overlapping promoters and pausing of RNAP outside of the promoter context might also sterically hinder DNA binding by an alternative RNAP and/or transcription factors, producing transcriptional interference and repression (30,31,33–35) (Figure 1C). Few examples of direct negative effect of σ have been reported so far (27–29,35) but the extensive overlap between σ factor binding sites recently reported in E. coli (36,37) is compatible with antagonisms between σ factors at the promoter DNA level. In particular, during the completion of this study, Cho et al. (37) reported that EσS and Eσ70 binding regions in the E. coli genome extensively overlap, and many σS binding promoters showed increased transcription and increased σ70 binding in a ΔrpoS strain. Interestingly, these data suggested that direct interference between EσS and Eσ70 might occur at the DNA level. However, it was not shown whether the decreased σ70 binding and transcription, observed in the wild-type strain compared to the ΔrpoS mutant, was a direct consequence of σS binding in the same promoter region (according to model C; Figure 1) or an indirect effect, due to σ competition for E (according to model A; Figure 1) and/or the synthesis of a negative effector under the control of σS (according to model B; Figure 1).
Figure 1.
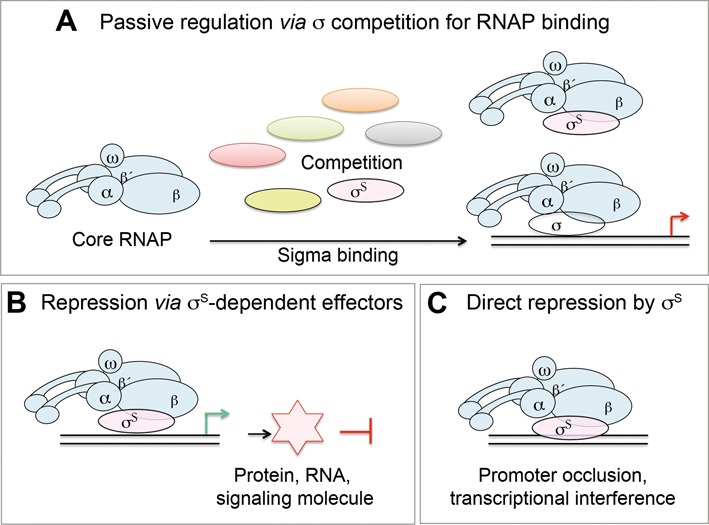
Schematic illustration of the main possible regulatory mechanisms of gene repression by σS. Graphical representation of the core RNA polymerase complex (in blue), σS (in light pink) and the other sigma subunits (different colors) is shown. Each box represents a possible σS repression mechanism. (A) Repression model based on competition of σ factors for RNAP core binding. (B) Indirect repression model via diverse effector molecules. (C) Direct repression model where EσS binding prevents the transcription by other Eσ holoenzymes. Models B and C require the DNA binding activity of σS. Note that combinations of different mechanisms are possible.
To shed light on the molecular mechanisms of negative regulation by σS, we first asked whether σS binding to E alone results in significant downregulation of gene expression by other σ. Since the competition model (Figure 1A) solely requires σS binding to E whereas alternative mechanisms (Figure 1B and C) also require the ability of σS to bind DNA, we characterized a σS variant proficient for EσS formation but impaired in binding to promoter DNA. Our data suggest that, under physiological conditions in stationary phase, σ competition for E binding alone does not result in detectable gene repression by σS, which instead relies on the ability of σS to bind DNA. Based on this finding and further analysis of the downregulation by σS of the sdh operon encoding succinate dehydrogenase, we propose that gene repression by σS relies on negative effects of EσS at the promoter DNA level and presumably on the action of EσS-dependent negative effectors.
MATERIALS AND METHODS
Bacterial strains, bacteriophage, plasmids and growth conditions
Strains and plasmids are listed in Supplementary Table S1. Bacteriophage P22HT105/1int was used to transfer mutations and lacZ fusions between Salmonella strains by transduction (38). Green plates, for screening for P22-infected cells or lysogens, were prepared as described previously (39). Bacteria were routinely grown in Luria-Bertani medium (LB) (40) at 37°C under aeration. To assess Salmonella growth at the expense of succinate and glucose (20 mM) as carbon sources, M9 minimal medium (40) was used. Stationary phase cultures were washed, resuspended in phosphate-buffered saline (40) to OD600 of 1.0 and 0.05, and 5 μl of each dilution was spotted onto plates that were incubated at 37°C for 48 h. Rdar morphotypes were analyzed on CR plates (LB agar without NaCl supplemented with Congo red 40 μg/ml and Coomassie brilliant blue R250 20 μg/ml), at 28°C as described (8). Antibiotics were used at the following concentrations (in μg per ml): carbenicillin (Cb), 100; chloramphenicol (Cm), 15 for the chromosomal resistance gene and 30 for the plasmid resistance gene; kanamycin, (Km) 50; and tetracycline (Tet) 20.
DNA manipulations, inactivation of chromosomal genes and construction of chromosomal lacZ fusions
Standard molecular biology techniques were used (8,40). Oligonucleotides were obtained from Sigma-Aldrich and are listed in Supplementary Table S2. DNA sequencing was performed by Beckman Coulter Genomics. Plasmids pVF9551, pVF9647 were obtained by site-directed mutagenesis of plasmid pUCC52-2922K using the QuikChange II Site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. pVF9793 was obtained by site-directed mutagenesis of pVF9551. Plasmids pQE30rpoSR141S, pQE30rpoSA157T and pQE30rpoSdb (Supplementary Table S1) were obtained by cloning polymerase chain reaction (PCR) amplified DNA fragments from strains VF9682 VF9676 and VF9849, respectively, between the BamHI and HindIII sites of pQE30 using primers HK1 and HK2 as described previously for the wild-type rpoS gene in pQE30rpoS (17). All plasmids were verified by DNA sequencing. Chromosomal deletions in Salmonella ATCC14028 were generated using PCR-generated linear DNA fragments (Supplementary Table S2) and the λ-Red recombination method as described by Datsenko and Wanner (41). Because the ΔrssB mutants were sick and might accumulate compensatory mutations, they were constructed freshly for each experiment by transduction using a P22 lysate prepared on strain VF8293. When required, the resistance cassette was eliminated using a temperature-sensitive helper plasmid pCP20, which encodes the FLP recombinase (41). Point mutations in the sdh promoter region were introduced on the chromosome by a two-step Red-recombinase-based recombineering procedure (42). The procedure involves (i) replacement of the wild-type sequence by a tetAR module produced by PCR (12) and (ii) replacement of the tetRA module by a DNA fragment obtained by PCR and carrying the desired mutations (Supplementary Table S2) through positive selection of tetracycline-sensitive recombinants (43). The presence of desired mutations in all strains was confirmed by DNA sequencing. Tn5B21 insertions creating lacZ fusions in σS-dependent genes have been previously isolated (16,44). Single-copy transcriptional lacZ fusions were constructed in ompD and ynfM using conditional plasmids containing promoterless lacZY genes and the FLP recognition target site as described (45). PCR assays were used to ensure integration of the plasmids in the correct location and to exclude the presence of multiple plasmid integrants (using standard test primers, such as those described in (45)). We also used flanking locus-specific primers to amplify junction fragments that were subsequently analyzed by DNA sequencing. When required, the KmR cartridge in chromosomal lacZY gene fusions was replaced with a CmR cartridge as described previously (16).
