Abstract
Epigenetic silencing of transgene expression represents a major obstacle for the efficient genetic modification of multipotent and pluripotent stem cells. We and others have demonstrated that a 1.5 kb methylation-free CpG island from the human HNRPA2B1-CBX3 housekeeping genes (A2UCOE) effectively prevents transgene silencing and variegation in cell lines, multipotent and pluripotent stem cells, and their differentiated progeny. However, the bidirectional promoter activity of this element may disturb expression of neighboring genes. Furthermore, the epigenetic basis underlying the anti-silencing effect of the UCOE on juxtaposed promoters has been only partially explored. In this study we removed the HNRPA2B1 moiety from the A2UCOE and demonstrate efficient anti-silencing properties also for a minimal 0.7 kb element containing merely the CBX3 promoter. This DNA element largely prevents silencing of viral and tissue-specific promoters in multipotent and pluripotent stem cells. The protective activity of CBX3 was associated with reduced promoter CpG-methylation, decreased levels of repressive and increased levels of active histone marks. Moreover, the anti-silencing effect of CBX3 was locally restricted and when linked to tissue-specific promoters did not activate transcription in off target cells. Thus, CBX3 is a highly attractive element for sustained, tissue-specific and copy-number dependent transgene expression in vitro and in vivo.
INTRODUCTION
The genetic modification of pluripotent and multipotent stem cells offers a myriad of opportunities in regenerative medicine. In most cases self-inactivating lentiviral vectors (SIN-LV) are used for the genetic modification of stem cells, as stable gene transfer by SIN-LVs has been shown to be less mutagenic and genotoxic than other viral-based gene transfer vectors (1–5). However, even advanced lentiviral vectors remain subject to a certain degree of silencing and are influenced by position effects leading to variegated transgene expression (position effect variegation, PEV). These limitations of SIN-LVs become particularly obvious in pluripotent stem cells (PSCs). These cells, similar to other stem cell entities, have a strong epigenetic defense against foreign DNA (6–10), and lentiviral vectors suffer from massive epigenetic silencing in PSCs, especially during their differentiation which is associated with extensive chromatin remodeling (11,12).
To counteract silencing of transgene expression and PEV, various genetic elements such as insulators, scaffold attachment regions, origins of replication, or CpG islands have been investigated (13–16). Here, the most commonly used insulator, the chicken hypersensitive site 4 element (cHS4), has been demonstrated to reduce the spread of repressive histone modifications and DNA methylation towards the expression cassette, when incorporated into the viral long terminal repeat (LTR) (17,18). However, the element has been shown to cause a marked reduction in viral titers and its effects have been reported to be context dependent (19,20). We and others have recently shown that ubiquitous chromatin opening elements (UCOEs) represent promising tools to avoid silencing and sustain transgene expression in a wide variety of cellular models including cell lines, multipotent hematopoietic stem cells, as well as PSCs and their differentiated progeny (21–25). These elements are characterized by unmethylated CpG islands spanning dual, divergently transcribed promoters of housekeeping genes. Moreover, their chromatin structure is highly permissive, marked by hyperacetylation of histones H3 and H4, histone H3 lysine 4 trimethylation (H3K4me3) and the lack of histone H1 (26). The best studied UCOE is a 1.5 kb sequence derived from the human HNRPA2B1-CBX3 locus (A2UCOE) (22,27). Different variants of the A2UCOE have been successfully employed to sustain transgene expression, counteract epigenetic silencing, and prevent PEV (21,27). However, the bidirectional promoter activity of these elements inherently carries the risk of read-through transcripts initiated at the reverse oriented promoter, in most cases the HNRPA2B1 promoter, and thus do have the potential to deregulate the expression of neighboring cellular genes (28). Moreover, the same transcript can result in the formation of an antisense RNA during virus production and reduction of virus titers. As the HNRPA2B1 promoter is methylated in embryonic carcinoma cells (27), we hypothesized that this moiety of the bidirectional promoter might be dispensable for the anti-silencing function of the element.
Here, we studied the properties of an A2UCOE fragment lacking the HNRPA2B1 promoter and document almost complete preservation of the anti-silencing properties of the resulting minimal 0.7 kb UCOE (CBX3-UCOE) in multipotent and pluripotent stem cells and as well as in combination with viral and tissue-specific promoters. Moreover, we demonstrate that the anti-silencing activity of this minimal element is associated with characteristic changes in promoter CpG-methylation and histone modification generating a transcriptionally permissive chromatin environment. Importantly, we show that the chromatin opening capability of CBX3-UCOE is locally restricted and does not override the specificity of tissue-specific promoters linked to it.
MATERIALS AND METHODS
Cell culture
Murine P19 cells were cultivated in α-MEM medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum (PAN Biotech, Aidenbach, Germany), 2 mM glutamine and penicillin/streptomycin (100 U ml−1 each) (all Life technologies, Carlsbad, CA, USA). Human PLB985 and Jurkat cells were kept in RPMI (Life technologies) containing 2 mM glutamine, penicillin/streptomycin (100 U ml−1 each) and 10% fetal calf serum.
Murine Lin− cells were isolated from bone marrow samples harvested from the femurs of B6.SJL-PtprcaPepcb/BoyCrl mice (Ly5.1) using the Miltenyi Lineage Cell Depletion Kit (Miltenyi, Bergisch Gladbach, Germany). Isolated cells were cultured in StemSpan serum-free medium (STEMCELL technologies, Vancouver, Canada), supplemented with penicillin/streptomycin (100 U ml−1 each), 2 mM glutamine 10 ng ml−1 mSCF, 20 ng ml−1 mTPO, 20 ng ml−1 mIGF-2 and 10 ng ml−1 hFGF1 (all Peprotech, Hamburg, Germany).
The mESC line CCE (29) was cultured on mitomycin C-treated MEF feeder cells in ESC medium (knockout DMEM, 15% ES-tested FCS, 1 mM L-glutamine, 0.1 mM nonessential amino acids, penicillin/streptomycin (100 U ml−1 each) (all Invitrogen), 100 μM β-mercaptoethanol and 1 μg ml−1 leukemia inhibitory factor (LIF) (kindly provided by the Institute of Technical Chemistry, Hannover Medical School, Hannover, Germany). Murine ESCs were passaged every 2–3 days using Trypsin (Invitrogen, Carlsbad, CA, USA).
The hiPSC line hCD34iPSC11 was previously generated from mobilized peripheral blood CD34+ cells using a polycistronic lentiviral vectors over-expressing OCT4, SOX2, KLF4, c-MYC and a dTomato-reporter (24), and was cultured on irradiated CF1-MEF feeder cells in ESC medium (knockout DMEM, 20% knock out serum replacement, 1 mM L-glutamine, 1% NEAA, penicillin/streptomycin (100 U ml−1 each) (all Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich) and 40 ng ml−1 fibroblast growth factor-basic (bFGF, kindly provided by the Institute of Technical Chemistry, Hannover Medical School, Hannover, Germany). Human iPSC were passaged weekly using 2 mg ml−1 collagenase V (STEMCELL technologies).
Generation and production of lentiviral vectors
The lentiviral vectors CBX3EW and CBX3MEW containing CBX3-UCOE were generated by excision of the A2 moiety from the vector UrEW (Christian Brendel, unpublished) and UrMEW (27) by enzymatic digestion with SmaI and EcoRV and subsequent ligation. The vector CBX3SEW was cloned by excision of the MRP8 promoter from CBX3MEW and insertion of SFFV. Canonical and cryptic splice sites in CBX3 were deleted by site directed mutagenesis to generate CBX3*.
Lentiviral vector supernatants were produced by transient cotransfection of 293T cells using polyethylenimine (PEI) or calcium phosphate precipitation according to standard protocols (30,31). 48 h after transfection supernatants were collected and concentrated 100-fold by ultracentrifugation at 4°C. The titers were analyzed by transduction of PLB985 cells in limiting dilution and analysis of reporter gene expression.
Transduction
Cell lines were transduced in 24-well plates by adding concentrated viral supernatant to 2 × 105 cells in 500 μl medium in the presence of protamine sulphate (6 μg ml−1) and spinoculation (1000 × g, 1 h, 32°C). Transduction of murine lin− cells isolated from bone marrow cells was done with the same protocol after 24 h prestimulation at a multiplicity of infection of 20–50.
For transduction, 1 × 105 human or murine PSC were seeded as single cells onto Matrigel- (Beckton & Dickinson, Heidelberg, Germany) or gelatine-coated 12-well plates in standard medium containing, respectively. The next day, cells were transduced with lentiviral vectors (MOI 10–50) in the presence of 10 μg ml−1 protamine sulphate (Sigma Aldrich). After 3–4 days cells were transferred to MEF cells and cultured as described above.
Generation of PLB985 clones
PLB985 cells were transduced with vector CBX3MEW at low and high MOI and eGFP expression was analyzed by flow cytometry 5 days later. Both cell populations were used for the generation of cell clones via limiting dilution. After several weeks in culture the MFI was analyzed for each clone by flow cytometry and the vector copy number (VCN) was determined by quantitative polymerase chain reaction (qPCR).
