Abstract
The pentatricopeptide repeat (PPR) protein family is a large family of RNA-binding proteins that is characterized by tandem arrays of a degenerate 35-amino-acid motif which form an α-solenoid structure. PPR proteins influence the editing, splicing, translation and stability of specific RNAs in mitochondria and chloroplasts. Zea mays PPR10 is amongst the best studied PPR proteins, where sequence-specific binding to two RNA transcripts, atpH and psaJ, has been demonstrated to follow a recognition code where the identity of two amino acids per repeat determines the base-specificity. A recently solved ZmPPR10:psaJ complex crystal structure suggested a homodimeric complex with considerably fewer sequence-specific protein–RNA contacts than inferred previously. Here we describe the solution structure of the ZmPPR10:atpH complex using size-exclusion chromatography-coupled synchrotron small-angle X-ray scattering (SEC-SY-SAXS). Our results support prior evidence that PPR10 binds RNA as a monomer, and that it does so in a manner that is commensurate with a canonical and predictable RNA-binding mode across much of the RNA–protein interface.
INTRODUCTION
The pentatricopeptide repeat (PPR) superfamily (1) is characterized by a degenerate 35 amino acid repeat and is specific to eukaryotes although most numerous and varied in the plant kingdom (2), with over 400 identified in Arabidopsis thaliana (3). PPR proteins function in RNA stabilization (4–6), editing (7), maturation and post-transcriptional modifications (8,9) and are essential for organelle biogenesis (3) and translational control (10–14). Mutations can result in cytoplasmic male sterility (15,16), impaired seed development (17) and diverse embryonic defects (18).
PPR proteins can be broadly divided into two families, those consisting of tandem arrays of canonical P-class motifs and those composed of repeats of triplets of P-, L- and S-class motifs (3). Regardless of repeat class, conservation of key amino acids from each motif has been correlated with the sequence of known RNA targets to establish a statistically and experimentally validated predictive code for RNA recognition by PPR proteins (19–21). This code is consistent with a modular interaction where a base is coordinated by the sixth residue of one PPR repeat, and the first residue of the next repeat. Such a modular, predictable, mode of single-stranded RNA binding makes PPR proteins a potential scaffold for the design of biotechnologically useful proteins (22,23).
Predictions of an α-solenoid tertiary structure for PPR tracts (1,24) have been confirmed recently by a number of crystal structures including a segment of the mitochondrial RNA polymerase (25), Arabidopsis proteinaceous ribonuclease P (26), the P-class PPR protein THA8L (27) and a synthetic consensus P-class PPR protein (28). Crystal structures of two PPR proteins, Brachypodium THA8 (29) and Zea mays PPR10 (ZmPPR10) (30), in complex with RNA have partially confirmed the modular RNA-binding mode, but revealed non-modular and idiosyncratic interactions that had not been anticipated based on prior data. In addition, these structures implicate a dimeric quaternary structure in RNA binding.
ZmPPR10 localizes to plastids where it binds cis-elements of two polycistronic transcripts (6,13), the minimal binding sites identified as 17 and 18 nucleotide tracts in the atpI-atpH and psaJ-rpl33 intercistronic regions. PPR10 binding was shown to stabilize the RNA in vivo, protecting the transcripts from exoribonucleases in either direction, and to activate mRNA translation by remodeling local RNA structure (6). Analytical ultracentrifugation experiments unambiguously demonstrated the association of PPR10 monomers into dimers at high concentration and dissociation of PPR10 dimers into monomers on binding an oligonucleotide representing atpH RNA, resulting in a 1:1 protein:RNA stoichiometry (19), with a dissociation constant that has been measured at ca. 1 nM (6).
In the recent crystal structure of ZmPPR10 in complex with an 18 nucleotide RNA fragment of psaJ (PDB 4M59 (30)), the protomer consists of a continuous array of 20 P-class PPR repeats forming a right-handed superhelix. A dimer is generated by the association of two anti-parallel molecules, and one RNA molecule is bound at each end of the dimer by elements of both monomers. Notably, reported attempts to crystallize PPR10 with atpH resulted in apparent dissociation of the dimer and failure to crystallize, suggesting the possibility of a different, higher-affinity, mode of RNA binding compared to psaJ, for which a dissociation constant of >200 μM has been measured (13). Here we use size-exclusion chromatography-coupled synchrotron small-angle X-ray scattering (SAXS) (31) to produce clear structural evidence for a monomeric PPR10:atpH complex in solution, demonstrating a potential structural rearrangement with respect to the PPR10:psaJ crystal structure, that is more consistent with the proposed PPR–RNA recognition code and with other prior data concerning PPR10–RNA interactions.
