Abstract
The Krüppel-associated box (KRAB) domain is a transcription repression module from the largest family of transcriptional regulators encoded by higher vertebrates. We developed a drug-controllable regulation system based on an artificial KRAB-containing repressor (tTS) that targets the endogenous Hprt gene to explore the regulatory mechanism and molecular basis of KRAB-containing regulators within the context of an endogenous gene in vivo. We show that KRAB can mediate irreversible and reversible regulation of endogenous genes in mouse that is dependent on embryonic developmental stage. KRAB-induced stable DNA methylation within the KRAB binding region during the early embryonic stage, resulting in irreversible gene repression. In later stages, KRAB mainly induced de-acetylation and methylation of histone, resulting in reversible gene repression. Thus, we have characterized the KRAB-mediated regulation system within the context of an endogenous gene and multiple spatiotemporal ranges, thereby providing a basis for identifying the function of KRAB-containing regulators and aiding development of novel KRAB-based gene regulation tools in vivo.
INTRODUCTION
There are ∼400 zinc finger proteins containing Krüppel-associated box (KRAB) domains in both human and mouse (1–3). Zinc finger proteins are the largest family of transcriptional regulators encoded by higher vertebrate genomes. After binding to DNA through zinc finger motifs, these transcription factors recruit KAP1/TIF1β corepressor complexes via the KRAB domain, and act as potent transcriptional repressors (4–6). KAP1 acts as a scaffold to further recruit factors associated with DNA methylation and formation of repressive chromatin, such as heterochromatin protein 1 (HP1), histone deacetylases and histone methyltransferases (7–10).
Recently, the potent transcriptional repressor activity of the KRAB domain was used to develop a conditional gene regulation system (11). In this system, the KRAB domain was fused to the DNA binding domain of the Escherichia coli tetracycline repressor (tetR), forming an artificial transcriptional silencer, tTS (also known as tTRKRAB) (11). In turn, tTS binds to tetracycline response elements (TRE; also known as ‘tetO’, tetracycline operator) that are inserted close to a target promoter and recruit corepressors, thereby causing transcriptional repression of the target gene. Repression can be released by doxycycline (dox) administration, enabling dox-controlled exogenous gene expression and endogenous gene knockdown (11). This system also provides an excellent platform to explore the mechanism(s) of the KRAB/KAP1 system in regulating gene expression. By decreasing histone H3-acetylation and increasing H3 lysine 9 trimethylation at the cell level, KRAB/KAP1 mediates reversible and long-range transcriptional repression through heterochromatin spreading (12). In an earlier report, transgenic mice expressing tTRKRAB and tetO-controlled green fluorescent protein (GFP) were generated using lentiviral vectors. GFP expression was irreversibly silenced with tTRKRAB binding to tetO sequences during early embryogenesis. This irreversible silencing was due to KRAB-induced de novo DNA methylation of the promoter (13). However, a recent study suggested that tethering of KRAB to an endogenous gene body does not contribute to irreversible gene silencing, even under the same circumstances as previously described, as KRAB binding to gene bodies did not induce stable DNA promoter methylation (14). It is worthy to note that in these experiments, tetO-controlled GFP was inserted into the genome using a lentivirus vector, and in some cases, KAP1 has silenced retroviruses in embryonic stem (ES) cells and early embryos (15–18). Thus, there is reasonable uncertainty as to whether irreversible silencing is due to KRAB binding or cooperation between KRAB binding and retrovirus elements.
The tTS conditional gene regulation system has also been used to regulate endogenous genes. In the limited examples, the tTS system has been effective in regulating endogenous gene expression (19–23), and could be a potentially versatile genetic tool that is advantageous over conventional gene targeting methods as it can be widely used to induce reversible regulation of endogenous genes in mice. Nevertheless, it is still unclear if irreversible silencing is induced by KRAB binding within the vicinity of an endogenous gene promoter. Furthermore, its application has been hampered because the advantages, limitations and potential solutions are not fully understood.
In the current study, we explored the regulatory mechanisms of KRAB in vivo to address application of the tTS system. Hypoxanthine guanine phosphoribosyl transferase (Hprt), a housekeeping gene responsible for recycling purines and located on the X chromosome (24), was selected as the target gene for tTS system regulation. We characterized repression performance of the KRAB domain in different tissues and stages, demonstrating that the KRAB domain induces irreversible and reversible regulation modes depending on the developmental stage. Tethering of KRAB within the vicinity of a gene promoter induced irreversible regulation during early embryogenesis, while reversible regulation was mediated by dox administration at other developmental stages. Our data identifies the potential regulatory mechanism of KRAB-containing zinc finger proteins in vivo, and suggests that they may play an important role in establishing DNA methylation patterns during early embryogenesis. Our results also further demonstrate the power of the tTS system for controlling endogenous gene expression.
MATERIALS AND METHODS
Animals
Mice (tTS transgenic and HprtTRE knockin) were housed in a pathogen-free facility and maintained under controlled conditions (21°C–24°C; 12-h light-dark periods). As described previously (22), for inducible and reversible experiments, doxycycline hydrochloride (dox) (Sigma-Aldrich, St. Louis, MO, USA) was administered in drinking water at a concentration of 2 mg/ml supplemented with 5% sucrose, and was refreshed every 2 days. All animal study protocols were reviewed and approved by the Institutional Animal Care and Use Committee in Shanghai Research Center for Model Organisms (No. 2010–0007).
Generation of HprtTRE knockin mice
Construction of targeting vectors has been described previously (22). The TRE site was amplified from pLVCT-tTRKRAB (Addgene, Cambridge, MA, USA). The final targeting construct contained a 2.469 kb 5′-homology arm, 2.302 kb knockin fragment (TRE-loxp-FRT-PGK-Neo-FRT-loxp) and 4.046-kb 3′-homology arm. The targeting vector was linearized by NotI digestion. The ES cell targeting and screening procedure has been described previously (22). Surviving ES cell colonies were polymerase chain reaction (PCR) amplified using primer sets I/II and III/IV (Figure 1B) and confirmed by sequencing. Positive ES cell clones were expanded and injected into C57BL/6J blastocysts to generate chimeric offspring. Chimeric mice were mated with C57BL/6J mice. Targeted F1 offspring were genotyped from tail genomic DNA.
