Abstract
CRISPR-Cas systems are RNA-based immune systems that protect prokaryotes from invaders such as phages and plasmids. In adaptation, the initial phase of the immune response, short foreign DNA fragments are captured and integrated into host CRISPR loci to provide heritable defense against encountered foreign nucleic acids. Each CRISPR contains a ∼100–500 bp leader element that typically includes a transcription promoter, followed by an array of captured ∼35 bp sequences (spacers) sandwiched between copies of an identical ∼35 bp direct repeat sequence. New spacers are added immediately downstream of the leader. Here, we have analyzed adaptation to phage infection in Streptococcus thermophilus at the CRISPR1 locus to identify cis-acting elements essential for the process. We show that the leader and a single repeat of the CRISPR locus are sufficient for adaptation in this system. Moreover, we identified a leader sequence element capable of stimulating adaptation at a dormant repeat. We found that sequences within 10 bp of the site of integration, in both the leader and repeat of the CRISPR, are required for the process. Our results indicate that information at the CRISPR leader-repeat junction is critical for adaptation in this Type II-A system and likely other CRISPR-Cas systems.
INTRODUCTION
To survive in natural environments, prokaryotes have evolved numerous defense systems to battle foreign nucleic acids from potentially harmful invaders such as viruses/phages and plasmids (1,2). In bacteria and archaea, recently discovered CRISPR-Cas systems provide adaptive immunity against these invasive genetic elements (3–12). Approximately half of bacteria and almost all archaea whose genomes have been sequenced possess one or multiple CRISPR-Cas systems (13,14). CRISPR-Cas systems are diverse and are classified to three major groups, Types I-III, with multiple subtypes in each group, based primarily on Cas protein composition (13).
A CRISPR (clustered regularly interspaced short palindromic repeat) locus consists of a DNA control element called the ‘leader’ followed by an array of alternating units of an identical direct repeat sequence (repeats) and variable invader-derived sequences (spacers) (7,15–17) (Figure 1A). A group of co-functional, CRISPR-associated (Cas) protein-coding genes are often (but not exclusively) located adjacent to the CRISPR locus (5,7,16–19). In the primary event that enables CRISPR-Cas immunity, appropriate short DNA sequences identified in the invader (protospacers) are integrated into the host CRISPR array (with concurrent generation of an additional repeat) (20–23). The process of acquiring novel spacers into a CRISPR locus is termed ‘spacer acquisition’ or ‘adaptation’. Selection of protospacers depends in part on the presence of short (3–5 bp) neighboring sequences within the invader called protospacer adjacent motifs (PAMs), which differ for distinct CRISPR-Cas systems (11,24,25). CRISPR locus transcripts are processed to generate individual mature CRISPR RNAs (crRNAs) that each contains a sequence complementary to the invader from which they were originally derived (5,26,27). Degradation of invader nucleic acids by crRNA-guided Cas protein complexes effects resistance to the invader and is termed ‘interference’ or ‘defense’ (5,7,8,10,16–19).
Figure 1.
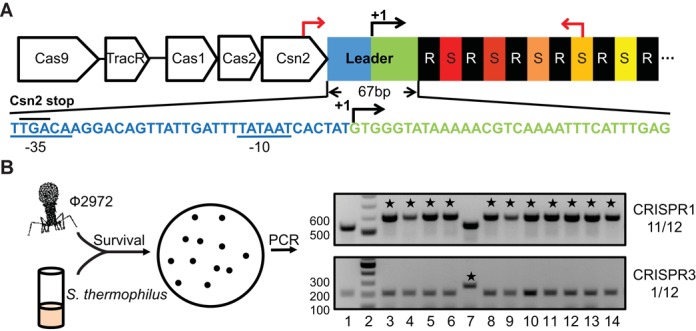
Adaptation occurs predominantly at CRISPR1 in Streptococcus thermophilus DGCC7710. (A) The CRISPR1 leader is divided into promoter region (blue) and transcribed region (green), with a right arrow and ‘+1’ indicating the mapped transcription start site (43). CRISPR1 contains 32 repeat (‘R’, black)-spacer (‘S’, colored) units, only some of which are illustrated. Nucleotide sequence of the leader is displayed. Predicted −35 and −10 elements of the promoter (43) are underlined and the stop codon of csn2 (TGA) is upper-lined. Primers used to amplify the CRISPR1 region are indicated by red arrows. (B) Sth cells were infected by phage 2972 and 12 surviving colonies (lane 3–14) were analyzed for spacer acquisition via colony PCR using primers specific for CRISPR1 (top panel) and CRISPR3 (bottom panel). Lane 1 is the control (no phage) of wild-type Sth and Lane 2 is the DNA ladder. PCR products consistent with acquisition of a new spacer are indicated with asterisks. Numbers on the right indicate adaptation-positive colonies/total colonies in this representative experiment. Selected molecular weight markers are labeled.
While our understanding of the mechanisms governing crRNA biogenesis and CRISPR-Cas-mediated defense against foreign nucleic acids has expanded rapidly in recent years, the adaptation process is still poorly understood. Although biochemical and structural studies of Cas proteins have uncovered activities consistent with potential roles in adaptation (e.g. nuclease and nucleic acid binding) (28–35), the activities of these proteins observed in vitro lack specificity toward substrates resembling PAM-containing invaders or CRISPR arrays. Genetic studies have shown that adaptation in Escherichia coli relies on only two Cas proteins, Cas1 and Cas2, which are the only Cas proteins common to the multiple CRISPR-Cas systems, indicating that they are likely of universal importance in mediating adaptation (23,34,36,37). Increased expression of Cas1 and Cas2 (above endogenous levels) is required for adaptation to be detected in E. coli under laboratory conditions (23,36–38).