Animals
Congenic B6.SJL-Ptprca Pepcb/BoyCrl (Ly5.1) and C57BL/6N mice were obtained from Charles River (Wilmington, MA, USA). All experimental procedures were performed in compliance with the local animal experimentation guidelines. Animal experiments were approved by the regional council (Regierungspräsidium, Darmstadt, Germany).
Transplantation
Lin− cells isolated from bone marrow of B6.SJL-PtprcaPepcb/BoyCrl mice (Ly5.1) were washed 1 day after transduction and resuspended in PBS. 5 × 105 to 1 × 106 cells were transplanted into lethally irradiated mice (9.5 Gy) via tail vain injection. Transplanted mice were kept in individually ventilated cages and drinking water was supplemented with 1.6 g l−1 neomycin (Carl Roth, Karlsruhe, Germany) for 2 weeks.
Hematopoietic differentiation of murine ESC
Hematopoietic differentiation of murine ESC was carried out as previously described (32). In brief, miPSCs were seeded for embryoid body (EB) formation in suspension cultures. On day 5 of differentiation, the medium was changed and supplemented with 30 ng ml−1 murine stem cell factor (mSCF) and 10 ng ml−1 murine interleukin (IL)-3 (both Peprotech). EBs were harvested on day 8, dissociated using Collagenase IV (STEMCELL technologies) and stained for CD41 expression.
Hematopoietic differentiation of human iPSC
For hematopoietic differentiation, human iPSC were subjected to an established hematopoietic differentiation protocol (33) with minor modifications. In brief, formation of EBs was induced in suspension culture. On day 5 EBs were transferred to adherent plates and cultured in differentiation medium containing 25 ng ml−1 human IL-3 and 50 ng ml−1 human granulocyte colony-stimulating factor (G-CSF). Medium was changed twice per week and myeloid cells were harvested from the supernatant from day 20 onwards and further differentiated in RPMI-medium containing 100 ng ml−1 G-CSF for 3–5 days.
Antibodies and FACS analysis
The following antibodies were used for the detection of hematopoietic subpopulations: Gr1-Vioblue (clone RB6–8C5), CD11b-APC (clone M1/70), CD45.1-PE-Cy7 (clone A20), CD45.2-PerCP-Cy5.5 (clone 104), CD3e-V500 (clone 145–2C11 or 500A2), B220-PE (clone RA3–6B2), Sca1-PE-Cy7 (clone D7) and cKit-APC (clone 2B8). Data acquisition was performed on a FACSCanto II flow cytometer and analyzed using the FACSDiva 6.0 software (all Beckton & Dickinson). Dead cells were excluded by staining with eFluor780 (eBioscience, San Diego, CA, USA). Sorting of cells for subsequent genomic DNA (gDNA) isolation was performed on a FACSAria II flow cytometer (Beckton & Dickinson) running with FACSDiva 6.0 software. For flow cytometric analysis of murine and human PSCs cells the following antibodies were used: mSSEA1-APC, mCD41-APC, hTra-1–60-PE, hCD45-APC, hCD11b-APC; isotype-controls: mouse-IgG1a- APC, mouse-IgM-PE (all eBioscience). Data acquisition was performed on a FACScalibur (Beckton & Dickinson) and raw data were analyzed using the software FlowJo (TreeStar, Ashland, OR, USA).
Quantitative PCR
Quantitative PCRs for the determination of VCNs in transduced cells were performed in a Roche LightCycler480 machine as duplex reactions. For this genomic DNA was isolated at least 5 days after transduction using the DNeasy extraction kit (Qiagen, Hilden, Germany). 20 ng of gDNA was used as template and mixed with Roche LC480 Probes Master mix (Roche, Basel, Switzerland) and primers and probes specific for eGFP (Primerdesign, Southampton, UK) and an internal control gene.
As a reference for human-derived samples, gDNA isolated from a PLB985 clone harboring a single vector integration was used, and for murine samples a Baf3 clone harboring a single vector integration was used. The primer sequences used are listed in Supplementary Table S1.
cLAM PCR
A cDNA based LAM (cLAM) protocol was used for the analysis of RNA transcripts as described in (34,35). Briefly, total RNA from a UrMgpsW transduced PLB-XCGD single cell clone was isolated with the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. This RNA was used for synthesis of double-stranded cDNA by the RETROscript Reverse Amplification Kit (Life technologies). Transcripts starting at the CBX3 promoter were linearly amplified using the biotinylated Primer CBX3 LAM1. After immobilization of the target cDNA on beads, the second strand was generated by using random hexanucleotide primers and Klenow polymerase. Subsequent to digestion with FatI, a restriction site complementary linker was ligated, enabling the amplification of CBX3 transcripts by nested PCR with primers CBX3 LAM2 and CBX3 LAM3 and LC1 and LC2. PCR products were analyzed on agarose gel and subcloned for sequencing. Sequences were aligned to the human genome (GRCh37/hg19, February 2009) using blat search genome (http://genome.ucsc.edu/). The primers used are listed in Supplementary Table S1.
Bisulfite conversion and sequencing
About 1 μg of isolated gDNA were used for bisulfite conversion with the EpiTect Bisulfit Kit (Qiagen) according to the manufacturer's protocol. The product was used as template for a semi-nested PCR amplification of promoter sequences. PCR reactions were shotgun cloned into the TOPO-TA plasmid (Invitrogen) and the plasmid preparation from clones was individually sequenced. The primers used are listed in Supplementary Table S1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to the Abcam-X-ChIP protocol with modifications as described in (36). For immunoprecipitation 2–10 μg of the following antibodies were used: H3K4me3 (ab1012 or ab12209, Abcam, Cambridge, UK). H3K9me3 (ab8898, Abcam), H3K27me3 (ab6002, Abcam), RNA polymerase II CTD repeat YSPTSPS (phospho S5) (ab5131, Abcam). As control immunoglobulin G (IgG) was used: IgG mouse (sc-2025, Santa-Cruz, Dallas, TX, USA). Purification of ChIP-DNA was performed using DNA purification columns (ChIP DNA Clean and ConcentratorTM, Zymo Research, Irvine, CA, USA). ChIP-DNA was eluted with 30 μl of buffer and analyzed by SYBR green (Thermo Fisher Scientific, Waltham, MA, USA) based quantitative PCR using 1 μl of chromatin. Primers used are listed in Supplementary Table S1.
RESULTS
Generation of a minimal A2UCOE
To identify a minimal A2UCOE fragment lacking divergent promoter activity but still preventing transgene silencing, we deleted the reverse-oriented HNRPA2B1 promoter in the 1.5 kb A2UCOE (5'-HNRPA2B1-CBX3–3′). The remaining 674 bp, uni-directional CBX3 core promoter fragment, comprises the two alternative first exons of the CBX3 gene and a CpG-rich intragenic region between the CBX3 and HNRPA2B1 promoters (Figure 1A). This minimal 0.7 kb UCOE (CBX3-UCOE) was then introduced into a variety of lentiviral vector configurations either upstream of the viral spleen focus forming virus (SFFV-) or the myeloid specific MRP8-promoter (also known as calcium-binding protein A8; S100A8, NM_002964) or used alone to drive expression of an eGFP reporter gene (Figure 1B). As the 1.5 kb A2UCOE is known to reduce titers of lentiviral vector preparations, most likely due to formation of antisense transcripts, we tested first the influence of the CBX3-UCOE on the titer of lentiviral vector preparations. To this end we produced lentiviral vectors from the constructs shown in Figure 1B by transient transfection of 293T cells using standard protocols and titrated those on HL60 cells. Indeed the titer of CBX3-containing vectors were significantly higher when compared to their counterparts containing the full 1.5 kb A2UCOE (1.8 × 109 versus 2.7 × 108 TU ml−1 for CBX3MEW versus UrMEW, and 2.4 × 109 versus 1.9 × 108 TU ml−1 for CBX3SEW versus UrSEW, respectively; P ≤ 0.05) and within the range of the MEW and SEW lentiviral vector preparations (Figure 1C).
Figure 1.