P-class PPR repeats forming a right-handed superhelix. A dimer is generated by the association of two anti-parallel molecules, and one RNA molecule is bound at each end of the dimer by elements of both monomers. Notably, reported attempts to crystallize PPR10 with atpH resulted in apparent dissociation of the dimer and failure to crystallize, suggesting the possibility of a different, higher-affinity, mode of RNA binding compared to psaJ, for which a dissociation constant of >200 μM has been measured (13). Here we use size-exclusion chromatography-coupled synchrotron small-angle X-ray scattering (SAXS) (31) to produce clear structural evidence for a monomeric PPR10:atpH complex in solution, demonstrating a potential structural rearrangement with respect to the PPR10:psaJ crystal structure, that is more consistent with the proposed PPR–RNA recognition code and with other prior data concerning PPR10–RNA interactions.
MATERIALS AND METHODS
Protein expression and purification
The ZmPPR10:pMAL-TEV plasmid generated as described previously (13) was used to transform E. coli Rosetta™ 2 (DE3) (Novagen). Cells were grown in 2YT media containing 1% (w/v) D-glucose, 50 mM Tris-HCl at pH 7.5 with kanamycin at 50 μg ml−1 and chloramphenicol at 50 μg ml−1. The culture was grown at 37ºC to an optical density (600 nm) of 0.6 and then cooled on ice for 5 min. Expression was then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside and the culture shaken overnight at 16ºC for protein expression. The bacterial pellet was resuspended in Buffer A (50 mM Tris-HCl at pH 8.0, 500 mM KCl, 10% (v/v) glycerol, 1 mM dithiothreitol) supplemented with 0.13 mM phenylmethanesulfonyl fluoride (Roche), 1 mini Complete protease inhibitor tablet (Roche), 0.5 μl Benzonase (Sigma-Aldrich) and 1 mg ml−1 lysozyme, and lysed under high pressure using a Emulsiflex C5 homogenizer (Avestin). Lysate supernatant was batch loaded onto amylose resin (GE healthcare) and left at 4ºC for 1 h with agitation. The column was washed with buffer A, and fusion protein eluted with buffer A supplemented with 50 mM maltose. Fusion protein was cleaved overnight at 4ºC with tobacco etch virus protease and then subjected to size-exclusion chromatography (BioLogic DuoFlow, Bio-Rad) with a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) developed in buffer A. Peak fractions were confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (NuPage®Novex 4–12%, Bis-Tris gel, Invitrogen) stained with Coomassie brilliant blue. Purified proteins were concentrated in a Vivaspin® 20 (MWCO 30 000) centrifugal concentrator (GE) and quantified by absorption at 280 nm (Nanodrop Lite, ThermoScientific). RNA concentration was analysed by absorption at 260 nm. PPR10:atpH complex was prepared by briefly incubating a 1:1 molar ratio of purified protein with RNA (5′-GUAUCCUUAACCAUUUC-3′ [IDT]).
Circular dichroism analysis
Purified PPR10 protein at a concentration of 0.1 mg ml−1 was dialysed into 10 mM potassium phosphate at pH 8.0, 100 mM KF, and analysed at 20ºC in a 1 mm path length quarz cuvette on a JASCO J-810 spectropolarimeter. Measurements from 260 to 185 nm with wavelength steps of 1 nm were conducted in triplicate to facilitate the generation of error bars denoting standard deviation. Estimations of α-helix content were made using characteristic signals at 208 and 222 nm:%α208 = [(|[Θ]208|−4000) / (33 000–4000)] x 100 and %α222 = [(|[Θ]222|−3000) / (36 000–3000)] x 100 (32,33).
Small-angle X-ray scattering analysis
SAXS measurements were conducted at the SAXS/WAXS beamline of the Australian Synchrotron with continuous data collection on a 1 M Pilatus detector (34). SEC-SY-SAXS was controlled by a Shimadzu HPLC system. For the PPR10:atpH complex, scattering data were collected from 50 μl of a 6 mg ml−1 sample loaded onto a Superdex200 Precision column (GE healthcare) at 0.1 ml min−1 in 50 mM Tris-HCl at pH 7.3, 250 mM NaCl, 10% glycerol, 5 mM β -mercaptoethanol. For the MBP-PPR10:atpH complex, 90 μl of a 6 mg ml−1 sample was run through the Precision column at 0.5 ml min−1 in Buffer A. For experiments where timed shutter opening was critical, the delay between the FPLC UV detector and X-ray detector was calibrated using glucose isomerase solution as a standard. Scattering data from 2 s exposures were background corrected and averaged using scatterBrain. Data were corrected for fouling by aggregates while the shutter was open, by using linear interpolation of background from averaged frames prior to the peak to averaged frames after the peak, in an analogous approach to that used in US-SOMO (35). A high-q cutoff of 0.21 was used due to increasing noise beyond this resolution. The radius of gyration (Rg), maximum dimension (Dmax) and P(r) distribution plots of the samples were determined with the ATSAS software (36) using PRIMUS (37) and GNOM (38) respectively. Initially, the automated Dmax determination by GNOM was used. However, we chose to manually increase Dmax as it was underestimated judging by the appearance of the P(r) plot (discussed below).
