Abstract
The replicative DNA polymerase Polδ consists of a catalytic subunit POLD1/p125 and three regulatory subunits POLD2/p50, POLD3/p66 and POLD4/p12. The ortholog of POLD3 in Saccharomyces cerevisiae, Pol32, is required for a significant proportion of spontaneous and UV-induced mutagenesis through its additional role in translesion synthesis (TLS) as a subunit of DNA polymerase ζ. Remarkably, chicken DT40 B lymphocytes deficient in POLD3 are viable and able to replicate undamaged genomic DNA with normal kinetics. Like its counterpart in yeast, POLD3 is required for fully effective TLS, its loss resulting in hypersensitivity to a variety of DNA damaging agents, a diminished ability to maintain replication fork progression after UV irradiation and a significant decrease in abasic site-induced mutagenesis in the immunoglobulin loci. However, these defects appear to be largely independent of Polζ, suggesting that POLD3 makes a significant contribution to TLS independently of Polζ in DT40 cells. Indeed, combining polη, polζ and pold3 mutations results in synthetic lethality. Additionally, we show in vitro that POLD3 promotes extension beyond an abasic by the Polδ holoenzyme suggesting that while POLD3 is not required for normal replication, it may help Polδ to complete abasic site bypass independently of canonical TLS polymerases.
INTRODUCTION
Polδ and Polϵ are the major eukaryotic replicative polymerases. Polδ has been shown to be predominantly responsible for lagging DNA strand synthesis during replication (1) but is also required for base and nucleotide excision repair (2–4), break induced replication and homologous recombination (HR) (5,6). Vertebrate Polδ consists of four subunits: POLD1/p125, the catalytic subunit, POLD2/p50, POLD3/p66 and POLD4/p12, which serve regulatory functions (7,8). Consistent with the central role of Polδ in genome replication and repair, POLD1 and POLD2 are both essential for cell proliferation (9–11). However, while the POLD3 ortholog in the fission yeast Schizosaccharomyces pombe is essential (12), the ortholog in the budding yeast Saccharomyces cerevisiae, Pol32, is not (11). Pol32-deficient S. cerevisiae exhibit defects in Polζ-dependent mutagenesis as well as in double-strand break repair by break-induced DNA replication (5–6,13–15). Recent reports have explained the role of Pol32 in mutagenesis by showing that, along with Pol31 (the yeast ortholog of POLD2) it is an integral component of the specialized translesion synthesis (TLS) polymerase Polζ (16,17). Moreover, POLD2 and POLD3 are common subunits of both Polδ and Polζ in mammalian cells (16–19), interacting with the iron–sulphur clusters found in the C-terminus of both Polδ and Polζ (19). The current model of TLS suggests that when replicative polymerases encounter DNA lesions they hand off DNA synthesis to one of a number of specialized translesion polymerases, which are able to insert nucleotides opposite the lesion. The resulting mismatch is further extended by Polζ, creating a conventional 3′ end from which the replicative polymerases can continue DNA synthesis (20,21). It has been suggested that POLD2 and POLD3, being subunits of both Polδ and Polζ, could contribute to this switch (18) although there is currently no direct evidence for this idea.
The in vivo role of POLD3 in vertebrate replication and TLS has not yet been defined. Here we have generated POLD3-deficient cells (pold3 cells) from the chicken DT40 B lymphocyte line. DT40 cells provide an opportunity for precisely examining in vivo TLS across abasic sites by sequencing the constitutively diversifying immunoglobulin light-chain variable gene (IgL). During avian IgL diversification, the action of activation induced deaminase (AID) leads to conversion of dC to dU, which is then efficiently excised by uracil DNA glycosylase to yield a high frequency of abasic sites (22,23), the most common spontaneously-arising lesion in the chromosomal DNA (24), within a defined window of around 500 bp. In DT40 these abasic sites can induce gene conversion with a set of homeologous upstream IgL pseudogenes, resulting in templated mutagenesis or can be bypassed directly by TLS resulting in nontemplated mutagenesis at dC/dG basepairs (22,25–28). Here we show that pold3 cells replicate with essentially normal kinetics but exhibit a 4.5-fold decrease in the nontemplated immunoglobulin mutagenesis, indicating that POLD3 is required for efficient TLS over abasic sites. Further, pold3 cells exhibit a marked defect in maintaining replication fork progression following UV irradiation. Neither of these phenotypes is observed in cells lacking both Polη and Polζ, and combined loss of Polη, Polζ and POLD3 results in lethality. Together, these results are consistent with POLD3 playing a critical role in TLS in DT40 cells that is substantially independent of Polζ.
MATERIALS AND METHODS
Disruption of POLD3 in DT40 cells
Constructs for targeting and disrupting the chicken POLD3 locus were generated from genomic polymerase chain reaction (PCR) products combined with puroR and bsrR selection-marker cassettes (Figure 1A). Genomic DNA sequences were amplified using primers 5′-GGGGCTCGAGCTGCAGCAAGTGAGTTGGTGTTCCTTC-3′ and 5′-GGGGAATTCCTGTGAGCTCTGCTTAGCAGCTGTAGGACC-3′ for the 5′-arm of the targeting construct and 5′-GGGACTAGTGTCAACTCAGTGCCTGGGCAGCCAAAGGAGG-3′ and 5′-GGGGCGGCCGCCCTTCCTCATCACTGCTGTCAGACTCC-3′ for the 3′-arm of the targeting construct. The resulting 3.6 kb 3′-arm and 1.5 kb 5′-arm were cloned into the XhoI–EcoRI and SpeI–NotI sites, respectively, of pBlueScript SK. puroR and bsrR selection cassettes, flanked by loxP sequences, were inserted into the BamHI site to generate pold3-puroR and pold3-bsrR targeting constructs. To generate pold3 cells, wild-type DT40 cells were transfected sequentially with pold3-puroR and pold3-bsrR. Targeted integration of the constructs was detected by Southern blotting of EcoRV- digested genomic DNA and a 0.70 kb probe generated by PCR from the genomic POLD3 locus with the primers 5′-GGCAGCCACGTGGGTCAGGGGCC-3′ and 5′-CTGGAGCTGTACACTGCACTGTTGG-3′. Targeting efficiency for the first and second allele was 12.5% (5/40) and 2% (2/100) respectively.
Figure 1.
