Abstract
Upstream open reading frames (uORFs) are often found in the 5′-leader regions of eukaryotic mRNAs and can negatively modulate the translational efficiency of the downstream main ORF. Although the effects of most uORFs are thought to be independent of their encoded peptide sequences, certain uORFs control translation of the main ORF in a peptide sequence-dependent manner. For genome-wide identification of such peptide sequence-dependent regulatory uORFs, exhaustive searches for uORFs with conserved amino acid sequences have been conducted using bioinformatic analyses. However, whether the conserved uORFs identified by these bioinformatic approaches encode regulatory peptides has not been experimentally determined. Here we analyzed 16 recently identified Arabidopsis thaliana conserved uORFs for the effects of their amino acid sequences on the expression of the main ORF using a transient expression assay. We identified five novel uORFs that repress main ORF expression in a peptide sequence-dependent manner. Mutational analysis revealed that, in four of them, the C-terminal region of the uORF-encoded peptide is critical for the repression of main ORF expression. Intriguingly, we also identified one exceptional sequence-dependent regulatory uORF, in which the stop codon position is not conserved and the C-terminal region is not important for the repression of main ORF expression.
INTRODUCTION
Nascent peptides with certain specific sequences cause ribosome stalling during mRNA translation and thereby regulate gene expression (1–5). In prokaryotes, small open reading frames (ORFs) located in the 5′-leader regions of several genes encode regulatory nascent peptides that cause ribosome stalling in the middle of or at the stop codon of the small ORFs under specific conditions. Ribosome stalling in these small ORFs induces expression of the downstream cistron by destabilizing the secondary structure to make the Shine–Dalgarno sequence accessible (1,6–10) or by inhibiting transcriptional termination before the downstream cistron (11). In eukaryotes, most documented regulatory nascent peptides are encoded by upstream open reading frames (uORFs), which are located in the 5′ untranslated regions (5′-UTRs). Although uORFs often negatively modulate the translational efficiency of the downstream main ORF, most uORFs exert their effects in a sequence-independent manner (12). By contrast, certain uORFs control translation of the main ORF in a peptide sequence-dependent manner (1–5,13). In the previously characterized sequence-dependent regulatory uORFs, such as those in the cytomegalovirus gpUL4, fungal arg-2 and CPA1, mammalian AdoMetDC, and Arabidopsis thaliana AdoMetDC1 genes, the uORF-encoded peptides cause ribosome stalling at the stop codon of the uORF (14–18). The stalled ribosome prevents other scanning ribosomes from reaching the start codon of the main ORF, resulting in translational repression of the main ORF. In many of these uORF peptide-mediated translational regulations, a metabolite acts as an effector molecule. For example, arginine induces ribosome stalling in the arg-2 and CPA1 uORFs (16,17), and polyamine induces ribosome stalling in the mammalian AdoMetDC and A. thaliana AdoMetDC1 uORFs (15,18). In both cases, the effector molecule is a metabolite produced by the pathway involving the main ORF-encoded enzyme (15–18). Therefore, the translational repression mediated by these uORF-encoded peptides acts as feedback regulation.
To date, only a limited number of regulatory nascent peptides have been found in prokaryotes and eukaryotes, and the prevalence of nascent peptide-mediated gene regulation is yet to be determined. In attempts to identify uORF-encoded regulatory peptides on a genome-wide scale, exhaustive searches for uORFs encoding conserved amino acid sequences, which are referred to as ‘conserved peptide uORFs (CPuORFs)’ (19), have been conducted in various organisms, such as mammals (20), plants (19,21) and insects (22), using comparative genomic analyses. In plants, Hayden and Jorgensen identified 26 homology groups of CPuORFs by comparing uORF sequences between A. thaliana and rice homologous genes or among A. thaliana paralogous genes (19). Recently, Vaughn et al. additionally identified four homology groups of CPuORFs by comparing uORF sequences between A. thaliana, cotton, orange, soybean, grape and tobacco (21). For a more comprehensive identification of CPuORFs, we developed a BLAST-based program, BAIUCAS, which permits comparisons of uORF sequences of certain species to those of any other species in the expressed sequence tag databases. Using BAIUCAS, we identified 13 additional homology groups of A. thaliana CPuORFs that are conserved beyond Brassicales (23). These bioinformatic analyses revealed that a plant genome contains more than 40 homology groups of CPuORFs. However, it has not been assessed how many of the identified CPuORFs encode regulatory peptides that control main ORF translation.
In our previous report, we classified the recently identified plant CPuORFs into two classes based on the conservation pattern of their encoded amino acid sequences (23). In class I CPuORFs, the C-terminal amino acid sequence and the stop codon position are evolutionarily conserved. In class II CPuORFs, the amino acid sequence is conserved entirely or the N-terminal and/or middle region is conserved, but the stop codon position is not conserved. Cryo-electron microscopy studies by Bhushan et al. revealed that the C-terminal regions of the gpUL4 and arg-2 uORF-encoded nascent peptides interact with components of the ribosomal exit tunnel when ribosome stalling occurs at the uORF stop codon (24). Additionally, genetic and biochemical studies have revealed that interaction between a regulatory nascent peptide and the exit tunnel components is important for ribosome stalling (7,9,25–31). Considering these observations, class I CPuORFs may be more likely to encode a regulatory nascent peptide. However, the relationship between the conservation patterns of the CPuORF-encoded peptide sequences and the abilities as regulatory nascent peptides has not been addressed.
In this study, to address what percentage and type of CPuORFs encode regulatory peptides, we selected 16 recently identified A. thaliana CPuORFs, all of which belong to distinct homology groups, and analyzed the effects of their peptide sequences on the expression of the main ORF. From this analysis, we identified five novel peptide sequence-dependent regulatory uORFs that repress main ORF expression. Additionally, we found that CPuORFs belonging to class I have an increased tendency to encode regulatory peptides compared with class II CPuORFs. However, we identified one sequence-dependent regulatory uORF from class II CPuORFs, and found that it has an exceptional feature in that the C-terminal region of the encoded peptide is not important for the repression of the main ORF expression.
MATERIALS AND METHODS
Plant material and growth condition
Arabidopsis thaliana MM2d suspension cells (32) were cultured in modified Linsmaier and Skoog (LS) medium (33) at 26°C in the dark with orbital shaking at 130 rpm. Cells were transferred to fresh medium every week.
Plasmid construction
For cloning of the 5′-UTRs of At1g67480 and At3g55050, poly(A)+ RNA was prepared from A. thaliana (Col-0 ecotype) flower buds and opened flowers using a Qiagen Plant RNeasy Mini Kit (Qiagen) and a GenElute mRNA Miniprep Kit (Sigma-Aldrich). For cloning of the 5′-UTRs of the other genes, poly(A)+ RNA was prepared from A. thaliana (Col-0 ecotype) seedlings using the same kits. cDNA of the 5′-UTRs was amplified from poly(A)+ RNA using the OneStep RT-PCR Kit (Qiagen) and the primers listed in Supplementary Table S1. The sense and antisense primers contained XbaI and SalI restriction endonuclease sites, respectively.
Plasmid pIE0, which harbors the cauliflower mosaic virus 35S RNA (35S) promoter, the 5′-UTR of the A. thaliana AdoMetDC1 gene, the Renilla luciferase (RLUC) coding sequence and the polyadenylation signal of the Agrobacterium tumefaciens nopaline synthase (NOS) gene in pUC19, was used as the cloning vector for the amplified 5′-UTRs. To construct this vector, we first digested plasmid pSY209 (18), which contains the AdoMetDC1 5′-UTR and the RLUC coding sequence in the pSP64 Poly(A) vector (Promega), with XbaI and SmaI at sites downstream of RLUC, treated the digested DNA with a T4 DNA polymerase to fill in the XbaI end, and then ligated the blunt-ended XbaI site to the SmaI site to remove the XbaI and SmaI sites. The modified pSY209 was then digested with HindIII, treated with the Klenow fragment to fill in the digested ends, and digested with SacI. The HindIII–SacI fragment of the modified pSY209 containing the AdoMetDC1 5′-UTR and the RLUC coding sequence was inserted between the 35S promoter and the NOS polyadenylation signal of plasmid pMI4(WT) (34,35) to generate pIE0, after pMI4(WT) was digested with XbaI, treated with the Klenow fragment, and digested with SacI.