Coordinates were manipulated with PDB-MODE (39). Rigid-body modeling was performed using SASREF (40). The starting model was derived from a single chain of PPR10 extracted from PDB 4M59 (30), broken into seven ‘domains’ each consisting of three PPR motifs. RNA was included in the domain definition by placing the appropriate trinucleotide adjacent to the predicted RNA-binding residues of each domain. For the MBP-PPR10 data, PDB entry 1MBP was used as an additional protein domain. Two distance constraints per domain were added to ensure physically reasonable modeling, as the PPR motifs in PPR10 form a continuous array. CRYSOL (41) and MOLEMAN2 (42) were used to evaluate model quality by quantifying how representative the atomic models are of the data and P(r) distribution plots. Molecular graphics were created with PyMOL (Schroedinger LLC).
RESULTS
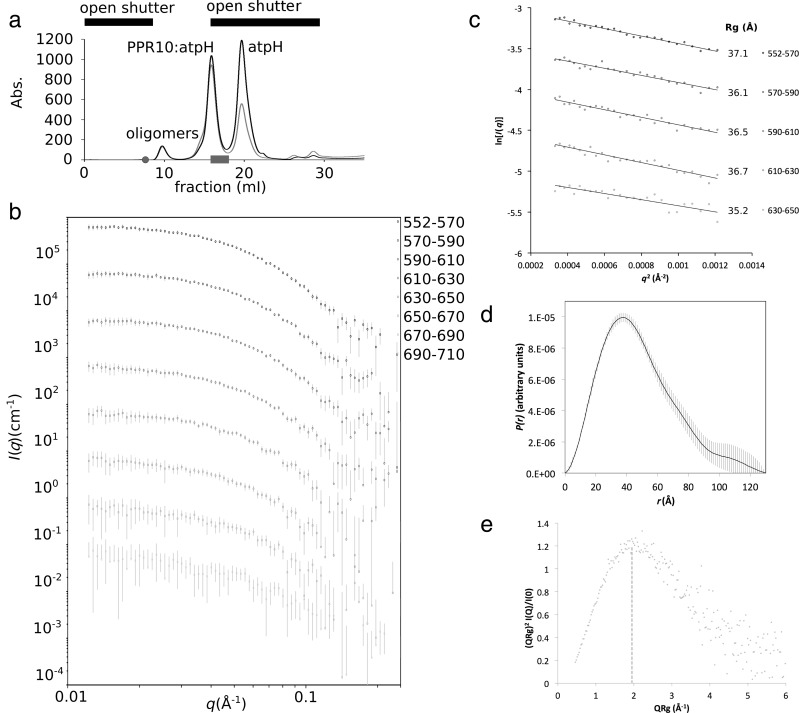
We have generated a plausible model of the solution structure of ZmPPR10 in complex with the 17 nucleotide minimal binding site of the maize atpH transcript informed by a combination of SAXS data, circular dichroism spectroscopy, and pre-existing bioinformatic, biochemical and crystallographic data. The secondary structure content of PPR10 was estimated to be predominantly α-helical (84%) from circular dichroism as expected for the anti-parallel α-helical topology of a PPR motif, and comparable with calculations based on the crystal structures (71% α-helix). Preliminary SAXS measurements on PPR10 alone and after incubation with atpH RNA yielded complex uninterpretable scattering profiles due to significant aggregation. We therefore employed size-exclusion chromatography-coupled synchrotron small-angle X-ray scattering (SEC-SY-SAXS; Figure 1). This method allowed us to separate the various complexes inline, identify their content from their A260/A280 ratios and examine the solution structure of specific SEC fractions (Figure 1a). A significant challenge to this method was encountered due to the earliest eluting aggregated proteins adhering to the sample capillary at the point of X-ray exposure, thus tainting all further images. By carefully timing the opening of the X-ray shutter to only measure regions of interest and appropriate background segments it was possible to measure high quality SAXS on the protein:RNA complex. Scattering data were obtained for a monodisperse peak with an A260/A280 ratio and molecular size commensurate with a monomeric PPR10:atpH complex (Figure 1b). Data collection and analysis are summarized in Tables 1 and 2, respectively.
Figure 1.