Figure 1.
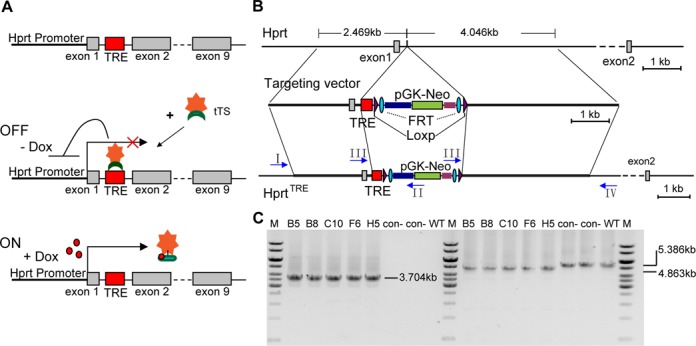
Schematic diagram of the genetic modification system for regulation of the endogenous Hprt gene and generation of HprtTRE ES clones. (A) Schematic representation of the model principle for regulating endogenous Hprt expression. (B) Structures of wild type Hprt, targeting vector and HprtTRE. In HprtTRE, a TRE-loxp-FRT-Neo-FRT-loxp cassette was inserted into the first intron of the murine Hprt gene between sites 198 and 199. (C) Representative PCR identifying HprtTRE from targeted HprtTRE ES cell clones using primers I-IV (arrows in panel B). Primer sets I and II, which identified the 5′-homology arm (left half of panel C), yielded a single fragment (3.074 kb) in positive clones and no product in negative clones (top). Primer sets III and IV, which identified the 3′-homology arm (right half of panel C), separately yielded single 4.863 and 5.386 kb fragments in positive and negative clones, respectively. B5, B8, C10, F6 and H5: positive ES cell clones; con-: a negative clone; WT: wild type ES cell as a negative control; M: 1 kb DNA ladder from Fermentas.
Mouse embryonic fibroblast (MEF) isolation and treatment
Isolation and propagation of MEFs has been previously described (25). For drug-based assays, MEFs were plated at an appropriate density in medium with or without dox (1 μg /ml). For analyzing Hprt and neomycin expression, MEFs were treated with 6-thioguanine (6-TG, 8 μg/ml; Sigma) or geneticin (G418, 600 μg/ml; Life Technologies, Grand Island, NY, USA) for 4 days before obtaining images. To investigate epigenetic mechanisms involved in gene regulation, MEFs were treated with trichostatin A (TSA, 300 nM; Sigma) or 5-Aza-2′-deoxycytidine (5-Aza, 7.5 μM; Sigma) for the indicated time before analysis (13).
Quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and cDNA synthesized using the PrimeScript RT reagent kit (Takara, Dalian, China). qPCR was performed using the Mastercycler Realplex2 detection system (Eppendorf, Hamburg, Germany) and SYBR Premix Ex Taq mixture (Takara). Primer sequences were: Hprt forward: 5′-GCTGGTGAAAAGGACCTCT-3′, reverse: 5′-CACAGGACTAGAACACCTGC -3′; Actin forward: 5′-CCTGTATGCCTCTGGTCGTA-3′, reverse: 5′-CCATCTCCTGCTCGAAGTCT-3′; Kap1 forward: 5′-CCTCGGCGGCCTCTGGTAG-3′, reverse: 5′-TGGCTGGGCATTATCTTCACA-3′. For expression analysis, all samples were normalized to actin signal.
Western blotting
MEFs were harvested and lysed in Radioimmunoprecipitation assay buffer (RIPA buffer) containing protease inhibitor cocktail (Sigma). The cerebral cortex was homogenized in RIPA buffer containing protease inhibitor cocktail using a glass homogenizer. Lysates were incubated for 10 min on ice and centrifuged at 13 000 × g for 10 min at 4°C. Supernatants were collected, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Amersham Biosciences, Piscataway, USA). Membranes were blocked with Tris-Buffered Saline and Tween 20 (TBST) buffer containing 5% fat-free milk powder for 1 h at room temperature, and then incubated overnight at 4°C with primary antibodies against Hprt (1:5000; Abcam, Cambridge, UK) and β-Actin (1:2000; Santa Cruz, CA, USA). After washing, membranes were incubated with fluorescent-conjugated secondary antibody for 1 h (1:10000; LI-COR Biosciences, Lincoln, USA). Protein bands of interest were analyzed using the Odyssey Infrared Imaging System (LI-COR).
DNA methylation analysis
Bisulfite sequencing was performed using the EpiTect bisulfite kit (Qiagen, Hilden, Germany), according to the manufacturer's recommendations. Methylation levels of two CpG islands located in the promoter and intron 1 of the Hprt gene were assayed. Converted DNA was amplified by PCR. PCR reaction conditions were: 94°C for 4 min, followed by 34 cycles of 94°C for 30 s, 57°C for 30 s and 72°C for 40 s. PCR amplified products were cloned into a T vector (Takara). At least 10 positive clones were sequenced. Primer sequences for amplification of the Hprt promoter were: forward 5′-TTTTTGAGTTATTGTTGAGG-3′; reverse 5′-AATCCCCTTAACTCACCAC-3′. To amplify the Hprt intronic region, a nested PCR reaction was performed. The first set of primer sequences were: forward 5′-GTGGGGATGTTTTTTTAGTGAGTT-3′; reverse 5′-CCAAACCTAAACATACCCTCTCATA-3′. The second set of primer sequences were: forward 5′-GTGGGGATGTTTTTTTAGTGAGTT-3′; reverse 5′-ATAGAGAGAGAGGGTAGGTTG-3′.