The amplified 5′-UTR cDNA fragments containing the CPuORFs analyzed in this study were digested with XbaI and SalI and ligated between the XbaI and SalI sites of pIE0 to generate 35S::UTR:RLUC reporter plasmids. The deletion and insertion mutations and the codon changes were introduced into the CPuORFs using the overlap extension polymerase chain reaction (PCR) method (36) with primers listed in Supplementary Tables S2–S8. In all of the constructs, sequence analysis confirmed the integrity of the PCR-amplified regions.
The 35S::UTR:FLUC reporter plasmids carry the 5′-UTR of the ANAC082, CIPK6, At3g15430, At5g27920 or OTLD1 genes between the 35S promoter and a firefly luciferase (FLUC) coding sequence. To construct these plasmids, oligonucleotides LUCSalIF (5′-TCCTCTAGATATCAATCTCTTCTCAAAAGATGGCGTCGACCATGGAAGC-3′) and LUCSalIR (5′-GCTTCCATGGTCGACGCCATCTTTTGAGAAGAGATTGATATCTAGAGGA-3′) were annealed, digested with XbaI and NcoI and ligated into the XbaI and NcoI sites of pMI21(WT) (37) to yield pMT61. pMT61 was then digested with SalI and SacI, and the SalI-SacI fragment containing the FLUC coding sequence was ligated into the SalI and SacI sites of the 35S::UTR:RLUC reporter plasmids to yield the 35S::UTR:FLUC reporter plasmids.
Transient expression assay
In the transient expression experiments, plasmid DNAs were introduced into MM2d protoplasts by electroporation or polyethylene glycol (PEG) treatment. To prepare protoplasts, MM2d cells were collected by centrifugation on the third day after transfer to fresh media and suspended in modified LS medium containing 1% (w/v) cellulase Onozuka RS (Yakult Pharmaceutical Industry), 0.5% (w/v) pectolyase Y23 (Seishin Pharmaceutical) and 0.4 M mannitol, and incubated at 26°C with gentle shaking until the suspension became turbid with protoplasts (∼3 h). The protoplasts were then washed five times with wash buffer (0.4 M mannitol, 5 mM CaCl2 and 12.5 mM NaOAc, pH 5.8).
For electroporation, protoplasts were suspended in electroporation buffer (5 mM morpholinoethanesulfonic acid, 70 mM KCl and 0.3 M mannitol, pH 5.8). Ten micrograms each of a 35S::UTR:RLUC reporter plasmid and the 35S::FLUC internal control plasmid, 221-LUC+ (34,38), were mixed with 1.5 × 106 protoplasts in 500 μl of electroporation buffer in an electroporation cuvette with a 4-mm electrode distance. Electroporation was carried out using a BTX Electro Cell Manipulator 600 with voltage, capacitance and resistance settings of 190 V, 100 μF and 480 Ω, respectively. The protoplasts were kept on ice for 30 min and then incubated at 25°C for 5 min, centrifuged (60 × g, 2 min at 25°C) and resuspended in 1 ml of the modified LS medium containing 0.4 M mannitol.
For PEG-mediated transfection, protoplasts were suspended in MaMg solution (5 mM morpholinoethanesulfonic acid, 15 mM MgCl2 and 0.4 M mannitol, pH 5.8). Five micrograms each of a 35S::UTR:RLUC reporter plasmid, a 35S::UTR:FLUC reporter plasmid and the 35S::GUS internal control plasmid, pBI221 (Clontech), which carries an Escherichia coli β-glucuronidase (GUS) coding sequence under control of the 35S promoter, were mixed with 3 × 105 protoplasts in 100 μl of MaMg solution and 110 μl of PEG solution (40% PEG4000, 0.5 M CaCl2, 0.4 M mannitol). This mixture was incubated for 15 min at room temperature, and diluted by adding 800 μl of wash buffer. The protoplasts were centrifuged (60 × g, 2 min at room temperature) and resuspended in 1 ml of the modified LS medium containing 0.4 M mannitol.
After 48 h of incubation at 23°C in the dark, cells were harvested and disrupted in 200 μl of extraction buffer [100 mM(NaH2/Na2H)PO4, 5 mM dithiothreitol, pH 7] by sonication on ice with a Branson Sonifier 250. A Dual-LUC Reporter Assay Kit (Promega) was used to measure the RLUC and FLUC activities. GUS activities were determined as described by Jefferson (39) with 4-methylumbelliferyl-β-D-glucuronide as the substrate, using a spectrofluorimeter (Hitachi, Fluorescence Spectrophotometer F-2500).
RESULTS
Identifying sequence-dependent regulatory uORFs
Among the recently identified CPuORFs, we selected 16 CPuORFs for analysis of their sequence-dependent effects on main ORF expression. Ten of the CPuORFs (in the ANAC082, ANAC096, ARF4, ATMPK20, CIPK6, At1g67480, At3g15430, At3g55050, At5g02480 and At5g27920 genes) belong to class I, whereas six (in the CIPK23, DIC1, OTLD1, At4g10170, At4g12790 and At5g63640 genes) belong to class II (23).
To investigate the regulatory functions of these CPuORFs, the 5′-UTR of each gene was amplified by reverse transcription PCR (RT-PCR) using primers designed based on full-length cDNA sequence information available at the Arabidopsis Information Resource (TAIR) website (http://arabidopsis.org/) (Figure 1, Supplementary Figure S1, Table S1). According to the current TAIR gene models (TAIR10), splice variant forms of the 5′-UTRs exist for some of the selected genes. In fact, multiple bands were detected when the 5′-UTRs of some genes were amplified by RT-PCR. In all such cases, the most abundant RT-PCR product was cloned and sequencing confirmed the presences of the CPuORFs in the cloned 5′-UTRs (Supplementary Figure S1). Each cloned 5′-UTR was fused to the RLUC coding sequence and placed under the control of the 35S promoter (Figure 2A).
Figure 1.
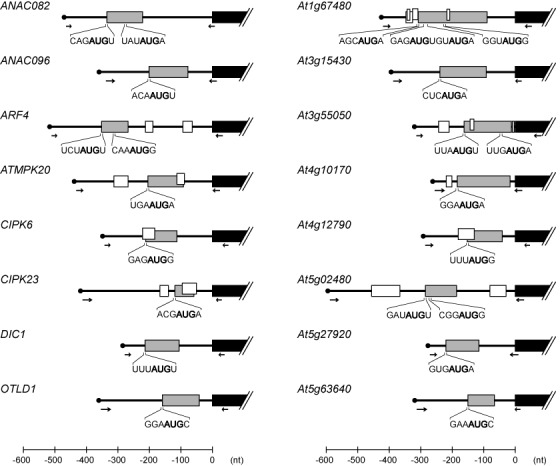
Schematic representation of the 5′-UTRs containing CPuORFs analyzed in this study. Shaded, open and closed boxes represent the CPuORFs, the other uORFs and the main ORFs, respectively. Arrows indicate the positions of primers used to clone the 5′-UTRs. The translation initiation contexts of the first and internal in-frame AUG codons of each CPuORF are shown.
Figure 2.