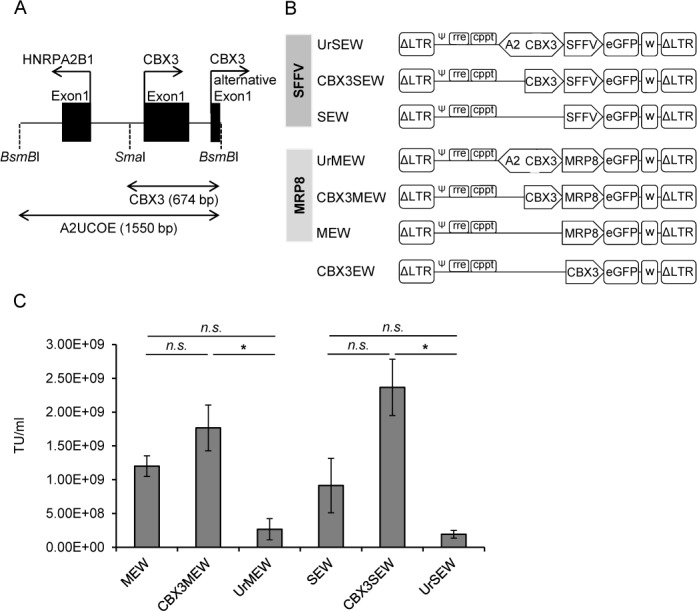
Schematic representation of lentiviral vectors used in this study. (A) Illustration of the human HNRPA2B1-CBX3 (heterogeneous nuclear ribonucleoproteins A2/B1/chromobox protein homolog 3) locus and the A2UCOE (1.55 kb) spanning the HNRPA2B1 and the CBX3 promoter. To generate the minimal 0.7 kb UCOE the HNRPA2B1 moiety was removed from A2UCOE by using a SmaI restriction site located upstream of the CBX3 promoter. The resulting fragment (CBX3) is 674 bp in size and spans the two alternative first exons of CBX3. (B) The 1.5 kb A2UCOE and the 0.7 kb CBX3-UCOE were cloned into self-inactivating (SIN) lentiviral vector backbones in combination with either the spleen focus forming virus (SFFV-) or myeloid-related protein 8 (MRP8) promoters to drive expression of an enhanced green fluorescent protein (eGFP). In CBX3EW, the 0.7 kb CBX3-UCOE was cloned directly in front of eGFP. (Abbreviations: ΔLTR, self-inactivating (SIN) long-terminal repeats; Ψ, extended encapsidation signal; cPPT, central polypurine tract; RRE, rev responsive element; SFFV, spleen focus-forming virus; UCOE, ubiquitous chromatin opening element; wPRE, woodchuck hepatitis virus post-transcriptional element). (C) Titers of lentiviral vector preparations determined 5 days after transduction of HL60 cells (mean ± SEM, n = 3; *, P < 0.05 by Student's t-test).
Stabilized transgene expression in P19 embryonic carcinoma cells by the CBX3-UCOE
To test the functionality of the CBX3-UCOE, we first transduced P19 embryonic carcinoma cells with lentiviral vector constructs containing the SFFV promoter either alone or linked to the 1.5 kb A2UCOE or the CBX3-UCOE (SEW, UrSEW, or CBX3SEW, respectively, Figure 1B) at a multiplicity of infection (MOI) of 5 to achieve comparable VCNs and a single vector integration per cell. In addition, a CBX3EW vector containing just the CBX3-UCOE driving eGFP expression was similarly tested. The percentage of eGFP expressing cells was monitored for a total of 6 weeks using flow cytometry. Transduction efficiency ranged between 30% and 55% five days after transduction at VCNs of 0.1 to 0.5 copies per cell (Supplementary Figure S1A).
As reported previously, transgene expression from the SFFV promoter was rapidly silenced in this CpG-methylation prone cell line and within 42 days eGFP expression declined to less than 20% of the initial value (Figure 2A). In contrast, when the SFFV promoter was linked to either the A2UCOE or the CBX3-UCOE the drop in eGFP expressing cells was less pronounced, resulting in stable eGFP expression in 66.2 ± 18.5% and 54.1 ± 16.8% of the cells for UrSEW and CBX3SEW, respectively, at day 42 post-transduction. No drop in the percentage of eGFP expressing cells was observed for the CBX3EW construct (Figure 2A). The levels of eGFP expression, as measured by the mean fluorescence intensity (MFI), decline for all vectors to 50–65% of the initial values. The MFI of eGFP in CBX3SEW expressing cells was close to 85% of that seen in UrSEW transduced cells at day 42 of culture (Supplementary Table S2). Given nearly constant VCNs throughout the follow-up for all four constructs (Supplementary Figure S1A), these data are consistent with substantial silencing of SFFV-driven transgene expression during culture, which is markedly reduced by both UCOEs.
Figure 2.
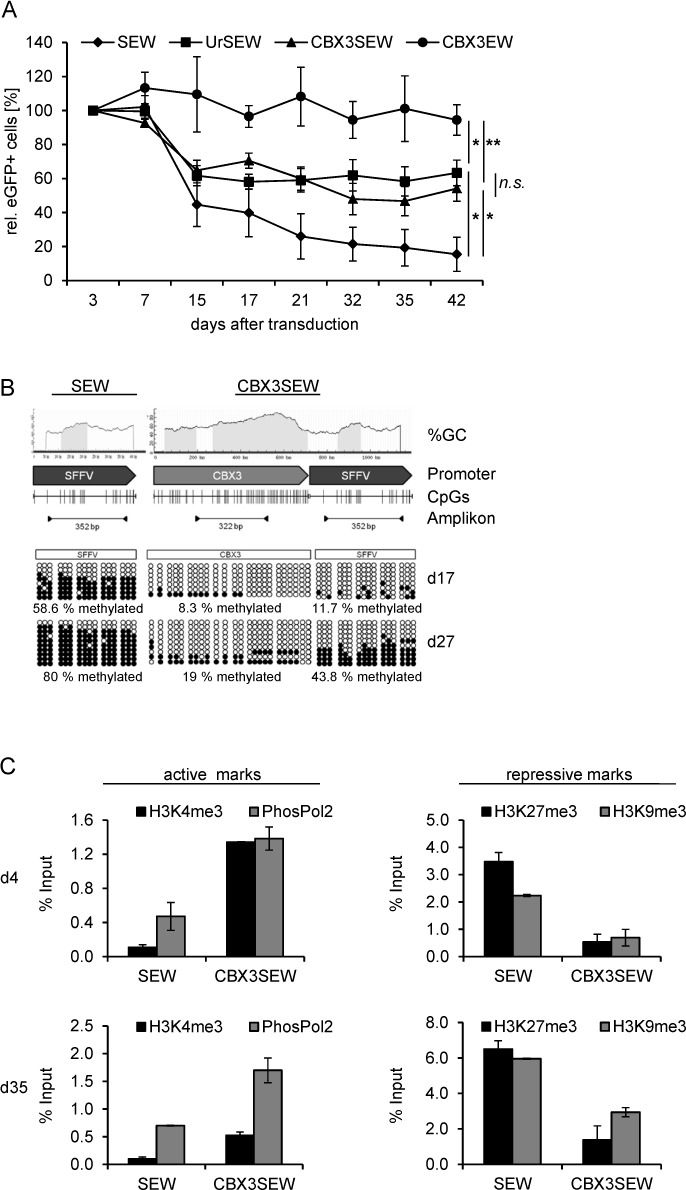
The CBX3-UCOE stabilizes transgene expression in the murine embryonic carcinoma cell line P19. (A) P19 cells were transduced with SEW, UrSEW, CBX3SEW and CBX3EW and eGFP expression was monitored over time by flow cytometry. To compensate different initial transduction efficiencies, eGFP expression was normalized to percentage of eGFP positive cells at day 3 after transduction (mean ± SEM, n = 6; *, P < 0.05, **, P < 0.01, both by Student's t-test). (B) CpG methylation at the SFFV and CBX3 promoters in SEW and CBX3SEW transduced cells was quantified at day 17 and 27 after transduction by PCR after bisulfite conversion. (C) Active (H3K4me3, phosphorylated Polymerase 2 (PhosPol2)) and repressive (H3K9me3, H3K27me3) chromatin modifications at the SFFV promoter in SEW and CBX3SEW transduced P19 cells were determined 4 and 35 days after transduction by ChIP (representative results shown, mean ± SD).
To correlate sustained transgene expression by the CBX3-UCOE with DNA-methylation, we analyzed CpG-methylation within the promoter regions of CBX3 and SFFV by bisulfite sequencing on samples taken at days 17 and 27 after transduction. As expected, in SEW transduced cells the SFFV promoter was hypermethylated already at day 17 (59% CpG methylation), and almost completely methylated 10 days later (80% CpG methylation; Figure 2B). In contrast, the degree of DNA methylation was significantly reduced to 11.7% (P = 0.008) at day 17 when the SFFV promoter was linked to the CBX3-UCOE, corresponding to an 80% reduction in CpG methylation when compared to the SFFV promoter alone. This extensive protection from CpG methylation is equivalent to that seen in a similar construct but containing the 1.5 kb A2UCOE instead of CBX3 (85% reduction in CpG methylation, (37)). At day 27 still only 44% of the CpGs were methylated, representing a significant improvement compared to the SEW construct (P = 0.012). Of note, the CBX3 region remained almost completely hypomethylated throughout the entire observation period.
In order to gain further insights into the epigenetic mechanisms underlying the anti-silencing effect of the CBX3-UCOE, we analyzed histone modifications at the SFFV promoter by ChIP experiments in SEW and CBX3SEW transduced cells at days 4 and 35 after transduction (Figure 2C, Supplementary Figure S1B). Already at day 4 the histone modification H3K4me3 as well as phosphorylated RNA polymerase 2 (PhosPol2), both associated with active transcription, were nearly absent along the SFFV-promoter in SEW transduced cells. In contrast, when combined with the CBX3 element the SFFV-promoter was highly enriched in these active chromatin marks which were partially maintained after prolonged cell culture (day 35). Consistent with these findings, substantially lower levels of repressive chromatin marks (H3K9me3 and H3K27me3) were detected along the SFFV-promoter when fused to the CBX3-UCOE at both time points. Importantly, the CBX3 moiety, in accordance with its hypomethylated status, was characterized by an open chromatin configuration comparable to that present at the control Gapdh promoter (Supplementary Figure S1B).