Construction of isogenic S. Typhimurium strains carrying the ΔrpoS and rpoSdb alleles
The rpoS allele in S. Typhimurium ATCC14028 was replaced by the rpoSR141S, rpoSA157T and rpoSdb alleles using the strategy previously described for rpoS allelic exchange (16,17). Briefly, pUCC52-2922K contains the rpoS and downstream sequences including a gene encoding a putative decarboxylase (named STM2922 in S. Typhimurium strain LT2 and STM14_3524 in ATCC14028) into which a Km cartridge has been inserted. When introduced into ATCC14028 by electroporation this plasmid was unstable. Recombination of the Km cartridge into the host genome with simultaneous loss of pUCC52-2922K resulted in the isolation of KmR CbS recombinants containing the STM2922::Km mutation. This mutation was then transduced into ATCC14028 and its ΔrpoS::Cm derivative VF7928, and the resulting strains (VF7969 and VF7975, respectively) were checked by PCR and DNA sequencing. The same strategy was applied with the pUCC52-2922K derivatives containing the rpoS mutations C421A, G469A and C421A-G469A (yielding to σS substitutions R141S, A157T and R141A-A57T, respectively). These plasmids were electroporated in the ATCC14028 strain containing the rpoS420::Cm mutation (VF9579) to facilitate screening of strains into which simultaneous recombination of the STM2922::Km and rpoS point mutations occurred. CbS recombinants that were KmR but CmS were selected, and recombination of the STM2922::Km mutation and simultaneous replacement of the rpoS420::Cm mutation by the mutated rpoS alleles were confirmed by PCR and DNA sequencing. The mutated rpoS alleles and STM2922::Km were then co-transduced into a fresh ΔrpoS::Cm background (VF7928) and the resulting KmR CmS strains (VF9849, VF9676 and VF9682) were checked by DNA sequencing for the presence of the mutated rpoS alleles. The resistance cassette in VF7975 was eliminated to yield strain VF9356 using a temperature-sensitive helper plasmid pCP20, which encodes the FLP recombinase (41). The rpoSdb mutation was also introduced in ATCC14028 using the two-step Red-recombinase-based recombineering procedure (42) mentioned above (Supplementary Table S1).
Isolation of total RNA from S. Typhimurium, cDNA library preparation, sequencing and analysis of sequences
Total RNA from three biological replicates of strains VF7969, VF9356 and VF9849 was isolated from late stationary phase cultures (18 h growth in LB at 37°C) and the rRNA depleted fraction was used for construction of strand specific single end cDNA libraries as described recently (12). Libraries were sequenced using an Illumina Hiseq2000 sequencer (multiplexing 3 samples per lane). One replicate of each strain was sequenced in each lane. Analysis of sequences and statistical analyses were performed as described (12).
Quantitative real-time PCR
Total RNA was extracted from cells grown to stationary phase in LB and reverse-transcribed as described (12). Quantitative real-time PCR was performed using Applied Biosystems 7300 Real-Time PCR system and methods recently detailed (12). Three biological replicates were analyzed in duplicate each. The rpoZ gene was used as a reference (12). Gene expression levels were calculated using the comparative Ct method (2−ΔΔCT) as previously described (12,46).
Electrophoresis and immunoblot analysis of proteins
Whole-cell extracts were prepared and sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described (16,17). For detection of σS proteins during growth, exponential-phase cultures of Salmonella in LB at 37°C were diluted into LB prewarmed at 37°C to prolong the exponential phase and aliquots were removed during the exponential phase and stationary phase. The amount of protein in whole-cell lysates was determined using the DC Protein Assay kit (Bio-Rad). Equal amounts of protein were loaded in each slot. The molecular sizes of the proteins were estimated using Precision Plus Protein Standard (Bio-Rad). Proteins were transferred to nitrocellulose blotting membranes (Hybond ECL membranes, GE Healthcare) and incubated with rabbit antibody against the σS protein of S. enterica serovar Typhimurium as previously described (16,17,47). Bound antibodies were detected using a secondary anti-rabbit antibody linked to peroxidase (NA934, GE Healthcare) and the Pierce ECL2 western blotting substrate (Thermoscientific).
Fractionation of free and RNAP-bound σS from cellular extracts and immunoblot analysis
Gel filtration of cellular extracts from stationary phase cultures of strains VF7969 and VF9849 was performed as previously described (20). Crude cell extracts were obtained using a Cell Disrupter (Constant Systems, Daventry, UK) in 10-mM Tris-HCl pH 7.9, 0.1-mM DTT, 0.1-mM ethylenediaminetetraacetic acid, glycerol 5%, 300-mM NaCl supplemented with antiprotease (Roche). A total of 20 μl of the supernatant was applied to a gel filtration column (Superdex 200 PC3.2/30, GE Healthcare) using the EttanLC System (GE Healthcare). Elution was performed at a flow rate of 0.04 ml/min at room temperature, gathering fractions of 50 μl. The proteins in the elution fractions were analyzed by SDS-PAGE and electroblotted onto nitrocellulose membranes (Hybond ECL GE Healthcare). Immunoblot analysis of the eluted fractions was carried out using the σS rabbit antibody (47) as described above and monoclonal antibodies against the β′ and α subunits of RNAP (WP001 and WP003, Neoclone) and a secondary anti-mouse antibody linked to peroxidase (A4416, Sigma-Aldrich).
Preparation of outer membrane proteins
Salmonella strains were grown for 18H in LB at 37°C. Cells were centrifuged and washed with 10 mM, Tris-HCl pH 8.0 and crude cell extracts were obtained using a Cell Disrupter (Constant System, Daventry, UK). Outer membrane protein preparations were obtained as described by Lobos and Mora (48).
Overproduction and purification of His6-σS variants
His6-σS wild-type and variants were purified from JM109 carrying plasmids pQErpoS, pQErpoSR141S, pQErpoSA157T and pQErpoSdb as described (17).
KMnO4 reactivity
The assays were performed as described in (49) with the following modifications: the katN promoter fragment was incubated with 60 nM reconstituted EσS (σS: E = 10) for 1 h at 30°C before KMnO4 addition.
Band shift analysis
Escherichia coli core and S. Typhimurium His6-σS (wild-type and/or mutants) in 6-μl buffer (40-mM Hepes pH 8.0, 10-mM MgCl2, 100-mM K-glutamate, 2-mM DTT containing 500-μg/ml bovine serum albumin) were incubated for 10 min at 37°C. Three microliter of [32P]-labeled katN fragment prepared as in (49) was then added and incubated for 20 min at 37°C. After addition of 3 μl loading buffer (buffer A containing 50% sucrose, 0.025% xylene cyanol blue and 150 μg/ml of heparin) the mixture was loaded onto a 5.5% native polyacrylamide gel run in TG buffer (25-mM Tris, 192-mM Glycine pH 8.5) at 10 V/cm.
Enzymatic assays
β-galactosidase activity was measured as described by Miller (50) and is expressed in Miller units.
RESULTS AND DISCUSSION
Construction of rpoS mutants affected in σS promoter DNA binding
To assess the molecular basis of σS mediated gene repression, we constructed an rpoS mutant specifically deficient in promoter DNA binding. Sigma factors from the σ70 family, including σS, are composed of globular domains divided into functional regions (Figure 2A). They direct the RNAP holoenzyme to promoter elements -10 and -35, recognized by domains σ2 and σ4, respectively (1–3,51,52). σ2 regions 2.3–2.4 are critical for recognition and melting of the -10 element, the most highly conserved and essential promoter motif (1–3,51,52). In contrast, most σS-dependent promoters display a poorly conserved -35 element and it is not clear how EσS uses the -35 element (7,53).
Figure 2.