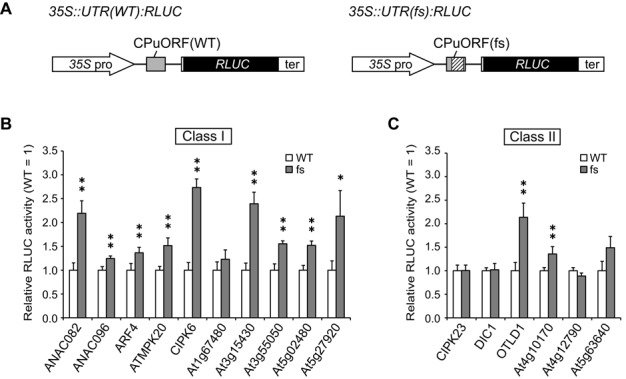
Search for sequence-dependent regulatory uORFs. (A) Schematic representation of the WT (35S::UTR(WT):RLUC) and fs mutant (35S::UTR(fs):RLUC) reporter constructs. The hatched box in the fs mutant CPuORF shows the frame-shifted region. Although only a single uORF is depicted in each construct, the actual 5′-UTRs of some genes have multiple uORFs. See Figure 1 and Supplementary Figure S1 for the exact 5′-UTR structure of each gene and the exact positions of the fs mutations in each CPuORF. The polyadenylation signal of the Agrobacterium tumefaciens NOS gene is designated as ‘ter’. (B and C) Transient expression studies of class I (B) and class II (C) CPuORFs. The reporter plasmids containing the WT or fs mutant CPuORF of each gene were co-transfected with the 35S::FLUC internal control plasmid into MM2d protoplasts by electroporation. After 48 h of incubation, the transfected cells were harvested and disrupted for luciferase assay. RLUC activity was normalized to FLUC activity, and the relative activity to that of the corresponding WT construct was calculated. Means ± S.D. of at least three biological replicates are shown. Each graph is representative of two or more separate experiments using independently prepared protoplasts. Single and double asterisks indicate significant differences between the WT and fs constructs at P < 0.05 and P < 0.01 by t-test, respectively.
To assess the sequence-dependence of the effect of each CPuORF on main ORF expression, frameshift (fs) mutations were introduced to alter the amino acid sequence of each CPuORF. A +1 or −1 fs mutation was introduced upstream or in the conserved region of each CPuORF, and another fs mutation was introduced before the stop codon to shift the reading frame back to the original frame (Supplementary Figure S1). In the AT3G55050 CPuORF, two sets of fs mutations were introduced to avoid changing the length of ORF beginning at the internal Met codon, Met-40 (Supplementary Figure S1K). In the ANAC096, OTLD1 and AT4G12790 CPuORFs, introduction of the fs mutations generated an in-frame premature stop codon. Therefore, an additional nucleotide change was made to replace the premature stop codon by an amino acid-encoding codon (Supplementary Figure S1B, H and M). In the ATMPK20, CIPK6, CIPK23, AT1G67480, AT3G55050 and AT4G12790 genes, another uORF overlaps with the CPuORF (Figure 1, Supplementary Figure S1D, E, F, I, K and M). In these cases, fs mutations were introduced leaving the presence and length of the overlapping uORF unaltered, because alteration in the presence or length of other uORFs may affect the main ORF expression.
The 35S::UTR:RLUC reporter plasmid containing the wild-type (WT) or fs mutant version of each CPuORF was introduced into protoplasts prepared from A. thaliana MM2d suspension cultured cells. After 48 h of incubation, cells were harvested and disrupted for analysis of RLUC activity. As shown in Figure 2, among the class I CPuORFs, the fs mutants of the ANAC082, CIPK6, At3g15430 and At5g27920 CPuORFs exhibited a more than two-fold increase in RLUC activity level compared with the corresponding WT (Figure 2B). In five of the remaining class I CPuORFs, the fs mutations had weaker effects, with 1.2- to 1.5-fold increases. By contrast, the At1g67480 uORF mutant showed no significant effect (Figure 2B). Among the class II CPuORFs, the fs mutations of the OTLD1 and At4g10170 CPuORFs significantly enhanced the RLUC activity by 2.1- and 1.4-fold, respectively (Figure 2C). The fs mutations of the other class II CPuORFs had no significant effect (Figure 2C). These results suggest that nine class I CPuORFs and two class II CPuORFs may have a sequence-dependent inhibitory effect on main ORF expression. We further analyzed the five CPuORFs whose fs mutation caused a more than two-fold increase in reporter expression level.
Peptides translated from the five CPuORFs repress main ORF expression
In the fs mutants, only a few nucleotide changes were introduced into each CPuORF, whereas most amino acid residues in the frameshifted region were altered. However, it is possible that those nucleotide changes affected main ORF expression independently of CPuORF-encoded peptide function; for example, by altering the secondary structure of the mRNA. To address this possibility, we eliminated the start codon of the ANAC082, CIPK6, At3g15430, At5g27920 and OTLD1 CPuORFs by changing them to an AAG codon, and tested if the fs mutations of these uORFs affected main ORF expression even in the absence of the uORF start codon by the transient expression assay. In the ANAC082 CPuORF, the internal Met codon, Met-20, was also replaced by an AAG codon. In all of the CPuORFs tested, eliminating the start codon elevated RLUC activity, indicating that translation of these CPuORFs have an inhibitory effect on main ORF expression (Figure 3). However, the fs mutations caused no further increase in RLUC activity in the absence of the CPuORF start codon (Figure 3). These results indicate that the effects of the fs mutations of the five CPuORFs depend on translation of the CPuORFs, and suggest that the peptides encoded by these CPuORFs are responsible for the sequence-dependent inhibitory effect on main ORF expression.
Figure 3.
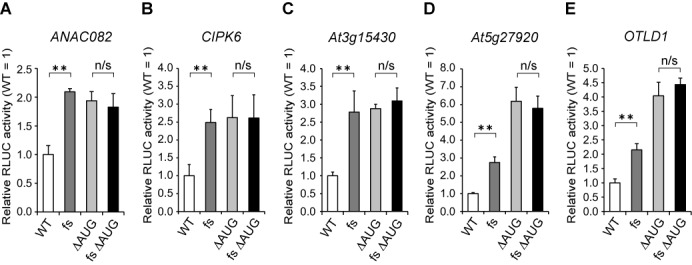
Effect of the fs mutation in the presence and absence of the uORF start codon. (A–E) Transient expression studies of the ANAC082 (A), CIPK6 (B), At3g15430 (C), At5g27920 (D) and OTLD1 (E) CPuORFs. The 35S::UTR:RLUC reporter plasmids carrying the WT CPuORF, the mutant CPuORF lacking the start codon (ΔAUG), the fs mutant CPuORF or fs ΔAUG double mutant CPuORF of each gene was co-transfected with the 35S::FLUC internal control plasmid into MM2d protoplasts by electroporation, and the reporter activities were analyzed as in Figure 2. Means ± S.D. of at least three biological replicates are shown. Each graph is representative of two or more separate experiments using independently prepared protoplasts. Double asterisks indicate a significant difference between two constructs (P < 0.01 by t-test), whereas ‘n/s’ indicates non-significant difference (P ≥ 0.05 by t-test).
The newly identified regulatory uORF peptides act in cis
If the peptides encoded by the five CPuORFs function as regulatory nascent peptides to repress main ORF expression, as do the previously characterized regulatory uORF peptides, they should act in cis to exert their effects only on the downstream main ORF on the same mRNA. To address this hypothesis, we tested whether the five CPuORFs acted in cis or in trans to repress main ORF expression. For this analysis, the 5′-UTR containing the WT or fs version of each CPuORF was fused to the FLUC reporter gene and placed under control of the 35S promoter to yield 35S::UTR:FLUC reporter plasmids (Figure 4A). The 35S::UTR:FLUC reporter plasmid harboring the WT or fs version of each CPuORF was co-transfected into MM2d protoplasts with the 35S::UTR:RLUC reporter plasmid carrying the WT or fs version of the corresponding CPuORF. As shown in Figure 4, for all the CPuORFs analyzed, the reporter activities of each reporter plasmid were not significantly different, regardless of whether it was co-transfected with the WT or fs version of the other reporter plasmid. These results indicate that neither the WT nor the fs mutant CPuORF affected the reporter activity of the other reporter plasmid, and suggest that the peptides encoded by these five CPuORFs act in cis to repress main ORF expression.
Figure 4.