Specific sequence elements within the CRISPR are expected to direct spacer acquisition, however these elements have not yet been defined. Sequences within the leader elements of CRISPR loci can be hypothesized to be important given that novel spacers are introduced immediately adjacent to the leader in several systems (3,23,36–42). However, leader elements are generally poorly defined and their functionality not known. Typical reported leader sequences range in size from ∼100–500 bp in length and often include adenine/thymine (A/T)-rich sequence elements (15,41,43–45). Several studies have demonstrated that transcriptional promoters for the CRISPR loci are embedded within leaders (43,44,46–48). In an E. coli (Type I-E) CRISPR-Cas system, it was found that the leader and a single repeat are sufficient for adaptation and that the promoter within the leader was dispensable (23,36–38). A partially matching spacer was found to be necessary for detectable adaptation by a Type I-B system in Haloarcula hispanica (42,49), however this requirement may reflect the role of the spacer in the special form of adaptation (termed ‘primed’ adaptation) that is promoted by the presence of pre-existing spacer(s) that partially match the invading DNA (37,38,41,42,45,49–52).
Adaptation was first observed with Type II CRISPR-Cas systems in Streptococcus thermophilus (Sth). Indeed, the causal relationship between CRISPR spacer acquisition and survival of lytic phage infection was first demonstrated in this system (3). The widely employed Sth strain DGCC7710 has two Type II-A systems (CRISPR1 and CRISPR3) that can be observed to acquire novel spacers from phages and plasmids (3,39,40,53,54), with CRISPR1 being the dominant system in terms of frequency of new spacer acquisition (39,55,56) (This Sth strain also contains a Type III-A and a Type I-E system (CRISPR2 and CRISPR4, respectively) for which spacer acquisition has not been observed (39,43,54)). Sth adaptation is active under laboratory conditions without genetic manipulation or Cas protein overexpression (3,40). While the universally conserved Cas proteins, Cas1 and Cas2, are likely critical for adaptation of the Type II systems, a protein unique to Type II-A systems, Csn2, also appears to be required; a csn2 gene insertion mutant was found to be incapable of acquiring spacers in response to phage infection (3).
Here, we have identified the cis-acting elements that are required for adaptation by the Type II-A system at Sth CRISPR1. We have analyzed an extensive series of constructs of CRISPR elements and mutants for the ability to mediate adaptation in phage infection assays. We found that the leader and a single repeat are sufficient for adaptation, and that adaptation can be induced at a downstream repeat by introduction of a short leader sequence within the CRISPR. Moreover, we identified sequence elements immediately flanking the leader-repeat junction that play an essential role in acquisition of new spacers within the CRISPR locus.
MATERIALS AND METHODS
Strains and plasmids
Streptococcus thermophilus DGCC7710 (Sth) and phage 2972 were kindly provided by Dr. Sylvain Moineau. Sth was maintained in M17 medium (Oxoid) supplemented with 0.5% lactose (LM17) (3). Sth cultures were grown at 37° for overnight, 42° during the day and 30° for strains harboring temperature sensitive plasmids derived from pINTRS (57). E. coli Top10 was used for cloning and plasmid maintenance. When needed, erythromycin was supplemented at 150 μg/ml and 15 μg/ml for E. coli and Sth, respectively. Strains used in this study are listed in Supplemental Table S1.
DNA manipulation
We followed standard procedures for cloning. Phusion polymerase, restriction enzymes and T4 DNA ligase from New England Biolabs (NEB) were used for cloning. Taq polymerase (with crimson taq buffer) (NEB) was used for colony polymerase chain reaction (PCR). Zymoclean™ Gel DNA Recovery Kit (Zymo Research) was used for gel extraction. QIAprep Spin Miniprep Kit (Qiagene) was used for plasmid preparations.
Construction of CRISPR1 variants
The methods developed by Renye and Somkuti using plasmid pINTRS were used to construct CRISPR1 variants on the Sth chromosome (57). Plasmid pINTRS contains upstream and downstream homologous regions of a pseudogene locus that encodes truncated components of the glucose phosphoenolpyruvate-dependent phosphotransferase system (PTS locus) (57). Constructs inserted into PTS locus (Figures 4 and 5) were first cloned into pINTRS. Plasmids with correct inserts were confirmed by sequencing and transformed into Sth via electroporation (40). Transformants harboring desired plasmids were first grown at 30° under selection (15 μg/ml erythromycin). Plasmid integration into the genome was selected by shifting the growth temperature to 37° in the presence of erythromycin for 8 h and subsequently plated on LM17 (with 1% agar) to achieve single colonies. Colonies with plasmid integrated were grown in LM17 broth at 37° with selection. Excision of the plasmid and plasmid loss were allowed to occur by shifting the growth temperature to 30° without selection for 4–5 days with two subcultures (1:500 dilution) per day. Cells that had lost the plasmid were identified through patching of single colonies on LM17 plates with and without selection. Correct mutations were confirmed via PCR amplification and sequencing of the PTS region.
Figure 4.