SEC-SY-SAXS of PPR10:atpH. (a) Size exclusion chromatography trace of PPR10:atpH showing peaks corresponding to oligomers, PPR10:atpH complex and unbound atpH RNA. Black bars indicate the part of the chromatogram for which SAXS was collected by opening the beamline shutter. The grey circle indicates position where the first data frame was recorded, and the bar, the region corresponding to frames 552–710. (b) Scattering profiles of the PPR10:atpH complex averaged across 20 frame segments, corresponding to the second half of the PPR10:atpH peak (grey bar in (a)). (c) Guinier analysis of the low-q scattering data for the five most strongly scattering 20 frame segments indicates a radius of gyration of 36 Å. (d)P(r) distribution for frames 552–710 indicates a maximum dimension of 125 Å. (e) Dimensionless Kratky plot is diagnostic of a globular but somewhat extended protein.
Table 1. SAXS data collection and refinement statistics.
| PPR10:atpH | MBPPPR10:atpH | |
|---|---|---|
| Data collection | ||
| Instrument | Australian synchrotron SAXS/WAXS beamline | Australian synchrotron SAXS/WAXS beamline |
| Beam geometry (μm) | 120 | 120 |
| Wavelength (Å) | 1.033 | 1.033 |
| q range (A−1) | 0.01–0.21 | 0.01–0.21 |
| Exposure per frame (s) | 2 | 2 |
| Flow-rate (ml min−1) | 0.1 | 0.5 |
| Concentration (mg ml−1) | 6–0 | 6–0 |
| Temperature (K) | 288 | 288 |
| Structural parameters | ||
| I(0) (cm−1) [from P(r)] | 0.007 ± 0.00 | 0.01 ± 0.00 |
| Rg (Å) [from P(r)] | 36.19 ± 1.08 | 63.2 ± 0.01 |
| I(0) (cm−1) (from Guinier) | 0.008 ± 0.00 | 0.009 ± 0.00 |
| Rg (Å) (from Guinier) | 37.39 ± 2.30 | 61.20.78 ± 1.87 |
| Dmax (Å) | 125 | 250 |
| Molecular-mass determination | ||
| Porod invariant (Vp)(Å3) | 165,328 | 203,698 |
| Estimated Mr from Vp (Da) | 97,251 | 161,900 |
| Calculated Mr (Da) | 87,882 | 135,607 |
Table 2. Guinier and Porod analysis of peak fractions from PPR10:atpH and MBP-PPR10:atpH SEC-SY-SAXS.
| Frame no. | I(0) | Radius of gyration, Rg (Å) | Guinier range (q) | Maximum dimension, Dmax (Å) | P(r) Range (q) | Porod invariant, Vp (Å3) | Molecular mass, Mr, from Vp (Da) | Oligomeric state from Mr |
|---|---|---|---|---|---|---|---|---|
| 552–570 | 0.007 ± 0.000 | 37.394 ± 2.299 | 0.014–0.034 | 130.1 | 0.014–0.25 | 165,328 | 97,251 | 1.11 |
| 570–590 | 0.006 ± 0.000 | 36.055 ± 3.811 | 0.014–0.034 | 126.2 | 0.014–0.25 | 162,893 | 95,819 | 1.09 |
| 590–610 | 0.005 ± 0.000 | 36.456 ± 3.967 | 0.014–0.034 | 127.6 | 0.014–0.25 | 165,752 | 97,501 | 1.11 |
| 610–630 | 0.004 ± 0.000 | 36.773 ± 5.734 | 0.014–0.034 | 126.4 | 0.014–0.25 | 158,006 | 92,944 | 1.06 |
| 630–650 | 0.003 ± 0.000 | 35.034 ± 9.020 | 0.014–0.034 | 123.9 | 0.014–0.25 | 187,843 | 110,495 | 1.26 |
| 650–670 | 0.002 ± 0.000 | 38.532 ± 5.436 | 0.014–0.034 | 130.0 | 0.014–0.25 | 162,642 | 95,671 | 1.09 |
| 215–220 | 0.012 ± 0.000 | 67.459 ± 6.188 | 0.012–0.025 | 250.0 | 0.012–0.35 | 210,905 | 124,061 | 0.94 |
| 221–226 | 0.012 ± 0.000 | 61.199 ± 1.868 | 0.012–0.025 | 250.0 | 0.012–0.35 | 203,698 | 135,607 | 1.03 |
| 227–233 | 0.007 ± 0.000 | 55.053 ± 1.385 | 0.012–0.025 | 250.0 | 0.012–0.35 | 204,390 | 143,800 | 1.09 |
| 234–239 | 0.005 ± 0.000 | 54.975 ± 2.248 | 0.012–0.025 | 250.0 | 0.012–0.35 | 184,072 | 129,100 | 0.98 |
| 240–245 | 0.003 ± 0.000 | 55.947 ± 3.515 | 0.012–0.025 | 250.0 | 0.012–0.35 | 160,763 | 112,200 | 0.85 |
Forward scatter,I(0), radius of gyration, Rg determined using PRIMUS. Maximum dimension, Dmax, and Porod volume,Vp, and molecular mass, Mr, determined using GNOM. Oligomeric state (n) determined from the ratio of Mr(Vp) to Mr (theoretical).