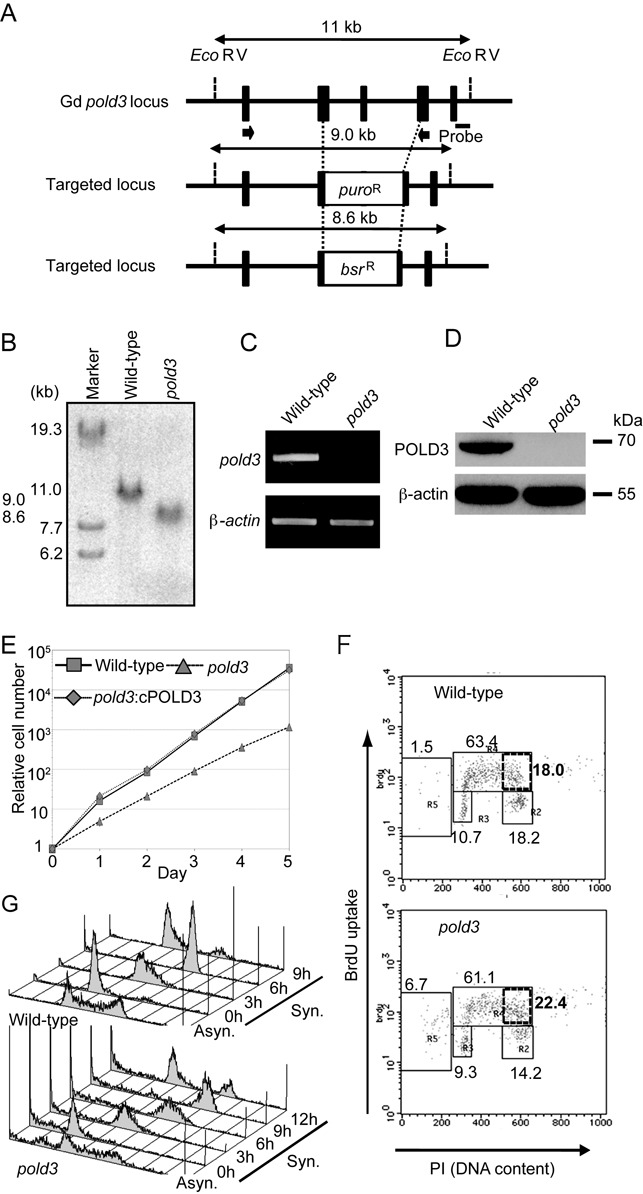
pold3 cells exhibit a prolonged S-phase. (A) POLD3 disruption in DT40 cells. The wild-type chicken POLD3 locus from exon 6 to exon 8 was replaced by a puro or bsr selection-marker gene. Targeted loci (middle and bottom) are shown and compared with the relevant chicken POLD3 genomic sequences (top). Solid boxes indicate the position of the exons. Relevant EcoRV sites and the position of the probe used in the Southern blot analysis are indicated. Black arrows indicate the position of primers used for RT-PCR in (C). (B) Disruption of POLD3 was confirmed by Southern blot. (C and D) Depletion of POLD3 mRNA and POLD3 protein in pold3 cells was confirmed by RT-PCR (C) and western blot (D). β-actin was used as an internal control. (E) Relative growth rate plotted for the indicated genotypes. (F) Representative cell-cycle distribution for the indicated genotypes. The top of the box, and the lower left, lower right, and left-most gates correspond to cells in the S, G1 and G2/M phases, and the sub-G1 fraction, respectively. The sub-G1 fraction represents dying and dead cells. The percentage of cells in each gate is indicated. The box outlined with bold lines corresponds to cells in the late S phase, with the bolded number indicating the percentage of cells. (G) Cells of the indicated genotypes were synchronized at the G1 phase with elutriation and released into culture. Cell-cycle progression profiles after release are shown.
RT-PCR
The loss of POLD3 transcript was confirmed by RT-PCR using primers 5′-ACTGCAGCAAGTTCAGTGCC-3′ and 5′-CTCTACTATGCTATTAGGAG-3′. β-actin transcripts were analyzed as a positive control for the RT-PCR analysis using primers 5′-CATTGCTGACAGGATGCAGAAGG-3′ and 5′-TGCTTGCTGATCCACATCTGCTGG-3′.
Construction of POLD3 cDNA-expression vector
Chicken POLD3 cDNA was isolated by PCR amplification using primers 5′-GACTGAGCGGCCGCATGGAGGACGAGCTGTACCTCGA-3′ and 5′-CTGACTCCAGCACACTGGTCATTTCTTCTGACAGAAGCCCATG-3′ and inserted into a pIRES2–EGFP expression vector (Invitrogen, Carlsbad, CA, USA).
Assessment of sensitivity of cells to genotoxic agents
Sensitivity to γ-rays, UV light, cisplatin and camptothecin was measured as a fraction of surviving colonies. A colony-formation assay was performed as described previously (29). Briefly, to analyze sensitivity to γ-rays, cells were plated into medium containing methylcellulose and subsequently irradiated with a 137Cs γ-ray source. For exposure of cells to UV light, 3 × 105 cells were suspended in 0.5 ml of PBS (phosphate-buffered saline) containing 1% FCS (fetal calf serum) in 6-well plates and irradiated with UVC (254 nm wavelength). For exposure of cells to cisplatin (Nihon-Kayaku, Tokyo, Japan), 1 × 105 cells were treated for 1 h at 39.5°C in 1 ml of complete medium containing cisplatin. Sensitivity to camptothecin, Methyl methansulfonate (MMS) and H2O2 was measured as a fraction of living cells after proliferation in liquid culture. Cells were exposed to camptothecin continuously. For exposure of cells to MMS or H2O2, 2 × 105 cells were treated for 1 h at 39.5°C in 1 ml of PBS containing 1% FCS and MMS or complete medium containing H2O2. Two hundred cells were seeded into 384-well white plates (#6007680 Perkin Elmer Life Sciences, Waltham, MA, USA) with 40 μl of medium per well. Plates were incubated at 39.5°C for 72 h. Cell survival was determined using the ATPlite 1-step kit (PerkinElmer). Briefly, 40 μl ATPlite solution was added to each well. After 5 min, luminescence was measured by Envision 2104 Multilabel Reader (PerkinElmer).
Flow-cytometric analysis of cell-cycle progression
To analyze cell-cycle progression, 1 × 106 cells were exposed for 10 min to 20 μM 5-bromo-2’-deoxyuridine (BrdUrd; Nacalai Tesque, Kyoto, Japan), then harvested and fixed with 70% ethanol. Fixed cells were incubated with 2 M HCl containing Triton X-100 at 0.5%, treated with mouse anti-BrdUrd monoclonal antibody (BD PharMingen, San Diego, CA, USA) and then with FITC-conjugated anti-mouse IgG antibody (Southern Biotechnology Associates, Birmingham, AL, USA). Cells were resuspended in PBS containing propidium iodide (PI) at 5 μg/ml for subsequent analysis using FACScaliber (BD Biosciences). To measure the progression of the cell cycle, cells were synchronized at the G1 phase using centrifugal counterflow elutriation (Hitachi Industrial, Tokyo, Japan). The cell suspension (∼5 × 107 cells) was loaded at a flow rate of 11 ml/min into an elutriation chamber running at 2000 rpm. Cell synchrony was confirmed by Fluorescence-activated cell sorting (FACS) analysis.
Dynamic molecular combing and immunofluorescent detection
Asynchronously growing DT40 cells were sequentially labelled for 15 min with 25 μM IdU and for 15 min with 25 μM CIdU. UV treated cells were irradiated at 20 J/m2 just before the CldU treatment. At the end of the labeling period (30 min), cells were placed in ice cold 1× PBS (1 volume of cells for 2 volumes of 1× PBS) and centrifuged at 250 g for 5 min at 4°C, washed in ice-cold PBS, and resuspended in PBS to a final concentration of 1 × 106 cells/ml. Three microliter of the cell suspension was spotted onto clean glass Superfrost slides and lysed with 7 μl of 0.5% sodium dodecyl sulphate (SDS) in 200 mM Tris–HCl (pH 5.5) and 50 mM ethylenediaminetetraacetic acid (EDTA) (5 min, at room temperature). Slides were tilted at 15° to horizontal, allowing the DNA to run slowly down the slide. Slides were then air dried and fixed in 3:1 methanol/acetic acid and stored at 4°C before immunolabeling. IdU, CldU, DNA revelations and analysis were performed as described (30), with minor modifications: the DNA was denatured for 30 min in 2.5 N HCl, and CldU was detected using rat anti BrdU (ABD Serotec, Raleigh, NC, USA) at 1/750. A stretching factor of 2.6 for conversion from μm to kb was applied, as previously described for the method in (31). Slides were mounted in 10% 1× PBS and 90% glycerol, kept at −20°C and imaged using a Nikon C1-si confocal microscope.