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the EZ-CHIP kit (Millipore, MA, USA) according to the manufacturer's recommendations but with minor modification. Approximately 107 MEFs were crosslinked using 1% formaldehyde at room temperature for 10 min. The crosslinked reaction was quenched by adding glycine and cells washed on ice with phosphate buffered saline. Cells were harvested by scraping the dish, resuspended in 200 μl lysis buffer and sonicated on ice to shear genomic DNA. Sonicated supernatants were divided into 100 μl aliquots per tube. One aliquot was directly de-crosslinked and used as the total input reference in subsequent quantitative analysis. The other aliquots were diluted with 900 μl dilution buffer and then pre-cleared using Protein G agarose. The antibodies for immunoprecipitation were added and incubated overnight at 4°C: anti-acetyl-Histone H4, anti-trimethyl H4 (Lys20), anti-trimethyl-Histone H3 (Lys9), ChIPAb+Acetyl-Histone H3 (Lys9) (2 μg for 106 cells; Millipore) and anti-Histone H3 antibody-Chip Grade (2 μg for 106 cells; Abcam). Antibody/antigen/DNA complexes were captured by Protein G agarose, washed and eluted with elution buffer. Crosslinked protein/DNA complexes were reversed, and captured chromatin harvested using spin columns. DNA fragments pulled down by antibodies were detected by qPCR using a primer pair targeting the Hprt promoter (5′-CCTTTCTTGGTAGCTGGGCAT-3′ and 5′- GCCCTCTGTCGTCTCCCAGA-3).
Statistical analysis
Data from the same litters was analyzed. Data are expressed as mean ± SEM. Statistical differences between groups were analyzed using one-way analysis of variance, with P < 0.05 considered significant for all statistical analyses.
RESULTS
Mode of tTS regulating endogenous Hprt gene expression
Figure 1A outlines tTS-dox system regulation of endogenous Hprt gene expression. A TRE site was artificially inserted into the vicinity of the endogenous Hprt promoter. In the absence of dox, tTS binds to the TRE site and recruits co-repressors to inhibit transcriptional activity of the Hprt gene promoter, therefore, the status of the Hprt gene will be ‘off’. In the presence of dox, tTS binds to dox and not the TRE site, therefore, the Hprt gene will be ‘on’ with normal expression.
Construction of HprtTRE mouse
To reduce the risk of interfering with normal Hprt transcription by TRE site insertion, the TRE site was inserted into the first intron, which had less potential transcription factor binding sites identified by JASPAR database analysis. To generate HprtTRE knockin mice (HprtTRE), a TRE-loxP-Flippase Recognition Target (FRT)-neomycin resistance gene (Neo)-FRT-loxP cassette was inserted into the first intron of the murine Hprt gene (Figure 1B). HprtTRE positive ES cells were identified by PCR (Figure 1C) and confirmed by DNA sequencing. Three positive ES cell clones were used to generate chimeric HprtTRE founder mice. F1 offspring of these founder mice were genotyped by tail genomic DNA to detect heterozygous HprtTRE mice. As Hprt is located on the X chromosome, homozygous HprtTRE female and male mice were separately named as HprtTRE/TRE and HprtTRE/Y mice. HprtTRE/? mice did not have their gender distinguished.
Reversible dox-dependent on/off regulation of Hprt expression in MEFs
To test tTS performance in regulating Hprt expression, HprtTRE mice were crossed with tTS transgenic mice, as described previously (22), generating HprtTRE/Y:tTS mice and HprtTRE/TRE littermates. Offspring were genotyped by PCR. MEFs from HprtTRE/Y:tTS and HprtTRE/TRE mice that had developed with dox exposure from conception to embryonic day 13.5 (E13.5) were harvested and cultured with or without dox. Tight and rapid regulation of Hprt gene expression was observed by qPCR (Figure 2A). After removing dox for 2 days, Hprt expression levels sharply decreased to 2.3% of normal expression. Increasing the time of dox removal to 4 days resulted in only 0.04% Hprt expression (relative to normal expression) (Figure 2A, red). After adding 1 μg/ml dox to the culture medium, Hprt expression levels returned to normal in only 1 day (Figure 2A, black). High dox responsiveness was also shown using a range of doses for dox treatment, and 0.0004–0.002 μg /ml dox could switch Hprt expression status from ‘off to on’ (Figure 2B, black bars) or ‘on to off’ within 2 days (Figure 2B, gray bars). Hprt expression status (‘on’ or ‘off’) could be freely switched by adding or removing dox (Figure 2C and D). Thus, these cells are in a ‘reversible’ condition. Dox-induced on or off regulation of Hprt expression was confirmed by the phenotype of MEFs exposed to 6-thioguanine (6-TG) (Figure 2E). In the presence of Hprt, 6-TG is converted to a guanine analog that is incorporated into DNA and induces cell apoptosis (26). In the presence of dox, Hprt was normally expressed and 6-TG cytotoxic to MEFs. In the absence of dox, Hprt was silenced and 6-TG not converted into the guanine analog, resulting in MEF survival. The capacity and effectiveness of tTS-mediated gene regulation was also shown by the phenotype of MEFs exposed to geneticin (G418), as tTS also synchronously regulated Neo expression (Figure 2E).
Figure 2.
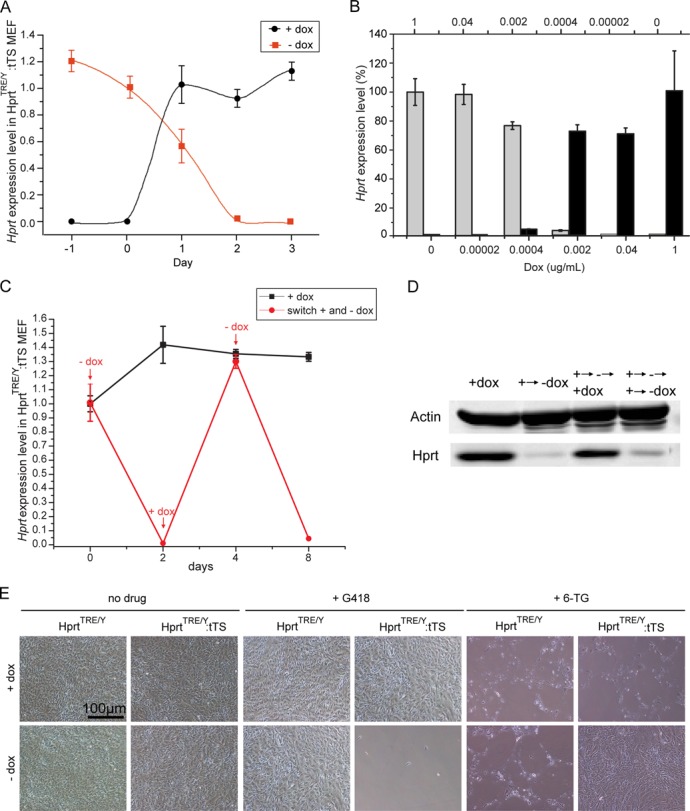
Reversible dox-dependent ‘on/off’ regulation of Hprt expression in MEFs isolated following dox exposure from conception to E13.5. (A) qPCR analysis of Hprt expression levels in HprtTRE/Y:tTS MEFs after removing (red) or adding (black) dox at the indicated time points. (B) Gray bars showed Hprt expression levels in HprtTRE/Y:tTS MEFs using the indicated dox concentrations (above) to maintain Hprt expression for 2 days before qPCR analysis. Black bars showed Hprt expression levels using the indicated dox concentrations (below) to switch on Hprt expression. (C) Reversible regulation of Hprt mRNA expression by adding or removing dox at the indicated time points. (D) Reversible regulation of Hprt protein expression by adding or removing dox at the indicated time points. (E) MEF survival status after 6-TG or G418 treatment in presence or absence of dox.