PPR10:atpH displayed a stable scattering profile for a globular protein throughout the SEC peak. Guinier analysis of the low-q data indicated a consistent radius of gyration (Rg) close to 36 Å for the most concentrated fractions (Table 2, Figure 1c). An upturn at low-q became evident in the scattering profile during later fractions, possibly due to adhesion of protein on the capillary (Figure 1b), rendering Guinier analysis of these lower concentration samples impossible. Thus, data from the peak fraction (frames 552–570) were averaged and used for subsequent analysis.
The P(r) distribution (Figure 1d) is indicative of a globular structure with an extended tail: an asymmetric distribution with a maximum at 40 Å with a shoulder at 20 Å and a steady decline through the longer distances with shoulders at 80 and 110 Å and a Dmax of ca. 125 Å. A dimensionless Kratky plot contains a peak at 1.95 with a height of around 1.2 indicating a somewhat elongated, but generally globular protein (with reference to (43)) that is suitable for Porod analysis (Figure 1e). The molecular mass (Mr) of the scattering particle, determined from the Porod invariant (Vp), (44), corresponds to a monodisperse, 1:1 PPR10:atpH complex (Tables 1 and 2).
As the monomeric state of the complex as determined by AUC, REMSA and now SAXS contrasts with the reported dimeric crystal structure, we next sought to build a molecular model—taking into account the limitations of data quality—that best reflects the scattering data. We first examined the crystal structures of ZmPPR10 in the absence (4M57) and presence (4M59) of psaJ RNA. Initially, we generated candidate models of the observed dimers, and monomers extracted from the crystal structures, and compared their minimized fit with the observed scattering profiles, and compared their distance distributions with the observed P(r) distribution (Figure 2). The relatively high χ values combined with the lack of common features in the distance distribution (e.g. a maximum at 60 Å compared to 40 Å) led us to conclude that there were notable differences between the solution structure and the crystallographic models.
Figure 2.
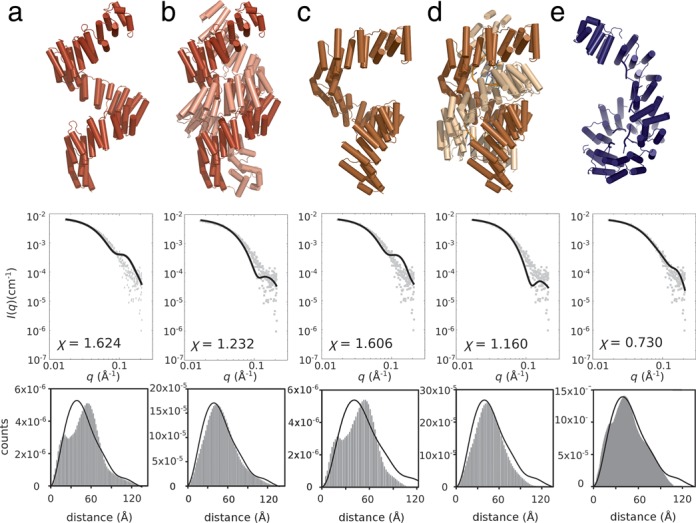
Validation of candidate molecular models against PPR10:atpH scattering data. Top row: Molecular model. Middle row: Optimized fit to data. Bottom row: Reconstructed pair distribution plots from the respective models (solid histogram) compared with the experimentally derived P(r) distribution (black line). (a) apoPPR10 monomer extracted from PDB 4M57; (b) apoPPR10 dimer from PDB 4M57; (c) PPR10:psaJ monomer extracted from PDB M459; (d) PPR10:psaJ dimer from PDB 4M59; (e) The best-fitting SASREF model. Fitting parameters indicate a considerably better fit for the SASREF model.
Armed with this knowledge we generated rigid body models from PPR10 broken into seven domains, with RNA fragments attached, and refined against the scattering data. Highly consistent results were obtained over multiple independent modeling runs for PPR10:atpH (Figure 2e) presented as a monomeric complex. The model has an excellent χ fitting parameter (0.73). The best-fitting model is a right-handed superhelical array with the C-terminal region forming a toroid. However, the pseudosymmetric nature of the PPR10 molecule—a superhelical array—made it impossible to objectively assign either end as the N- or C-terminus.