The CBX3-UCOE stabilizes transgene expression during the hematopoietic differentiation of murine ES cells
Given that the differentiation of PSCs into specialized cell types involves massive epigenetic changes and is frequently associated with silencing of integrated lentiviral vectors, we evaluated the CBX3-UCOE also in this model. We have recently demonstrated that the 1.5 kb A2UCOE protects the SFFV promoter from epigenetic silencing in murine ES cells and their differentiated progeny (24). Here, we transduced murine ES cells (CCE) with SEW, CBX3SEW, UrSEW and CBX3EW at an MOI of 50, resulting in 78.6% to 95.6% eGFP positive cells 3 days after transduction (Figure 3A). When these ES cells were monitored in culture for 35 days by flow cytometry we observed a slight reduction in the percentage of eGFP positive cells for SEW and UrSEW (75% of initial values) but constant values for CBX3EW and CBX3SEW, while the same moderate reduction in gene expression levels (MFI) was observed for all constructs (Figure 3A–C). In contrast, clear differences were observed upon hematopoietic differentiation of transduced cells. Employing a previously established EB-based differentiation protocol (32) which yields about 60% CD41+ early hematopoietic stem/progenitor cells after 8 days of differentiation (Supplementary Figure S2A), rapid and complete silencing of SFFV-mediated eGFP expression was observed, whereas both the CBX3-UCOE and A2UCOE were able to effectively stabilize eGFP expression from the SFFV promoter to a similar extend (6.1% versus 84.6 ± 8.7% and 68.8 ± 3.8% for SEW, CBX3SEW and UrSEW eGFP+ cells at day 8 of differentiation, respectively; Figure 3D and E, and Supplementary Figure S2B). As expected, the level of transgene expression (MFI of eGFP+ cells) after hematopoietic differentiation increases to a similar extent, most likely as a consequence of the activation of the SFFV promoter in differentiated cells (Supplementary Figure S2C). Also the CBX3 element alone (CBX3EW) was able to sustain 53.3 ± 13.9% of transgene expression after hematopoietic differentiation. Analysis of VCN revealed higher numbers of lentiviral integrations in SEW transduced cells (5.4 VCN/cell) when compared to CBX3SEW (0.98 VCN/cell), UrSEW (1.49 VCN/cell) and CBX3EW (3.63 VCN/cell) transduced cells.
Figure 3.
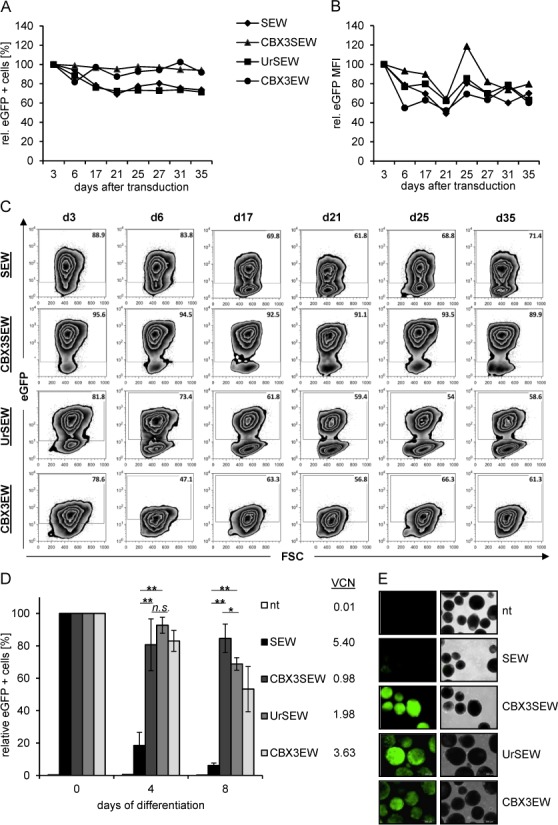
The CBX3-UCOE mediates stable transgene expression during hematopoietic differentiation of murine ES-cells. (A and B) Murine ESCs were transduced with SEW, CBX3SEW, UrSEW and CBX3EW. The percentage of eGFP positive cells (A) and the mean fluorescence intensity of eGFP expression in GFP+ cells (B) was analyzed over 35 days by flow cytometry in SSEA1 (stage specific embryonic antigen 1) positive cells. (C) Representative FACS analysis of eGFP expression in transduced mES-cells shown in A and B. (D) Transduced mESC were differentiated towards hematopoietic stem/progenitor cells following an EB-based protocol and transgene expression was analyzed by flow cytometry at day 4 and 8 of differentiation (mean ± SD, n = 4; *, P <0.05; **, P < 0.01 by Student's t-test). VCNs for the individual cell populations are given at the right. (E) Visualization of eGFP expression in murine ES cells after differentiation by fluorescence microscopy.
Again we analyzed the SFFV promoter for methylated CpG motifs. Despite several integrations of the lentiviral vector cassette, 41% methylated CpGs were detected in SEW transduced cells in the pluripotent state, which increased to 80% at day 8 after hematopoietic differentiation. In contrast, in CBX3SEW transduced cells only 1.3% methylated CpGs were observed in the pluripotent status and 11% after hematopoietic differentiation (Supplementary Figure S2D). Thus, the CBX3-UCOE effectively protects heterologous promoters from silencing in murine ES cells before and after differentiation.
The CBX3-UCOE confers vector copy-dependent expression and prevents transgene silencing without disturbing the physiological regulation of the myeloid specific MRP8 promoter
Since cell type-specific promoters hold great potential for gene therapeutic applications as they reduce both, genotoxicity and phenotoxicity, we asked if the CBX3-UCOE would preserve the specificity of tissue-restricted promoters. For this we combined the CBX3 element with the myeloid biased MRP8 promoter to generate the lentiviral vector CBX3MEW (Figure 1B). First, this vector was tested in the P19 cell line, as we have previously shown that the MRP8 promoter is inactive in this cell line (27). In agreement with this, we did not observe eGFP expression from the MRP8 promoter in P19 cells (Figure 4A). To our surprise, however, significant levels of eGFP expression were detectable when the MRP8 promoter was linked to the minimal CBX3 element. As previous experiments with the full length 1.5 kb A2UCOE had revealed aberrant gene expression as a consequence of transcripts initiated at the CBX3 promoter region and spliced into cellular exons or a cryptic acceptor site in the 3' end of the MRP8 promoter (Figure 4B and C and (28)), we mutated the canonical donor splice site and a cryptic splice acceptor site present within the CBX3-UCOE to generate the construct CBX3*MEW (Supplementary Figure S3). No eGFP expression was observed from this construct in P19 cells after 20–25 days, arguing for maintenance of cell type specificity by MRP8 even when linked to CBX3*. Lack of transgene expression in P19 cells from the MEW construct correlated with the absence of active (H3K4me3 and PhosPol2) and presence of repressive histone marks (H3K27me3 and H3K9me3) at the MRP8 promoter (Supplementary Figure S4A). When linked to the CBX3-UCOE, PhosPol2 was slightly enriched at the MRP8 promoter, whereas the repressive histone mark H3K27me3 was markedly reduced, consistent with our previous observations at the SFFV promoter in CBX3SEW transduced P19 cells (Figure 2C). In strong contrast, the CBX3 promoter in CBX3MEW transduced cells was enriched for H3K4me3 and PhosPol2, suggesting a transcriptional active state (Supplementary Figure S4B).
Figure 4.
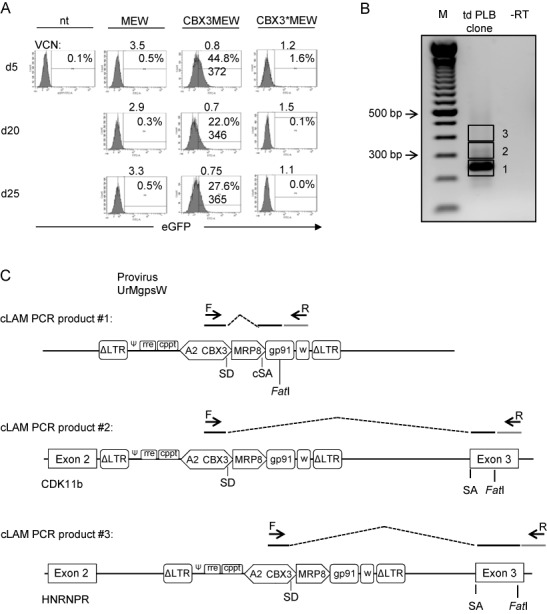
eGFP expression in P19 cells transduced with MRP8 containing vectors is caused by transcripts initiated at CBX3 and not by aberrant activation of the MRP8 promoter. (A) P19 cells were transduced with MEW, CBX3MEW and CBX3*MEW. eGFP expression was monitored over time by flow cytometry and VCNs were analyzed by quantitative PCR. (B) cDNA based linear amplification-mediated (cLAM)-PCR analysis of transcripts arising from the CBX3 promoter in UrMgpsW transduced PLB985 cells. Three major regions in the agarose gel were excised and DNA fragments analyzed. (C) Schematic representation of cLAM-PCR products as analyzed by sequencing. CBX3 transcripts fused to cellular exons were mapped according to the vector integration sites (see text for details).