Irreversible regulation of Hprt expression in MEFs
In a previous study (13), tTS-induced irreversible silencing of lentivirus-mediated exogenous gene expression during early embryogenesis. To determine if tTS induces irreversible regulation of an endogenous gene, MEFs from HprtTRE/Y:tTS and HprtTRE/TRE mice were harvested and cultured with or without dox. HprtTRE/Y:tTS MEFs lost responsiveness to dox and Hprt expression was silenced, even if the culture time was extended to 5 days in the presence of dox (Figure 3A and B). This was also confirmed by the survival status of MEFs under 6-TG and G418 treatment (Figure 3C), showing these cells are in an ‘irreversible’ condition.
Figure 3.
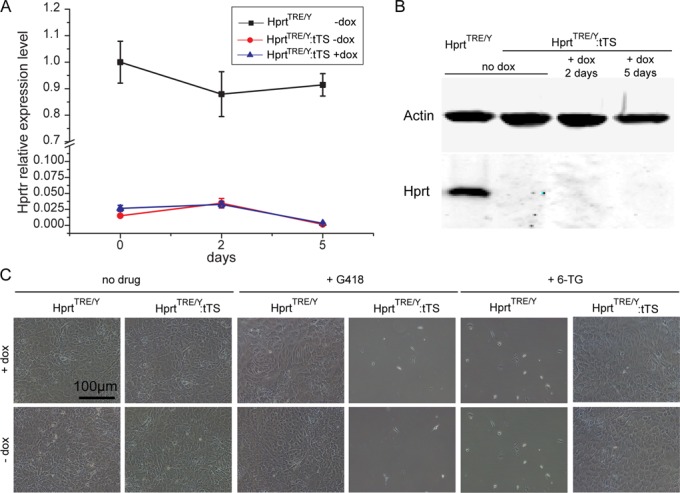
Irreversible regulation of Hprt expression in MEFs isolated following dox exposure from conception to E13.5. qPCR (A) and western blotting (B) analysis of Hprt expression levels in HprtTRE/Y and HprtTRE/Y:tTS MEFs after adding dox at the indicated time points. (C) MEF survival status after 6-TG or G418 treatment in presence or absence of dox.
Reversible dox-dependent on/off regulation of Hprt expression in vivo
To test performance of this system in vivo, Hprt expression was analyzed by qPCR in E10.5 embryos. In E10.5 embryos without dox exposure, Hprt expression from HprtTRE/?:tTS mice was less than 0.8% (P < 10−7) levels observed in HprtTRE/? mice (Figure 4A). In HprtTRE/?:tTS embryos exposed to dox since E0.5, Hprt expression was not significantly different between genotypes (Figure 4A).
Figure 4.
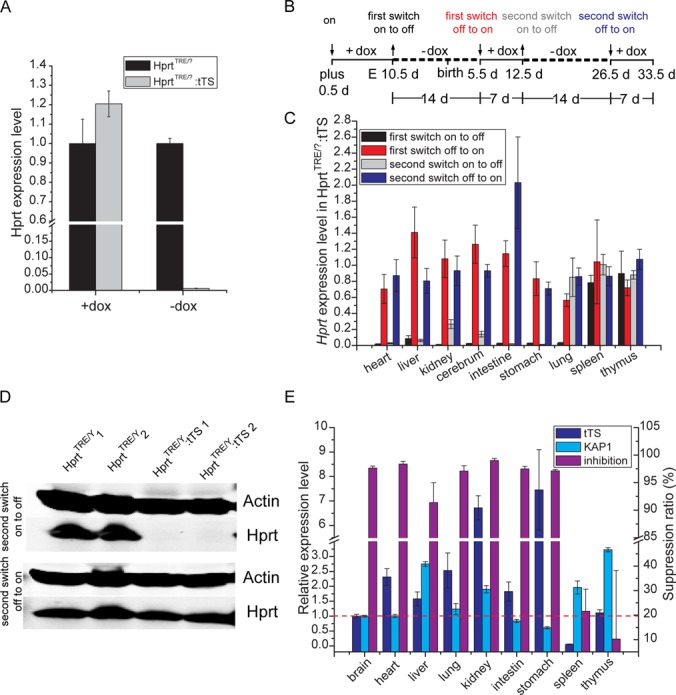
tTS-induced irreversible regulation of Hprt expression in vivo. (A) qPCR of Hprt expression in HprtTRE/? and HprtTRE/?:tTS E10.5 embryos. Pregnant mice were exposed to dox or no dox-containing drinking water for 10 days from E0.5. (B) Schedule of dox introduction or removal from drinking water of pregnant mice. The symbols ‘↓’ and ‘↑’ together with the terms ‘on’, ‘switch off’ and ‘switch off to on’ indicate the action of adding and removing dox, respectively, at the corresponding time points and resulting effects. (C) qPCR of Hprt expression in HprtTRE/?:tTS mice at post-natal day 5.5, 12.5, 26.5 and 33.5 before and after dox re-treatment. Hprt expression levels in HprtTRE/?:tTS mice were normalized to littermate HprtTRE/? mice. Hprt expression in HprtTRE/?:tTS mice switched off after dox removal for 2 weeks, and switched ‘off to on’ after dox administration for 1 week. (D) In the brain of HprtTRE/?:tTS mice, western blotting detected reversible Hprt expression at the second ‘on to off’ and ‘off to on’ switches. (E) tTS and Kap1 expression levels, and the effect of repressing Hprt expression in different tissues at the first ‘on to off’ switch.