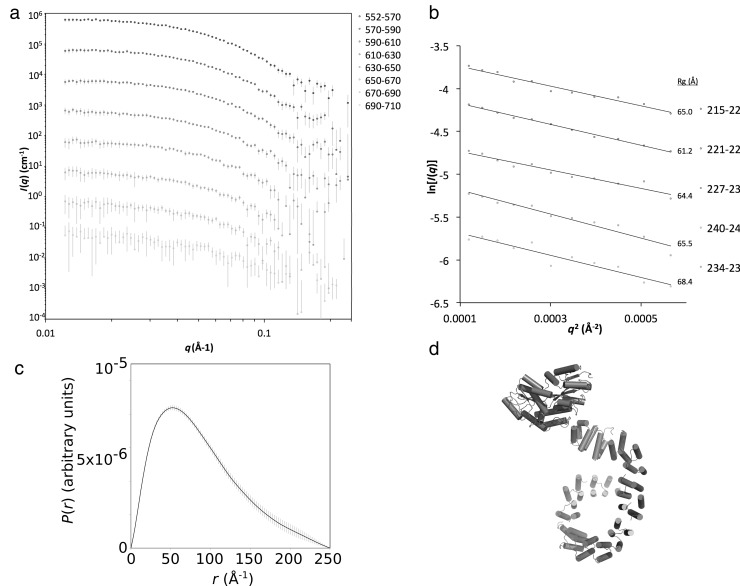
To address this problem, we repeated the data collection procedure with an N-terminal MBP-PPR10 fusion protein. Scattering data were analysed (Figure 3) revealing increases of Rg to 61 Å and Dmax to 250 Å, as expected from addition of an MBP domain. Rigid body modeling of this complex produced a satisfactory solution (χ = 0.78) for a protein with MBP at the N-terminus and a toroidal arrangement at the C-terminus, thus supporting our model of the untagged protein, which is used in the following comparative analysis.
Figure 3.
SEC-SY-SAXS analysis of MBP-PPR10 fusion protein:atpH complex. (a) Scattering profiles of averaged and background subtracted five-frame region of the peak fraction of MBP-PPR10:atpH. (b) Guinier analysis and (c)P(r) distribution for the 215–220 averaged scattering profile. (d) The best-fitting SASREF model for the MBP fusion protein.
Alignment of monomeric PPR10 structures from three different available structural contexts (a monomer extracted from the apo-PPR10 dimer (PDB 4M57); a monomer extracted from the psaJ-bound dimer (PDB 4M59) and the monomeric atpH-bound solution structure, this work) reveals two notable regions of structural rearrangement (Figure 4a) that are also discernable from the scattering profiles and distance distributions (Figure 2). While the C-terminal region of the apo- and psaJ-bound monomers aligns well, the four N-terminal PPR motifs are arranged differently. Conversely, the psaJ- and atpH-bound conformations only differ significantly around the four C-terminal PPR motifs.
Figure 4.

Global conformation of PPR10. (a) Dynamic structure of a PPR10 monomer:A single PPR10 molecule extracted from the RNA-free dimer structure (4M57 chain A, red cartoon), the PPR10:psaJ dimer (4M59 chain A, gold) and the monomeric PPR10:atpH solution structure (this work, blue) can be superimposed effectively (RMSD ranging from 7 to 11 Å including Cα atoms from most of the structure: black lines link superimposed Cα atoms between two structures). The N-terminal region (oriented at the top) differs between apo- and psaJ-bound proteins, while only the C-terminus differs significantly between psaJ- and atpH-bound proteins. Global differences between superposed structures are indicated by black arrows. (b)PsaJ RNA coordination by PPR10 motifs 6 and 7 in the crystal structure (4M59; chain A orange cartoon; chain B, beige/gray surface). Residues Asn284, Asp314 and Asn319, Asn349 (positions 6 and 1′ for motifs 6 and 7 respectively) are known to dictate RNA specificity, however Asn349 is displaced from RNA binding by the dimeric interface. (c) The solution structure of PPR10:atpH illustrates that the arrangement of the PPR motifs (colored blue to red from N-terminus) is conducive to ssRNA binding (black cartoon) as a monomer (Note: breaks in RNA chain are a consequence of the rigid body modeling approach used).
DISCUSSION
This study shows how the coupling of size-exclusion chromatography with synchrotron SAXS allows productive study of challenging macromolecular complexes, particularly when studying a well-characterized system with ample orthogonal information, from genomic, biochemical and crystallographic sources.