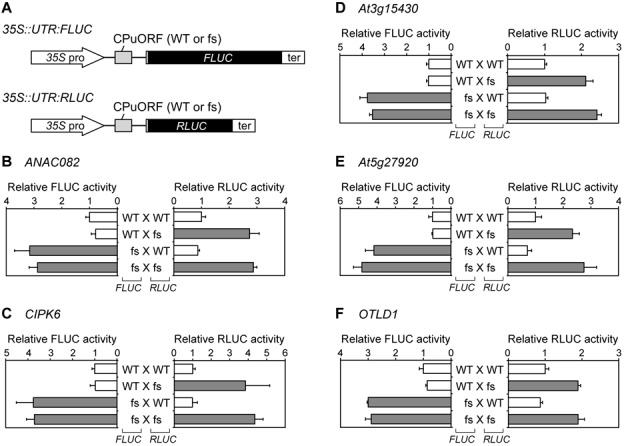
The peptides encoded by the five sequence-dependent CPuORFs act in cis. (A) Schematic representation of the 35S::UTR:FLUC and 35S::UTR:RLUC reporter constructs. (B–F) Transient expression studies of the co-transfected 35S::UTR:RLUC and 35S::UTR:FLUC reporter plasmids. MM2d protoplasts were co-transfected with three plasmids, the 35S::UTR:FLUC and 35S::UTR:RLUC reporter plasmids and the 35S::GUS internal control plasmid, by PEG treatment. The 35S::UTR:FLUC and 35S::UTR:RLUC reporter plasmids contained the WT or fs version of the ANAC082 (B), CIPK6 (C), At3g15430 (D), At5g27920 (E) or OTLD1 (F) CPuORFs. Co-transfection was carried out for all four combinations for each CPuORF, as indicated. After 48 h of incubation, the transfected cells were harvested and disrupted for luciferase and GUS assays. FLUC and RLUC activities were normalized to GUS activity, and the FLUC and RLUC activities relative to those in the experiment where both reporter plasmids had the WT CPuORF were calculated. Means ± S.D. of at least three biological replicates are shown. Each graph is representative of two or more separate experiments using independently prepared protoplasts. In each graph, bars with the same colors are not significantly different, whereas bars with different colors differ significantly (P < 0.05 by t-test).
Identification of critical residues of the uORF-encoded regulatory peptides
To identify critical amino acid residues of the five CPuORF-encoded peptides responsible for the inhibitory function, we next performed Ala scanning mutagenesis. Several amino acids of each CPuORF peptide were individually changed to Ala, and their effects on expression of the downstream RLUC reporter gene were examined by the transient expression assay. To compare the functional importance of the amino acid residues in the CPuORF peptides and their evolutionary conservation levels, conservation scores (40) were calculated based on the alignments shown in Supplementary Figure S2 and indicated below the graphs showing data of the transient expression assays in Figure 5.
Figure 5.
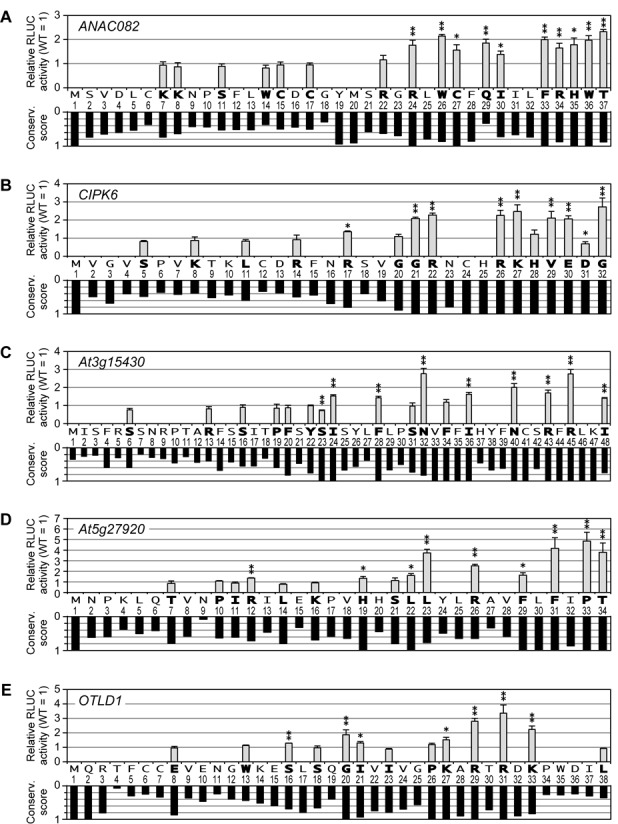
Alanine scanning of the five sequence-dependent CPuORFs. (A–E) Effects of Ala substitutions and conservation scores of amino acid residues in the ANAC082 (A), CIPK6 (B), At3g15430 (C), At5g27920 (D) and OTLD1 (E) CPuORFs. The 35S::UTR:RLUC reporter plasmid harboring a WT CPuORF or its mutant with an Ala substitution was co-transfected with the 35S::FLUC internal control plasmid into MM2d protoplasts by electroporation, and the reporter activities were analyzed as in Figure 2. Means ± S.D. of at least three biological replicates are shown. Each graph is representative of at least two separate experiments using independently prepared protoplasts. Single and double asterisks indicate significant differences from the corresponding WT at P < 0.05 and P < 0.01 by t-test, respectively. The amino acid sequence of each CPuORF is indicated below the graph. Amino acid residues analyzed in the Ala scanning mutagenesis are shown in bold. The numbers below the amino acid residues indicate the positions of the residues in each CPuORF peptide. Conservation scores of the amino acid residues of each CPuORF were calculated based on the alignments presented in Supplementary Figure S2, using the Scorecons server (40), and shown in the graph below the numbers indicating the positions of the amino acid residues.
In the ANAC082 CPuORF peptide, the C-terminal region comprising the 24th to the 37th amino acid residues is highly conserved (Figure 5A, Supplementary Figure S2A). As shown in Figure 5A, all the Ala substitutions tested in this region increased RLUC activity compared with the WT, suggesting that this region is crucial for the repression of main ORF expression. Even the Ala substitution of Q29, which shows a low conservation score, had a strong effect (Figure 5A). By contrast, the Ala substitutions introduced outside of the highly conserved region exhibited no significant effect. Although Y19 and M20 are highly conserved, we did not test Ala substitutions of these residues because they may also affect translation initiation from M20 and it would be difficult to assess their effects solely on the peptide function.
In the CIPK6 CPuORF peptide, the C-terminal region comprising the 20th to the 32nd amino acid residues is highly conserved (Figure 5B, Supplementary Figure S2B). As shown in Figure 5B, many of the Ala substitutions in this region affected RLUC activity. Of these, D31A had a stronger repressive effect than the WT. Among the Ala substitutions introduced outside of the highly conserved region, only R17A caused an increase in RLUC activity; however, its effect was much weaker than the Ala substitutions of Arg residues in the highly conserved region (R22 and R26). These results suggest that the C-terminal region comprising 12 amino acid residues is critical for the repression of main ORF expression and that R17 may have an accessory role.
The At3g15430 CPuORF peptide has a long highly conserved C-terminal region comprising the 19th to the 48th amino acid residues (Figure 5C, Supplementary Figure S2C). Of the first four amino acid residues in the highly conserved region, three residues showing relatively high conservation scores (P19, F20 and Y22) were individually replaced by Ala, and their effects were tested. As shown in Figure 5C, none of them exhibited a significant effect. By contrast, in the remaining C-terminal highly conserved region, many of the Ala substitutions did affect RLUC activity level. These results suggest that the C-terminal region comprising 26 amino acid residues is important for the repression of main ORF expression.
In the At5g27920 CPuORF peptide, the C-terminal region comprising the 19th to the 34th amino acids is relatively highly conserved, and the region comprising the 10th to 18th amino acid residues is weakly conserved (Figure 5D, Supplementary Figure S2D). In the highly conserved region, all the Ala substitutions tested increased RLUC activity except for S21A. By contrast, none of the Ala substitutions in the weakly conserved region showed a significant effect, except that R12A slightly upregulated RLUC activity (Figure 5D). These results suggest that the C-terminal highly conserved region is crucial for the repression of main ORF expression, and that R12 in the weakly conserved region may have an accessory role.
In the OTLD1 CPuORF peptide, the region comprising the 20th to 33th amino acid residues is highly conserved (Figure 5E, Supplementary Figure S2E). In addition, the region comprising the 16th to 18th amino acid residues is weakly conserved. In the highly conserved region, many of the Ala substitutions tested enhanced RLUC activity level. In the weakly conserved region, S16A slightly increased RLUC activity (Figure 5E). These results suggest that the highly conserved region is important for the repression of main ORF expression, and that S16 in the weakly conserved region may have an accessory role.