To determine performance of dox-dependent reversible regulation in vivo, pregnant mice were given dox-containing (2 mg/ml) drinking water for 10 days beginning at E0.5, followed by no exposure for 14 days, and then access to dox-containing drinking water again for 7 days, no dox for 14 days and dox again for 7 days (Figure 4B). During repeated ‘on–off’ switches, changes in Hprt expression levels in multiple tissues were monitored by qPCR, with different regulation efficiency observed in different tissues (Figure 4C). The expected dox-dependent reversible regulation was observed in heart, liver, kidney, cerebrum, intestine and stomach (Figure 4C and D). However, in spleen and thymus, no significant regulation of Hprt expression was observed after dox withdrawal (Figure 4C), and in lung, efficient regulation was lost at the second dox withdrawal (Figure 4C). To investigate the reason for distinct regulation efficiency in different tissues, expression of tTS and the corepressor, Kap1, were measured by qPCR in different tissues (Figure 4E). No obvious correlation between tTS or Kap1 expression and regulation efficiency was observed.
tTS-induced irreversible regulation of Hprt expression in vivo
To determine if tTS can induce irreversible regulation of an endogenous gene in vivo, pregnant mice were exposed to no dox-containing drinking water for 10 days from E0.5, followed by dox exposure for 14 days. Compared with littermate mice, Hprt expression was silenced in all tissues of HprtTRE/Y:tTS mice (Figure 5A). This silencing could not be reversed, even if the time the mice were exposed to dox was increased to 1 month (data not shown).
Figure 5.
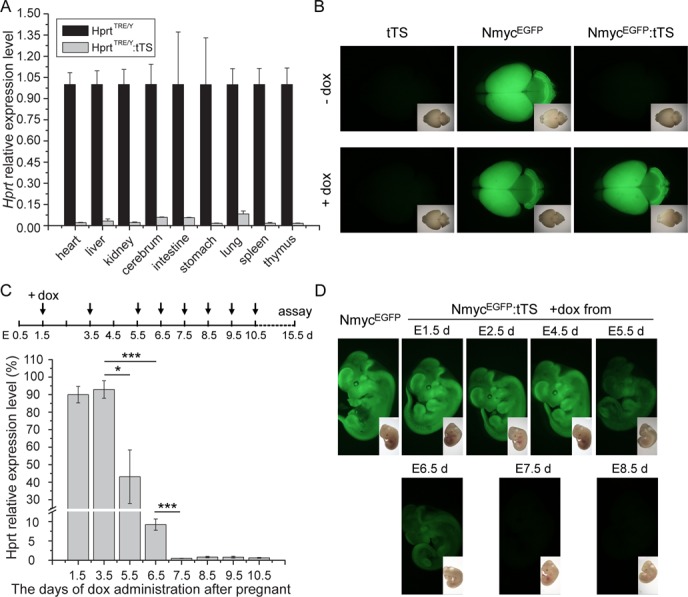
tTS-induced irreversible regulation of Hprt expression in vivo. (A) qPCR analysis of Hprt expression levels in different tissues from HprtTRE/Y and HprtTRE/Y:tTS mice at post-natal day 5. Pregnant mice were given no dox-containing drinking water for 10 days from E0.5, followed by 2 mg/ml dox exposure for 14 days. (B) tTS-induced irreversible silencing of endogenous N-myc promoter activity. EGFP fluorescence assays were performed using brain from post-natal day 7 mice. Top panels: dams were exposed to no dox-containing drinking water until birth, followed by dox exposure for 7 days. Bottom panels: dams were exposed to dox for 6 days from E0.5, followed by dox removal until birth and then dox added for 7 days. Insets show bright field images. (C) Analysis of the time window for inducing irreversible Hprt silencing. Schedule of dox introduction to drinking water of pregnant mice at the indicated time points (top). qPCR analysis of Hprt expression levels in brain of HprtTRE/Y:tTS mice at E15.5 (bottom). These embryos were exposed to dox as in the top schedule. Hprt levels in HprtTRE/Y:tTS mice were normalized to littermate HprtTRE/? mice (n ≥ 3 in each group). (D) Analysis of the time window for inducing irreversible silencing of N-myc promoter activity. EGFP fluorescence assays were performed using E11.5 embryos. Dox was separately introduced to drinking water of pregnant mice at the indicated time points. Insets show bright field images.
To determine if tTS-mediated irreversible silencing observed in HprtTRE:tTS mice is applicable to other tTS-regulated mouse models, a N-myc endogenous gene model was used. In this model (22), a TRE site is inserted in an intron near the promoter of the N-myc gene, and an EGFP cassette fused to the endogenous start codon of N-myc to monitor transcriptional activity of the N-myc promoter. Similar tTS-mediated irreversible silencing was observed using the N-myc mouse model (Figure 5B). Exposing the dam to no dox-containing drinking water during pregnancy caused irreversible EGFP expression silencing, even if the dam was subsequently exposed to dox for 7 days (Figure 5B).
To investigate the time window of irreversible silencing, pregnant mice were exposed to dox for different durations (Figure 5C), beginning from E1.5 to E10.5, and Hprt expression in brain was measured at E15.5. If dox was added at E3.5 or earlier, Hprt expression in HprtTRE/Y:tTS mice was normal. With delayed dox introduction, progressive and significant decreased Hprt expression was observed. Starting dox administration at E7.5 or later caused irreversible silencing of Hprt. These results demonstrate that KRAB domain-induced irreversible gene silencing occurs between E3.5 and E7.5. Time-dependent irreversible silencing was also observed in tTS-mediated irreversible N-myc endogenous promoter silencing. A similar dox introducing strategy as for Hprt regulation was performed (Figure 5D). Dox exposure at E4.5 or earlier resulted in normal EGFP expression in NmycEGFP:tTS mice, but if dox was introduced at E7.5 or later, no EGFP fluorescence was observed at E13.5 (Figure 5D).
Epigenetic mechanism of tTS-induced Hprt reversible and irreversible regulation
To investigate the potential epigenetic mechanism behind tTS-induced Hprt reversible and irreversible regulation, reversible or irreversible MEFs were treated with inhibitors of DNA methyltransferases (5-Aza) and histone deacetylases (TSA) with or without dox.