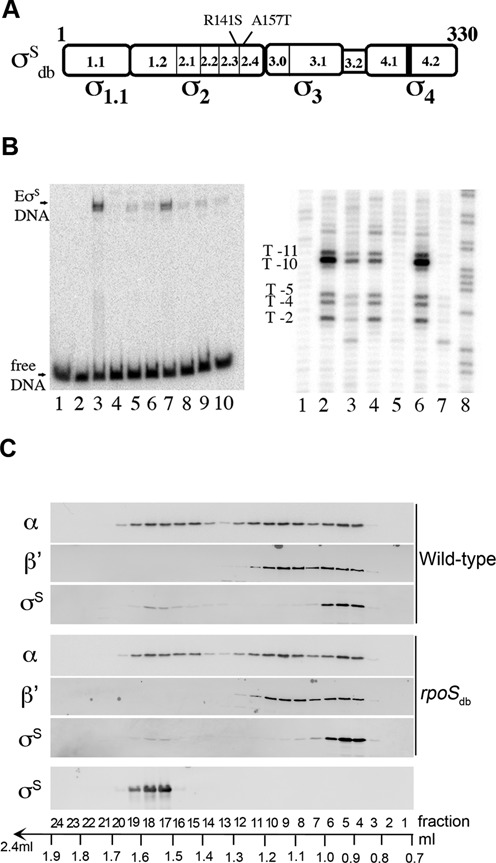
σSdb is impaired in DNA promoter binding but not in RNAP core binding. (A) Schematic representation of the four regions of the σS protein (17,51) and location of the amino-acid substitutions in σSdb in regions involved in binding and melting the -10 promoter element (54,55). (B) The σS variants containing one or both substitutions are deficient in DNA promoter binding. When bound to core RNA polymerase (E), σS makes sequence-specific promoter contacts and plays a crucial role in DNA melting (54–56). Left panel: electrophoretic mobility shift assay indicating binding of EσS, but not EσSdb, to the katN promoter region. The [32P]-labeled katN promoter fragment was incubated for 20 min at 37°C with buffer (lane 1), E (lane 2) or holoenzymes reconstituted with His6-σS wild-type (lanes 3 and 7), His6-σSR141S (lanes 4 and 8), His6-σSA157T (lanes 5 and 9) and His6-σSdb (lanes 6 and 10) before heparin challenge. The E:σS ratios were 1:33 (lanes 3–6) and 1:4 (lanes 7–10). Right panel: KMnO4 probing of the EσS holoenzymes (E:σS of 1:10) on the 5′-[32P]-labeled template strand katN fragment. KMnO4 preferentially oxidizes exposed unstacked thymines of RNAP–promoter complexes and gives rise to marked KMnO4 reactivity at the katN promoter as previously reported (49). The positions of the reactive Ts with respect to the transcription start are indicated (T-11, T-10, T-5, T-4 and T-2). Lane 1: control with no protein; lanes 2 and 6: E His6-σS wild-type; lane 3: E His6-σSR141S; lane 4: E His6-σSA157T; lane 5: E His6-σSdb; lane 7: E only. Lane 8 is a G+A reaction performed on the same DNA fragment according to Maxam and Gilbert (58). (C) Distribution of σS between free and holoenzyme forms in the wild-type and rpoSdb strains. Whole cell lysates from wild-type strain VF7969 and rpoSdb mutant VF9849 were fractionated by size exclusion chromatography and the relative concentration of RNAP subunits was subsequently analyzed in the fractions by immunoblot using monoclonal antibodies against the β’ and α subunits of RNA polymerase and a polyclonal anti-σS antibody. Purified σS was used to pinpoint fractions containing free σS. The percentage of total σS in fractions corresponding to free σS was very low (and appeared slightly lower in rpoSdb than in wild-type) suggesting that most σS molecules are associated with RNAP in stationary phase. Two independent experiments were performed with similar results (the elution profiles were similar for the wild-type strain and the rpoSdb mutant, and the percentages of total σS in fractions 4–8 (bound σS) were 69 and 83% for the wild-type strain and 83 and 93% for the rpoSdb mutant, calculated using the IMAGEJ software).
Few biochemical studies have been performed on EσS (7,53–56) and the three-dimensional structure of σS is unknown. Although the overall sequence of the σS and σ70 DNA-binding regions is well conserved (51,54), the corresponding holoenzymes are distinct in some mechanistic features and residues important for DNA recognition by EσS appear to be significantly different from those of Eσ70 (5,7,53–56). Amino-acid substitutions R141S and A157T (Figure 2A) have been shown to impair σS promoter binding but not E binding (54). These residues are not conserved in σ70 (Supplementary Figure S1A). However, the corresponding residues in σ70 are located in a DNA binding α helix, and residue K426, corresponding to R141 in σS (Supplementary Figure S1A), interacts with nucleotides in the -10 element of the promoter (57). The rpoS alleles encoding σSR141S, σSA157T (54) and σSR141SA157T (named σSdb) were generated and introduced in the chromosome of S. Typhimurium ATCC14028 as described in the Materials and Methods section. The mutant derivatives, rpoSR141S and rpoSdb (encoding σSR141S and σSdb, respectively) and to a lesser extent rpoSA157T (encoding σSA157T), were unable to display the σS-dependent rdar morphotype (8,9) of Salmonella wild-type (Supplementary Figure S2A). They were also impaired in expression of a transcriptional lacZ fusion to katN (Supplementary Figure S2B), a well-characterized σS-dependent gene (16,49,58). Since σSR141S, σSdb and to a lesser extent σSA157T were produced in increased amounts compared to wild-type σS (Supplementary Figure S2C), these variant σS proteins were likely impaired in their activity. Consistent with this hypothesis, RNAP holoenzymes containing the σS variants were affected for binding at the katN promoter, as assessed by band shift assays and potassium permanganate reactivity footprinting (Figure 2B). The σSdb variant was more strongly affected than the σSR141S and σSA157T variants, especially in vivo (Supplementary Figure S2B), and the corresponding mutant was thus selected for further studies.
σSdb is not impaired in RNAP core binding
As mentioned above, Lee and Gralla established that substitutions R141S and A157T do not affect the ability of σS to bind E (54). Since we used a combination of these two substitutions in σSdb, and given that in vivo modulators of RNAP formation (2–5,52) might differentially affect the interaction between E and the σS and σSdb variants, it was important to compare the ability of σS and σSdb to form EσS holoenzyme in vivo.
Separation by size exclusion chromatography and immuno-detection in crude cell extracts of free and E-associated σ has been used previously to assess the effect on the assembly of Eσ70 and EσS holoenzymes of ppGpp (20), Crl (59) and the ω subunit of RNAP (60). Here we used the same methodology to separate and compare levels of free σS from σS bound to RNAP in the wild-type strain and its rpoSdb derivative in stationary phase (Figure 2C). Two populations of σS were found in both strains, the major one (fractions 4–8) co-eluted with the β and α subunits of core RNAP and was interpreted to represent holoenzyme-associated σS, while the other, in fractions 17–19, represented free (unbound) σS. Indeed, free σS used as a control eluted in fractions 17–19 only (Figure 2C). The elution profiles were similar for the wild-type and rpoSdb strains, showing that σSdb was not impaired in RNAP binding, compared to wild-type σS. Thus, the defect of σSdb in gene activation was likely due to its inability to bind the promoter DNA.
σS, but not σSdb, downregulates expression of sdh and ompD genes
To further characterize the activity of σSdb on different gene targets besides katN, a collection of 17 σS-activated lacZ gene fusions was used. Expression of all the fusions was downregulated in the rpoSdb mutant, compared to the wild-type strain (Figure 3A), even though it is worth noting that σSdb retained basal activity at some promoters (e.g. yeaG and yahO). The cloned rpoS gene restored expression of the lacZ fusions as shown for katE and katN and of the rdar morphotype in the rpoSdb mutant (Figure 3B and C, respectively).
Figure 3.
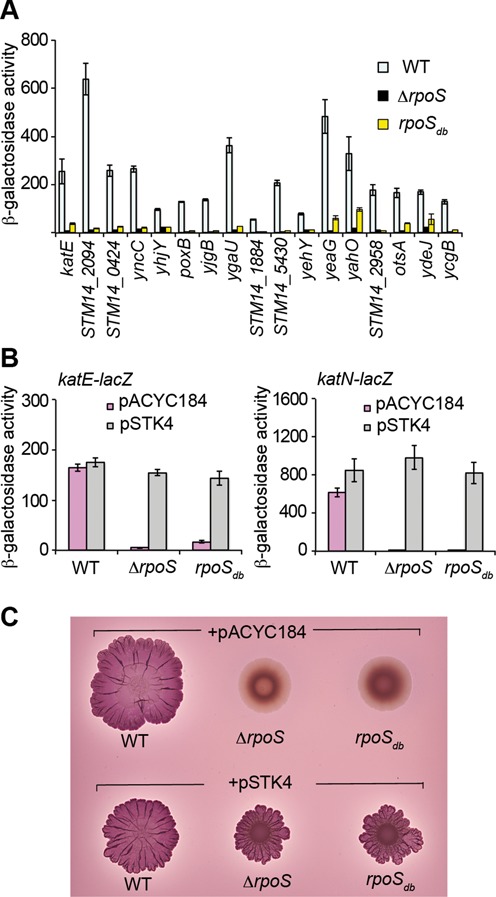
Effects of the ΔrpoS and rpoSdb mutations on σS-activated genes and the rdar morphotype. (A, B) Expression of transcriptional lacZ fusions in σS-dependent genes in the wild-type (WT) strain VF7969 and its rpoS derivatives VF9356 and VF9849, grown for 18 h in LB at 37°C. Bar graphs represent the mean β-galactosidase activity and error bars represent standard deviation of at least three independent experiments. (C) Development of the red dry and rough (rdar) morphotype of the wild-type strain and rpoS mutants carrying the plasmid indicated was visualized on CR plates at 28°C. Empty vector pACYC184 and plasmid pSTK4 carrying the rpoS gene were used in complementation experiments (B, C).