We have determined that the solution structure of ZmPPR10 complexed with a fragment of atpH is not compatible with the dimeric form observed in the PPR10:psaJ crystal structure, thus confirming previous analytical ultracentrifugation analyses. The nature of recognition of atpH and psaJ has been well characterized (6,13,19), and the PPR–RNA recognition code has been validated biochemically for the N-terminus/5′ end of the PPR–RNA duplex by mutating residues Asn284, Asp314 and Asn319, Asn349 in PPR motifs 6 and 7, and observing a predictable change in binding specificity (19). The dimeric conformation observed in the psaJ-bound PPR10 structure is not fully compatible with this result, as one of the key residues in motif 7 does not engage the RNA (Figure 4b). Indeed the structure of the helix bearing Asn349 is highly disturbed, presenting considerably different positions in the two subunits of the crystal structure: in both cases Asn319 is well over 10 Å distant from the RNA. It seems likely that the presence of the other subunit of the dimer close to this region is a cause of the disruption of this binding site (Figure 4b).
In the SAXS structure, the more highly overwound superhelix of the C-terminal region would sterically hinder dimerisation of the protein (Figure 4c). In this tight, toroid-like part of the structure there are intramolecular interactions between residues of the C-terminal repeats and repeats earlier in the sequence, which in the dimeric crystal structure are involved in intermolecular interactions instead. Our results are consistent with the 1:1 stoichiometry established by analytical ultracentrifugation for a PPR10-atpH complex (19). That a similar architecture would apply to the PPR10-psaJ complex in solution is supported by the comigration of PPR10-atpH complexes and PPR10-psaJ complexes in native gels and by the extensive sequence conservation across the length of the psaJ and atpH RNAs.
A question is raised about whether all of these unambiguously observed configurations can be correct. The answer may lie in the necessarily convoluted dynamic process that is required for PPR10 to dissociate from a dimeric state to a monomeric state and to bind sequence specifically to RNA in a cellular context involving other binding partners and RNA secondary structures (6).
It is unclear whether the dimeric PPR10:psaJ structure observed crystallographically represents an intermediate state in the dynamic process of RNA recognition by PPR10, or if it represents a dead-end complex formed at the high concentrations required for crystallisation. At a late point in our analysis, a paper became available that raises further doubt regarding the dimeric nature of the complex (45). Here, the authors of the PPR10 crystal structures determine that an N-terminal truncation and a point mutation that were essential for crystallogenesis in fact drive dimerization. Our wild-type PPR10 protein makes a more physiologically relevant model for the interaction between PPR10 and target RNA, taking into account the miniscule cellular concentration of PPR10 (with respect to the micromolar dissociation constant for dimerisation) and the large excess of ‘non-specific’ single-stranded RNA. While it remains plausible that dimeric apoPPR10 binds to an RNA transcript in low-affinity mode causing a structural rearrangement that dissociates the dimer and allows the high-affinity monomeric complex to form, it is likely that in the cell a monomeric PPR10 interacts with bulk single-stranded RNA before exchanging partners to achieve high-affinity binding to its target RNA motif as a monomeric protein:RNA complex similar to the solution structure presented here.
Despite their well-described modularity and relatively predictable RNA-binding specificity, this example of a native PPR protein reveals a potentially convoluted process of RNA binding. If the promise for PPR proteins to be applied as ssRNA-binding tools in biotechnology is to be maximized, a fuller understanding of the detailed structural changes that occur on RNA binding is essential.
Footnotes
Present address: Will A. Stanley, Northern Institute for Cancer Research, University of Newcastle upon Tyne, UK.