In irreversible MEFs without dox, 5-Aza slightly but significantly elevated Hprt expression. With dox for 3 days, 5-Aza progressively and significantly increased Hprt expression by 548-fold. Additionally, TSA also significantly increased Hprt expression by 12- to19-fold compared with no dox (Figure 6A). These results suggest that relative to histone deacetylation, DNA methylation plays a major role in silencing Hprt expression in irreversible MEFs.
Figure 6.
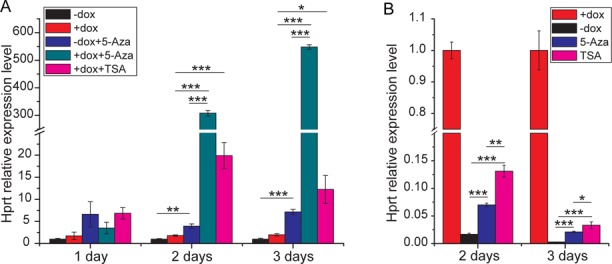
Epigenetic analysis of tTS-induced Hprt reversible and irreversible regulation. (A) qPCR analysis of Hprt expression levels in 5-Aza or TSA-treated HprtTRE/Y:tTS MEFs. MEFs were isolated from embryos that developed with (A) or without (B) dox exposure from conception to E13.5d, and cultured without (A) or with dox (B). In (A), Hprt expression was normalized to the ‘-dox’ group at the same time point. In (B), Hprt expression was normalized to the ‘+dox’ group at the same time point. The start time of 5-Aza or TSA treatment was designated point 0.
In reversible MEFs, both 5-Aza and TSA attenuated Hprt expression switching after dox removal (Figure 6B), suggesting that both histone deacetylation and DNA methylation play important roles in tTS-induced reversible repression.
To support the role of DNA methylation in irreversible and reversible regulation of Hprt expression, bisulfite sequencing analysis of CpG islands within the Hprt promoter and intron1 was performed. In irreversible MEFs, bisulfite sequencing revealed a very high proportion of CpG methylation in the Hprt promoter and intron 1 region. No significant changes in methylation were observed in the presence or absence of dox (Figure 7A, right and C, right). In contrast, in reversible MEFs, low levels of promoter methylation were observed. Similarly, significantly low levels of intron methylation were observed without dox (Figure 7A, left and C, left). Although removing dox increased the average methylation level in reversible MEFs, the increase was not significant. Statistical graphs were shown in Figure 7B and D. Overall, these findings demonstrate that tTS binding during the early embryonic period induces a high level of DNA methylation within the binding region, resulting in irreversible gene silencing. Our results also indicate that binding or removing of tTS does not change DNA methylation levels in reversible MEFs.
Figure 7.
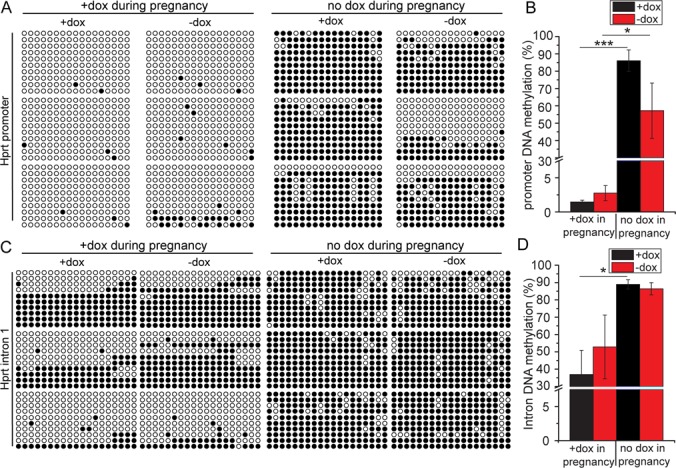
DNA methylation analysis of tTS-induced Hprt reversible and irreversible regulation. Bisulfite sequencing analysis of Hprt promoter (A) and intron 1 (C) methylation levels in HprtTRE/Y:tTS MEFs. Quantification of (A) and (C) are presented in (B) and (D). MEFs were isolated from six mice. Three mice were exposed to dox during development from E0.5, while the other three mice received no dox. MEFs were cultured in presence or absence of dox. DNA methylation levels of 17 and 19 CpG dinucleotides within the Hprt promoter and intron 1, respectively, were assayed. Ten clones amplified from each mouse were sequenced and are presented. Each circle represents one CpG dinucleotide, with empty and full circles representing unmethylated and methylated CpGs, respectively (*P < 0.05; **P < 0.01; ***P < 0.001).
To investigate the role of histone modifications in irreversible and reversible regulation of Hprt expression, histone modifications within the Hprt promoter region, namely, histone H3-acetylation (H3Ac), H3 lysine 9 trimethylation (H3K9me3), histone H3-acetylation (H4Ac) and H4 lysine 20 trimethylation (H4K20me3) were examined by ChIP (Figure 8A and B). In both irreversible and reversible MEFs, dox removal caused decreased H3Ac and H4Ac levels, and increased H3K9me3 and H4K20me3 levels upon tTS-binding. Compared with reversible MEFs, lower H3Ac and H4Ac levels, and higher H3K9me3 and H4K20me3 levels were observed with or without dox. Differences in histone modification levels between irreversible and reversible MEFs were particularly notable with H4Ac levels. These results demonstrate that tTS binding results in a heterochromatin status of increased histone methylation and decreased histone acetylation.
Figure 8.