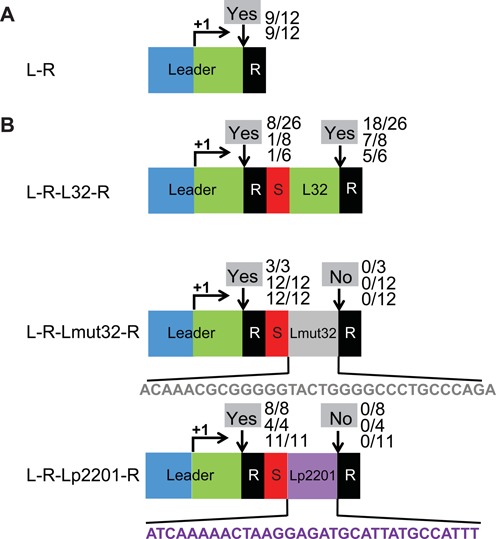
Leader sequence directs adaptation. (A) A minimal CRISPR1 comprised of the leader (157 bp upstream of the repeat) and a repeat sequence (‘R’ black) was introduced at an ectopic locus in Sth. The promoter region and transcribed region of the leader are indicated in blue and green, respectively. A right arrow and ‘+1’ indicate the transcription start site. (B) Variants of CRISPR1 were engineered at the ectopic site with various sequences inserted between the spacer and second repeat of an L-R-S-R locus as indicated. The inserts are: 32 bp transcribed leader region (green, denoted ‘L32’) in ‘L-R-L32-R’, mutated transcribed leader region (gray, denoted ‘Lmut32’) in ‘L-R-Lmut32-R’ and +1 to +32 of the transcribed region of pSTH2201 (purple, denoted ‘Lp2201’) in ‘L-R-Lp2201-R’. Experimentally observed adaptation events are indicated at the downward arrows at each site as in Figure 2.
Figure 5.
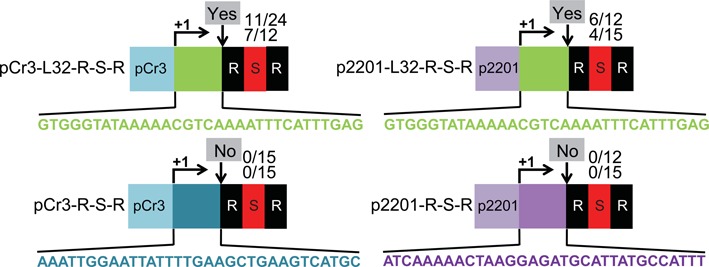
Adaptation does not depend on specific leader promoter sequences. Variants of CRISPR1 were engineered at the ectopic site with various substitutions of the promoter and transcribed regions of the leader of an L-R-S-R locus as indicated. The leaders of ‘pCr3-L32-R-S-R’ and ‘pCr3-R-S-R’ contain the promoter region of CRISPR3 (light blue) and the transcribed region of either CRISPR1 (green) or CRISPR3 (dark blue), respectively as indicated. The leaders of ‘p2201-L32-R-S-R’ and ‘p2201-R-S-R’ contain the promoter region of pSTH2201 (light purple) and the transcribed region of either CRISPR1 (green) or pSTH2201 (dark purple), respectively as indicated. Sequences of the transcribed regions are shown. Experimentally observed adaptation events are indicated at the downward arrows at each site as in Figure 2.
CRISPR1 variants at the native locus were similarly constructed (Figures 2, 6 and 7), with a few alterations. We first constructed a derivative of pINTRS by replacing the PTS homologous regions with a multiple cloning site, yielding plasmid pINTRS-MCS. Seven hundred to eight hundred upstream and downstream homologous regions of CRISPR1 locus were PCR amplified, combined to a single fragment by overlap PCR, cloned into pINTRS-MCS and sequenced to yield pINTRS-Cr1. Mutations of the CRISPR1 leader and the repeat were achieved via quikchange mutagenesis on pINTRS-Cr1. The above protocol for chromosomal insertions into PTS locus was followed to accomplish deletions or mutations at the native CRISPR1 locus.
Figure 2.
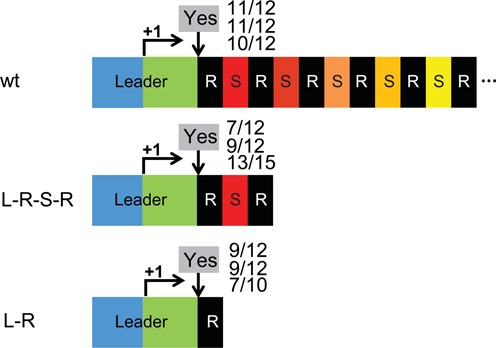
The leader and a single repeat are required for adaptation. Adaptation was examined at various truncated CRISPR1 loci as described in Figure 1. ‘L-R-S-R’ retains the first spacer (‘S’, red) with upstream and downstream repeats (‘R’, black). ‘L-R’ includes a single repeat. The leader is divided into promoter region (blue) and transcribed region (green), with transcription start site indicated with a right arrow and ‘+1’. Downward arrow indicates the leader-repeat junction where new spacer acquisition generally occurs. Occurrence of adaptation (detected by PCR in multiple independent experiments) is indicated by ‘Yes’ or ‘No’. Numbers of adaptation events observed among total survivors examined (by PCR in independent experiments) are indicated.
Figure 6.