Overall, the Ala scanning mutagenesis revealed that, in the newly identified regulatory uORF peptides, the regions comprising 12 to 26 amino acid residues in the highly conserved regions have a pivotal role in the repression of main ORF expression, and that, in some of the uORF peptides, the weakly conserved region located upstream of the highly conserved region may have an accessory role. In the ANAC082 CPuORF, Q29A exhibited a strong effect despite its low conservation, whereas G20A and H28A in CIPK6, F34A in At3g15430 and P26A in OTLD1 showed no significant effect despite their high conservation. These results suggest that changes of these amino acid residues to certain specific amino acids are tolerated, but changes to the other amino acids are not.
Synonymous codon changes in the critical region
Although the analysis in Figure 3 suggested that the peptides encoded by the five CPuORFs are responsible for main ORF repression, to further confirm the peptide sequence-dependence of the effects of the five CPuORFs, we investigated the effects of synonymous codon changes in the crucial region of the five CPuORFs. For this analysis, we introduced synonymous changes in the Arg codons in the crucial region of each CPuORF, because the Ala substitutions of the Arg residues showed relatively strong effects in many cases (Figure 5) and two of three nucleotides in an Arg codon can be altered synonymously. We examined the effects of the synonymous codon changes in the five CPuORFs on expression of the downstream RLUC reporter gene by the transient expression assay. As shown in Figure 6, in all five CPuORFs, none of the synonymous changes tested affected RLUC activity, in contrast to the effects of the Ala substitutions of the same codons. These results confirmed that the amino acid sequences of the five CPuORFs are responsible for the sequence-dependent inhibitory effects on main ORF expression.
Figure 6.
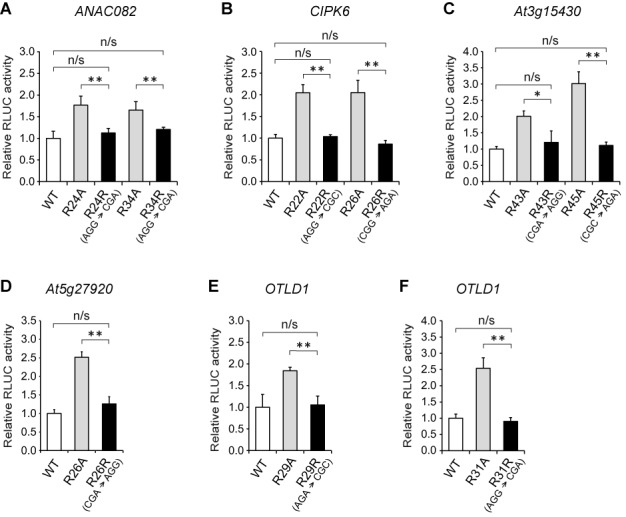
Effects of synonymous codon changes in the five CPuORFs. (A–F) Transient expression studies to compare the effects of Ala substitutions and synonymous codon changes in the ANAC082 (A), CIPK6 (B), At3g15430 (C), At5g27920 (D) and OTLD1 (E and F) CPuORFs. The 35S::UTR:RLUC reporter plasmid harboring a WT CPuORF or its mutant with an Ala substitution or a synonymous codon change was co-transfected with the 35S::FLUC internal control plasmid into MM2d protoplasts by electroporation, and the reporter activities were analyzed as in Figure 2. Means ± S.D. of at least three biological replicates are shown. Each graph is representative of three separate experiments using independently prepared protoplasts. Single and double asterisks indicate significant differences between two constructs at P < 0.05 and P < 0.01 by t-test, respectively, whereas ‘n/s’ indicate a non-significant difference. In (A) and (D), the data for the R24A, R34A and R26A mutants are the same as those presented in Figure 5A and D.
The 3′ non-conserved region and the stop codon position of the OTLD1 uORF are not important
The C-terminal amino acid sequence and the stop codon position of the OTLD1 CPuORF are not evolutionarily conserved (Supplementary Figure S2E), implying that the C-terminal region of the OTLD1 CPuORF-encoded peptide is not essential for the repression of main ORF expression. To address this possibility, we generated a deletion series of the 3′ non-conserved region of the OTLD1 CPuORF (Figure 7A). The changes in the uORF length by these deletions may affect gene expression, regardless of the function of the uORF peptide, because the efficiencies of translational reinitiation following uORF translation and uORF-induced nonsense-mediated mRNA decay depend on uORF size (41,42). Therefore, to assess the effects of the deletions solely on the uORF peptide function, we also constructed an fs version (fs2) of each deletion mutant (Figure 7A) and compared the effects of the deletion mutant OTLD1 CPuORFs on expression of the downstream RLUC reporter gene with those of their corresponding fs versions, using the transient expression assay. As shown in Figure 7B, even in the presence of any of these deletions, the fs2 mutation showed a similar effect to that in their absence. These results suggest that the C-terminal non-conserved region of the OTLD1 CPuORF-encoded peptide is not essential for the repression of main ORF expression. Additionally, these results also suggest that the stop codon position of the OTLD1 CPuORF is not important for the repression, because the position of the stop codon was moved four or five codons upstream in the deletion mutants.
Figure 7.
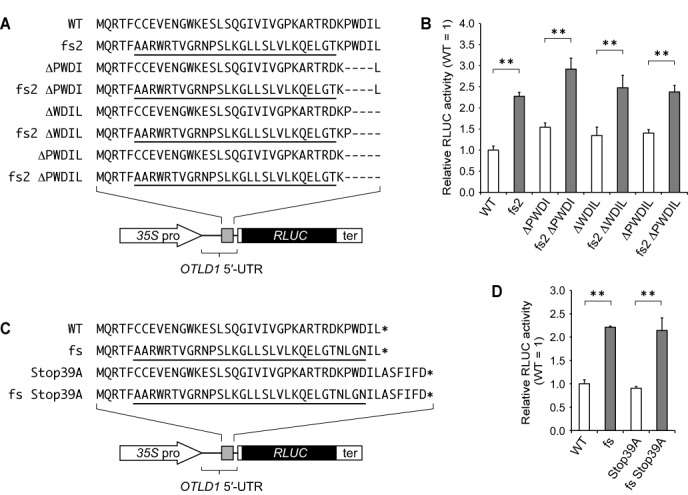
The 3′-non-conserved region and the stop codon of the OTLD1 CPuORF are not important for the peptide sequence-dependent repressive effect. (A) Amino acid sequences of the WT OTLD1 CPuORF and its mutants with a deletion (ΔPWDI, ΔWDIL or ΔPWDIL) and/or a fs (fs2) mutation. The frameshifted region in the fs2 mutant is underlined. Hyphens indicate the deleted amino acid residues. (B) Transient expression assay to test the effect of the C-terminal deletion series. (C) Amino acid sequences of the WT OTLD1 CPuORF and its mutants with the Ala substitution of the stop codon (Stop39A) and/or a fs mutation. Asterisks represent stop codons. The frameshifted region in the fs mutant is underlined. (D) Transient expression assay to examine the effect of uORF stop codon elimination. In (B) and (D), the 35S::UTR:RLUC reporter plasmids containing the WT or mutant OTLD1 CPuORF whose sequence is presented in (A) and (C), respectively, was co-transfected with the 35S::FLUC internal control plasmid into MM2d protoplasts by electroporation, and the reporter activities were analyzed as in Figure 2. Means ± S.D. of four and three biological replicates are shown in (B) and (D), respectively. Each graph is representative of two or more separate experiments using independently prepared protoplasts. Double asterisks indicate a significant difference between two constructs (P < 0.01 by t-test).
To further confirm the unimportance of the stop codon position of the OTLD1 CPuORF, we next substituted an Ala codon for the stop codon (Stop39A) (Figure 7C). This Ala substitution moved the stop codon position six codons downstream, because there is another in-frame stop codon six codons downstream of the original stop codon (Supplementary Figure S1H). We compared the effect of the CPuORF carrying the Stop39A mutation on the RLUC gene expression with that of its fs version using the transient expression assay. As shown in Figure 7D, even in the presence of the Stop39A mutation, the fs mutation exhibited a similar effect to that in its absence. This result suggests that the stop codon position of the OTLD1 CPuORF is not critical for the repression of main ORF expression.