We previously showed that, in stationary phase of growth in rich medium, σS downregulates most S. Typhimurium genes involved in the TCA cycle (12). For example, the sdhCDAB operon encoding succinate dehydrogenase, a membrane bound complex that directly connects the TCA cycle with the respiratory electron transport chain, is downregulated by σS in Salmonella (12) and E. coli K-12 (10). Interestingly, sdhB transcript levels were strongly increased in both the rpoSdb and ΔrpoS mutants, compared to that in the wild-type strain (Figure 4A). Consistently, the ΔrpoS and rpoSdb strains grew better than wild-type in minimal medium with succinate and this phenotype was complemented by wild-type rpoS expressed from plasmid pSTK4 (Figure 4B). The ability of Salmonella to grow on succinate was also improved by the single rpoS mutation rpoSR141S and to a lesser extent rpoSA157T (Supplementary Figure S2D). The ompD (nmpC) gene is another target of negative regulation by σS (12), encoding a major porin of Salmonella and a target of a protective antibody response (61). In the rpoSdb and ΔrpoS strains, the levels of ompD-lacZ fusion expression and OmpD production were higher than in the wild-type strain (Figure 4C and D). Overall, these results suggested that σS binding to DNA is required for downregulation of sdh and ompD.
Figure 4.
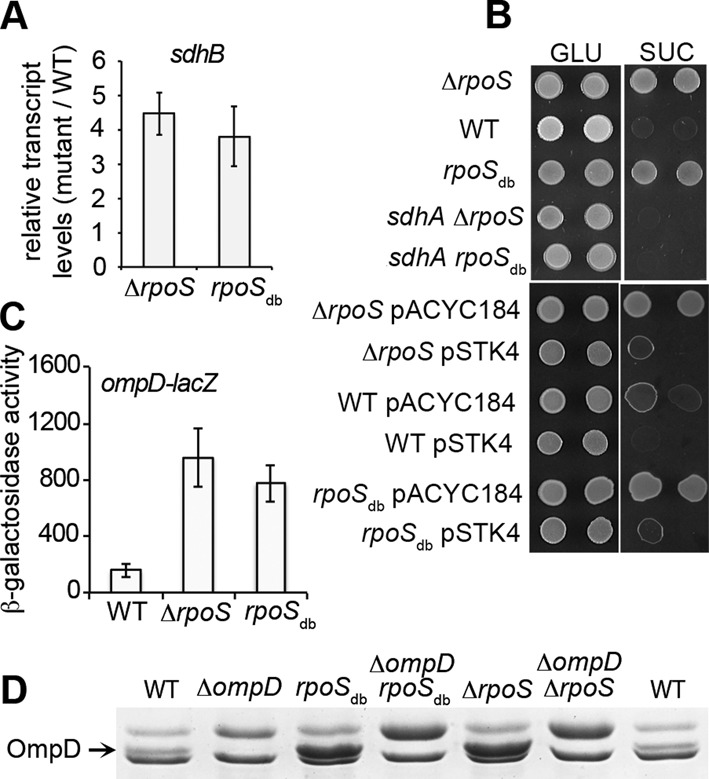
Effects of the ΔrpoS and rpoSdb mutations on sdh and ompD expression. (A) sdhB transcript levels using quantitative real-time PCR in the ΔrpoS and rpoSdb mutants, VF9356 and VF9849, respectively, were compared to that in the wild-type strain VF7969. Strains were grown 18 h in LB at 37°C. Three biological replicates were used. Results shown are the mean and standard deviation. (B) The wild-type (WT) and rpoS strains carrying the mutations and/or plasmid indicated were spotted on minimal medium with succinate (SUC) and glucose (GLU) as the sole carbon source (5 μl of cultures diluted to OD600 of 1.0 and 0.05 were spotted). The rpoS mutations were complemented by the rpoS gene on pSTK4 but not by the empty vector pACYC184. (C) ompD-lacZ expression in the rpoS mutants compared to the wild-type strain. Strains were grown 18 h in LB at 37°C. Results shown are the mean and standard deviation of at least three independent experiments. (D) Detection of the OmpD porin in membranes from the wild-type and rpoS strains carrying the indicated mutations (Supplementary Table S1) and grown for 18 h in LB at 37°C.
The rpoSdb and ΔrpoS mutations alleviate growth restriction of a ΔrssB mutant
The levels of σS are low in exponential phase due to proteolysis by the ClpXP protease and to RssB, a protein required for σS recognition by ClpXP (4,5) (Figure 5A). Consistently, the levels of σSdb and σS were higher in the ΔrssB mutant than the wild-type strain (Figure 5A). In ΔrssB mutants, the accumulation of σS in exponential phase resulted in growth defects that were alleviated by the rpoSdb and ΔrpoS mutations (Figure 5B). Thus, in contrast to wild-type σS, σSdb did not impair Salmonella growth when produced to high levels in exponential phase. The increased levels in stationary phase of σS variants with reduced activity, compared to that of wild-type σS (Figure 5A and Supplementary Figure S2C), might result from the inactivation of auto-regulatory circuits. Indeed, σS controls transcript levels of numerous genes that modulate its expression (5,7,12). The increased level of σSdb compared to that of σS (Figure 5A) might contribute to the observed residual activity of σSdb at some promoters in stationary phase (Figure 3A and see below Table 1).
Figure 5.
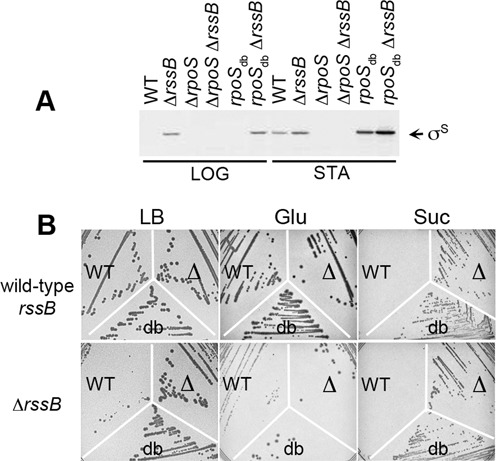
Effect of a ΔrssB mutation on σS expression and Salmonella growth. (A) Immunodetection of σS and σSdb in exponential (LOG, OD600 of 0.3) and stationary phase (STA, OD600 of 4.5) LB cultures of the wild-type (WT) strain VF7969 and its rpoS mutants VF9356 and VF9849, carrying wild-type and ΔrssB alleles. (B) Growth, on LB and minimal medium with succinate (Suc) and glucose (Glu) as a carbon source at 37°C, of the wild-type and ΔrssB strains carrying a wild-type (WT) or a mutated rpoS allele (Δ, deletion of rpoS; db, rpoSdb).