AID overexpression by retrovirus infection
Wild-type and pold3 cells were inoculated in 96-well plates to obtain single colonies (Supplementary Figure S4). Several single colonies were picked up and genomic DNA was extracted. After the DNA sequence of the V(D)J locus was analyzed, clones without any mutation or gene conversion in the V(D)J locus were obtained. AID overexpression was carried out by infection of retrovirus containing the AID gene and the internal ribosomal entry site (IRES) followed by the Green fluorescent protein (GFP) gene (Supplementary Figure S4), as described previously (27,32). The efficiency of infection was about 70%, as assayed by GFP expression. The per division rate of Ig-gene conversion and hypermutation (events/IgV segment/division) was calculated assuming doubling times of 7.6 and 11 h for wild-type and pold3 cells respectively.
Analysis of Ig Vλ diversification
Genomic DNA was extracted at 14 days post infection. Using primers 5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′, in the Vλ leader intron and 5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′, in the J-Cλ intron, the rearranged Vλ segments were PCR amplified, with Prime Star DNA polymerase (Takarabio, Shiga, Japan), a high fidelity thermostable polymerase, cloned into pTOPO-Zeroblunt (Invitrogen) and sequenced with the M13 forward (−20) primer. Sequence alignment with DNASIS-MAC v3.3 (Hitachi Solutions, Tokyo, Japan) allowed identification of changes from the parental sequences in each clone. The method for classification of gene conversion and non-templated point mutations was as previously described (33,34,35).
Creation of the anti-POLD3 antibody
Chicken POLD3 was amplified from DT40 cDNA with the primers 5′-ATAGGATCCATGGAGGACGAGCTGTAC-3′ and 5′-GGCGAATTCTCATTTCTTCTGACAGAAG-3′ and cloned into pGEX-6P1 (Amersham, Buckinghamshire, UK) for expression as in GST-tagged form in Escherichia coli [BL21 STAR (DE3) (Invitrogen)]. Cells were induced with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultured for a further 4 h at 22°C. Cells were harvested, disrupted by sonication and the lysate cleared by ultracentrifugation. The lysate was passed over a glutathione sepharose 4B column and the GST-POLD3 protein eluted with 50 mM Tris–HCL pH 8.0, 150 mM NaCl, 20 mM glutathione. The eluted protein was then applied to a 1 ml HiTrap Q XL column (Amersham) and eluted with a NaCl gradient. GST-POLD3 eluted at 750 mM NaCl. For antibody production, two rabbits were immunized (Eurogentec, Seraing, Belgium) with 200 μg GST-POLD3. Serum was collected 4 weeks after the final immunization. The antibody was purified against a his-tagged POLD3 immobilized on an agarose column.
Protein purification and primer extension assays
The human Polδ holoenzyme with or without POLD3, with N-terminal His-tagged p50, was expressed using a baculovirus vector (pBacPAK9, Clontech, Palo Alto, CA, USA) in insect cells (High Five, Life Technologies, Palo Alto, CA, USA), as described previously (36). The concentrations and purity of the proteins was estimated from the intensity of protein bands in an SDS-polyacrylamide gel (Supplementary Figure S6). For primer-extension analysis, DNA synthesis was carried out with 0.06 pmol 32P-labeled M17 primer (AGCTATGACCATGATTA) annealed with a 49-mer template oligo DNA (AGCTACCATGCCTGCACGAATXAAGCAATTCGTAATCATGGTCATAGCT), where X can be an abasic site. The assay was carried out in a reaction mixture (5 μl) containing 30 mM HEPES-NaOH (pH 7.4), 7 mM MgCl2, 8 mM NaCl, 0.5 mM dithiothreitol and 10 μM each dNTP in the presence of 2 nM of Polδ and 50 nM of PCNA for 15 min at 37°C. Used dNTP concentration reflects that in human cycling cells (37). At the end of the reaction, the products were denatured with formamide and loaded onto 15.6% polyacrylamide gels containing 7 M urea in Tris, Boric acid, EDTA (TBE) buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). After electrophoresis, radioactivity was measured with a Fuji Image analyzer, FLA2500 (Fujifilm, Tokyo, Japan).
RESULTS
DT40 cells lacking POLD3 are viable but exhibit a moderate S phase delay
To explore the role of POLD3, we disrupted the POLD3 gene in the chicken DT40 B cell line, creating pold3 cells (Figure 1A and B). Loss of POLD3 expression was verified by RT-PCR and western blot analysis (Figure 1C and D). pold3 cells are able to proliferate although their doubling time was 1.5× longer than that of wild-type cells (Figure 1E) due to a delay in late S-phase and a higher frequency of apoptosis (the left-most R5 gate, Figure 1F). To confirm this, we monitored cell cycle progression after release from synchronization in G1. Wild-type and pold3 cells completed S-phase at 6 and 9 h after release respectively and returned to the next G1-phase at 9 and 12 h (Figure 1G). Thus, completion of DNA replication is delayed in the absence of POLD3.
pold3 cells have a normal velocity of DNA synthesis but an increase in unidirectional fork movement
To understand the nature of the S phase delay, we examined the kinetics of unperturbed DNA replication using molecular combing (Tables 1 and 2). Strikingly, the replication rate for wild-type and pold3 cells in unperturbed conditions was not significantly different at 1.36 ± 0.05 kbp/min and 1.28 ± 0.03 kbp/min respectively (P > 0.34). The average inter-origin distance was also comparable between the two lines. These findings indicate that loss of POLD3 does not attenuate global progression of DNA replication forks on undamaged DNA templates (Table 1). However, pold3 cells did exhibit an increase (20% as opposed to 13% in wild-type cells) in the proportion of unidirectional forks, in which one of the two forks heading away from a presumed origin is missing (Table 2, Supplementary Figure S1). Thus, POLD3 might be required for releasing replication forks that stall at spontaneously arising DNA damage on genomic DNA.
Table 1. Replication profiles of mutants in this study with and without UV treatment.
| Cell | UV | ΣIdU/ΣDNA (%) | Fork density (per Mb) | Average inter origins distances (kb) | Fork speed (kb/min) | Fiber length (kb) | Mb of DNA analyzed/number of fiber |
|---|---|---|---|---|---|---|---|
| Wild-type | − | 47.05 | 8.63 | 81 ± 7 | 1.36 ± 0.05 | 368 ± 25 | 36.7/111 |
| + | 44.21 | 8.00 | 76 ± 5 | 1.27 ± 0.06 | 360 ± 24 | 24.6/75 | |
| pold3 | − | 46.58 | 10.42 | 81 ± 4 | 1.28 ± 0.03 | 345 ± 17 | 62.4/190 |
| + | 31.03 | 7.35 | 88 ± 7 | 1.14 ± 0.04 | 359 ± 23 | 28.8/89 | |
| polζ/polη | − | 41.53 | 8.54 | 78 ± 5 | 1.40 ± 0.04 | 312 ± 14 | 42.9/153 |
| + | 41.96 | 8.51 | 82 ± 4 | 1.24 ± 0.03 | 367 ± 28 | 48.2/149 | |
| polζ/polη | − | 43.98 | 8.63 | 91 ± 8 | 1.40 ± 0.06 | 383 ± 24 | 21.9/58 |
| pold3/tet-ON | + | 46.18 | 8 | 78 ± 7 | 1.41 ± 0.08 | 326 ± 28 | 15.1/49 |
| polζ/polη | − | 49.57 | 6.94 | 85 ± 6 | 1.43 ± 0.06 | 341 ± 25 | 31.7/104 |
| pold3/tet-OFF | + | 53.27 | 8.32 | 73 ± 5 | 1.13 ± 0.05 | 304 ± 21 | 26.8/97 |
Profile of the DNA replication kinetics in indicated cells with or without UV irradiation was analysed by molecular combing.