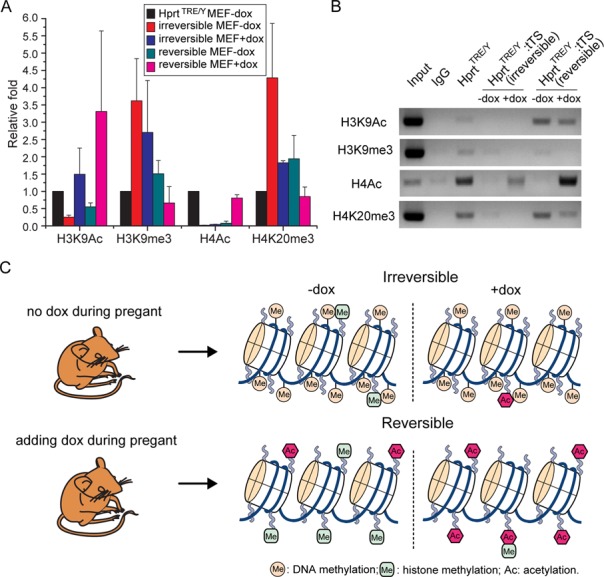
Schematic representation of the KRAB-mediated irreversible and reversible regulation model. (A) ChIP analysis of histone modifications within the Hprt promoter region. MEFs isolated from embryos that developed without or with dox exposure were named ‘irreversible MEFs’ or ‘reversible MEFs’, respectively. Chromatin immunoprecipitated DNA was analyzed by qPCR. Input was used as the control. Modification levels in HprtTRE/Y:tTS MEFs were denoted 1, and used to normalize data from the other samples. (B) Chromatin immunoprecipitated DNA analysis by semi-quantitative gel electrophoresis within the Hprt promoter region. (C) Schematic representation of the KRAB-mediated irreversible and reversible regulation model.
In conclusion, we have described two distinct regulatory modes mediated by the KRAB domain for regulation of endogenous genes in vivo (Figure 8C). Depending on the developmental time point, KRAB-mediated regulation is divided into irreversible or reversible regulation modes. If a KRAB-containing regulator acts during the early embryonic development, irreversible gene silencing is mediated mainly by stable DNA methylation. Otherwise, reversible gene regulation induced by rewriteable acetylation and methylation of histone H3 and H4 occurs.
DISCUSSION
In this study, we have shown that a KRAB-containing regulator can induce irreversible or reversible regulation of endogenous genes in vivo by mediating different chromatin states dependent on developmental stage. Tethering the KRAB domain to chromosomal DNA during early mouse embryonic development induces irreversible endogenous gene silencing mainly through stable DNA methylation near the binding region. Previously, two different types of KRAB-mediated regulation were reported using lentivirus-mediated transgenes. In one study, KRAB induced irreversible exogenous gene silencing by KRAB-induced de novo DNA methylation of the promoter (13). Alternatively, in the other study, KRAB induced reversible heterochromatization of the endogenous Kif2A gene in vivo, and not stable DNA promoter methylation (14). In contrast to these previous studies (13,14), we used homologous recombination to insert a TRE site, and thereby eliminated the possibility of lentivirus-induced DNA methylation. We have shown that gene silencing is mainly due to stable de novo DNA methylation, consistent with the gene silencing role of DNA methylation. However, methylation and deacetylation of histone H3 or H4 are also involved in gene silencing, as indicated by TSA treatment and ChIP analysis. Although the maximum range of KRAB-mediated DNA methylation is still unknown, it is reasonable to speculate that such methylation occurs within the vicinity of the KRAB binding site (27). In our study, within at least a 2-kb radius distal from the binding site is hypermethylated. The limited size of the methylated region may explain why KRAB did not induce irreversible gene silencing at the early developmental stage in the previous study, as KRAB was bound to the gene body and 5 kb distal to the promoter (14).
Furthermore in our current study, a definite time window during which KRAB induces irreversible gene silencing was identified using two endogenous gene regulation models. Before E3.5, KRAB binding does not induce irreversible gene silencing, and results in progressive repression from E3.5 to E6.5. If KRAB binding persists until E7.5, irreversible gene silencing occurs. This time course is consistent with the genomic demethylation-remethylation process in mouse development. During early embryonic development, genomic DNA methylation inherited from the gametes is erased, reaching its lowest point in the blastocyst at E3.5 (28,29). DNA methylation is then progressively restored and established, and in which, KRAB-containing transcription factors may play a key role.
KRAB-mediated reversible regulation of endogenous genes in vivo
The KRAB domain can recruit KAP1 (5,6), which acts as a scaffold to further recruit HP1, histone methyltransferase SETDB1 and the NuRD histone deacetylase complex to mediate transcriptional repression through heterochromatin spreading (7,8,12,30). In the KRAB-KAP1 repression system, SETDB1 has histone H3 lysine 9 (H3K9)-specific methyltransferase activity and mediates H3K9 methylation (8,31). HP1 can bind to methylated H3K9 residues and recruit more histone methyltransferase for a second round of H3K9 methylation/HP1 binding (12,32–35). Therefore, H3K9 methylation is a hallmark of KARB-KAP1-mediated gene repression, with H3Ac widely reported (12,14,15,23,32). In our study, increased H3K9me3 and decreased H3Ac were observed after dox removal and KRAB tethering to DNA. Of note, increased H4K20me3 and decreased H4Ac were also observed. Consistent with our findings, increased H4Ac at the 5′ UTR of intracisternal A-type particles was shown in KAP1 knockout ES cells (15). H4K20me3 enrichment at the proximal regulatory region was also reported in KAP1-mediated repression at the cell level (36).
Compared with uniform gene silencing efficiency in different tissues, tight control of Hprt expression was not detected during reversible regulation in spleen and thymus after dox removal. A similar lack of complete repression was also observed in thymus for the tTS-based mouse model targeting the N-myc gene (unpublished data). Normal tTS and higher Kap1 expression were observed in thymus, and does not explain the low levels of tTS and Kap1. Nevertheless, these results suggest that KRAB may mediate different regulatory mechanisms in thymus and spleen compared with other tissues. Results from the Kap1 conditional knockout mouse model provides further support for this (37,38). In contrast with the role of KRAB/KAP1 in transcriptional repression, genome-wide chromatin studies in T cells have shown that markers of active transcription, including the histone acetyltransferase CBP and TFIIB, are significantly enriched at Kap1 binding sites (37,38), although this hypothesis needs further verification. Moreover, the reason for repression failure in the spleen and thymus needs to be further examined. Differences in repression efficiency between irreversible and reversible regulation also reflect differences in regulatory mechanisms.