To confirm the myeloid expression specificity of the MRP8 promoter when juxtaposed to the CBX3-UCOE, we transduced the myelomonocytic cell line PLB985 and the T cell line Jurkat with MEW, CBX3MEW, CBX3EW and CBX3*MEW. After transduction with the CBX3-containing vectors 34–36% of the PLB985 cells expressed eGFP, whereas eGFP expression was hardly detectable in Jurkat cells (Figure 5A). Almost every single vector copy in CBX3*MEW and CBX3MEW transduced PLB985 cells expressed the transgene, while the percentage of silent proviruses in MEW transduced PLB985 cells was 10-fold higher (Figure 5A). Moreover, linking CBX3* to the MRP8 promoter resulted in a 2.6-fold increase in expression levels in PLB985 cells but not in Jurkat cells as determined by the median fluorescent intensity of eGFP expressing cells (Figure 5B). The myeloid specificity of MRP8 in CBX3*MEW was maintained as the level of eGFP expression was 4.3-fold higher in PLB985 cells than in Jurkat cells. This was not due to a lack of promoter activity of the CBX3 element, as eGFP expression was clearly seen in PLB985 and Jurkat cells transduced with CBX3EW (Figure 5A and B).
Figure 5.
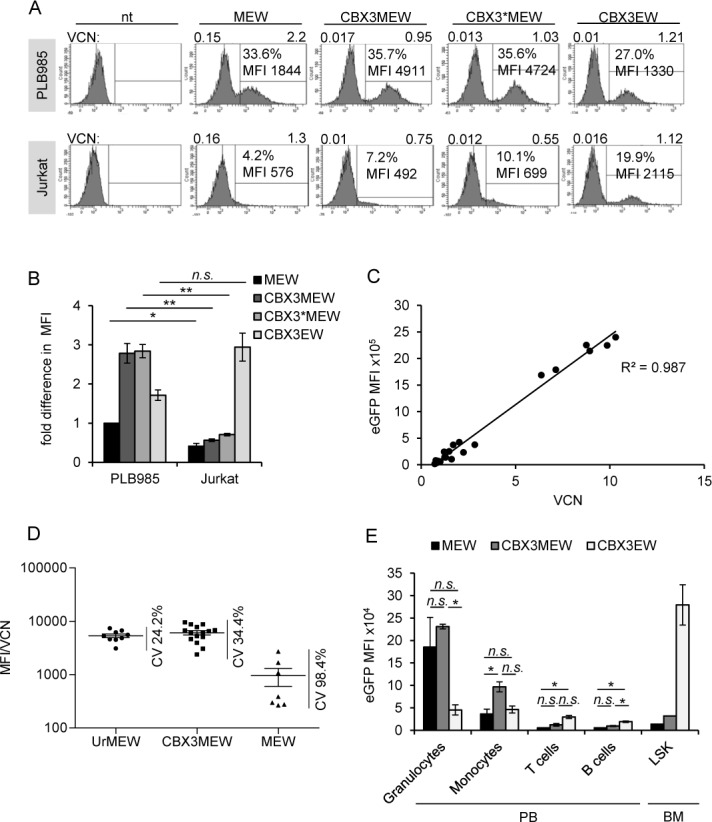
The CBX3MEW vector mediates high transgene expression levels in myeloid cell lines and in vivo. (A) Myeloid (PLB985) and T (Jurkat) cells were transduced with MEW, CBX3MEW, CBX3*MEW and CBX3EW and cultured for 5 days. Median fluorescent intensity (MFI) as well as VCN was determined in eGFP+ and eGFP− sorted cells. (B) Comparison of the relative MFI of MEW CBX3MEW, CBX3*MEW and CBX3EW vectors in PBL 985 and Jurkat cells normalized to the level measured in MEW transduced PLB985 cells (n = 3, mean ± SD). (C) Correlation between VCN and MFI in single cell clones obtained from CBX3MEW transduced PLB985 cells. (D) Variation in eGFP expression in PLB985 cell clones containing 1 to 3 copies of UrMEW (n = 9), CBX3MEW (n = 16) or MEW (n = 7). The coefficient of variation (CV) in the MFI of the different clones with respect to the mean MFI is shown after correction for VCNs. (E) Murine lineage negative cells derived from SJL mice (CD45.1) were transduced with MEW, CBX3MEW and CBX3EW and transplanted into lethally irradiated C57BL/6 mice (CD45.2). EGFP expression in donor-derived cells (CD45.1+) was analyzed by FACS in peripheral blood and bone marrow subpopulations 24 weeks after transplantation (n = 3, except for LSK n = 2, mean ± SEM). *, P <0.05; **, P < 0.01 by Student's t-test.
We generated single cell clones from CBX3MEW-transduced PLB985 cells and correlated the VCN to the MFI of eGFP expression (Figure 5C). As expected for a non-silenced construct, a direct correlation between expression levels and VCNs was observed (R2 = 0.99). The expression levels of eGFP in A2UCOE and CBX3-containing clones with similar VCNs was homogeneous as reveled by the coefficient of variation in eGFP expression among the different clones (24.2% and 34.5% for UrMEW and CBX3MEW, respectively; Figure 5D). In contrast, the coefficient of variation in eGFP expression for MEW clones was almost 100% reflecting a strong PEV.
The myeloid specificity of the CBX3MEW vector was also confirmed in vivo. For this purpose lineage negative bone marrow cells (CD45.1+) were transduced with MEW, CBX3MEW and CBX3EW vectors and transplanted into lethally irradiated recipient mice (CD45.2+). Transgene expression was assessed by flow cytometry in peripheral blood cells and bone marrow cells 24 weeks later. As depicted in Figure 5E mice transplanted with MEW- or CBX3MEW-transduced cells exhibited high levels of transgene expression in myeloid cells and particularly in granulocytes, whereas expression in T and B cells was low. In contrast, relatively uniform eGFP expression levels in all mature subpopulations analyzed was observed for the CBX3 promoter when used alone. EGFP expression in murine progenitor (LSK) cells was also low when the CBX3 element was juxtaposed to the MRP8 promoter in contrast to the CBX3 alone, an observation with significance for the assessment of the safety of this element in clinical applications (Figure 5E).
The CBX3 element supports myeloid-specific expression of the MRP8 promoter during hematopoietic differentiation of human iPS cells
Additionally, we analyzed the expression pattern of CBX3*MRP8 during myeloid differentiation of human (h)iPSC together with the associated epigenetics marks at the MRP8 promoter. To this end human iPSCs derived from mobilized peripheral blood CD34+ cells were transduced with the MEW, CBX3*MEW and UrMEW vectors at a MOI of 10 and cultured for several weeks before inducing differentiation. While eGFP expression was low to undetectable in cells transduced with MEW or CBX3*MEW (1.0 ± 0.4% and 5.9 ± 2.6% eGFP+ cells, respectively), high levels of eGFP expression were observed in UrMEW transduced iPSC (28.5 ± 5.7% eGFP+ cells, n = 3–4; Figure 6A and B, Supplementary Figure S5A). For myeloid differentiation of hiPSC we employed a previously established EB-based differentiation protocol, which allows for the continuous generation of myeloid cells for several months (33). After differentiation into CD45+/CD11b+ myeloid cells (Supplementary Figure S5B) eGFP expression was markedly upregulated in MEW, CBX3*MEW and UrMEW transduced cells (Figure 6A–C). However, the fraction of eGFP expressing cells was significantly higher when the CBX3-UCOE or the 1.5 kb A2UCOE were included in front of the MRP8 promoter (MEW 27.8 ± 3.4%, CBX*MEW 54.0 ± 7.3%, UrMEW 56.2 ± 5.8% eGFP+ cells, n = 3–4; Figure 6B). These results were obtained despite a 2.5-fold higher VCN in the MEW-transduced control group (MEW: 2.5 VCN/cell, CBX3*MEW: 0.96 VCN/cell, UrMEW: 0.88 VCN/cell). Importantly, also here the effect of the CBX3-UCOE was similar to that of the full length 1.5 kb A2UCOE. To confirm the myeloid-restricted expression pattern of the CBX3*MEW vector we also analyzed CD45 negative, non-hematopoietic cells generated during the differentiation process for eGFP expression. Again, only minimal transgenic eGFP expression was observed in MEW and CBX3*MEW transduced cells (1.8 ± 0.6% and 2.7 ± 0.48% eGFP+ cells, respectively). In contrast, considerable eGFP expression was observed when the 1.5 kb A2UCOE was used (28.5 ± 5.7% eGFP+ cells) most likely as a consequence of read through transcripts and aberrant splicing (Figure 6A and B, and (28)). No major differences in transgene expression levels (MFI) in myeloid cells were observed for the three vector constructs, though there was a tendency towards higher expression from the CBX3*MEW construct (Figure 6C).
Figure 6.