Table 2. Proportion of unidirectional forks.
| Cell | Wild-type -UV | Wild-type +UV | pold3 -UV | pold3 +UV |
|---|---|---|---|---|
| Unidirectional fork% | 13 | 18 | 20 | 26 |
| Compared pair | P-values of chi-square test for pairwise comparison | |||
| Wild-type − UV/Wild-type + UV | 0.0206 | |||
| pold3 − UV/pold3 + UV | 0.0201 | |||
| Wild-type − UV/pold3 − UV | 0.0006 | |||
| Wild-type + UV/pold3 + UV | 0.0116 | |||
Proportion of unidirectional fork in indicated cells was examined as illustrated in Supplementary Figure S1.
pold3 cells exhibit sensitivity to a wide range of DNA damaging agents
pold3 cells exhibited sensitivity to a broad range of DNA damaging agents, including ionizing radiation (γ-rays), UV irradiation, the methylatingagent (methyl methane sulphonate, MMS), H2O2 and the chemical crosslinker (cisplatin) (Figure 2). This hypersensitivity to DNA damage was reversed by the ectopic expression of chicken POLD3 (Figure 2).
Figure 2.
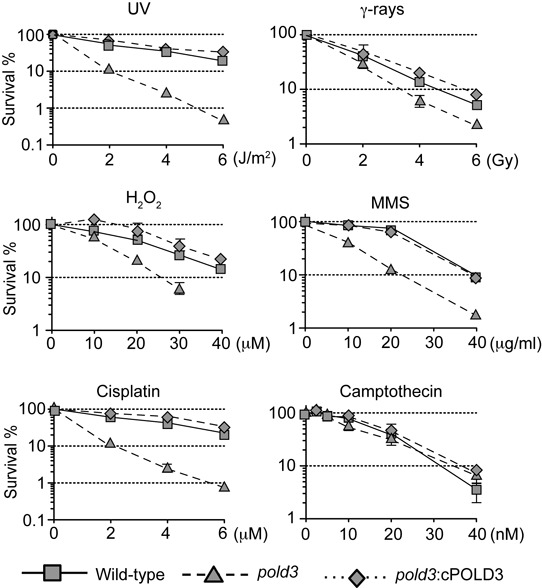
The POLD3 deficient cells are sensitive to a wide range of DNA-damaging agents. pold3 cells exhibit hypersensitivity to various types of DNA damage. Cells with the indicated genotype were exposed to the indicated genotoxic agents. The dose of the genotoxic agent is displayed on the x-axis on a linear scale, while the percentage fraction of surviving cells is displayed on the y-axis on a logarithmic scale. Error bars show the SD of mean for three independent assays.
Since DNA polymerase δ and ζ are involved in a number of DNA repair and damage tolerance pathways (38), we next sought to define the pathways affected by loss of POLD3. The sensitivity of cells to the methylating agent, MMS and oxidising agent, H2O2 prompted us to test whether pold3 cells exhibit a defect in base excision repair. However, gap filling during base-excision repair (39) occurred normally in pold3 cells (Supplementary Figure S2). To investigate the involvement of POLD3 in nucleotide excision repair, we performed epistasis analysis of POLD3 and XPA, an essential protein of nucleotide excision repair (40). Inactivation of the XPA and POLD3 genes had a synergistic impact on cellular sensitivity to UV, suggesting that POLD3 contributes to cellular tolerance of UV independently of nucleotide excision repair (Supplementary Figure S3). To investigate the involvement of POLD3 in HR, we measured cellular sensitivity to the topoisomerase I poison, camptothecin and gene targeting efficiency. pold3 cells are not hypersensitive to camptothecin (Figure 2) and are proficient in gene targeting (Supplementary Table S1). Taken together, hypersensitivity of the pold3 cells to various DNA damaging agents is not attributable to defects in the excision repair pathways or in HR.
POLD3 is required for efficient Ig Vλ gene diversification by TLS
The wide-ranging hypersensitivity of pold3 cells to DNA-damaging agents is reminiscent of the phenotype of TLS-deficient cells (41–43). To examine the role of POLD3 in TLS, we took advantage of the diversification of the Ig Vλ light chain locus of DT40 cells, described in the Introduction and summarized in Supplementary Figures S4 and S5.
To examine Ig Vλ diversification, we ectopically overexpressed AID and isolated at least three subclones each of wild-type and pold3 cells (Supplementary Figure S4). Following two weeks clonal expansion, we compared the frequency and spectrum of mutations in the IgL variable segment in wild-type and pold3 clones (Supplementary Figure S4). pold3 cells exhibited a 4.5-fold reduction in the rate of non-templated mutations (Figure 3A and B). This reduced frequency of TLS was accompanied by a two-fold increase in the frequency of Ig gene conversion events suggesting that the failure of POLD3-dependent TLS results in more lesions being available to trigger Ig gene conversion and showing that POLD3 is not required for robust intragenic HR. We next looked in detail at the mutation spectrum of abasic site bypass. Under conditions of AID over-expression, 62% of the TLS in wild-type cells involves insertion of dC opposite the abasic sites created on both strands, leading to dC to dG transversions. This reaction is largely dependent on the deoxycytidyl transferase activity of REV1 (44,45). Following disruption of POLD3, the frequency of mutagenesis is decreased but 78% of the remaining mutations indicate insertion of dC opposite the abasic site. Intriguingly, few remaining mutations at dC in the pold3 mutant appear to be highly strand biased. However, confirming this result will require a very much more substantial dataset.
Figure 3.