Table 1. Genes differentially expressed in the rpoSdb and ΔrpoS mutants (P < 0.001).
| Name | Normalized read countsa | Product | ||
|---|---|---|---|---|
| WT | rpoSdb | ΔrpoS | ||
| STM14_0422 | 1203 | 25 | 3 | Cytochrome BD2 subunit I |
| yahO | 961 | 112 | 15 | Hypothetical protein |
| ybgS | 4003 | 116 | 6 | Hypothetical protein |
| yeaG | 5730 | 352 | 26 | Putative serine protein kinase |
| STM14_1559b | 589 | 30 | 1 | Hypothetical protein |
| osmC | 453 | 26 | 4 | Putative envelope protein |
| otsA | 865 | 161 | 11 | Trehalose-6-phosphate synthase |
| otsB | 832 | 99 | 6 | Trehalose-6-phosphate phosphatase |
| yohC | 1820 | 79 | 8 | Putative transport protein |
| STM14_2680c | 415 | 23 | 3 | Hypothetical protein |
| ygdI | 1871 | 229 | 23 | Putative lipoprotein |
| yghA | 716 | 18 | 1 | Oxidoreductase |
| STM14_5096d | 1830 | 117 | 2 | Putative cytoplasmic protein |
| yjbJ | 8161 | 571 | 25 | Putative stress-response protein |
| STM14_5129e | 9767 | 195 | 21 | Hypothetical protein |
| ecnB | 3219 | 114 | 12 | Entericidin B membrane lipoprotein |
| osmY | 2396 | 333 | 17 | Hypothetical protein |
| STM14_5481 | 419 | 46 | 3 | Hypothetical protein |
| oat | 6006 | 288 | 35 | Putrescine-2-oxoglutarate aminotransferase |
| ynfM | 190 | 13 | 220 | Putative transport protein |
Global gene expression in wild-type, ΔrpoS and rpoSdbSalmonella strains
The above results suggested that σS binding to DNA is required for downregulation of sdh and ompD genes. To assess regulation of additional gene targets, and attempt to identify a subset of genes downregulated by σS primarily via σ competition for E binding (Figure 1A), the activity of σSdb was determined at the genome level. Transcript levels of the wild-type strain and the rpoSdb and ΔrpoS mutants were measured by directional RNA-seq using three biological replicates of strains grown to stationary phase in LB medium. We recently reported the comparative analysis of expression profiles between the wild-type and ΔrpoS strains (12). Six hundred and seven genes were differentially expressed in the wild-type strain and the ΔrpoS mutant with a high probability (P < 0.001), of which 145 were repressed by σS (12), including many genes also repressed by σS in E. coli (5,10). It is worth noting that whereas some σS-activated genes are directly regulated through binding of σS to σS-dependent promoters, others are likely regulated indirectly by σS. When the expression profile of the rpoSdb mutant was compared to that of the wild-type and ΔrpoS control strains, the rpoSdb and ΔrpoS mutants showed similar expression profiles (Figure 6 and Supplementary data set S1). Besides rpoS, only 19 genes showed expression levels higher in the rpoSdb mutant than in the ΔrpoS strain (P < 0.001; Figure 6 and Table 1). These σS-activated genes exhibited high expression levels in the wild-type strain and strong σS-dependency (12) (Table 1). Consistent with this finding, promoter sequences of those genes (62,63) show typical features of σS-promoters (7,53,56) (Figure 7A and B and Supplementary Figure S3). In particular, the conserved -12T, -11A and -7T are of paramount importance for promoter recognition and use by EσS (7,53,56). In contrast, the data did not show any conserved nucleotides corresponding to a -35 region (Figure 7A), in agreement with previous findings that EσS promoters display low level of sequence conservation near -35 (7,53,56). It is likely that these σS-dependent promoters are functional to some extent with EσSdb (Table 1). Basal expression levels in the rpoSdb mutant of yahO, yeaG and otsA were also detected using lacZ fusions (Figure 3A).
Figure 6.
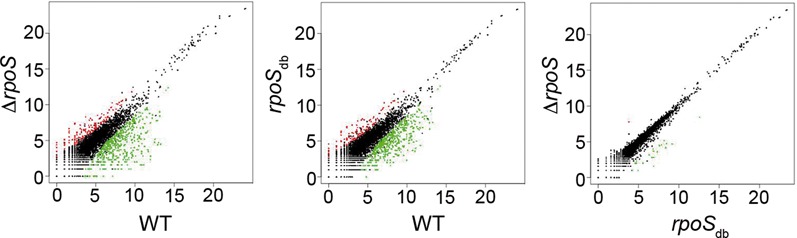
Transcriptome profile of wild-type and rpoS Salmonella strains. Scatterplot was used to compare gene expression (log2-transformed normalized read counts) in the rpoSdb mutant and the wild-type (WT) and ΔrpoS strains grown to stationary phase in LB. Transcriptome profiles of the wild-type and ΔrpoS strains, used as controls in the experiment, have been recently reported (12). Red and green dots represent genes differentially expressed (P < 0.001) and black dots represent genes not differentially expressed.
Figure 7.
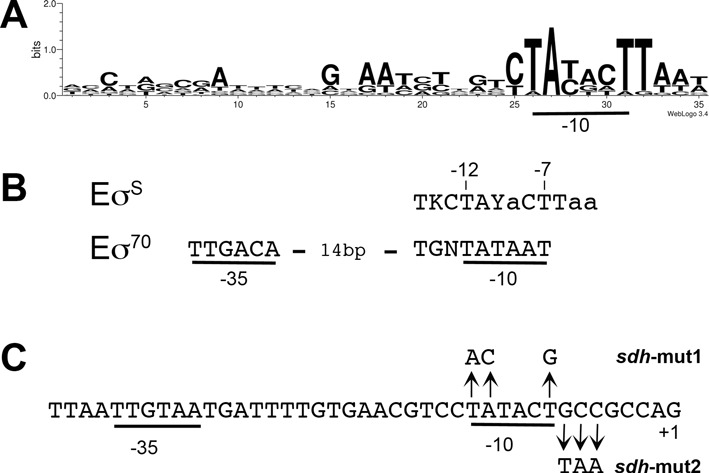
Promoter sequences and features. (A) Sequence logo generated with promoter sequences of genes in Table 1 (Supplementary Figure S3) and the WebLogo application (http://weblogo.threeplusone.com/create.cgi). (B) Possible consensus sequences for the -10 element of promoters preferentially recognized and transcribed by Eσ70 and EσS (7,53). The -35 sequence of Eσ70-dependent promoters is also indicated. The -35 element is less conserved in EσS-dependent promoters and is variable in its sequence and location (7,53). The most conserved nucleotides are indicated in capital letters. Y denotes a pyrimidine (T/C). K stands for T/G. (C) DNA sequence of the sdh promoter (Psdh) region and base substitutions generated in the chromosome of Salmonella to yield sdh-mut1 and sdh-mut2.
One gene only, ynfM, was upregulated in the ΔrpoS strain compared to the rpoSdb mutant (Table 1), even when a P-value cut-off of 0.05 was used (Supplementary data set S1). The ynfM gene was poorly transcribed under the growth condition used (Table 1 and Supplementary Figure S4A). Surprisingly, its transcript levels were not significantly different in the ΔrpoS and wild-type strains (12) (Table 1), whereas expression of a transcriptional ynfM-lacZ fusion was downregulated in the ΔrpoS and rpoSdb strains, compared to the wild-type strain (Supplementary Figure S4A). Complementation of the ΔrpoS and rpoSdb mutations was observed when σS was produced in trans from plasmid pSTK4 (Supplementary Figure S4B), confirming that ynfM-lacZ transcription was activated by σS. Activation of the ynfM promoter by σS might be masked, at the ynfM transcript level, by compensatory negative effects of σS on ynfM mRNA elongation and/or stability, resulting in a complex regulatory pattern.