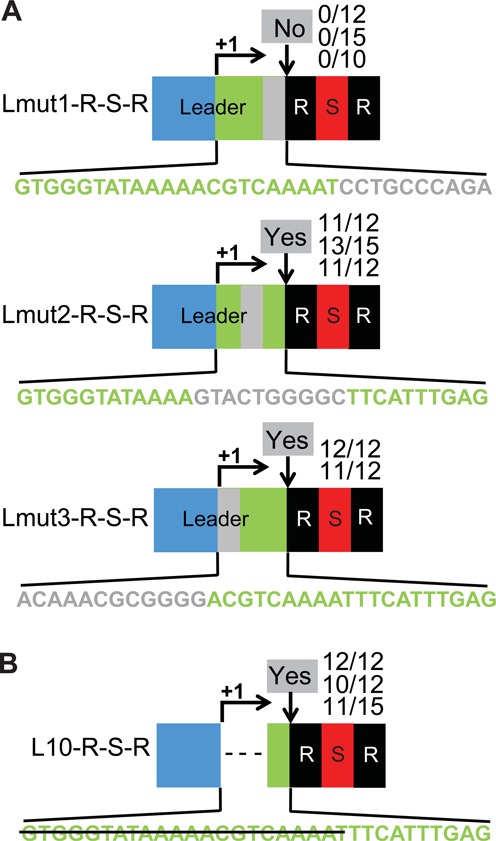
Leader sequences essential for adaptation are found in 10 bp region adjacent to repeat. (A) Three segments of the 32 bp transcribed region of the CRISPR1 leader (green) were mutated in the context of the L-R-S-R locus (Figure 2). Substitutions (adenine<−>guanine, cytosine<−>thymine) were made individually in the repeat-proximal 10 bp (gray in ‘Lmut1-R-S-R’), central 10 bp (gray in ‘Lmut2-R-S-R’) and repeat-distal 12 bp (gray in ‘Lmut3-R-S-R’) of the transcribed region. Sequences of the transcribed regions are shown. (B) The ‘L10-R-S-R’ locus includes just the repeat-proximal 10 bp of the CRISPR1 leader transcribed region (green); the first two segments (22 bp) of the transcribed region are deleted (strikethrough). Experimentally observed adaptation events are indicated at the downward arrows at each site as in Figure 2.
Figure 7.
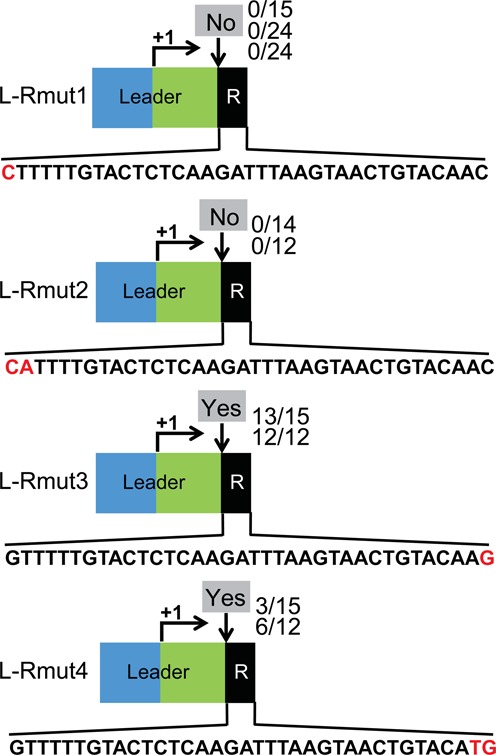
Repeat sequence adjacent to the leader is critical for adaptation. Constructs with mutations in the repeat element of the minimal CRISPR1 locus L-R (Figure 2) were examined. One or two nucleotides at the leader-proximal end (‘L-Rmut1’ and ‘L-Rmut2’) or at the leader-distal end (‘L-Rmut3’ and ‘L-Rmut4’) of the repeat were substituted, respectively as indicated. Sequences of the repeats are shown. Experimentally observed adaptation events are indicated at the downward arrows at each site as in Figure 2.
Phage infection and adaptation analysis
Overnight cultures of Sth strains were diluted into fresh media (1:100) and grown for 2–3 h at 42° until the optical densities (A600) reached ∼0.3. Phage infection was performed with phage 2972 at multiplicity of infection of 0.3 (3,40). Survivors were tested for spacer acquisition by using specific primers (listed in Supplemental Table S2) for the CRISPR locus of interest.
RESULTS
Adaptation at Sth CRISPR1
Among the four CRISPR systems in Sth, only the two Type II-A systems (CRISPR1 and CRISPR3) in Sth DGCC7710 have been observed to incorporate new spacers during phage infection under laboratory conditions (3,39,40). CRISPR1 has been reported to be the dominant system for adaptation (39,55,56). We adopted the phage infection model established in earlier studies with Sth lytic phage 2972 (Figure 1B) (3,39,40,55,56). Following phage infections, bacterial survivors were analyzed for adaptation events at both CRISPR1 and CRISPR3. Consistent with previous findings (39,55,56), PCR amplification of leader-proximal regions of the respective CRISPRs revealed that the majority of the survivors (e.g. 11 out of 12 in the representative example shown) had a new spacer incorporated at CRISPR1, indicated by a repeat-spacer unit-longer PCR product compared to the control, while a small fraction (e.g. 1 out of 12) of the survivors had new spacers incorporated at CRISPR3 (Figure 1B). PCR products of selected survivors were sequenced (data not shown) and found to match phage 2972 genome sequences with adjacent PAM sequences 5′-NNAGAAW-3′ and 5′-NGGNG-3′ expected for CRISPR1 and CRISPR3, respectively (39). Here, we utilize the lytic phage infection model described above to study cis-acting elements required for novel spacer acquisition in Sth CRISPR1 locus.
Adaptation requires a single CRISPR repeat downstream of the leader
Sth CRISPR1 contains 32 repeat-spacer units (each with a 36 bp repeat and 30 bp spacer) and a terminal repeat ((3); see Figure 1A). We investigated the minimum number of repeats required for adaptation by truncating the CRISPR1 array to two repeats (Leader-Repeat-Spacer-Repeat or L-R-S-R) and one repeat (L-R) (Figure 2). Survivors of Sth phage 2972 infection were analyzed via PCR. Wild-type Sth with a full CRISPR array was used as a control (wt, Figure 2). In strains with a CRISPR1 locus with either a single repeat (L-R) or two repeats (L-R-S-R), the majority of the survivors successfully acquired spacers in CRISPR1 (Figure 2; rates of integration observed in independent experiments are indicated). The results indicate that a CRISPR array with a leader and single repeat is functional for adaptation and that additional downstream elements are dispensable.