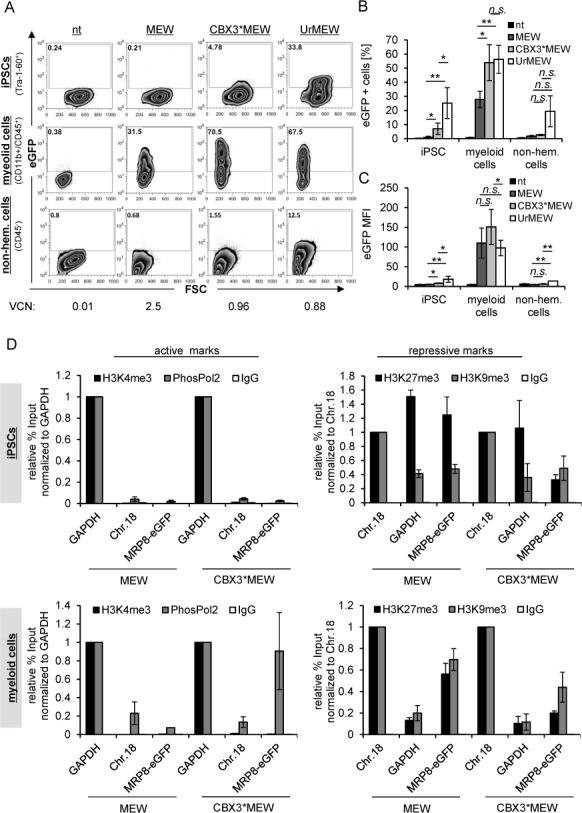
The CBX3-UCOE stabilizes transgene expression while maintaining tissue-specificity of the MRP8 promoter during myeloid differentiation of hiPSC. (A) Human iPSC were transduced with MEW, CBX3*MEW and UrMEW and differentiated towards myeloid cells following an EB-based protocol. EGFP expression was analyzed by flow cytometry in pluripotent iPSC, iPSC-derived myeloid cells (CD45+/CD11b+) and non-hematopoietic (CD45−) cells. A representative experiment is shown. VCNs were determined in (h)iPSCs before differentiation. The percentage of eGFP positive cells (mean ± SD) obtained from 3–4 independent experiments is given in (B). (C) Analysis of of eGFP expression levels (MFI) in pluripotent iPSC, iPSC-derived myeloid cells (CD45+/CD11b+) and non-hematopoietic (CD45−) cells (n = 3–4, mean ± SD). (D) Chromatin structure at the MRP8 promoter in MEW and CBX3*MEW transduced cells was investigated in pluripotent iPSC and differentiated myeloid cells by ChIP (mean ± SD). Active and repressive histone marks together with phosphorylated Polymerase 2 (PhosPol2) were quantified in transduced cells. The actively transcribed GAPDH promoter and a repressed locus on chromosome 18 (Chr.18) were used as controls for the ChIP experiments. Values are given as percentage of input and normalized to the percentage of input for GAPDH (active chromatin marks) or Chr.18 (repressive chromatin marks). *, P <0.05; **, P < 0.01 by Student's t-test.
To decipher the regulatory mechanisms underlying the myeloid restricted expression pattern of the MRP8 promoter when fused to the CBX3-UCOE, we analyzed the epigenetic status of the MRP8 promoter with or without CBX3* in hiPSCs before and after myeloid differentiation by ChIP experiments. No enrichment of the active histone marks H3K4me3 and PhosPol2 was detected at the MRP8 promoter in the pluripotent state, irrespectively if the CBX3 moiety was present or not (Figure 6D), reflecting the epigenetic status of the endogenous MRP8 locus in hiPSCs (Supplementary Figure S5C). While H3K4me3 remained absent from the MRP8 promoter upon granulocytic differentiation, PhosPol2 occupancy at the MRP8 promoter increased during differentiation, particularly when MRP8 was linked to CBX3*, reaching levels similar to those observed at the actively transcribed GAPDH promoter (Figure 6D).
The repressive histone mark H3K27me3 was moderately enriched at all loci analyzed in (h)iPSCs, reflecting the bivalent chromatin structure of pluripotent cells (Figure 6D and (38)). However, H3K27me3 marks at the MRP8 promoter were clearly reduced in CBX3*MEW transduced cells as compared to MEW and to the endogenous promoter in (h)iPSC (Figure 6D and Supplementary Figure S5C). Following myeloid differentiation the H3K27me3 mark at the endogenous MRP8 locus and within the MEW vector sequence decreased to similar levels (0.7 relative to the repressed control locus on chromosome 18). In the presence of CBX3, H3K27me3 levels at the MRP8 promoter were further reduced and were similar to that observed at the GAPDH promoter which is constitutively active in these cells.
The level of another histone repressive mark, H3K9me3, at the MRP8 promoter in transduced pluripotent cells remained unchanged, irrespectively of the presence of CBX3 and was similar to the H3K9me3 levels at the endogenous MRP8 promoter (Figure 6D and Supplementary Figure S5C). In comparison to GAPDH, the H3K9me3 levels at the MRP8 promoter did not decrease after myeloid differentiation in MEW transduced cells, but were strongly reduced when the MRP8 promoter was linked to CBX3.
In aggregate, our data suggest that the CBX3 element protects the MRP8 promoter from repressive epigenetic marks in stem cells and their differentiated progeny thus providing a permissive chromatin environment for transcription. The MRP8 promoter becomes transcriptionally active once the appropriate myeloid-specific transcription factors are expressed.
DISCUSSION
We and others have effectively utilized the A2UCOE to overcome epigenetic silencing and stabilize transgene expression in genetically manipulated hematopoietic as well as PSCs (reviewed in (21)). Although the utility of the A2UCOE as a protective element against silencing is well documented, side-effects associated with the use of this element have only recently been addressed. In particular the existence of aberrant splice products arising from transcripts initiated at the dual divergent promoters within the A2UCOE was recently recognized (28). In the context of gene therapy applications, aberrant splice products should be avoided as aberrant splicing was linked to clonal outgrowth in a gene therapy trial for β-thalassemia (39). Consequently, splicing-defective versions of the A2UCOE with an improved genotoxicity profile and maintenance of regulatory activity have been generated (28). Also shorter versions of the A2UCOE were generated aiming for a reduction in DNA fragment size, as the originally described 2.2 kb A2UCOE was rather large, limiting the size of the transgene cassette that could be included in the A2UCOE-containing retro- and lentiviral vectors (23,40). A 1.5 kb A2UCOE, which still includes the HNRPA2B1-CBX3 divergent promoter has been shown to retain all properties of the original 2.2 kb fragment including protection against silencing and VCN dependent transgene expression in hematopoietic and pluripotent stem cells including their differentiated progeny (25,37). Moreover, this fragment when linked to tissue restricted promoters provided protection against CpG methylation and copy-dependent expression without interfering with promoter specificity (27,41). Interestingly, protection against silencing was best when the CBX3 promoter of the A2UCOE was juxtaposed to a heterologous promoter, suggesting functional heterogeneity within the 1.5 kb A2UCOE (27,37). Indeed during our studies, we observed profound CpG methylation at the HNRPA2B1 promoter in P19 cells when the A2UCOE was placed upstream of a myeloid-specific promoter in a SIN-LV construct, whereas the CBX3 region of the A2UCOE remained mostly unmethylated (27). Therefore, we hypothesized that the HNRPA2B1 promoter might be dispensable for the anti-silencing effect of the element, and we now demonstrate that a 0.7 kb minimal UCOE devoid of the HNRPA2B1 promoter part of the element can also stabilize SIN-LV-driven transgene expression in murine P19 embryonic carcinoma cells as well as murine ESCs and their hematopoietic progeny. This minimal 0.7 kb CBX3-UCOE provided protection against CpG-methylation dependent silencing to the SFFV promoter in murine ES, human iPSCs and their hematopoietic differentiated progeny and sustained gene expression from the SFFV and MRP8 promoters over time in a vector copy-dependent manner in vitro and in vivo. In addition, the 0.7 kb CBX3-UCOE did not only protect heterologous promoters from silencing, but retained full promoter activity in pluripotent cells when linked directly to eGFP. In contrast to the CBX3-UCOE described here, other A2UCOE-derived DNA-fragments have failed to provide extensive protection against methylation to heterologous promoters. For example, Uchiyama et al. described a 631 bp A2UCOE-derived fragment containing the HNRPA2B1 promoter and 5'-flanking region but devoid completely of CBX3 sequences (42). While the authors claim sustained gene expression of eGFP and of the Wiskott-Aldrich syndrome protein gene (WAS) in hematopoietic cells in vitro and in vivo, no detailed epigenetic studies were presented. Similar HNRPA2B1-only promoter fragments lacking CBX3 have been shown previously by others to be transcriptionally unstable or to lack methylation-protective functions in hematopoietic cells (28,40). Another 0.7 kb UCOE fragment derived from a region within the first intron of CBX3 but devoid of promoter activity and thus different from the CBX3-UCOE described here was shown to partially sustain gene expression from the SFFV promoter in vitro (43). In contrast to the CBX3-UCOE, the 0.7 kb intronic UCOE fragment described by Bandaranayake et al. did not sustained the initial gene expression levels despite high VCNs and a reduction in MFI to less than 50% of the initial values was observed. Although the configuration of the different UCOE-containing constructs mentioned above differs from the one tested in our study and thus a direct comparison between the constructs in terms of performance is difficult, the novel CBX3-UCOE presented here is the only A2UCOE subfragment described to date that retains most if not all of the properties ascribed to the full length 1.5 kb A2UCOE in vitro and in vivo.