FUNDING
Australian Research Council (Discovery Project DP120102870 to I.D.S., A.B., C.S.B.) Pearl Technologies Ltd; University of Western Australia (scholarship for B.S.G.). Funding for open access charge: The University of Western Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Small I.D., Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 2.Barkan A., Small I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 3.Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B., et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2000;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choquet Y. 5′ and 3′ ends of chloroplast transcripts can both be stabilised by protein ‘caps’: a new model for polycistronic RNA maturation. EMBO J. 2009;28:1989–1990. doi: 10.1038/emboj.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 8.Williams-Carrier R., Kroeger T., Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delannoy E., Stanley W.A., Bond C.S., Small I.D. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 10.Davies S.M., Lopez Sanchez M.I., Narsai R., Shearwood A.M., Razif M.F., Small I.D., Whelan J., Rackham O., Filipovska A. MRPS27 is a pentatricopeptide repeat domain protein required for the translation of mitochondrially encoded proteins. FEBS Lett. 2012;586:3555–3561. doi: 10.1016/j.febslet.2012.07.043. [DOI] [PubMed] [Google Scholar]
- 11.Zoschke R., Kroeger T., Belcher S., Schottler M.A., Barkan A., Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72:547–558. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- 12.Zoschke R., Qu Y., Zubo Y.O., Borner T., Schmitz-Linneweber C. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 2013;126:403–414. doi: 10.1007/s10265-012-0527-1. [DOI] [PubMed] [Google Scholar]
- 13.Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkan A., Walker M., Nolasco M., Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentolila S., Alfonso A.A., Hanson M.R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akagi H., Nakamura A., Yokozeki-Misono Y., Inagaki A., Takahashi H., Mori K., Fujimura T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. App. Gen. 2004;108:1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez-Marcos J.F., Dal Pra M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., et al. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell. 2007;19:196–210. doi: 10.1105/tpc.105.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cushing D.A., Forsthoefel N.R., Gestaut D.R., Vernon D.M. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta. 2005;221:424–436. doi: 10.1007/s00425-004-1452-x. [DOI] [PubMed] [Google Scholar]
- 19.Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takenaka M., Zehrmann A., Brennicke A., Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8:e65343. doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8:e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipovska A., Rackham O. Pentatricopeptide repeats: modular blocks for building RNA-binding proteins. RNA Biol. 2013;10:1426–1432. doi: 10.4161/rna.24769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi Y., Nakamura T., Small I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014;78:772–782. doi: 10.1111/tpj.12377. [DOI] [PubMed] [Google Scholar]
- 24.Fujii S., Bond C.S., Small I.D. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringel R., Sologub M., Morozov Y.I., Litonin D., Cramer P., Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 26.Howard M.J., Lim W.H., Fierke C.A., Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16149–16154. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ban T., Ke J., Chen R., Gu X., Tan M.H.E., Zhou X.E., Kang Y., Melcher K., Zhu J.-K., Xu H.E. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J. Biol. Chem. 2013;288:31540–31548. doi: 10.1074/jbc.M113.496828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gully B.S., Shah K.R., Lee M., Shearston K., Smith N.M., Sadowska A., Blythe A.J., Bernath-Levin K., Stanley W.A., Small I.D., et al. The design and structural characterisation of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D. 2015;71 doi: 10.1107/S1399004714024869. doi:10.1107/S1399004714024869. [DOI] [PubMed] [Google Scholar]
- 29.Ke J., Chen R.Z., Ban T., Zhou X.E., Gu X., Tan M.H., Chen C., Kang Y., Brunzelle J.S., Zhu J.K., et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 2013;20:1377–1382. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 30.Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W., et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 31.Beckham S.A., Brouwer J., Roth A., Wang D., Sadler A.J., John M., Jahn-Hofmann K., Williams B.R.G., Wilce J.A., Wilce M.C.J. Conformational rearrangements of RIG-I receptor on formation of a multiprotein:dsRNA assembly. Nucleic Acids Res. 2013;41:3436–3445. doi: 10.1093/nar/gks1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenfield N.J., Fasman G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 33.Morrisett J.D., David J.S.K., Pownall H.J., Gotto A.M. Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12:1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 34.Kirby N.M., Mudie S.T., Hawley A.M., Cookson D.J., Mertens H.D.T., Cowieson N., Samardzic-Boban V. A low-background-intensity focusing small-angle X-ray scattering undulator beamline. J. Appl. Crystallogr. 2013;46:1670–1680. [Google Scholar]
- 35.Brookes E., Perez J., Cardinali B., Profumo A., Vachette P., Rocco M. Fibrinogen species as resolved by HPLC-SAXS data processing within the UltraScan Solution Modeler (US-SOMO) enhanced SAS module. J. Appl. Crystallogr. 2013;46:1823–1833. doi: 10.1107/S0021889813027751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 38.Semenyuk A.V., Svergun D.I. GNOM - a program package for small-angle scattering data processing. J. Appl. Crystallogr. 1991;24:537–540. [Google Scholar]
- 39.Bond C.S. Easy editing of Protein Data Bank formatted files with EMACS. J. Appl. Crystallogr. 2003;36:350–351. [Google Scholar]
- 40.Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svergun D., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 42.Kleywegt G. J., Jones T. A. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 43.Durand D., Vives C., Cannella D., Perez J., Pebay-Peyroula E., Vachette P., Fieschi F. NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 2010;169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 44.Fischer H., de Oliveira Neto M., Napolitano H.B., Polikarpov I., Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010;43:101–109. [Google Scholar]
- 45.Li Q., Yan C., Xu H., Wang Z., Long J., Li W., Wu J., Yin P., Yan N. Examination of the dimerization states of the single-stranded RNA recognition Protein Pentatricopeptide Repeat 10 (PPR10) J. Biol. Chem. 2014;289:31503–31512. doi: 10.1074/jbc.M114.575472. [DOI] [PMC free article] [PubMed] [Google Scholar]