Leader sequence conservation
Repeat sequences are conserved among related CRISPR-Cas systems (58). We analyzed the leader sequences associated with CRISPR-Cas systems harboring identical or highly similar repeat sequences to that of Sth CRISPR1 (Figure 3). The leader sequences identified are all associated with Type II CRISPR-Cas systems. As expected, the −10 and −35 regions of the promoters of the leaders are highly conserved, consistent with a conserved function in driving crRNA transcription (blue boxes). In addition, an adenosine-rich region close to the transcription start site is weakly conserved (yellow box). Another region of high sequence conservation is found immediately adjacent to the repeat (green box); a 5′-ATTTGAG-3′ sequence is identical among the analyzed CRISPRs (Figure 3).
Figure 3.
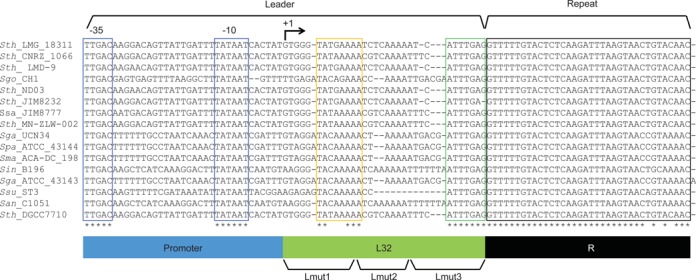
Conservation in leaders from CRISPR systems with repeat sequences related to Sth CRISPR1. CRISPR systems with repeat sequences identical or highly similar to Sth CRISPR1 were identified using CRISPRs Finder (CRISPRdb). Leader sequences and the first repeats were retrieved and aligned. Highly conserved regions are boxed in blue, yellow, green (in the leaders) or black (in the repeats). Identical nucleotides in the alignment are indicated with asterisks. Specific elements are illustrated at the bottom of the alignment. The promoter region, transcribed region of the leader (L32), and the repeat are indicated in blue, green and black, respectively. Three regions that are mutated in Figure 6 (Lmut1, Lmut2 and Lmut3) are indicated. Sequences analyzed are all from Type II CRISPR-Cas systems and are from (accession numbers in parenthesis): Streptococcus thermophilus (Sth) LMG 18311 (NC_006448_1), S. thermophilus CNRZ1066 (NC_006449_1), S. thermophilus LMD-9 (NC_008532_2), Streptococcus gordonii (Sgo) str. Challis substr. CH1 (NC_009785_2), S. thermophilus ND03 (NC_017563_1), S. thermophilus JIM 8232 (NC_017581_1), Streptococcus salivarius (Ssa) JIM8777 (NC_0, 17595_3), S. thermophilus MN-ZLW-002 (NC_017927_1), Streptococcus gallolyticus (Sga) UCN34 (NC_013798_1), Streptococcus pasteurianus (Spa) ATCC_43144 (NC_015600_1), Streptococcus macedonicus (Sma)ACA-DC_198 (NC_016749_1), Streptococcus intermedius (Sin) B196 (NC_022246_1), S. gallolyticus (Sga) subsp.gallolyticus_ATCC_43143 (NC_017576_1), Streptococcus suis (Ssu)ST3 (NC_015433_2), Streptococcus anginosus (San) C1051 (NC_022244_4) and S. thermophilus DGCC7710 (subject of this study) (NZ_AWVZ01000000).
Identification of a leader region capable of stimulating adaptation at a dormant repeat
It has been shown in various types of CRISPR-Cas systems, including CRISPR1 of Sth, that acquisition of new spacers occurs predominantly at the leader-proximal repeat of the CRISPR array (3,23,37,38,41,42,45,52). The leader contains the transcriptional promoter for the CRISPR and this could play a role in the site of adaptation, i.e. adaptation could preferentially occur at the repeat closest to the promoter/transcription start site. Alternatively, the preference could reflect the existence of other specific sequences in the leader that direct adaptation at the neighboring repeat.
Because the stop codon of the upstream csn2 gene overlaps the promoter within the CRISPR leader (Figure 1), we established a minimal CRISPR1 with a single repeat (L-R) at an ectopic chromosomal location (a pseudogene locus that encodes truncated components of the glucose phosphoenolpyruvate-dependent phosphotransferase system (PTS locus) (57)) to eliminate any potential for secondary effects of manipulations on the Csn2 protein in initial experiments. Spacer expansion was observed at the ectopic single repeat CRISPR in response to phage challenge (Figure 4A, L-R), indicating that the element functions effectively at the ectopic locus and also that spacer acquisition does not require close proximity between the Cas protein-coding genes and the CRISPR array.
We then engineered and examined multiple variants of CRISPR1 at the ectopic locus, aiming to dissect the function of the leader in adaptation. The transcription start site of CRISPR1 is located 32 bp upstream of the first repeat (43). We refer to this 32 bp region of the leader that is proximal to the first repeat as L32 (green segment, Figures 1B, 3 and 4A). We placed an additional L32 region upstream of the second repeat element in a L-R-S-R locus (L-R-S-L32-R in Figure 4B). Phage infections were performed with the strain containing the ectopic L-R-S-L32-R locus (Figure 4B). Sth survivors were first pre-screened for spacer expansion at the ectopic locus and then further analyzed for spacer expansion at each leader-repeat junction using specific primers. We found that adaptation occurred at both leader elements (Figure 4B), suggesting that the 32 bp of leader sequence nearest the first CRISPR repeat (L32) is capable of directing adaptation at an immediately downstream dormant repeat, and that proximity to the promoter is not critical (within the range tested). Interestingly, for unknown reasons, adaptation at the second repeat occurred at a higher frequency than at the first (upstream) repeat.