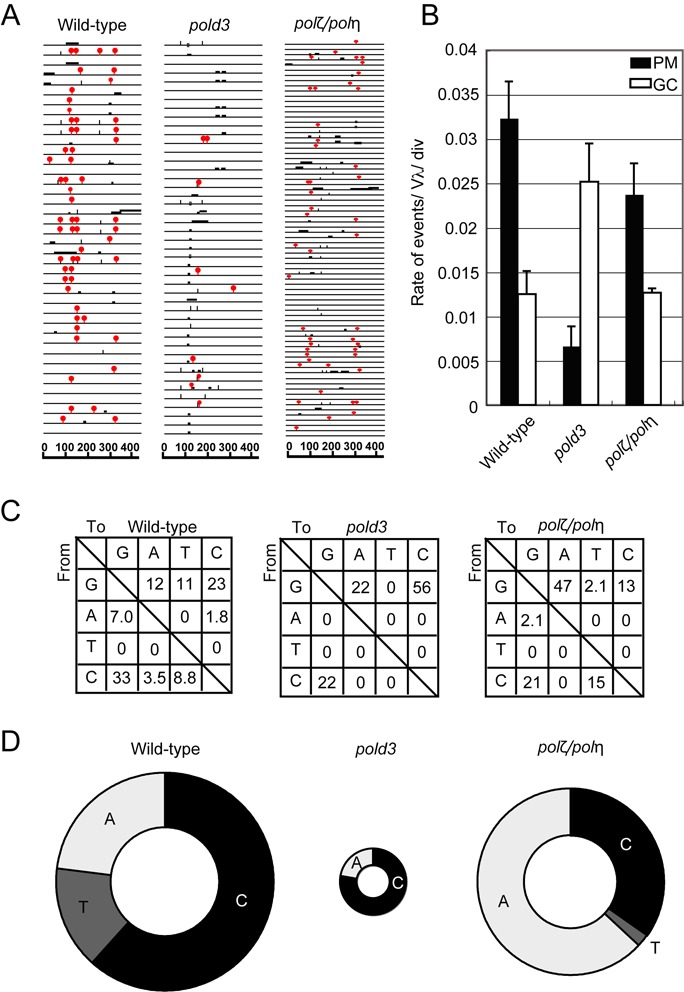
The important role of POLD3 in TLS past abasic sites during Ig Vλ hypermutation. (A) Ig Vλ segments isolated from indicated cells, clonally expanded for two weeks. Horizontal lines represent the rearranged Ig Vλ (450 bp), with hypermutation (red (gray in print) lollipop shapes), gene conversion (horizontal bars), single-nucleotide substitutions that could be the result of hypermutation or gene conversion (vertical bars) and single-base deletion (boxes) determined as described previously (34,35). More than three clones were expanded for two weeks and analyzed for each dataset. (B) The rates of gene conversion (GC) and hypermutation (PM) are indicated with standard error. (C) Pattern of point mutation in wild-type, pold3 and polζ/polη cells. Tables showing the pattern of mutation in each line, given a percentage of mutations observed. (D) Frequency of mutagenic base insertion of C, T or A opposite C on either strand, corresponding to mutation from C to G, A and T, respectively. The size of the pie charts reflects the frequency of overall point mutation within mutated sequences, while the segments reflect the relative use of C, T or A in bypass.
Cells lacking Polζ alone do not tolerate AID overexpression, dying within a few days of retroviral infection (20). However, cells deficient in both Polη and Polζ are able to tolerate AID overexpression and exhibit a level of gene conversion and point mutagenesis comparable to wild-type cells, but with a marked increase in dA incorporation opposite the AID-induced abasic sites (20) (Figure 3). Thus, the immunoglobulin mutagenesis in cells lacking POLD3 and those lacking both Polη and Polζ is quite distinct. This suggests that while POLD3 contributes to TLS in the immunoglobulin loci, it does so substantially independently of Polζ. Moreover, polζ cells displayed significantly higher sensitivity to the ectopic AID expression in comparison with pold3 cells, further suggesting functional independence of POLD3 and Polζ.
Cells deficient in POLD3, Polη and Polζ are inviable
To further test whether POLD3 acts independently of the Polη/Polζ TLS axis, we conditionally disrupted POLD3 in polη/polζ double mutant cells by covering the deletion of POLD3 with a chicken POLD3 transgene, the expression of which was under a tetracycline-repressible promoter (Figure 4A). polη/polζ/pold3 triple mutant cells expressing the cPOLD3 transgene proliferated slightly more slowly than polη/polζ/pold3+/− cells (Figure 4B). Upon addition of doxycycline, POLD3 expression was significantly reduced by 24 h and lost completely by 48 h. By 3–4 days the cells had stopped proliferating. This observation further supports the idea that POLD3 plays a role that is independent of Polη and Polζ.
Figure 4.
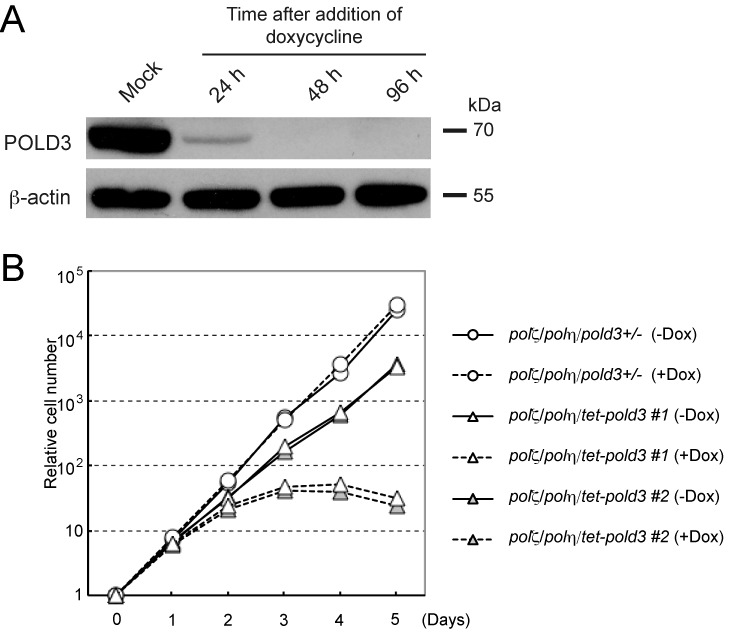
Synthetic lethality of pold3 and polζ/polη. (A) Western blot for probing existence of POLD3 protein was shown. Protein samples from indicated cells were analyzed. β-actin was analyzed as a loading control. (B) Growth curves of the indicated cells. The transcription of tet-POLD3 was active without doxycycline (−Dox) and inhibited upon addition of doxycycline (+Dox).
POLD3 is required to maintain replication fork progression on UV damaged DNA but not to promote the filling of post-replicative gaps
Previous work in DT40 cells has revealed temporally separated modes of lesion bypass. One mechanism is responsible for timely filling of postreplicative gaps at UV-damage sites, while another operates at or very close to stalled replication forks and maintains normal fork progression on UV-damaged DNA (46). These modes of damage bypass can be assessed by alkaline sucrose gradient sedimentation of newly replicated DNA (Figure 5A) and DNA molecular combing (Tables 1 and 2, and Figure 5B and C), respectively.
Figure 5.