Altogether, these results strongly suggested that, under the conditions used, the negative effects of σS on gene expression require its binding to DNA and are unlikely to result solely from competition between σS and other σ factors for E binding. Our finding, that σS binding to E alone did not result in detectable gene repression by σS, might be explained if promoters are saturated, i.e. they bind Eσ efficiently but display a low rate of transcription initiation, so that they are occupied by Eσ most of the time and are weakly affected by σ competition. It is also possible that, in stationary phase under physiological conditions (as opposed to conditions where a σ factor is over-expressed), σ competition for E is weakened and/or its effects on gene transcription are not detected by the methods used here. In stationary phase, the stop of transcription of ribosomal RNA increases the availability of E (the activities of these promoters sequester 60–70% of the transcriptional machinery during rapid growth in rich media; (2)). In addition, molecules produced in stationary phase (such as anti-σ, 6SRNA, ppGpp and metabolites) also alter σ competition by sequestering σ and/or modulating Eσ formation (2,52). Variations in the efficiency of promoter escape and transcript elongation in stationary phase may also alter the distribution of E, σ and Eσ and thus σ competition. To determine whether elimination of σS favors the formation of Eσ70 in the conditions of our study, levels of σ70 bound to RNAP were compared in the wild-type strain and the ΔrpoS and rpoSdb mutants, by size exclusion chromatography of crude extracts and immuno-detection of σ70 and the β′ subunit of RNAP. The percentage of total σ70 co-eluting with the β′ subunit of core RNAP was 94–98% and similar for the three strains, suggesting that most σ70 molecules were associated with RNAP in stationary phase (Supplementary Figure S5). A more detailed study and the use of complementary techniques, to assess the concentrations of the different holoenzymes at different time points upon entry to stationary phase, in different media and bacterial genetic backgrounds, are required to carefully address the relevance of the σ competition for E model during stationary phase. Nevertheless, these data reinforced our conclusion that the extensive gene repression by σS in stationary phase (Supplementary data set S1 and Figure 6) (12) does not rely on the regulatory model depicted in Figure 1A.
σS and σ70 factor antagonism at the sdh promoter
The sdhCDAB succinate dehydrogenase operon is negatively controlled at the post-transcriptional level by the RyhB sRNAs (22,64). Expression of the Salmonella homologous RyhB1 and RyhB2 sRNAs is positively controlled by σS in the growth conditions used in the present study (i.e. late stationary phase in rich medium) (12,22), thus making these sRNAs possible intermediates in the downregulation of sdh expression by σS (according to the regulatory model in Figure 1B). However, sdhB transcript levels were similar in the wild-type strain and the ryhB1/2 strain containing mutations in both ryhB genes (Supplementary Table S1) and were increased to similar levels in the ΔrpoS and ΔrpoS ryhB1/2 strains, compared to that in wild-type (Figure 8A). These data suggested the existence of a RyhB-independent mechanism of negative regulation of sdh by σS.
Figure 8.
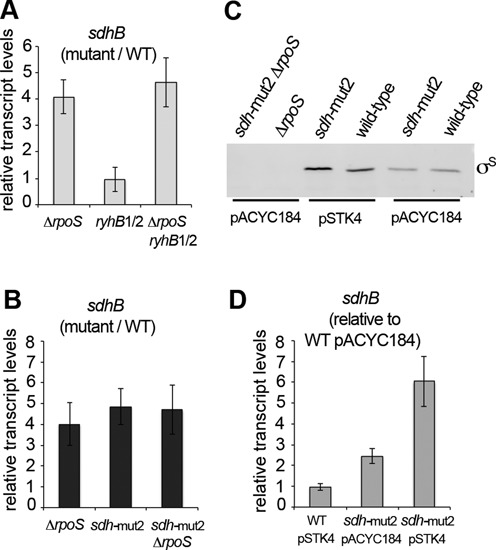
Role of the RyhBs sRNAs and discriminator region of Psdh in sdhB gene repression by EσS. (A) sdhB transcript levels in the ΔrpoS, ryhB1/2 and ΔrpoS ryhB1/2 strains (VF8158, VFB867 and VFB868, respectively; Supplementary Table S1), relative to sdhB transcript levels in the wild-type strain ATCC14028. (B) sdhB transcript levels in the ΔrpoS, sdh-mut2 and ΔrpoS sdh-mut2 strains (VF8158, VFD984 and VFD987, respectively; Supplementary Table S1), relative to sdhB transcript levels in the wild-type strain ATCC14028. (C) Immunodetection of σS in stationary phase LB cultures of wild-type and mutant strains carrying the vector pACYC184 and rpoS on pSTK4. (D) sdhB transcript levels in strains containing the plasmid vector pACYC184 or the rpoS gene on pSTK4, relative to sdhB transcript levels in the wild-type strain containing pACYC184. Three biological replicates were used for qRT-PCR experiments in panels (A), (B) and (D), and results shown are the mean and standard deviation.
A single promoter has been identified upstream of sdhC in wild-type Salmonella (62,63) (Figure 7C). Chromosomal mutations were introduced in the −10 element of this promoter (Psdh) at positions -12T, -11A and -7T that are of paramount importance for promoter recognition by both Eσ70 and EσS (7,52,53,55) (sdh-mut1, Figure 7C). These mutations strongly reduced sdhC, sdhD sdhA and sdhB mRNAs levels in the wild-type strain and in the ΔrpoS mutant (Figure 9A) and impaired the ability of these strains to grow on succinate (Figure 9B), suggesting that sdh expression in both wild-type and ΔrpoS strains is driven mainly from Psdh. In vitro, Eσ70 and EσS were both able to bind the sdh promoter region (data not shown). This finding was not unexpected since (i) the DNA sequence specificity of σS is very similar to that of σ70 and many promoters are bound in vitro by both holoenzymes (5,7,53), and (ii) the −10 sequence of Psdh is identical to that of promoters preferentially recognized by EσS and Eσ70 (7,53,55) (Figure 7C). In vivo however, variable combinations of intrinsic promoter features and regulatory proteins determine whether a promoter is recognized and transcribed by EσS or Eσ70 (5,7,53). Interestingly, genome-wide mapping of Eσ70 and EσS binding sites in E. coli (37) shows that both Eσ70 and EσS bind the sdh promoter region, in vivo as well. These findings raised the possibility that EσS binding to the sdh promoter in vivo competes with Eσ70-dependent transcription initiation (as proposed in the regulatory model in Figure 1C).
Figure 9.
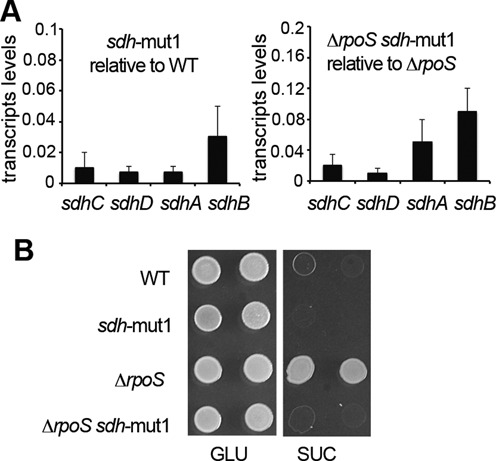
Mutational analysis of the -10 Psdh promoter element. (A) Relative transcript levels of sdhC, sdhD, sdhA and sdhB in wild-type (left) and ΔrpoS (right) strains carrying the sdh-mut1 mutations (Figure 7C and Supplementary Table S1). Three biological replicates were used and results shown are the mean and standard deviation. sdh transcripts levels were reduced at least 10-fold by the sdh-mut1 mutations and were close to the background level of detection, leading to variations in the values. (B) Effect of sdh-mut1 on the ability of the wild-type strain and ΔrpoS mutant to grow at the expense of glucose (GLU) and succinate (SUC) as a sole carbon source (5 μl of cultures diluted to OD600 of 1.0 and 0.05 were spotted).
A striking feature of Psdh is the presence of a GC-rich region just downstream the −10 box (Figure 7C), in the discriminator region of the promoter (3,52,57). In contrast, the discriminator of EσS-dependent promoters is frequently TA rich (7,53,55) (Figure 7B). Interestingly, a TA motif is conserved in the discriminator region of promoters retaining basal activity in the rpoSdb mutant (Figure 7A), and may favor residual activity of σSdb. When the GCC sequence in Psdh was substituted by TAA (sdh-mut2; Figure 7C), sdhB transcript levels were upregulated in the wild-type strain (Figure 8B). In stark contrast, sdh-mut2 had no significant effect on sdhB transcript levels in the ΔrpoS strain (Figure 8B), indicating a σS-specific effect of the TAA motif. These data suggested that sdh-mut2 mutations eliminate the negative effects of σS on sdhB transcription (Figure 8B) and are consistent with a model in which EσS binding to Psdh poises the promoter due to an unfavorable discriminator region and disrupts normal transcription by competing with Eσ70 binding.