To address the specificity of this finding, we examined constructs in which the additional L32 element was mutated via transitions (purine–purine, pyrimidine–pyrimidine, denoted Lmut32) or replaced by the first 32 bp of the transcribed region of an unrelated constitutive promoter pSTH2201 (denoted Lp2201) (59) (Figure 4B). Both alterations resulted in loss of adaptation specifically at the second junction (Figure 4B). These results suggest that sequences within the last 32 bp of the leader are necessary and sufficient to direct adaptation at the junction with an adjacent downstream CRISPR repeat element in the context of a CRISPR array.
Upstream leader sequences are not important for adaptation
The results shown in Figure 4 suggest that the L32 region of the leader nearest the repeat is sufficient to mediate adaptation, however, to more rigorously determine if sequences in the upstream region of the leader play a role in adaptation (e.g. in recruiting factors required for the process to the region), we replaced the upstream region with other promoter sequences. We replaced the CRISPR1 promoter with the promoter of CRISPR3 (pCr3) or a non-CRISPR, constitutive promoter pSTH2201 (p2201) (59) upstream of the last 32 bp of the leader (L32) and the repeat-spacer-repeat unit (pCr3-L32-R-S-R and p2201-L32-R-S-R, Figure 5). Phage infections and Sth survivor analyses were performed and adaptation occurred for both promoter swap constructs (Figure 5). However, when the last 32 bp of the leader (L32) was replaced by the sequences immediately downstream of each of the respective alternate promoters, adaptation did not occur (pCr3-R-S-R and p2201-R-S-R, Figure 5). These results indicate that the leader sequence upstream of the 32 bp region adjacent to the first repeat is not required for adaptation. We note that we were unable to test for any requirement for transcription per se since the assay requires crRNA production to survive phage infection.
Leader sequences within 10 bp of the repeat are crucial for adaptation
Further mutational analysis revealed that sequences in the 10 bp of the leader closest to the leader-repeat junction are essential for the process of adaptation (Figure 6). We made substitutions in the L32 region of an L-R-S-R unit at the native CRISPR1 locus (Figure 6 and see results for wild-type L-R-S-R construct in Figure 2). Transition mutations were introduced in the repeat-proximal 10 bp (Lmut1-R-S-R), central 10 bp (Lmut2-R-S-R) and repeat-distal 12 bp (Lmut3-R-S-R) of the L32 region of the leader (Figure 4) and Sth survivors of phage infection were analyzed for successful adaptation at the mutant loci via PCR (Figure 6A). Mutation of the distal and central regions of L32 did not disrupt adaptation (Figure 6A). Furthermore, deletion of these two regions did not significantly affect adaptation (Figure 6B), indicating that the first 22 bp of L32, including the weakly conserved A-rich region (Figure 3), are dispensable for adaptation. However, mutation of the repeat-proximal region (Lmut1-R-S-R) that includes the highly conserved ATTTGA sequence (Figure 3) resulted in loss of acquisition of spacers (Figure 6A). The results indicate that the leader sequence within 10 bp of the leader-repeat junction plays a key role in CRISPR adaptation.
Repeat sequences near the leader are crucial for directing adaptation
A single repeat is sufficient for adaptation (Figure 2). The ability to further analyze repeat sequences important in adaptation observed in response to phage infection, however, will be limited to the extent that the repeat sequences are also important in the downstream processes of crRNA processing and phage silencing that are required for survival (and therefore for detection of the adaptation event) (3–12). We tested the effect of mutations in the two nucleotides at each end of the repeat in the L-R construct at the native CRISPR1 locus (Figures 2 and 7). Single nucleotide (G to C) or double nucleotide (GT to CA) mutations were introduced at the leader-proximal end of the repeat (L-Rmut1 and L-Rmut2, respectively, Figure 7). Likewise, single nucleotide (C to G) or double nucleotide (AC to TG) mutations were made at the leader-distal end of the repeat (L-Rmut3 and L-Rmut4, respectively, Figure 7). Phage infections were performed and the resulting Sth survivors were analyzed. The strains containing repeat mutations at the leader-proximal end (L-Rmut1 and L-Rmut2) failed to acquire spacers at the mutant locus (Figure 7). In contrast, adaptation occurred at the locus harboring mutations in the leader-distal end of the repeat (L-Rmut3 and L-Rmut4; note that the double mutation appeared to acquire new spacers at a lower efficiency than the single mutation, Figure 7).
Because the adaptation assay depends on production of functional crRNAs for survival of phage infection as noted above, we tested for the ability of a crRNA from both the L-Rmut1 and L-Rmut2 constructs to guide defense. The leader region of CRISPR1 gives rise to a crRNA (43), so we transformed a plasmid containing the leader target sequence and a PAM into the L-Rmut1 and L-Rmut2 strains and found that transformation efficiency was greatly reduced (relative to control plasmids lacking the target sequence (no target) and to a control strain lacking a repeat (L)), indicating that the repeat sequence changes in L-Rmut1 and L-Rmut2 do not interfere with defense (Supplemental Figure S1) and specifically affect adaptation. Mutation of the equivalent terminal nucleotide in the E. coli Type I-E system is tolerated (60), suggesting a difference between the sequence requirements of the systems. We also note that, as expected, sequence analysis revealed that the mutated repeat sequences were duplicated when spacers were acquired in the L-Rmut3 and L-Rmut4 strains (data not shown). In summary, the results of our experiments indicate that at least the first nucleotide of the repeat adjacent to the leader is critical for acquisition of novel spacers in Type II-A CRISPR-Cas systems.