DISCUSSION
Previously, ∼10 peptide sequence-dependent regulatory uORFs have been reported in eukaryotes (2–3,43–46). In plants, although five CPuORFs have been reported to be involved in the regulation of main ORF expression (44,46–51), to date, their peptide sequence-dependence has only been shown in two of them (44,46). In the present study, we analyzed 16 A. thaliana CPuORFs for their effects on main ORF expression, and identified five novel regulatory uORFs that control main ORF expression in a peptide sequence-dependent manner.
Identification of peptide sequence-dependent regulatory uORF
Of the CPuORFs analyzed in this study, the fs mutations of the ANAC082, CIPK6, At3g15430, At5g27920 and OTLD1 CPuORFs conferred a more than two-fold increase in main ORF expression compared with their corresponding WT constructs (Figure 2). The effects of the fs mutations were abolished in the absence of the uORF start codon (Figure 3). These results indicated that translation of the CPuORFs is required for the fs mutations to exert their effects, and suggested that the effects of the fs mutations are caused by amino acid sequence alterations of the CPuORFs rather than by nucleotide sequence changes. Another possibility to explain the dependence of the fs mutations' effects on uORF translation is that the fs mutations affected the translation initiation efficiencies of the CPuORFs, and that the reduced translation efficiencies of the CPuORFs resulted in increased main ORF translation. However, this possibility is unlikely for the following reasons: firstly, in the five CPuORFs, the fs mutations were introduced more than eight nucleotides downstream of the uORF start codon (Supplementary Figure S1A, E, H, J and O), and, therefore, the fs mutations did not change the translation initiation context sequence (52,53). Secondly, in all five CPuORFs, some Ala substitutions introduced at positions further away from the uORF initiation codon and in different positions from the fs mutations showed similar effects to the corresponding fs mutation (Figure 5). Furthermore, synonymous codon changes introduced at codons whose Ala substitution elevated main ORF expression showed no significant repressive effect (Figure 6). These observations strongly suggest that the peptides encoded by these five CPuORFs function to repress main ORF expression.
Of these five CPuORFs, only the OTLD1 CPuORF belongs to class II, whereas the other four CPuORFs belong to class I. Among the remaining class I CPuORFs analyzed, the fs mutations of the ATMPK20, At3g55050 and At5g02480 CPuORFs conferred ∼1.5-fold increases in the main ORF expression level compared with their corresponding WT (Figure 2B), suggesting that these CPuORFs may have a modest sequence-dependent repressive effect, although further mutational analyses are necessary to establish the peptide sequence-dependence of these uORFs. The remaining class I CPuORFs (At1g67480, ANAC096 and ARF4 uORFs) exhibited little or no significant sequence-dependent effect on main ORF expression (Figure 2B). The amino acid sequence conservations of these three CPuORFs are relatively low in A. thaliana compared with those of other plant orthologs (23). Therefore, little or no significant sequence-dependent effects of these CPuORFs are likely because of the low sequence conservation. By contrast, in only two of the six class II CPuORFs, the fs mutations exhibited a significant effect on main ORF expression. In particular, the CIPK23 and DIC1 CPuORFs showed no sequence-dependent effect, despite their highly conserved amino acid sequences (Figure 2C) (23). These observations suggest that class I CPuORF peptides tend to possess a regulatory function that controls main ORF expression, compared with class II CPuORF peptides. However, in all the transient assays performed in this study, the protoplasts were cultured under normal culture conditions; therefore, we cannot rule out the possibility that the CPuORFs that showed little or no sequence-dependent effect may exert their effects under certain specific conditions. In fact, many of the previously reported sequence-dependent regulatory uORFs repress main ORF translation in response to metabolites, such as polyamine, arginine and sucrose (15–18,46). Therefore, it is possible that the CPuORFs that showed little or no sequence-dependent effect in this study may require a metabolite as an effector molecule to exert their effects, and that the cellular concentration of the metabolite was not sufficient under our conditions.
Possible mechanisms of gene expression control by the newly identified regulatory uORF peptides
The co-transfection assays in Figure 4 revealed that the peptides encoded by the five newly identified sequence-dependent uORFs act in cis to repress main ORF expression, suggesting that these uORF peptides function as regulatory nascent peptides. The most likely underlying mechanism for the repression is that the nascent peptides encoded by these uORFs cause ribosome stalling. Genetic and biochemical studies have revealed that some regulatory nascent peptides cause ribosome stalling by interacting with components of the ribosomal exit tunnel (7,9,25–31). Cryo-electron microscopy studies have observed how regulatory nascent peptides interact with exit tunnel components. In prokaryotes, several of the C-terminal 16 and 11 amino acid residues in the TnaC and SecM nascent peptides interact with exit tunnel components when ribosome stalling occurs (54,55). In eukaryotes, several of the C-terminal 16 and 19 amino acid residues in the gpUL4 and arg-2 uORF-encoded nascent peptides interact with exit tunnel components (24). Ala scanning mutagenesis in this study revealed that, in the five newly identified regulatory uORF peptides, the regions comprising 12 to 26 amino acid residues in the highly conserved regions have a pivotal role for the repression of main ORF expression (Figure 5). Thus, the lengths of the crucial regions in these uORF peptides are roughly consistent with those of the regions interacting with the exit tunnel components in the previously characterized regulatory nascent peptides. In some of the newly identified uORF peptides, an Ala substitution in the weakly conserved region located upstream of the highly conserved region showed a weak effect on reporter expression (Figure 5). This suggested that the weakly conserved regions might have an accessory role in ribosome stalling. Alternatively, because a eukaryotic ribosomal exit tunnel holds 30–40 amino acid residues (56,57) and, therefore, the weakly conserved regions in the nascent uORF peptides should be inside the exit tunnel when ribosome stalling occurs, the Ala substitutions in the weakly conserved regions may have affected the structure or position of the crucial region in the nascent peptide to some extent, resulting in a slight impairment of the uORF peptide function.
The Ala scanning mutagenesis also indicated that, in the peptides encoded by ANAC082, CIPK6, At3g15430 and At5g27920 CPuORFs, all of which belong to class I, the crucial regions are located at the C-terminus. By contrast, the deletion analysis of the OTLD1 CPuORF, which belongs to class II, revealed that the C-terminal five amino acid residues are essentially dispensable for the repression of main ORF expression (Figure 7B). If the OTLD1 CPuORF-encoded peptide causes ribosome stalling by interacting with exit tunnel components, it is unlikely that the ribosome is stalled at the uORF stop codon as seen in the gpUL4 and arg-2 uORFs, because deletion of the C-terminal five amino acid residues, which would change the position of the crucial region of the uORF-encoded peptide in the exit tunnel if ribosome stalling occurs at the stop codon, had little effect (Figure 7B). Additionally, elimination of the stop codon, which changed the stop codon position to one six codons downstream from the original position, did not affect the sequence-dependent inhibitory effect of the OTLD1 CPuORF (Figure 7D). Therefore, it is more likely that ribosome stalling occurs at the translation elongation step before the 3′-terminal non-conserved region of the OTLD1 CPuORF. In prokaryotes, there are examples of nascent peptide-mediated translation elongation arrest, in which ribosomal stalling occurs in the middle of small ORFs (1,4). In eukaryotes, the regulatory nascent peptide encoded by the A. thaliana CGS1 gene causes ribosomal stalling in the middle of the main ORF (58,59). However, in all of the previously characterized regulatory uORFs in which the stall position has been determined (i.e. the gpUL4, arg-2, CPA1, mammalian AdoMetDC and A. thaliana AdoMetDC1 uORFs), ribosome stalling mainly occurs at the uORF stop codon (14–18,60), although the arg-2 uORF-encoded peptide can cause ribosomal stalling at the translation elongation step if the uORF stop codon is removed (61). Therefore, the OTLD1 CPuORF-mediated regulation may involve, at least in part, a distinct mechanism from the regulation mediated by the previously characterized native uORF peptides.