Altogether, the results suggest that σ competition at the promoter DNA plays an important role in gene repression by σS and open new field of investigation regarding the role of the promoter discriminator region in EσS activity. σ factors of the σ70 family are involved in promoter recognition to position RNAP, but also in the formation of the open promoter complex, in which melted DNA includes the −10 region and extends downstream to the transcription start site (3,52). In housekeeping RNAP, region 1.2 of domain 2 of sigma interacts with three non-template strand nucleotides immediately downstream the −10 element (GG and G of the discriminator sequence GGGA (3,52,57)). The strength of interaction between the discriminator and σ70 region 1.2 influences open complex formation/stability and its consequence on transcriptional output and regulation depends on the intrinsic kinetics of the promoter (3,52,65). Although the discriminator sequence likely influences σS-dependent transcription in concert with the -10 hexamer (7,53,66), its exact role in EσS-transcription is unknown. In addition, residues of σ70 interacting with the discriminator (57) are not all conserved in σS (Supplementary Figure S1B) raising the possibility that EσS and Eσ70 use different discriminator sequences. The nature of the Psdh discriminator may influence different steps in EσS-dependent transcription, such as positioning of EσS on the promoter DNA, formation/stability of the open complex and/or promoter clearance. As EσS binds duplex (unmelted) DNA promoters more weakly than Eσ70, AT-rich discriminator sequences may well optimize promoter melting near the transcription site (7,53,55). When the level of σS was increased in Salmonella by providing additional copies of rpoS in trans on plasmid pSTK4 (Figure 8C), sdhB transcript levels increased in the sdh-mut2 strain but not in wild-type (Figure 8D). Altogether, these data suggest that σS (i) represses transcription from the wild-type Psdh promoter, but not from the sdh-mut2 promoter (Figure 8B), and (ii) activates transcription from the sdh-mut2 promoter, but not from the wild-type Psdh promoter, at least when it is over-produced (Figure 8 CD). It is thus possible that the sdh promoter is poised by EσS engaged and having difficulties to escape and that sdh-mut2 allows to some extent poised EσS to escape into elongation mode.
The discriminator sequence has been implicated in the regulation of Eσ70-dependent promoters by ppGpp and its cofactor DksA (2,52,65). Since a GC-rich discriminator favors promoter repression by ppGpp (2,52,65), it would be interesting to determine whether ppGpp and DksA have a direct role in the regulation by σS of the sdh promoter. In a recent transcriptome analysis, the sdh promoter was shown to be upregulated in a relAspoT mutant of Salmonella deficient for the production of ppGpp (62). However, this effect may be indirect, via σS, since ppGpp has positive effects on rpoS transcription, σS stability and the formation/performance of EσS (2,4,20). Changing the GC-rich discriminator to a TA-rich one in sdh-mut2 had no significant effect on sdhB transcript levels in the absence of σS (Figure 8B), suggesting that this modification had no major impact on Eσ70 transcription in these conditions. It remains to be determined how it affects EσS transcription and whether ppGpp is involved.
Although additional experiments are required to highlight the mechanistic implication of the EσS-discriminator interaction in EσS-dependent transcription and how sdh-mut2 might influence transcriptional outputs at Psdh, our data suggest that negative regulation of sdh transcription by σS is mediated by competition between σS and σ70 at the DNA rather than E level, a finding consistent with data from experiments using σSdb. It must be emphasized that the presence of a perfect −10 σS-consensus followed by GC-rich region is not a general characteristic of promoters of genes negatively controlled by σS (data not shown) and the repression mechanism used by EσS likely adapts to promoter characteristics. Also, we cannot exclude a role for additional factors helping EσS to form unproductive complex at Psdh in vivo. Indeed, there are a few examples of poised RNAP generated by transcriptional regulators and/or inappropriate environmental condition (27,29,52,67).
CONCLUSION
Our study provides novel insights into mechanisms of downregulation of gene expression by σS by showing that, under physiological conditions in stationary phase, gene repression requires σS binding to DNA. Furthermore, we suggest that EσS can function as a transcriptional repressor. Our data are likely the tip of the iceberg and will inspire future studies to decipher the underlying molecular mechanisms. These mechanisms are likely to be specific to individual genes and environmental conditions and to rely on two main categories of regulatory process: direct effects of EσS at the promoter DNA level, as shown in the present study (Figure 1C), and the action of EσS-dependent negative effectors (Figure 1B).
Direct negative regulation of transcription by σ factors (Figure 1C) is not well documented and the possibility that EσS blocks active gene transcription as a repressor calls for further investigation. Our data suggest that EσS occludes the sdh promoter and this lowers transcription by competition with Eσ70 binding. Other genes downregulated by σS are transcribed by more than one promoter and EσS binding at one promoter or pausing might sterically hinder binding of Eσ70 or an alternative RNAP and/or transcription factors at a second promoter, resulting in promoter interference and gene repression. Due to the extensive overlap between promoter regions bound by housekeeping and alternative RNAP in vivo, especially Eσ70 and EσS (36,37), σ factor antagonisms at the promoter DNA level might be more frequent in vivo than initially thought.
A second main category of regulatory processes requiring the DNA binding activity of σS might endow σS with repressor functions: the involvement of σS-dependent effectors (Figure 1B). Future experiments will assess whether some of the σS-controlled secondary regulators (12) are intermediates in regulatory cascades (Figure 1B), or co-repressors in feed-forward regulatory loops (Figure 1C), to control gene transcription negatively. Another exciting issue will be to determine to which extent σS-dependent transcriptional and post-transcriptional control mechanisms are combined to allow for dynamic and flexible regulatory patterns and additional signal inputs. The RyhB sRNAs are another possible tool for σS to downregulate sdh expression. It is possible that the expression levels of these sRNAs and/or the growth conditions used in the present study are not appropriate to observe a significant impact of the ryhB mutations on sdhB transcript levels (Figure 8A). Experiments are underway to determine how the different regulatory components of the σS control cooperate to adjust the levels and dynamics of sdh expression in response to different environmental conditions, and whether these controls contribute to the cell fitness.
ACCESSION NUMBER
The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE46380 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rhedtgougouiuri&acc=GSE46380).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Olivera Francetic, Anthony Pugsley and all members of the laboratory for their kind support. We are very grateful to Bertil Gummesson for helpful discussions and to Nara Figueroa-Bossi for the gift of strain MA10200.
Author Contributions: J.Y.C. and F.N. designed research; C.L.M., V.M., O.S., A.K., S.S.D., M.B. and F.N. performed research; M.M. and B.D. contributed analytic tools; C.L.M., V.M., M.A.D., B.J., A.K., M.M. and F.N. analyzed data; and C.L.M., V.M. and F.N. wrote the paper.
Footnotes
These authors contributed equally to the paper as first authors.