DISCUSSION
The acquisition of invader-derived spacers into the host CRISPR locus upon an invader encounter is a requisite step for achieving CRISPR-Cas immunity against the invader. However, the mechanisms underlying this initial step of CRISPR action remain understudied and largely unresolved. Using a well-established Sth host/phage 2972 system (3), we have identified the cis-acting sequence requirements for capture of novel phage spacers into a CRISPR locus by a Type II-A CRISPR-Cas system. Our detailed genetic analyses demonstrate that the leader sequence and a single downstream repeat provide sufficient sequence information for directing adaptation (Figure 2). Importantly, our results show sequences immediately spanning the leader-repeat junction provide the crucial information required for spacer integration at CRISPR loci (Figures 4–6). Our results suggest that the defined leader-repeat junction sequences are likely recognition signals responsible for recruiting the adaptation machinery for spacer integration. A common role of leader-repeat sequences in mediating CRISPR adaptation in many or all CRISPR-Cas systems would provide a plausible explanation for why new spacer acquisition almost invariably occurs at the leader-proximal repeat rather than at identical repeat structures located elsewhere in the CRISPR array (3,23,36–42,52). Indeed, we demonstrated that introduction of a key leader sequence element adjacent to an internal repeat induces adaptation at the internal repeat (Figure 4).
Conserved role of leader sequences adjacent to the first CRISPR repeat in directing adaptation
The critical role of the repeat-proximal leader region for adaptation defined in our study (Figures 4–6) could be a general feature for (all) other CRISPR-Cas systems. Leaders are typically operationally defined as ∼100–500 bp sequences located adjacent to CRISPR arrays that do not contain open reading frames (15,23,41,43,44). One conserved function of CRISPR leaders appears to be to control expression of downstream crRNA transcripts. Leaders frequently contain core promoter sequences that drive crRNA expression (41,43–48) and we speculate that additional identified conserved leader elements may help regulate CRISPR array transcription (Figure 3 (yellow box) and (44)). A second conserved function of leaders, supported by this study, is in directing new spacer acquisition at the leader-proximal repeat. Our finding that the critical leader information for Sth CRISPR1 (Type II-A) adaptation is located within a 10-bp segment at the leader-repeat junction (Figure 6) that contains a 7-bp sequence element that is highly conserved among CRISPRs with similar repeat sequences (Figure 3) is consistent with the findings of mutational analysis of the E. coli CRISPR (Type I) leaders that narrowed the critical leader information to within 43 bp upstream of the repeat (23,36). Moreover, in silico comparisons of predicted leaders for over 50 sequenced Sulfolobales and for 6 Pyrococcales genomes indicate the presence of conserved leader sequences adjacent to other CRISPR repeat family types as well (44,61). Further experiments are needed to verify whether these conserved motifs are important for directing adaptation in the corresponding CRISPRs. Collectively, the findings to date indicate that repeat-proximal leader sequences play a role in conjunction with the adjacent repeat in facilitating CRISPR adaptation.
Mechanism for CRISPR array opening during spacer integration
CRISPR adaptation appears to involve multiple steps including: (i) PAM-dependent recognition and cutting of invader DNA into spacer fragments of defined size, (ii) opening of the CRISPR array, likely by staggered nicks at the end of the first repeat by Cas1 (in complex with Cas2) (33,34), (iii) end joining of the incoming spacer into the ‘opened’ repeat and (iv) gap filling by cellular machinery (polymerases), resulting in the addition of a spacer between duplicated repeats immediately adjacent to the leader. It has been proposed that the proximal nick, near the leader-repeat junction, relies on sequence-specific recognition and strand cleavage, while the repeat nicking event at the leader-distal end occurs a fixed distance from the first nick. In studies using Cas proteins and a CRISPR from different systems (Cas1 and Cas2 from E. coli K-12 and CRISPR from non-K12 E. coli), it was observed that the new repeat generated during adaptation included the last two nucleotides of the leader and correspondingly, the last two nucleotides of the original repeat were absent (36). The authors proposed that Cas1/Cas2 generated the first nick at an incorrect position due to faulty recognition of the non-cognate leader-repeat sequence and that the second nick is generated a repeat-length (ruler) downstream of the first nick (36). Our results also suggest that any sequence recognition involved in generation of the nicks would occur at the leader-repeat junction, in the vicinity of the proposed first nick. We note that it is highly likely that additional leader-proximal repeat sequences (in addition to the terminal nucleotide that we identified) are involved in this recognition. Cas1 and Cas2 are common to the diverse CRISPR-Cas systems. The proposed mechanism predicts co-evolution of Cas1/Cas2 with CRISPR leaders-repeat junction sequences (36).
Sth CRISPR1 is an active system for ‘naïve’ adaptation
The presence of pre-existing CRISPR spacers that partially match invader sequences dramatically increases spacer acquisition from the same invader (i.e. primed adaptation) (37,38,42). Adaptation has not been observed in response to invasion by phage or plasmid in the absence of pre-existing spacers in several systems (42,52). It has been speculated that the ability of Sth CRISPR1 to readily adapt to lytic phage 2972 may be due to the existence of two spacers partially matching the phage in Sth CRISPR1 (Spacer 14 and spacer 21 match the phage 2972 genome with one and three mismatches, respectively) (42). However, our results demonstrate that efficient adaptation does not depend on these partially matching spacers and can occur at a locus without spacers (Figure 2). Thus, adaptation by the Sth CRISPR1 system does not require priming and can occur upon a virgin encounter (‘naïve’ adaptation) with the phage.