The anti-silencing function of A2UCOE relies on a central 2.6 kb CpG island around the divergently transcribed HNRPA2B1 and CBX3 promoters generating a 5.0 kb genomic region of unmethylated DNA (26). This region extend at least 1.5 kb and 3.0 kb downstream of the CBX3 and HNRPA2B1 promoters, respectively, and thus it could be assumed that any DNA sequence placed within this distance to the A2UCOE promoters would be protected from transcriptional silencing. Indeed A2UCOE has been shown to confer protection from CpG-methylation to promoter sequences incorporated into the vicinity of the element. This property has been demonstrated in the context of SIN-LV as well as SIN γ-retroviral backbones and has been shown for viral as well as housekeeping promoters such as the SFFV, or CMV but also for the PGK, and EFS1a promoters (reviewed in (21)). In our work we have extended these studies and show that a subfragment of A2UCOE, CBX3-UCOE, retained most of the anti-silencing properties of A2UCOE. At the endogenous HNRPA2B1-CBX3 locus the lack of CpG methylation correlates with the presence of histones H3 and H4 acetylation as epigenetic marks for active chromatin regions. Likewise H3K4me3, another marker of active chromatin, is enriched at the CBX3 promoter region but absent at the HNRPA2B1 promoter (26). Remarkably, active chromatin marks were imposed by the CBX3-UCOE at the SFFV and MRP8 promoters in cells in which both native promoters were heavy methylated or not expressed. The SFFV promoter is well known to be rapidly silenced in stem cells and enriched in epigenetic marks correlated with closed chromatin. When combined with the CBX3-UCOE we observed a profound enrichment of the active chromatin mark H3K4me3 in combination with decreased levels of the repressive marks H3K9me3 and H3K27me3 along the SFFV promoter. The generation of an open chromatin environment by the CBX3-UCOE was also associated with increased levels of PhosPol2, correlating with the robust transgene expression observed in CBX3SEW transduced stem cells up to 35 days after transduction. More impressive is the chromatin remodeling at the MRP8 promoter in PSCs, as in the absence of CBX3-UCOE the MRP8 promoter is devoid of active chromatin marks but enriched in repressive marks, thus resembling the chromatin status of the endogenous promoter. In the presence of CBX3-UCOE, the MPRP8 promoter is still devoid of active chromatin marks in PSCs, but the levels of repressive marks, like H3K27me3, are markedly reduced. Despite a transcriptionally permissive chromatin environment, the MRP8 promoter remained transcriptionally inactive in stem cells, suggesting that CBX3-UCOE prevents heterochromatin spreading towards the expression cassette resulting in a reduced level of H3K9 and H3K27 trimethylation. Thus, the MRP8 promoter remains accessible to myeloid specific transcription factors once they become expressed, resulting in stable and vector copy number dependent transgene expression. Interestingly, CpG methylation at the MRP8 promoter was not prevented by the CBX3-UCOE or the A2UCOE (data not shown and (27)). The MRP8 promoter contains only a few CpG which are persistently methylated even in myeloid cells. The UCOEs do not override this epigenetic mark, which is essential for appropriate gene regulation, as the transcription factor C/EBPα, a master of myeloid differentiation, binds to methylated CpGs (44).
The remarkable properties of A2UCOE are most likely determined by chromatin remodeling factors, like the CCCTC-binding factor (CTCF). Indeed two CTCF binding sites have been experimentally determined in A2UCOE (http://www.biobase-international.com/product/transcription-factor-binding-sites). Both of them are retained in the CBX3-UCOE. CTCF organizes the 3D structure of chromatin by the formation of loop domains, defining thereby boundaries between heterochromatin und euchromatin (45). Moreover, CTCF actively target genes to transcriptional factories, defined as clusters within the nucleus were all components of the transcriptional machinery are highly concentrated (46). This might also explain the high enrichment for H3K4me3 and PhosPol2 we observed at the CBX3-UCOE in all analyzed cell types. Additionally a CFP1 (CXXC finger protein 1) binding site in mouse brain was detected by ChIP-seq in CBX3 (data accessible at NCBI GEO database, accession GSM462191). As this TF binds to unmethylated CpGs and induces the activating trimethylation of histone 3 at lysine 4 (H3K4me3) by recruiting the methyltransferase Setd1 (47) it might support the anti-silencing effect of the UCOE. Lastly, a high frequency of replication initiation events has been identified in the intergenic region between HNRPA2B1 and CBX3, which is partially conserved in the CBX3-UCOE (48). These ORIs have been described to confer an accessible chromatin environment that facilitates early transcription and are associated with histone acetylation patterns characteristics of euchromatin (16). Thus, most likely a combination of several factors regulates the function of the UCOE.
The independent, dominant chromatin remodeling function of UCOEs has been attributed to a large methylation-free CpG island in combination with the dual divergent promoter activity of the element (21). Our data now seem to implicate primarily the methylation-free CpG-island in UCO functionality, while the dual promoter structure seems to be dispensable. Nonetheless, we believe that a certain level of transcription is required for the CBX3-UCOE function, as the CBX3-derived 0.7 UCOE described by Bandaranayake et al., which lacks the CBX3 promoter, provides only partial protection against silencing when compared to the full active 1.5 kb A2UCOE (43). This notion is further supported by the fact that within the 1.5 kb A2UCOE CpG density is highest within the intragenic region between the two alternative first exons of the CBX3 gene, a region included in the CBX3-UCOE. Interestingly this area is missing in the 1.0 kb CBX3-NSD variant described by Knight et al., which failed to protect against transgene silencing in P19 cells (28). The CpG density in CBX3-NSD is 0.086, while that of CBX3-UCOE is 0.095 and increases to 0.154 when the intragenic region between the two alternative first exons of CBX3 is included, emphasizing the essential role of this region in anti-silencing activity.
In summary we here not only introduce a functional, 0.7 kb minimal CBX3-UCOE which can be combined with constitutive as well as tissue-specific promoters to counteract epigenetic silencing of transgene expression in multipotent and pluripotent stem cells, but also for the first time describe the epigenetic changes imposed by UCO-elements at juxtaposed heterologous promoters. Besides preventing promoter silencing, we here demonstrate the spread of the chromatin opening function of this element to adjacent heterologous DNA regions. This latest feature of the CBX3-UCOE could be of concern, as integrated CBX3-UCOE containing vectors could alter the chromatin status and therefore the expression pattern of cellular genes located nearby the integration site. However, the chromatin opening function of the endogenous CBX3 gene appears to be centered at the promoter region and spans a DNA region of approximately 1.5 kb, as defined by the transition from unmethylated to methylated DNA and the presence of active histone marks (26). Within lentiviral vectors, the distance of the CBX3-UCOE to the left border of the 5'-LTR is 1.7 kb. Therefore, at least in theory, the minimal distance of cellular sequences to the CBX3-UCOE should be beyond the distance covered by the chromatin opening effect of the element. Our data favor the use of CBX3-UCOE for future applications in cell and gene therapy. In contrast to vectors containing the 1.5 kb A2UCOE, lentiviral vectors containing CBX3-UCOE can be produced at high titer, emphasizing the potential of this fragment for its prospective incorporation into clinical vectors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Linping Chen-Wichmann, Joachim Schwäble and Tefik Merovci (GSH, Frankfurt, Germany), Manfred Schmidt and Simone Scholz (NCT, Heidelberg, Germany) and Doreen Lüttge (Hannover Medical School, Hannover, Germany) for technical assistance during this study.
Footnotes
These authors contributed equally to the paper as first authors.
Present address: Christian Brendel, Division of Hematology/Oncology, Boston Children's Hospital, Boston, MA, USA.