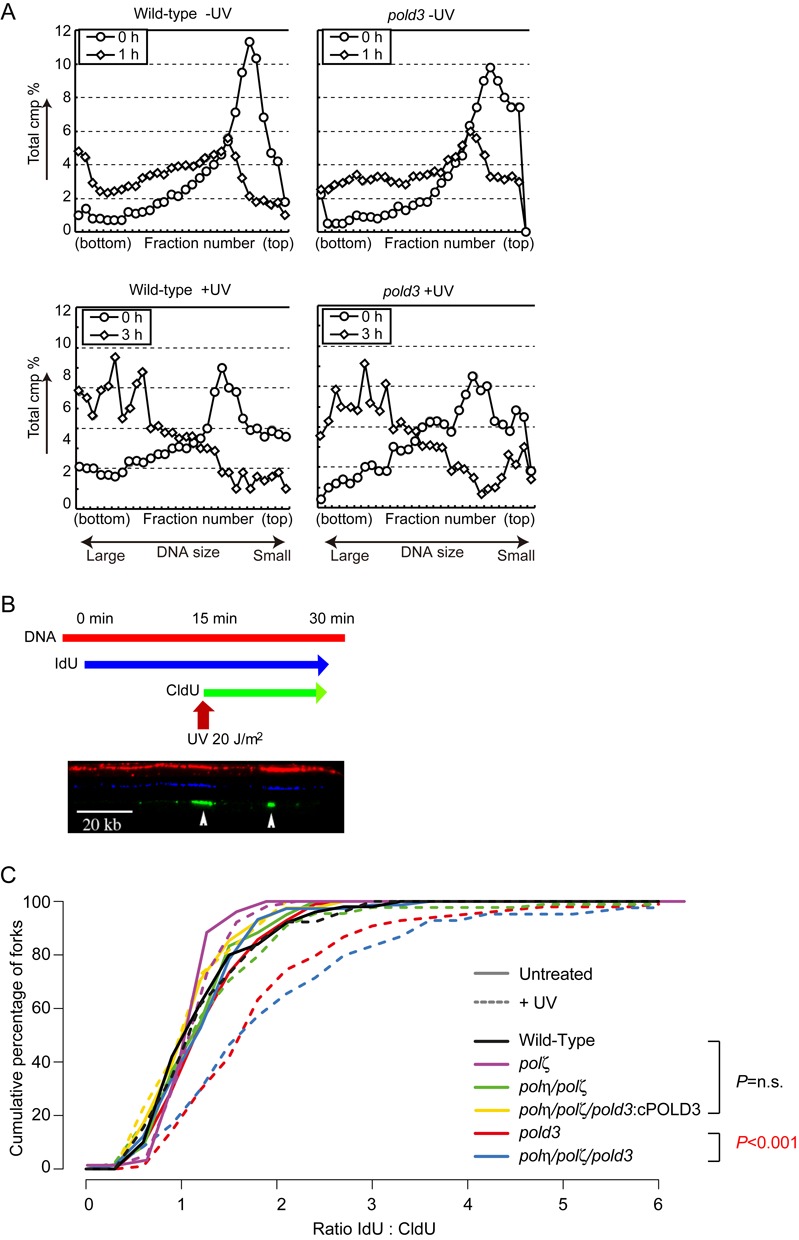
POLD3 but not Polζ is required to maintain replication fork progression on UV damaged DNA. (A) Intact postreplicative gap filling in pold3 cells. Indicated cells without UV (upper panel) or with UV (5 J/m2) (lower panel) were pulse-labeled with [methyl-14C] thymidine, then incubated a further in fresh medium containing 10 μM unlabeled thymidine for the indicated time. Samples were separated on 5–20% alkaline sucrose gradient sedimentation. (B) Representative image showing stained DNA fibres. DT40 cells were labelled sequentially with IdU and CldU with or without UV treatment after IdU labeling. (C) The data for cells carrying the indicated genotypes was plotted as a cumulative percentage (y-axis) of forks at each ratio (x-axis). The P-values of the Kolmogorov–Smirnov test for ratio distribution of each mutant for UV compared to wild-type are indicated. n.s.: not significant.
We first asked whether POLD3 is required for post-replicative gap filling. Without exposure to UV, the 14C thymidine label was incorporated into higher molecular weight DNA similarly in wild-type and pold3 cells (Figure 5A upper panel). This is consistent with the observation that POLD3 is not essential for unperturbed DNA replication (Table 1). However, pold3 cells are also able to reconstitute high molecular weight DNA during a 3-h chase period following UV irradiation (Figure 5A lower panel) with similar kinetics to wild-type cells. Similarly, polζ cells exhibited reconstitution of higher molecular weight DNA following UV irradiation that was indistinguishable from wild-type cells (42). These data contrast with the defective postreplicative gap filling seen in polη, rad18 and pcnaK164R/K164R DT40 cells (46). Thus, neither Polζ nor POLD3 play a critical role in postreplicative gap filling at UV damaged sites in DT40.
We next examined replication fork progression after UV irradiation. To do this, we labeled nascent strands with IdU for 15 min, UV irradiated the cells and then continued labeling the nascent strands with CldU (Figure 5B and Table 1). After DNA combing, we revealed the tracts of CldU and IdU and calculated the ratio between them to allow comparison of the total DNA synthesized before and after UV exposure on a fork-by-fork basis. We plotted the data as a cumulative percentage of forks at each ratio (Figure 5C). polζ cells exhibit a response indistinguishable from wild-type i.e. a modest reduction in the average length of DNA synthesized during the second labeling period following UV (Figure 5C). This is consistent with a previous report examining Polζ-deficient mouse cells (47). In contrast, pold3 cells exhibit a significant reduction in the length of DNA synthesized during the second labeling period after UV, consistent with a significantly increased proportion of forks stalling at UV lesions (P = 2.77 × 10−7, Supplementary Figure S6). In the time course of this experiment (15′ after 20 J/m2 UV), we estimate that forks would have about a 60% chance of encountering a UV dimer (based on (46), Supplementary Figure S1). Thus, POLD3 is required to maintain replication fork progression on a UV damaged template independently of Polζ. Taken together, these data indicate that POLD3 facilitates TLS independently of Polζ, leading us to speculate that it may facilitate TLS by Polδ itself.
POLD3 facilitates in vitro TLS past abasic sites by Polδ
To address the possibility that TLS by Polδ might be modulated by POLD3, we purified the intact human Polδ holoenzyme (Polδ (POLD3+)) and POLD3-deficient holoenzyme (Polδ (POLD3−)). We employed the human Polδ holoenzyme since the method for protein purification has been established (48). Polδ (POLD3+) and Polδ (POLD3−) were expressed and purified with the same efficiency (Supplementary Figure S7), indicating that the absence of POLD3 does not diminish the stability of the other three components of the holoenzyme. While the Proliferating cell nuclear antigen (PCNA) binding capability of POLD3 may increase the processivity of Polδ on a long circular template (49), to focus our analysis on the TLS capability of Polδ, we used a short (49-mer) linear template. As expected from the in vivo data shown in Table 1 and Figure 5A (upper panels), DNA synthesis by purified Polδ (POLD3+) and Polδ (POLD3−) on an intact template DNA strand was comparable (Figure 6A and C). However, bypass replication across an abasic site was significantly reduced in Polδ (POLD3−) (Figure 6B and C). The bypass is performed in two steps, the insertion of a nucleotide opposite to the lesion followed by extension of the resulting mismatch. Polδ (POLD3+) and Polδ (POLD3−) performed the insertion step with comparable efficiency, as the yield of product indicating an incorporation opposite the abasic site (‘0’ in Figure 6B) relative to that reflecting stalling at the −1 position was very similar with both Polδ (POLD3−) and Polδ (POLD3+). On the other hand, Polδ (POLD3+) was able to extend beyond the abasic site and complete DNA synthesis much more efficiently than Polδ (POLD3−) (Figure 6B and C). The reduced extension by Polδ (POLD3−) might be attributable to (i) higher exonuclease activity of Polδ (POLD3−) and/or to (ii) lower extension ability from the mismatched primer terminus than Polδ (POLD3+). To address these possibilities, we measured degradation and extension using a 28-mer primer, which carries dA in its 3′ terminus opposite the abasic site in template DNA. The ratio between intact and degraded primers in Polδ (POLD3+) and Polδ (POLD3−) was indistinguishable (Figure 6D). Moreover, the kinetics of the primer degradation in the absence of deoxynucleotides was also indistinguishable between Polδ (POLD3+) and Polδ (POLD3−) (Figure 6E). Thus, we propose that POLD3 is required for effective coupling of nucleotide insertion with subsequent extension thereby enhancing the overall processivity of Polδ during abasic site bypass.
Figure 6.