In the analysis of Figure 3, eliminating the start codon of the At5g27920 and OTLD1 CPuORFs caused stronger derepression than the corresponding fs mutation; whereas, in the ANAC082, CIPK6 and At3g15430 CPuORFs, start codon elimination and the fs mutation showed a similar derepressive effect. The presence of a uORF can have an inhibitory effect on main ORF translation, regardless of its peptide function, if the uORF is translated and ribosomes dissociate after translation. It is likely that the fs mutant version of the At5g27920 and OTLD1 CPuORFs had a repressive effect to some extent by this mechanism. By contrast, the results shown in Figure 3A–C suggest that, for ANAC082, CIPK6 and At3g15430, the presence of the fs mutant CPuORF had little or no effect on main ORF translation. One possibility to explain this observation is that translational initiation efficiencies of these CPuORFs are low and therefore most scanning ribosomes bypass these CPuORFs. Another possibility is that ribosomes that had translated these CPuORFs efficiently reinitiate translation at the main ORF. However, this latter possibility is unlikely, because reinitiation efficiency depends on the length of the uORF, and the ANAC082, CIPK6 and At3g1543 0 CPuORFs, whose sizes are 114, 99 and 147 nt, respectively, are too long for efficient reinitiation to occur (41). By contrast, the former possibility is more likely for the following reason. In A. thaliana, a purine (A or G) at position −3 and a guanine at position +4, where the A of AUG is defined as +1, are the optimal context for efficient translation initiation (62,63), as established in mammals by Kozak (52,53). The initiation contexts of the At5g27920 and OTLD1 CPuORFs are partially consistent with the optimal context, whereas those of the ANAC082 and At3g15430 CPuORFs are completely inconsistent with the optimal context (Figure 1). Although the initiation context of the CIPK6 CPuORF is consistent with the optimal context, another uORF overlaps with the CPuORF and has its start codon upstream of the CPuORF (Figure 1, Supplementary Figure S1E); therefore, it is likely that the presence of the overlapping uORF reduces translation initiation efficiency of the CIPK6 CPuORF. In addition, the former possibility is also supported by the observation that, in the Saccharomyces cerevisiae CPA1 uORF, whose translation initiation efficiency is low (64), a missense mutation impairing the regulatory function of the uORF peptide showed a similar derepressive effect to removal of the uORF start codon in yeast cells (65). It has been shown in vitro that scanning ribosomes frequently bypass the CPA1 uORF and reach the main ORF start codon, and that they are blocked when ribosome stalling occurs at the uORF in response to arginine (64).
The main ORFs regulated by the five newly identified sequence-dependent uORFs encode proteins involved in the control of gene expression or protein activity. ANAC082 encodes a NAC (NAM, ATAF1,2 and CUC2) domain-containing transcription factor. CIPK6 encodes a serine/threonine protein kinase that modulates the activity of a potassium channel, AKT2 (66), and is involved in the response to salt and osmotic stresses (67). At3g15430 encodes a protein related to the mammalian regulator of chromosome condensation, RCC1, which is the Ran guanine-exchange factor that regulates nuclear transport and mitotic spindle formation (68). At5g27920 encodes an F-box family protein. OTLD1 encodes an otubain-like histone deubiquitinase, which is involved in transcriptional repression via histone deubiquitination (69). Among the previously reported eukaryotic regulatory nascent peptides whose regulatory roles have been elucidated, many of them control the expression of metabolic enzyme genes and act in feedback regulation of the metabolic pathways (15–17,34,44). By contrast, the genes regulated by the uORF peptides identified in this study do not include any metabolic enzyme genes. The five newly identified regulatory uORF peptides had an inhibitory effect under normal culture conditions; therefore, these uORF peptides are likely to repress main ORF expression constitutively or in response to a metabolite that is present at a sufficient level in MM2d protoplast cells cultured under normal conditions. Even in the case where the uORF-encoded peptides always repress main ORF expression when the uORFs are translated, these uORFs can be involved in conditional regulation of gene expression if the translational initiation efficiencies of these uORFs are conditionally modulated. Such conditional modulation of translational initiation efficiencies of sequence-dependent regulatory uORFs has been reported in the human CHOP and A. thaliana AdoMetDC1 genes. In the CHOP gene, scanning ribosomes frequently bypass the sequence-dependent inhibitory uORF under endoplasmic reticulum stress condition, and thereby translation of the main ORF is enhanced (70). In the A. thaliana AdoMetDC1 gene, an overlapping uORF, whose start codon is located upstream of the sequence-dependent inhibitory uORF, was suggested to be involved in polyamine-responsive modulation of the translational initiation efficiency of the inhibitory uORF (44). As mentioned above, there is also an overlapping uORF upstream of the CIPK6 CPuORF (Figure 1, Supplementary Figure S1E). The size of the overlapping uORF and its position relative to the CPuORF are evolutionarily conserved. Additionally, it has been suggested that expression of the chickpea CIPK6 ortholog is regulated in response to salt stress at both the transcriptional and post-transcriptional level (71). Therefore, the CIPK6 uORFs might be involved in the post-transcriptional regulation in response to salt stress.
CONCLUSIONS
The present study showed that at least five of the 16 CPuORFs tested encode regulatory peptides. In A. thaliana, 43 homology groups of CPuORFs have been identified (19,21,23) and two of them have been reported to exert a peptide sequence-dependent effect on main ORF expression (44,46). If the remaining CPuORFs contain peptide sequence-dependent regulatory uORFs at the same ratio as seen in this study, there would be at least 15 homology groups of sequence-dependent regulatory uORFs in the A. thaliana genome. As mentioned above, it is possible that the CPuORFs that showed only weak or no significant effect in this study may exert a strong effect under certain conditions. If that is the case, even higher numbers of sequence-dependent regulatory uORFs would exist in the A. thaliana genome. In addition, the genes regulated by the peptide sequence-dependent uORFs identified in this study comprise a variety of regulatory genes. Thus, this study suggests that uORF peptide-mediated gene regulation is more prevalent than previously thought and is involved in the control of a wide variety of genes. Further studies on the regulatory roles of the identified uORFs will reveal novel roles of the uORF-encoded regulatory peptides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Ms Hitomi Sekihara, Ms Eriko Tanaka and Ms Saeko Yasokawa for skillful technical assistance and Ms Maki Mori for general assistance. We used the DNA sequencing facility of the Graduate School of Agriculture, Hokkaido University.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Present address:
Isao Ebina, Chugai Pharmaceutical Co., Ltd, Gotemba 412-8513, Japan.
Mariko Takemoto-Tsutsumi, SRD Corporation, Chuo-ku, Tokyo 104-0032, Japan.
Shun Watanabe, Wakou Shokuhin Co., Ltd, Otaru 047-0261, Japan.
Hiroaki Koyama, Nigata Prefectural Police, Nigata 950-8553, Japan.
Yayoi Endo, Tsu Agriculture, Forestry and Fisheries Office, Tsu 514-8567, Japan.