FUNDING
French National Research Agency [ANR-11-BSV3-009 to F.N.]; Institut Pasteur; Centre National de la Recherche Scientifique. Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 2.Österberg S., del Peso-Santos T., Shingler V. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 3.Feklistov A., Sharon B.D., Darst S.A., Gross C.A. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 4.Battesti A., Majdalani N., Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengge R. The general stress response in Gram-negative bacteria. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, DC: ASM Press; 2011. pp. 251–289. [Google Scholar]
- 6.Schellhorn H.E. Elucidating the function of the RpoS regulon. Future Microbiol. 2014;9:497–507. doi: 10.2217/fmb.14.9. [DOI] [PubMed] [Google Scholar]
- 7.Landini P., Egli T., Wolf J., Lacour S. SigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 2014;6:1–13. doi: 10.1111/1758-2229.12112. [DOI] [PubMed] [Google Scholar]
- 8.Robbe-Saule V., Jaumouillé V., Prévost M.-C., Guadagnini S., Talhouarne C., Mathout H., Kolb A., Norel F. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006;188:3983–3994. doi: 10.1128/JB.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Römling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patten C.L., Kirchhof M.G., Schertzberg M.R., Morton R.A., Schellhorn H.E. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Gen. Genomics. 2004;272:580–591. doi: 10.1007/s00438-004-1089-2. [DOI] [PubMed] [Google Scholar]
- 11.Notley-McRobb L., King T., Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lévi-Meyrueis C., Monteil V., Sismeiro O., Dillies M.A., Monot M., Jagla B., Coppée J-Y., Dupuy B., Norel F. Expanding the RpoS/ σS-network by RNA sequencing and identification of σS-controlled small RNAs in Salmonella. PLoS One. 2014;9:e96918. doi: 10.1371/journal.pone.0096918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong T., Yu R., Schellhorn H. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 2011;79:375–386. doi: 10.1111/j.1365-2958.2010.07449.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 15.Zambrano M.M., Siegele D.A., Almiron M., Tormo A., Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 16.Robbe-Saule V., Dias Lopes M., Kolb A., Norel F. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 2007;189:2976–2987. doi: 10.1128/JB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteil V., Kolb A., Mayer C., Hoos S., England P., Norel F. Crl binds to domain 2 of σS and confers a competitive advantage to a natural rpoS mutant of Salmonella enterica serovar Typhi. J. Bacteriol. 2010;192:6401–6410. doi: 10.1128/JB.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farewell A., Kvint K., Nyström T. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 19.Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition. Mol. Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 20.Jishage M., Kvint K., Shingler V., Nyström T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurie A.D., Bernardo L.M., Sze C.C., Skarfstad E., Szalewska-Palasz A., Nyström T., Shingler V. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 2003;278:1494–1503. doi: 10.1074/jbc.M209268200. [DOI] [PubMed] [Google Scholar]
- 22.Padalon-Brauch G., Hershberg R., Elgrably-Weiss M., Baruch K., Rosenshine I., Margalit H., Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fröhlich K.S., Papenfort K., Berger A.A., Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40:3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva I.J., Ortega A.D., Viegas S.C., García-Del Portillo F., Arraiano C.M. An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA. 2013;19:1253–1265. doi: 10.1261/rna.039537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber H., Polen T., Heuveling J., Wendisch V.F., Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maciag A., Peano C., Pietrelli A., Egli T., De Bellis G., Landini P. In vitro transcription profiling of the σS subunit of bacterial RNA polymerase: re-definition of the σS regulon and identification of σS-specific promoter sequence elements. Nucleic Acids Res. 2011;39:5338–5355. doi: 10.1093/nar/gkr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grainger D.C., Goldberg M.D., Lee D.J., Busby S.J.W. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 2008;68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 28.Boucher J.C., Schurr M.J., Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.J., Gralla J.D. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004;14:153–162. doi: 10.1016/s1097-2765(04)00202-3. [DOI] [PubMed] [Google Scholar]
- 30.Artsimovitch I. Post-initiation control by the initiation factor sigma. Mol. Microbiol. 2008;68:1–3. doi: 10.1111/j.1365-2958.2008.06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perdue S.A., Roberts J.W. σ70-dependent transcription pausing in Escherichia coli. J. Mol. Biol. 2011;412:782–792. doi: 10.1016/j.jmb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Reppas N.B., Wade J.T., Church G.M., Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 33.Palmer A.C., Ahlgren-Berg A., Egan J.B., Dodd I.B., Shearwin K.E. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendtsen K.M., Erdossy J., Csiszovszki Z., Svenningsen S.L., Sneppen K., Krishna S., Semsey S. Direct and indirect effects in the regulation of overlapping promoters. Nucleic Acids Res. 2011;39:6879–6885. doi: 10.1093/nar/gkr390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zafar M.A., Carabetta V.J., Mandel M.J., Silhavy T.J. Transcriptional occlusion caused by overlapping promoters. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1557–1561. doi: 10.1073/pnas.1323413111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade J.T., Castro Roa D., Grainger D.C., Hurd D., Busby S.J., Struhl K., Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 37.Cho B.K., Kim D., Knight E.M., Zengler K., Palsson B.O. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014;12:4. doi: 10.1186/1741-7007-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 39.Sternberg N.L., Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlach R.G., Jäckel D., Hölzer S.U., Hensel M. Rapid oligonucleotide-based recombineering of the chromosome of Salmonella enterica. Appl. Environ. Microbiol. 2009;75:1575–1580. doi: 10.1128/AEM.02509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochner B.R., Huang H-C., Schieven G.L., Ames B.N. Positive selection for loss of tetracycline resistance. J. Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibanez-Ruiz M., Robbe-Saule V., Hermant D., Labrude S., Norel F. Identification of RpoS (sigmaS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2000;182:5749–5756. doi: 10.1128/jb.182.20.5749-5756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellermeier C.D., Janakiraman A., Slauch J.M. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 46.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Coynault C., Robbe-Saule V., Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 48.Lobos S.R., Mora G.C. Alteration in the electrophoretic mobility of OmpC due to variations in the ammonium persulfate concentration in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Electrophoresis. 1991;12:448–450. doi: 10.1002/elps.1150120615. [DOI] [PubMed] [Google Scholar]
- 49.England P., Westblade L.F., Karimova G., Robbe-Saule V., Norel F., Kolb A. Binding of the unorthodox transcription activator, Crl, to the components of the transcription machinery. J. Biol. Chem. 2008;283:33455–33464. doi: 10.1074/jbc.M807380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller J.H. Experiments in Molecular Genetics. NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 51.Paget M.S., Helmann J.D. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haugen S.P., Ross W., Gourse R.L. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Typas A., Becker G., Hengge R. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- 54.Lee S.J., Gralla J.D. Promoter use by σ38 (rpoS) RNA polymerase. J. Biol. Chem. 2002;277:47420–47427. doi: 10.1074/jbc.M208363200. [DOI] [PubMed] [Google Scholar]
- 55.Lee S.J., Gralla J.D. Sigma38 (rpoS) RNA polymerase promoter engagement via -10 region nucleotides. J. Biol. Chem. 2001;276:30064–30071. doi: 10.1074/jbc.M102886200. [DOI] [PubMed] [Google Scholar]
- 56.Lee S.J., Gralla J.D. Open complex formation in vitro by sigma38 (rpoS) RNA polymerase: roles for region 2 amino acids. J. Mol. Biol. 2003;329:941–948. doi: 10.1016/s0022-2836(03)00369-3. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maxam A.M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 59.Typas A., Barembruch C., Possling A., Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geertz M., Travers A., Mehandziska S., Sobetzko P., Chandra-Janga S., Shimamoto N., Muskhelishvili G. Structural coupling between RNA polymerase composition and DNA supercoiling in coordinating transcription: a global role for the omega subunit. MBio. 2011;2 doi: 10.1128/mBio.00034-11. doi:10.1128/mBio.00034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gil-Cruz C., Bobat S., Marshall J.L., Kingsley R.A., Ross E.A., Henderson I.R., Leyton D.L., Coughlan R.E., Khan M., Jensen K.T., et al. The porin OmpD from nontyphoidal Salmonella is a key target for a protective B1b cell antibody response. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9803–9808. doi: 10.1073/pnas.0812431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramachandran V.K., Shearer N., Thompson A. The primary transcriptome of Salmonella enterica serovar Typhimurium and its dependence on ppGpp during late stationary phase. PLoS One. 2014;9:e92690. doi: 10.1371/journal.pone.0092690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kröger C., Dillon S.C., Cameron A.D., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hébrard M., Händler K., et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richards G.R., Vanderpool C.K. Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta. 2011;1809:525–531. doi: 10.1016/j.bbagrm.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gummesson B., Lovmar M., Nyström T.A. Proximal promoter element required for positive transcriptional control by guanosine tetraphosphate and DksA protein during the stringent response. J. Biol. Chem. 2013;288:21055–21064. doi: 10.1074/jbc.M113.479998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ojangu E.L., Tover A., Teras R., Kivisaar M. Effects of combination of different -10 hexamers and downstream sequences on stationary-phase-specific sigma factor sigma(S)-dependent transcription in Pseudomonas putida. J. Bacteriol. 2000;182:6707–6713. doi: 10.1128/jb.182.23.6707-6713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wade J.T., Struhl K. The transition from transcriptional initiation to elongation. Curr. Opin. Genet. Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