Application of KRAB-mediated endogenous gene regulation in vivo
In our previous study, we developed an inducible and reversible system based on an artificial KRAB-containing regulator (‘tTS’) to regulate endogenous genes in vivo (22). Although this system showed strong potential for regulating endogenous gene expression and could be a versatile tool to replace conditional targeting methods, application of this system was limited to a few examples, for example, Htr1a, Mlc1, N-myc and CD4 locus (20–23). Expression of these target genes was limited to some tissues or a specific period, which precluded general evaluation of the efficiency of this system. To overcome these limitations, we selected Hprt, a ubiquitously expressed housekeeping gene, for tTS system regulation in this study. We proposed that the tTS system would irreversibly silence Hprt in all studied tissues if no dox was added during early embryogenesis. Indeed, tTS-mediated reversible regulation of Hprt was achieved in most tissues examined, except spleen and thymus. Feasibility of tTS-mediated irreversible and reversible regulation of an endogenous gene in 10 different cells and tissues during multiple time windows in this study, suggests a potential application range for tTS-mediated endogenous gene regulation system in vivo. Furthermore, development of new genomic modification tools, such as zinc finger effectors, TAL effectors and Cas9, provides the potential for designing DNA binding effectors according to DNA sequence (39–41). Accordingly, KRAB has been fused with TAL effector or Cas9 to specifically repress transcription of an endogenous gene at the cell level (42). Our work will accelerate use of these artificial DNA binding effectors and KRAB to regulate endogenous genes in vivo.
Acknowledgments
We thank Prof. Shisan Bao for English manuscript editing. We also thank Ms Wenting Wu for schematic diagramming.
Footnotes
These authors contributed equally to the paper as first authors.
FUNDING
National Key Project [2010CB945501]; National Natural Science Foundation of China [81171300]; E-Institutes of Shanghai Municipal Education Commission [E03003]. Funding for open access charge: National Natural Science Foundation of China [81171300].
Conflict of interest statement. None declared.
REFERENCES
- 1.Looman C., Abrink M., Mark C., Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- 2.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emerson R.O., Thomas J.H. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S.S., Chen Y.M., O'Leary E., Witzgall R., Vidal M., Bonventre J.V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman J.R., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher F.J., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 6.Moosmann P., Georgiev O., Le Douarin B., Bourquin J.P., Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz D.C., Friedman J.R., Rauscher F.J., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szulc J., Wiznerowicz M., Sauvain M.O., Trono D., Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat. Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 12.Groner A.C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Denervaud N., Bucher P., Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiznerowicz M., Jakobsson J., Szulc J., Liao S., Quazzola A., Beermann F., Aebischer P., Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- 14.Groner A.C., Tschopp P., Challet L., Dietrich J.E., Verp S., Offner S., Barde I., Rodriguez I., Hiiragi T., Trono D. The Kruppel-associated box repressor domain can induce reversible heterochromatization of a mouse locus in vivo. J. Biol. Chem. 2012;287:25361–25369. doi: 10.1074/jbc.M112.350884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J., et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 16.Wolf D., Cammas F., Losson R., Goff S.P. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 2008;82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf D., Goff S.P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Wolf D., Goff S.P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallo M., Kanzler B., Ohnemus S. Reversible gene inactivation in the mouse. Genomics. 2003;81:356–360. doi: 10.1016/s0888-7543(03)00032-6. [DOI] [PubMed] [Google Scholar]
- 20.Richardson-Jones J.W., Craige C.P., Guiard B.P., Stephen A., Metzger K.L., Kung H.F., Gardier A.M., Dranovsky A., David D.J., Beck S.G., et al. 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron. 2010;65:40–52. doi: 10.1016/j.neuron.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K.F., Ahmari S.E., Leonardo E.D., Richardson-Jones J.W., Budreck E.C., Scheiffele P., Sugio S., Inamura N., Ikenaka K., Hen R. Flexible Accelerated STOP Tetracycline Operator-knockin (FAST): a versatile and efficient new gene modulating system. Biol. Psych. 2010;67:770–773. doi: 10.1016/j.biopsych.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun R., Zhao K., Shen R., Cai L., Yang X., Kuang Y., Mao J., Huang F., Wang Z., Fei J. Inducible and reversible regulation of endogenous gene in mouse. Nucleic Acids Res. 2012;40:e166. doi: 10.1093/nar/gks738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan M., Kaundal R., Huang H., Zhao J., Yang X., Chaiyachati B.H., Li S., Chi T. A general approach for controlling transcription and probing epigenetic mechanisms: application to the CD4 locus. J. Immunol. 2013;190:737–747. doi: 10.4049/jimmunol.1201278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C.L., Melton D.W. Production of a model for Lesch-Nyhan syndrome in hypoxanthine phosphoribosyltransferase-deficient mice. Nat. Genet. 1993;3:235–240. doi: 10.1038/ng0393-235. [DOI] [PubMed] [Google Scholar]
- 25.Michalska A.E. Isolation and propagation of mouse embryonic fibroblasts and preparation of mouse embryonic feeder layer cells. Curr. Protoc. Stem Cell Biol. 2007 doi: 10.1002/9780470151808.sc01c03s3. doi:10.1002/9780470151808.sc01c03s3. [DOI] [PubMed] [Google Scholar]
- 26.Waters T.R., Swann P.F. Cytotoxic mechanism of 6-thioguanine: hMutSalpha, the human mismatch binding heterodimer, binds to DNA containing S6-methylthioguanine. Biochemistry. 1997;36:2501–2506. doi: 10.1021/bi9621573. [DOI] [PubMed] [Google Scholar]
- 27.Quenneville S., Turelli P., Bojkowska K., Raclot C., Offner S., Kapopoulou A., Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 29.Santos F., Hendrich B., Reik W., Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 30.Lechner M.S., Begg G.E., Speicher D.W., Rauscher F.J., 3rd Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 32.Hathaway N.A., Bell O., Hodges C., Miller E.L., Neel D.S., Crabtree G.R. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 34.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 36.Sripathy S.P., Stevens J., Schultz D.C. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoni de Sio F.R., Barde I., Offner S., Kapopoulou A., Corsinotti A., Bojkowska K., Genolet R., Thomas J.H., Luescher I.F., Pinschewer D., et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012;26:4561–4575. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santoni de Sio F.R. Kruppel-associated box (KRAB) proteins in the adaptive immune system. Nucleus. 2014;5:138–148. doi: 10.4161/nucl.28738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L., Zhou R., Kuo Y.C., Cunniff M., Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandell J.G., Barbas C.F., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]