Taken together, our bioinformatic and genetic analyses indicate that the CRISPR leader-repeat junction plays a specific role in directing spacer adaptation in the Type II-A CRISPR-Cas system at Sth CRISPR1 (Type II-A), and likely in many CRISPR-Cas systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank members of the Terns lab and Claiborne Glover for helpful discussions. We would also like to thank Rodolphe Barrangou, Sylvain Moineau, Michael Federle, John Renye and George Somkuti for generously providing strains, plasmids and phages, and Denise Tremblay and Sylvain Moineau for protocols and invaluable technical assistance.
FUNDING
National Institutes of Health (NIH) [RO1 GM099876 to M.T. and R.T.]; NIGMS of the NIH [F32GM108263 to Y.W.]. Funding for open access charge: NIH [RO1 GM099876].
Conflict of interest statement. None declared.
REFERENCES
- 1.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 2.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terns M.P., Terns R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeks J., Naismith J.H., White M.F. CRISPR interference: a structural perspective. Biochem. J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 11.Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 12.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 16.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 17.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 18.Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 19.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 20.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 21.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 22.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 23.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 25.Shah S.A., Erdmann S., Mojica F.J., Garrett R.A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carte J., Wang R., Li H., Terns R.M., Terns M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiedenheft B., Zhou K., Jinek M., Coyle S.M., Ma W., Doudna J.A. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Beloglazova N., Brown G., Zimmerman M.D., Proudfoot M., Makarova K.S., Kudritska M., Kochinyan S., Wang S., Chruszcz M., Minor W., et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam K.H., Kurinov I., Ke A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J. Biol. Chem. 2011;286:30759–30768. doi: 10.1074/jbc.M111.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam K.H., Ding F., Haitjema C., Huang Q., Delisa M.P., Ke A. Double-stranded endonuclease activity in B. halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J. Biol. Chem. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo Y., Jung D.K., Bae E. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PloS One. 2012;7:e33401. doi: 10.1371/journal.pone.0033401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arslan Z., Wurm R., Brener O., Ellinger P., Nagel-Steger L., Oesterhelt F., Schmitt L., Willbold D., Wagner R., Gohlke H., et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013;41:6347–6359. doi: 10.1093/nar/gkt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Kasciukovic T., White M.F. The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PloS One. 2012;7:e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diez-Villasenor C., Guzman N.M., Almendros C., Garcia-Martinez J., Mojica F.J. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 2013;10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 38.Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J. CRISPR interference directs strand specific spacer acquisition. PloS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath P., Romero D.A., Coute-Monvoisin A.C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 41.Erdmann S., Garrett R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carte J., Christopher R.T., Smith J.T., Olson S., Barrangou R., Moineau S., Glover C.V., 3rd, Graveley B.R., Terns R.M., Terns M.P. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 2014;93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillestol R.K., Shah S.A., Brugger K., Redder P., Phan H., Christiansen J., Garrett R.A. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 45.Erdmann S., Le Moine Bauer S., Garrett R.A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 2014;91:900–917. doi: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 46.Bernick D.L., Cox C.L., Dennis P.P., Lowe T.M. Comparative genomic and transcriptional analyses of CRISPR systems across the genus Pyrobaculum. Front. Microbiol. 2012;3:251. doi: 10.3389/fmicb.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V., 3rd, Graveley B.R., Terns R.M., et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M., Wang R., Xiang H. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014;42:7226–7235. doi: 10.1093/nar/gku389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fineran P.C., Gerritzen M.J., Suarez-Diez M., Kunne T., Boekhorst J., van Hijum S.A., Staals R.H., Brouns S.J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erdmann S., Shah S.A., Garrett R.A. SMV1 virus-induced CRISPR spacer acquisition from the conjugative plasmid pMGB1 in Sulfolobus solfataricus P2. Biochem. Soc. Trans. 2013;41:1449–1458. doi: 10.1042/BST20130196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horvath P., Coute-Monvoisin A.C., Romero D.A., Boyaval P., Fremaux C., Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 55.Sun C.L., Barrangou R., Thomas B.C., Horvath P., Fremaux C., Banfield J.F. Phage mutations in response to CRISPR diversification in a bacterial population. Environ. Microbiol. 2013;15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- 56.Paez-Espino D., Morovic W., Sun C.L., Thomas B.C., Ueda K., Stahl B., Barrangou R., Banfield J.F. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 57.Renye J.A., Jr., Somkuti G.A. Insertion of a heterologous gene construct into a non-functional ORF of the Streptococcus thermophilus chromosome. Biotechnol. Lett. 2009;31:759–764. doi: 10.1007/s10529-009-9931-6. [DOI] [PubMed] [Google Scholar]
- 58.Kunin V., Sorek R., Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somkuti G.A., Solaiman D.K. STP2201, a chromosomal promoter sequence of Streptococcus thermophilus. Curr. Microbiol. 1997;35:180–185. doi: 10.1007/s002849900235. [DOI] [PubMed] [Google Scholar]
- 60.Goren M.G., Yosef I., Auster O., Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J. Mol. Biol. 2012;423:14–16. doi: 10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 61.Norais C., Moisan A., Gaspin C., Clouet-d'Orval B. Diversity of CRISPR systems in the euryarchaeal Pyrococcales. RNA Biol. 2013;10:659–670. doi: 10.4161/rna.23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