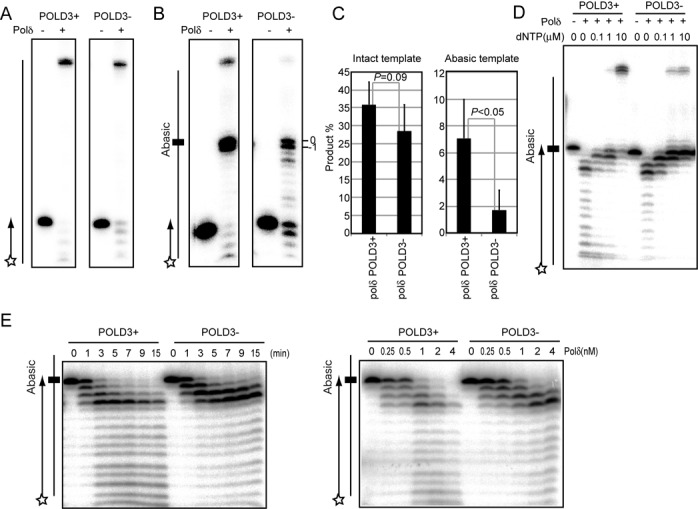
POLD3 is required for efficient Polδ-dependent TLS past abasic sites in vitro. (A and B) DNA synthesis reactions carried out with the indicated Polδ holoenzymes (2 nM each) on template and primer strands, which are schematically shown on the left. The 49-nucleotides template strands carry a dC (A) or abasic site (B) at the 25th nucleotide from the 3′ end. The 5′ end of the primer was 32P labeled as shown by a star. The DNA synthesis was done the presence of NaCl (8 mM), MgCl2 (7mM), PCNA (50 nM) and dNTP (10 μM), a concentration which reflects that in human cycling cells (37). After incubation at 37°C for 10 min, reaction products were separated on a 15.6% polyacrylamide gel containing 7 M urea and analyzed using a PhosphoImager. (B) The bands shown by −1 and +0 represent the DNA strand having stopped its extension one nucleotide before and at the abasic site, respectively. (C) The amounts of extension product (49 nucleotides) relative to the amount of input primer is shown as percentage. The experiment was performed at least three times, and averages are presented with SD and P-values. (D) Degradation and extension of the primer carrying a dA opposite the abasic site of the shown template DNA strand with the indicated Polδ holoenzymes (2 nM each) was examined at 15 min. The reaction was carried out with the indicated concentrations of dNTP. Less than 0.1 μm concentrations of dNTP significantly enhanced the proofreading exonuclease over polymerase activity. (E) Kinetics of the primer degradation in the absence of deoxynucleotides was examined. (Left) Reaction was carried out with 2 nM of the indicated Polδ holoenzymes for the indicated time. (Right) Reaction was carried out with the indicated concentrations of Polδ holoenzymes for 15 min.
DISCUSSION
Here we have shown that a vertebrate cell line is capable of proliferating and replicating undamaged DNA with apparently normal kinetics in the absence of the POLD3 subunit of Polδ. This suggests that POLD3 does not play a major role in the central function of Polδ as the main lagging strand replicase (1). This rather surprising finding contrasts with the essential function of the POLD3/Pol32 ortholog of the fission yeast S. pombe (12). Further, while Polδ is involved in excision repair (2–4) and recombination (5–6,50), we could not detect any gross defects in these repair pathways in pold3 cells. Rather, we found that POLD3 contributes significantly to cellular tolerance of a wide variety of DNA damaging agents by presumably promoting TLS.
It has been known for some time that Pol32 plays a role in TLS in S. cerevisiae and that genetically it played roles that are both dependent and independent of REV3, the catalytic subunit of Polζ, depending on the mutator examined (13–15,51–53). Recent biochemical studies have shown that Pol32 is not only a subunit of Polδ but is also an integral component of Polζ (16,17). This has led to the attractive suggestion that this association with Polζ explains the role of POLD3 in TLS and has led to the suggestion that Pol32 may facilitate switching between Polδ and Polζ (16–19). POLD3 and POLD2 are also subunits of human Polζ, interacting with the C terminal iron–sulphur clusters of REV3 in a similar manner to that reported in yeast (18,19). However, to date genetic data addressing the role played by vertebrate POLD3 in TLS have been lacking. The data we present here highlight striking phenotypic differences between cells lacking POLD3 and those lacking Polζ. Together with the lethality that results when both are disrupted the data suggest that the main role played by POLD3 in TLS in DT40 cells is independent of Polζ.
An important question is how POLD3 promotes TLS independently of Polζ. An obvious and attractive potential explanation is that POLD3 facilitates lesion bypass by Polδ itself. While Polδ is stalled by many lesions, there is good in vitro evidence that it is able to insert and extend over a number of single base lesions both with and without PCNA (48,54–56), including abasic sites (48,56–57). We show that purified human Polδ (POLD3+) is able to insert and extend over an abasic site at a physiological concentration of dNTP (Figure 6). Polδ preferentially inserts dA (57), an activity that could explain the bias toward the incorporation of dA we observed in the IgVλ gene in the absence of Polζ and Polη. How might POLD3 facilitate TLS by Polδ, given that it appears largely dispensable for general replication? We show that while Polδ (POLD3+) and Polδ (POLD3−) insert a single nucleotide opposite an abasic site with very similar efficiency, Polδ (POLD3+) is much more proficient at extension of the resulting mismatch (Figure 6B and C). This more efficient extension is attributable to higher processivity, but not lower proofreading exonuclease activity, in the presence of POLD3 (Figure 6D and E). It is not unlikely that Polδ (POLD3+) would cycle opposite the abasic site with nucleotide insertion balanced by exonucleolytic excision, with occasional extension events from the inserted nucleotide allowing for completion of the bypass of the abasic site. A possible explanation for the role of POLD3 would be that it helps prevent Polδ dissociating from the mismatched primer terminus, increasing the chance of a successful extension by POLD1.
Importantly, our data do not exclude cooperation between POLD3 and Polζ. Despite our data supporting Polδ being able to perform abasic site TLS on its own, independently of Polζ, it may remain the case that the efficiency of extension in vivo is facilitated by Polζ, which is able to complete bypass regardless of which polymerase mediated the insertion step. However, this role may become essential when the ability of Polδ to extend directly is compromised by loss of POLD3. Such a model would explain the synthetic lethality of cells lacking both POLD3 and Polζ and would suggest that the non-redundant function of the two proteins is in mediating the extension step of abasic site bypass.
Our study also indicates that POLD3 contributes to TLS operating at or close replication forks but not obviously to post-replicative gap filling. Previous studies both in DT40 cells (46) and mice (47) have shown that REV1 is also important for maintaining fork progression on damaged DNA and for effective Ig hypermutation (44,58). It is thus tempting to speculate that the two proteins may cooperate. Indeed, a physical interaction between Pol32 and REV1 has been described in S. cerevisiae (59), although this interaction has not been demonstrated in vertebrates. Nonetheless, it will be interesting to further explore the genetic and physical relationship between REV1 and POLD3 in vertebrates to examine whether POLD3 and REV1 collaborate to facilitate restart of replication by Polδ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank A. Noguchi for technical assistance. We also acknowledge the Radioisotope Research Center in Kyoto University, Kyushu University and Tokyo Metropolitan University for support in the use of isotopes.