FUNDING
Japan Society for the Promotion of Science [Grant-in-aid for Scientific Research (C) 25440122 to H.O.]; Ministry of Education, Culture, Sports, Science and Technology of Japan [Grant-in-aid for Scientific Research on Innovative Areas 21112501 to H.O., 22119006 to S.N.]. Funding for open access charge: Japan Society for the Promotion of Science [Grant-in-aid for Scientific Research (C) 25440122 to H.O.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lovett P.S., Rogers E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenson T., Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 4.Ito K., Chiba S. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Vera L.R., Sachs M.S., Squires C.L., Yanofsky C. Nascent polypeptide sequences that influence ribosome function. Curr. Opin. Microbiol. 2011;14:160–166. doi: 10.1016/j.mib.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Nakatogawa H., Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 7.Chiba S., Lamsa A., Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba S., Ito K. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol. Cell. 2012;47:863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez-Laslop N., Thum C., Mankin A.S. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Ramu H., Mankin A., Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 11.Gong F., Ito K., Nakamura Y., Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro) Proc. Natl. Acad. Sci. U.S.A. 2001;98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvo S.E., Pagliarini D.J., Mootha V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Arnim A.G., Jia Q., Vaughn J.N. Regulation of plant translation by upstream open reading frames. Plant Sci. 2014;214:1–12. doi: 10.1016/j.plantsci.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Cao J., Geballe A.P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law G.L., Raney A., Heusner C., Morris D.R. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z., Sachs M.S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Gaba A., Sachs M.S. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J. Biol. Chem. 1999;274:37565–37574. doi: 10.1074/jbc.274.53.37565. [DOI] [PubMed] [Google Scholar]
- 18.Uchiyama-Kadokura N., Murakami K., Takemoto M., Koyanagi N., Murota K., Naito S., Onouchi H. Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:1556–1567. doi: 10.1093/pcp/pcu086. [DOI] [PubMed] [Google Scholar]
- 19.Hayden C.A., Jorgensen R.A. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowe M.L., Wang X.Q., Rothnagel J.A. Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides. BMC Genomics. 2006;7:16. doi: 10.1186/1471-2164-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughn J.N., Ellingson S.R., Mignone F., Arnim A. Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA. 2012;18:368–384. doi: 10.1261/rna.031179.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden C.A., Bosco G. Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics. 2008;9:61. doi: 10.1186/1471-2164-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi H., Takahashi A., Naito S., Onouchi H. BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics. 2012;28:2231–2241. doi: 10.1093/bioinformatics/bts303. [DOI] [PubMed] [Google Scholar]
- 24.Bhushan S., Meyer H., Starosta A.L., Becker T., Mielke T., Berninghausen O., Sattler M., Wilson D.N., Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Nakatogawa H., Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 26.Wu C., Wei J., Lin P.J., Tu L., Deutsch C., Johnson A.E., Sachs M.S. Arginine changes the conformation of the arginine attenuator peptide relative to the ribosome tunnel. J. Mol. Biol. 2012;416:518–533. doi: 10.1016/j.jmb.2011.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz-Vera L.R., Rajagopal S., Squires C., Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Martinez A.K., Shirole N.H., Murakami S., Benedik M.J., Sachs M.S., Cruz-Vera L.R. Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function. Nucleic Acids Res. 2012;40:2247–2257. doi: 10.1093/nar/gkr1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez A.K., Gordon E., Sengupta A., Shirole N., Klepacki D., Martinez-Garriga B., Brown L.M., Benedik M.J., Yanofsky C., Mankin A.S., et al. Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res. 2014;42:1245–1256. doi: 10.1093/nar/gkt923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence M.G., Lindahl L., Zengel J.M. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 2008;190:5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yap M.N., Bernstein H.D. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menges M., Murray J.A. Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 2002;30:203–212. doi: 10.1046/j.1365-313x.2002.01274.x. [DOI] [PubMed] [Google Scholar]
- 33.Nagata T., Nemoto Y., Hasezawa S. Tobacco BY-2 cell line as the ‘Hela’ cell in the cell biology of higher plants. Int. Rev. Cytol. 1992;132:1–30. [Google Scholar]
- 34.Chiba Y., Ishikawa M., Kijima F., Tyson R.H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R.M., Naito S. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- 35.Ominato K., Akita H., Suzuki A., Kijima F., Yoshino T., Chiba Y., Onouchi H., Naito S. Identification of a short highly conserved amino acid sequence as the functional region required for posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. J. Biol. Chem. 2002;277:36380–36386. doi: 10.1074/jbc.M204645200. [DOI] [PubMed] [Google Scholar]
- 36.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 37.Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. S-Adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10225–10230. doi: 10.1073/pnas.1831512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuo N., Minami M., Maeda T., Hiratsuka K. Dual luciferase assay for monitoring gene expression in higher plants. Plant Biotechnol. 2001;18:71–75. [Google Scholar]
- 39.Jefferson R.A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 1987;5:387–405. [Google Scholar]
- 40.Valdar W.S.J. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 41.Rajkowitsch L., Vilela C., Berthelot K., Ramirez C.V., McCarthy J.E. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- 42.Nyikó T., Sonkoly B., Merai Z., Benkovics A.H., Silhavy D. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 2009;71:367–378. doi: 10.1007/s11103-009-9528-4. [DOI] [PubMed] [Google Scholar]
- 43.Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D., Fafournoux P. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 2001;29:4341–4351. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanfrey C., Elliott K.A., Franceschetti M., Mayer M.J., Illingworth C., Michael A.J. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J. Biol. Chem. 2005;280:39229–39237. doi: 10.1074/jbc.M509340200. [DOI] [PubMed] [Google Scholar]
- 45.Ivanov I.P., Loughran G., Atkins J.F. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahmani F., Hummel M., Schuurmans J., Wiese-Klinkenberg A., Smeekens S., Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai A., Hanzawa Y., Komura M., Yamamoto K.T., Komeda Y., Takahashi T. The dwarf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORF of a bHLH gene. Development. 2006;133:3575–3585. doi: 10.1242/dev.02535. [DOI] [PubMed] [Google Scholar]
- 48.Tabuchi T., Okada T., Azuma T., Nanmori T., Yasuda T. Posttranscriptional regulation by the upstream open reading frame of the phosphoethanolamine N-methyltransferase gene. Biosci. Biotechnol. Biochem. 2006;70:2330–2334. doi: 10.1271/bbb.60309. [DOI] [PubMed] [Google Scholar]
- 49.Alatorre-Cobos F., Cruz-Ramirez A., Hayden C.A., Perez-Torres C.A., Chauvin A.L., Ibarra-Laclette E., Alva-Cortes E., Jorgensen R.A., Herrera-Estrella L. Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J. Exp. Bot. 2012;63:5203–5221. doi: 10.1093/jxb/ers180. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X., Thalor S.K., Takahashi Y., Berberich T., Kusano T. An inhibitory effect of the sequence-conserved upstream open-reading frame on the translation of the main open-reading frame of HsfB1 transcripts in Arabidopsis. Plant Cell Environ. 2012;35:2014–2030. doi: 10.1111/j.1365-3040.2012.02533.x. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen R.A., Dorantes-Acosta A.E. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front. Plant Sci. 2012;3:191. doi: 10.3389/fpls.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 53.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seidelt B., Innis C.A., Wilson D.N., Gartmann M., Armache J.P., Villa E., Trabuco L.G., Becker T., Mielke T., Schulten K., et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhushan S., Hoffmann T., Seidelt B., Frauenfeld J., Mielke T., Berninghausen O., Wilson D.N., Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malkin L.I., Rich A. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J. Mol. Biol. 1967;26:329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- 57.Blobel G., Sabatini D.D. Controlled proteolysis of nascent polypeptides in rat liver cell fractions. I. Location of the polypeptides within ribosomes. J. Cell Biol. 1970;45:130–145. doi: 10.1083/jcb.45.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005;19:1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamashita Y., Kadokura Y., Sotta N., Fujiwara T., Takigawa I., Satake A., Onouchi H., Naito S. Ribosomes in a stacked array: elucidation of the step in translation elongation at which they are stalled during S-adenosyl-L-methionine-induced translation arrest of CGS1 mRNA. J. Biol. Chem. 2014;289:12693–12704. doi: 10.1074/jbc.M113.526616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J., Wu C., Sachs M.S. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol. 2012;32:2396–2406. doi: 10.1128/MCB.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang P., Spevak C.C., Wu C., Sachs M.S. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawaguchi R., Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005;33:955–965. doi: 10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugio T., Matsuura H., Matsui T., Matsunaga M., Nosho T., Kanaya S., Shinmyo A., Kato K. Effect of the sequence context of the AUG initiation codon on the rate of translation in dicotyledonous and monocotyledonous plant cells. J. Biosci. Bioeng. 2010;109:170–173. doi: 10.1016/j.jbiosc.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A.G., Sachs M.S. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaba A., Jacobson A., Sachs M.S. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 66.Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratge-Faillie C., Offenborn J.N., Lacombe B., Dreyer I., Thibaud J.B., et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011;21:1116–1130. doi: 10.1038/cr.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen L., Wang Q.Q., Zhou L., Ren F., Li D.D., Li X.B. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 2013;40:4759–4767. doi: 10.1007/s11033-013-2572-9. [DOI] [PubMed] [Google Scholar]
- 68.Hadjebi O., Casas-Terradellas E., Garcia-Gonzalo F.R., Rosa J.L. The RCC1 superfamily: from genes, to function, to disease. Biochim. Biophys. Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 69.Krichevsky A., Zaltsman A., Lacroix B., Citovsky V. Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11157–11162. doi: 10.1073/pnas.1014030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palam L.R., Baird T.D., Wek R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripathi V., Parasuraman B., Laxmi A., Chattopadhyay D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009;58:778–790. doi: 10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