FUNDING
European Union [FP7 integrated projects CELL-PID (HEALTH-2010–261387) and NET4CGD (HEALTH.2012.2.4.4-1) to M.G.]; Research Priority Program 1230 from the Deutsche Forschungsgemeinschaft (DFG; to M.G. and T.M.); LOEWE Center for Cell and Gene Therapy Frankfurt funded by the Hessische Ministerium für Wissenschaft und Kunst [HMWK; III L 4-518/17.004 (2010) to M.G.]. Further DFG funding included the REBIRTH cluster-of-excellence (Exc. 62/1) and individual grants MO886/4-1 and MO886/6-1 (all T.M.). M.A. was supported by a fellowship from the Hannover Biomedical Research School (HBRS; MHH-Hannover). The Georg-Speyer-Haus is supported by the Bundesministerium für Gesundheit and the Hessisches Ministerium für Wissenschaft und Kunst. Funding for open access charge: European Union [FP7 integrated projects CELL-PID (HEALTH-2010-261387) and NET4CGD (HEALTH.2012.2.4.4-1) to M.G.]; Research Priority Program 1230 from the Deutsche Forschungsgemeinschaft (DFG; to M.G. and T.M.); LOEWE Center for Cell and Gene Therapy Frankfurt funded by the Hessische Ministerium für Wissenschaft und Kunst [HMWK; III L 4-518/17.004 (2010) to M.G.]. Further DFG funding included the REBIRTH cluster-of-excellence (Exc. 62/1) and individual grants MO886/4-1 and MO886/6-1 (all T.M.). M.A. was supported by a fellowship from the Hannover Biomedical Research School (HBRS; MHH-Hannover). The Georg-Speyer-Haus is supported by the Bundesministerium für Gesundheit and the Hessisches Ministerium für Wissenschaft und Kunst.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 3.Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 4.Schambach A., Zychlinski D., Ehrnstroem B., Baum C. Biosafety features of lentiviral vectors. Hum. Gene Ther. 2013;24:132–142. doi: 10.1089/hum.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F., et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J., Yang Q., Chang L.J. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J. Virol. 2005;79:13497–13508. doi: 10.1128/JVI.79.21.13497-13508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotta A., Ellis J. Retroviral vector silencing during iPS cell induction: an epigenetic beacon that signals distinct pluripotent states. J. Cell Biochem. 2008;105:940–948. doi: 10.1002/jcb.21912. [DOI] [PubMed] [Google Scholar]
- 8.Minoguchi S., Iba H. Instability of retroviral DNA methylation in embryonic stem cells. Stem Cells. 2008;26:1166–1173. doi: 10.1634/stemcells.2007-1106. [DOI] [PubMed] [Google Scholar]
- 9.Herbst F., Ball C.R., Tuorto F., Nowrouzi A., Wang W., Zavidij O., Dieter S.M., Fessler S., van der Hoeven F., Kloz U., et al. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol. Ther. 2012;20:1014–1021. doi: 10.1038/mt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klug C.A., Cheshier S., Weissman I.L. Inactivation of a GFP retrovirus occurs at multiple levels in long-term repopulating stem cells and their differentiated progeny. Blood. 2000;96:894–901. [PubMed] [Google Scholar]
- 11.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 12.Mok H.P., Javed S., Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Ther. 2007;14:741–751. doi: 10.1038/sj.gt.3302923. [DOI] [PubMed] [Google Scholar]
- 13.Hejnar J., Hajkova P., Plachy J., Elleder D., Stepanets V., Svoboda J. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. U.S.A. 2001;98:565–569. doi: 10.1073/pnas.98.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park F., Kay M.A. Modified HIV-1 based lentiviral vectors have an effect on viral transduction efficiency and gene expression in vitro and in vivo. Mol. Ther. 2001;4:164–173. doi: 10.1006/mthe.2001.0450. [DOI] [PubMed] [Google Scholar]
- 15.Ramezani A., Hawley T.S., Hawley R.G. Performance- and safety-enhanced lentiviral vectors containing the human interferon-beta scaffold attachment region and the chicken beta-globin insulator. Blood. 2003;101:4717–4724. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- 16.Fu H., Wang L., Lin C.M., Singhania S., Bouhassira E.E., Aladjem M.I. Preventing gene silencing with human replicators. Nat. Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 17.Emery D.W., Yannaki E., Tubb J., Stamatoyannopoulos G. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery D.W. The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum. Gene Ther. 2011;22:761–774. doi: 10.1089/hum.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen T.T., Jakobsson J., Rosenqvist N., Lundberg C. Incorporating double copies of a chromatin insulator into lentiviral vectors results in less viral integrants. BMC Biotechnol. 2009;9:13. doi: 10.1186/1472-6750-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbinati F., Arumugam P., Higashimoto T., Perumbeti A., Mitts K., Xia P., Malik P. Mechanism of reduction in titers from lentivirus vectors carrying large inserts in the 3′LTR. Mol. Ther. 2009;17:1527–1536. doi: 10.1038/mt.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniou M.N., Skipper K.A., Anakok O. Optimizing retroviral gene expression for effective therapies. Hum. Gene Ther. 2013;24:363–374. doi: 10.1089/hum.2013.062. [DOI] [PubMed] [Google Scholar]
- 22.Williams S., Mustoe T., Mulcahy T., Griffiths M., Simpson D., Antoniou M., Irvine A., Mountain A., Crombie R. CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnol. 2005;5:17. doi: 10.1186/1472-6750-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., Thornhill S.I., Howe S.J., Ulaganathan M., Schambach A., Sinclair J., Kinnon C., Gaspar H.B., Antoniou M., Thrasher A.J. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann M., Lachmann N., Hartung S., Eggenschwiler R., Pfaff N., Happle C., Mucci A., Gohring G., Niemann H., Hansen G., et al. Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials. 2014;35:1531–1542. doi: 10.1016/j.biomaterials.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Pfaff N., Lachmann N., Ackermann M., Kohlscheen S., Brendel C., Maetzig T., Niemann H., Antoniou M.N., Grez M., Schambach A., et al. A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells. 2013;31:488–499. doi: 10.1002/stem.1316. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl Allen M., Antoniou M. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE) Epigenetics. 2007;2:227–236. doi: 10.4161/epi.2.4.5231. [DOI] [PubMed] [Google Scholar]
- 27.Brendel C., Muller-Kuller U., Schultze-Strasser S., Stein S., Chen-Wichmann L., Krattenmacher A., Kunkel H., Dillmann A., Antoniou M.N., Grez M. Physiological regulation of transgene expression by a lentiviral vector containing the A2UCOE linked to a myeloid promoter. Gene. Ther. 2012;19:1018–1029. doi: 10.1038/gt.2011.167. [DOI] [PubMed] [Google Scholar]
- 28.Knight S., Zhang F., Mueller-Kuller U., Bokhoven M., Gupta A., Broughton T., Sha S., Antoniou M.N., Brendel C., Grez M., et al. Safer, silencing-resistant lentiviral vectors: optimization of the ubiquitous chromatin-opening element through elimination of aberrant splicing. J. Virol. 2012;86:9088–9095. doi: 10.1128/JVI.00485-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiles M.V., Keller G. Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development. 1991;111:259–267. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda H., Kutner R.H., Bazan N.G., Reiser J. Simplified lentivirus vector production in protein-free media using polyethylenimine-mediated transfection. J Virol. Methods. 2009;157:113–121. doi: 10.1016/j.jviromet.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Lachmann N., Jagielska J., Heckl D., Brennig S., Pfaff N., Maetzig T., Modlich U., Cantz T., Gentner B., Schambach A., et al. MicroRNA-150-regulated vectors allow lymphocyte-sparing transgene expression in hematopoietic gene therapy. Gene Ther. 2012;19:915–924. doi: 10.1038/gt.2011.148. [DOI] [PubMed] [Google Scholar]
- 32.Pfaff N., Lachmann N., Kohlscheen S., Sgodda M., Arauzo-Bravo M.J., Greber B., Kues W., Glage S., Baum C., Niemann H., et al. Efficient hematopoietic redifferentiation of induced pluripotent stem cells derived from primitive murine bone marrow cells. Stem Cells Dev. 2012;21:689–701. doi: 10.1089/scd.2011.0010. [DOI] [PubMed] [Google Scholar]
- 33.Lachmann N., Happle C., Ackermann M., Luttge D., Wetzke M., Merkert S., Hetzel M., Kensah G., Jara-Avaca M., Mucci A., et al. Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am. J. Respir. Crit. Care Med. 2014;189:167–182. doi: 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- 34.Cesana D., Sgualdino J., Rudilosso L., Merella S., Naldini L., Montini E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J. Clin. Invest. 2012;122:1667–1676. doi: 10.1172/JCI62189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 36.Kolodziej S., Kuvardina O.N., Oellerich T., Herglotz J., Backert I., Kohrs N., Buscato E., Wittmann S.K., Salinas-Riester G., Bonig H., et al. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat. Commun. 2014;5:3995. doi: 10.1038/ncomms4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F., Frost A.R., Blundell M.P., Bales O., Antoniou M.N., Thrasher A.J. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol. Ther. 2010;18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 39.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K., et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoniou M., Harland L., Mustoe T., Williams S., Holdstock J., Yague E., Mulcahy T., Griffiths M., Edwards S., Ioannou P.A., et al. Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics. 2003;82:269–279. doi: 10.1016/s0888-7543(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 41.Talbot G.E., Waddington S.N., Bales O., Tchen R.C., Antoniou M.N. Desmin-regulated lentiviral vectors for skeletal muscle gene transfer. Mol. Ther. 2010;18:601–608. doi: 10.1038/mt.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchiyama T., Adriani M., Jagadeesh G.J., Paine A., Candotti F. Foamy virus vector-mediated gene correction of a mouse model of Wiskott-Aldrich syndrome. Mol. Ther. 2012;20:1270–1279. doi: 10.1038/mt.2011.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandaranayake A.D., Correnti C., Ryu B.Y., Brault M., Strong R.K., Rawlings D.J. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 2011;39:e143. doi: 10.1093/nar/gkr706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., Fitzgerald P., Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H.B., Ohno K., Gui H., Pirrotta V. Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies. PLoS Genet. 2013;9:e1003436. doi: 10.1371/journal.pgen.1003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A.R., Deaton A., Andrews R., James K.D., et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conner A.L., Aladjem M.I. The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses. Biochim. Biophys. Acta. 2012;1819:794–801. doi: 10.1016/j.bbagrm.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