FUNDING
Grant-in-Aid for Scientific Research on Innovative Areas [21114509, 24114509, 26116518], JSPS KAKENHI [20770147, 22687001, 25281021] and, grants from the Uehara Memorial Foundation and the Naito Foundation [to K.H.]; JSPS KAKENHI [20241012, 23221005 to S.T.]; National Institutes of Health Grants [P30 ES10126, P42 ES05948 to J.N.]; LMB from the Medical Research Council [U105178808] and Fanconi Anemia Research Fund [to G.G., L.G.P. and J.E.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank A., Kim B., Loeb L.A. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shivji K.K., Kenny M.K., Wood R.D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 4.Wood R.D., Shivji M.K. Which DNA polymerases are used for DNA-repair in eukaryotes. Carcinogenesis. 1997;18:605–610. doi: 10.1093/carcin/18.4.605. [DOI] [PubMed] [Google Scholar]
- 5.Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., Halazonetis T.D. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lydeard J.R., Jain S., Yamaguchi M., Haber J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 7.Podust V.N., Chang L.S., Ott R., Dianov G.L., Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 8.Lee M.Y., Zhang S., Lin S.H., Wang X., Darzynkiewicz Z., Zhang Z., Lee E.Y. The tail that wags the dog: p12, the smallest subunit of DNA polymerase delta, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression. Cell Cycle. 2014;13:23–31. doi: 10.4161/cc.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulet A., Simon M., Faye G., Bauer G.A., Burgers P.M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989;8:1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchimura A., Hidaka Y., Hirabayashi T., Hirabayashi M., Yagi T. DNA polymerase delta is required for early mammalian embryogenesis. PLoS One. 2009;4:e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerik K.J., Li X., Pautz A., Burgers P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 12.Bermudez V.P., MacNeill S.A., Tappin I., Hurwitz J. The influence of the Cdc27 subunit on the properties of the Schizosaccharomyces pombe DNA polymerase delta. J. Biol. Chem. 2002;277:36853–36862. doi: 10.1074/jbc.M202897200. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs P.E., McDonald J., Woodgate R., Lawrence C.W. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M.E., de Calignon A., Nicolas A., Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 15.Huang M.E., Rio A.G., Galibert M.D., Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson R.E., Prakash L., Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova A.V., Stodola J.L., Burgers P.M. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranovskiy A.G., Lada A.G., Siebler H.M., Zhang Y., Pavlov Y.I., Tahirov T.H. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netz D.J., Stith C.M., Stumpfig M., Kopf G., Vogel D., Genau H.M., Stodola J.L., Lill R., Burgers P.M., Pierik A.J. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota K., Sonoda E., Kawamoto T., Motegi A., Masutani C., Hanaoka F., Szuts D., Iwai S., Sale J.E., Lehmann A., et al. Simultaneous disruption of two DNA polymerases, Poleta and Polzeta, in Avian DT40 cells unmasks the role of Poleta in cellular response to various DNA lesions. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001151. doi:10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livneh Z., Ziv O., Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 22.Di Noia J., Neuberger M.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 23.Saribasak H., Saribasak N.N., Ipek F.M., Ellwart J.W., Arakawa H., Buerstedde J.M. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 2006;176:365–371. doi: 10.4049/jimmunol.176.1.365. [DOI] [PubMed] [Google Scholar]
- 24.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA repair and Mutagenesis. 2nd edn. ASM press; 2005. [Google Scholar]
- 25.Arakawa H., Hauschild J., Buerstedde J.M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 26.Kohzaki M., Nishihara K., Hirota K., Sonoda E., Yoshimura M., Ekino S., Butler J.E., Watanabe M., Halazonetis T.D., Takeda S. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J. Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saberi A., Nakahara M., Sale J.E., Kikuchi K., Arakawa H., Buerstedde J.M., Yamamoto K., Takeda S., Sonoda E. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol. Cell. Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sale J.E. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA Rep. 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 29.Okada T., Sonoda E., Yamashita Y.M., Koyoshi S., Tateishi S., Yamaizumi M., Takata M., Ogawa O., Takeda S. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 30.Guilbaud G., Rappailles A., Baker A., Chen C.L., Arneodo A., Goldar A., d'Aubenton-Carafa Y., Thermes C., Audit B., Hyrien O. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput. Biol. 2011;7:e1002322. doi: 10.1371/journal.pcbi.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 33.Sale J.E. Measurement of diversification in the immunoglobulin light chain gene of DT40 cells. Methods Mol. Biol. 2012;920:417–432. doi: 10.1007/978-1-61779-998-3_29. [DOI] [PubMed] [Google Scholar]
- 34.Nakahara M., Sonoda E., Nojima K., Sale J.E., Takenaka K., Kikuchi K., Taniguchi Y., Nakamura K., Sumitomo Y., Bree R.T., et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009;5:e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 36.Shikata K., Ohta S., Yamada K., Obuse C., Yoshikawa H., Tsurimoto T. The human homologue of fission Yeast cdc27, p66, is a component of active human DNA polymerase delta. J. Biochem. 2001;129:699–708. doi: 10.1093/oxfordjournals.jbchem.a002909. [DOI] [PubMed] [Google Scholar]
- 37.Jamburuthugoda V.K., Chugh P., Kim B. Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J. Biol. Chem. 2006;281:13388–13395. doi: 10.1074/jbc.M600291200. [DOI] [PubMed] [Google Scholar]
- 38.Prindle M.J., Loeb L.A. DNA polymerase delta in DNA replication and genome maintenance. Environ. Mol. Mutagen. 2012;53:666–682. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimura M., Kohzaki M., Nakamura J., Asagoshi K., Sonoda E., Hou E., Prasad R., Wilson S.H., Tano K., Yasui A., et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camenisch U., Nageli H. XPA gene, its product and biological roles. Adv. Exp. Med. Biol. 2008;637:28–38. doi: 10.1007/978-0-387-09599-8_4. [DOI] [PubMed] [Google Scholar]
- 41.Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonoda E., Okada T., Zhao G.Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N.S., van Gent D.C., Takata M., et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita Y.M., Okada T., Matsusaka T., Sonoda E., Zhao G.Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross A.L., Sale J.E. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Edmunds C.E., Simpson L.J., Sale J.E. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Jansen J.G., Tsaalbi-Shtylik A., Hendriks G., Verspuy J., Gali H., Haracska L., de Wind N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Rep. 2009;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Narita T., Tsurimoto T., Yamamoto J., Nishihara K., Ogawa K., Ohashi E., Evans T., Iwai S., Takeda S., Hirota K. Human replicative DNA polymerase delta can bypass T-T (6–4) ultraviolet photoproducts on template strands. Genes Cells. 2010;15:1228–1239. doi: 10.1111/j.1365-2443.2010.01457.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y., Meng X., Zhang S., Lee E.Y., Lee M.Y. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the MultiBac system. PLoS One. 2012;7:e39156. doi: 10.1371/journal.pone.0039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sneeden J.L., Grossi S.M., Tappin I., Hurwitz J., Heyer W.D. Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res. 2013;41:4913–4925. doi: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haracska L., Unk I., Johnson R.E., Johansson E., Burgers P.M., Prakash S., Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minesinger B.K., Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous polzeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939–1955. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebler H.M., Lada A.G., Baranovskiy A.G., Tahirov T.H., Pavlov Y.I. A novel variant of DNA polymerase zeta, Rev3DeltaC, highlights differential regulation of Pol32 as a subunit of polymerase delta versus zeta in Saccharomyces cerevisiae. DNA Rep. 2014;24:138–149. doi: 10.1016/j.dnarep.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi J.Y., Chowdhury G., Zang H., Angel K.C., Vu C.C., Peterson L.A., Guengerich F.P. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 55.Meng X., Zhou Y., Zhang S., Lee E.Y., Frick D.N., Lee M.Y. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt M.W., Matsumoto Y., Loeb L.A. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi J.Y., Lim S., Kim E.J., Jo A., Guengerich F.P. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson L.J., Sale J.E. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Acharya N., Johnson R.E., Pages V., Prakash L., Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


