Abstract
Emerging lines of evidence have shown that extracellular vesicles (EVs) mediate cell-to-cell communication by exporting encapsulated materials, such as microRNAs (miRNAs), to target cells. Endothelial cell-derived EVs (E-EVs) are upregulated in circulating blood in different pathological conditions; however, the characteristics and the role of these E-EVs are not yet well understood. In vitro studies were conducted to determine the role of inflammation-induced E-EVs in the cell-to-cell communication between vascular endothelial cells and pericytes/vSMCs. Stimulation with inflammatory cytokines and endotoxin immediately induced release of shedding type E-EVs from the vascular endothelial cells, and flow cytometry showed that the induction was dose dependent. MiRNA array analyses revealed that group of miRNAs were specifically increased in the inflammation-induced E-EVs. E-EVs added to the culture media of cerebrovascular pericytes were incorporated into the cells. The E-EV-supplemented cells showed highly induced mRNA and protein expression of VEGF-B, which was assumed to be a downstream target of the miRNA that was increased within the E-EVs after inflammatory stimulation. The results suggest that E-EVs mediate inflammation-induced endothelial cell-pericyte/vSMC communication, and the miRNAs encapsulated within the E-EVs may play a role in regulating target cell function. E-EVs may be new therapeutic targets for the treatment of inflammatory diseases.
Cell-to-cell communication is mediated by secreted bioactive molecules, such as short form peptides, proteins, lipids, and nucleic acids. These small molecules are commonly released by cells and bind to specific receptors on target cells, which induces intracellular signaling and changes the target cell's pathophysiological state. Extracellular vesicles (EVs), which include microparticles, microvesicles, and exosomes1,2,3,4, are released from different cell types, and emerging evidence suggests that EVs function as carriers of these bioactive molecules5,6,7,8.
Clinically, EVs are found in circulating blood, and the number of EVs is elevated in acute and chronic inflammatory diseases, such as sepsis, stroke, preeclampsia, atherosclerosis, diabetes mellitus, and metabolic syndrome9,10,11,12,13,14. Vascular endothelial cells are thought to be one of the major cell types that release EVs into the blood stream15. The number of endothelial-derived EVs (E-EVs) circulating in the blood stream correlates with the severity of disease; however, the pathophysiological significance of E-EVs is largely unknown12.
MicroRNAs (miRNAs) are small, single-stranded, non-coding RNAs that are transcribed in the nucleus. They are processed by the enzymes Drosha and Dicer, incorporated into RNA-induced silencing complexes, and mediate the translational inhibition or degradation of target mRNAs16,17. Many miRNAs have been shown to play key roles in pathophysiological processes18,19. In particular, the inflammation-related miRNAs, miR-101, miR-144, and miR-155, were reported to modulate protein biogenesis in lung epithelial and endothelial cells20,21. These miRNAs can be carried by E-EVs; however, their roles in E-EV-mediated cell-to-cell communication are not yet known.
Vascular endothelial cells and pericytes/vascular smooth muscle cells (vSMCs) are juxtapositioned to each other in blood vessels22. The interactions between these two cell types are important for the regulation of vascular integrity, and perturbation of their interaction has been observed in many diseases, including inflammatory diseases that cause vascular dysfunction, such as disturbance of the blood brain barrier (BBB) in cerebral blood vessels23,24,25,26. Here, we aimed to determine the involvement of EVs in cerebrovascular endothelial cell-pericyte communication in inflammatory disease. We found that the E-EVs induced by inflammatory stimuli carry numerous specific miRNAs and can induce pericyte responses to endothelial cells. These results suggest that E-EVs are an important mediator of vascular cell communication in inflammatory conditions.
Results
Induction of inflammatory responses in cerebrovascular endothelial cells
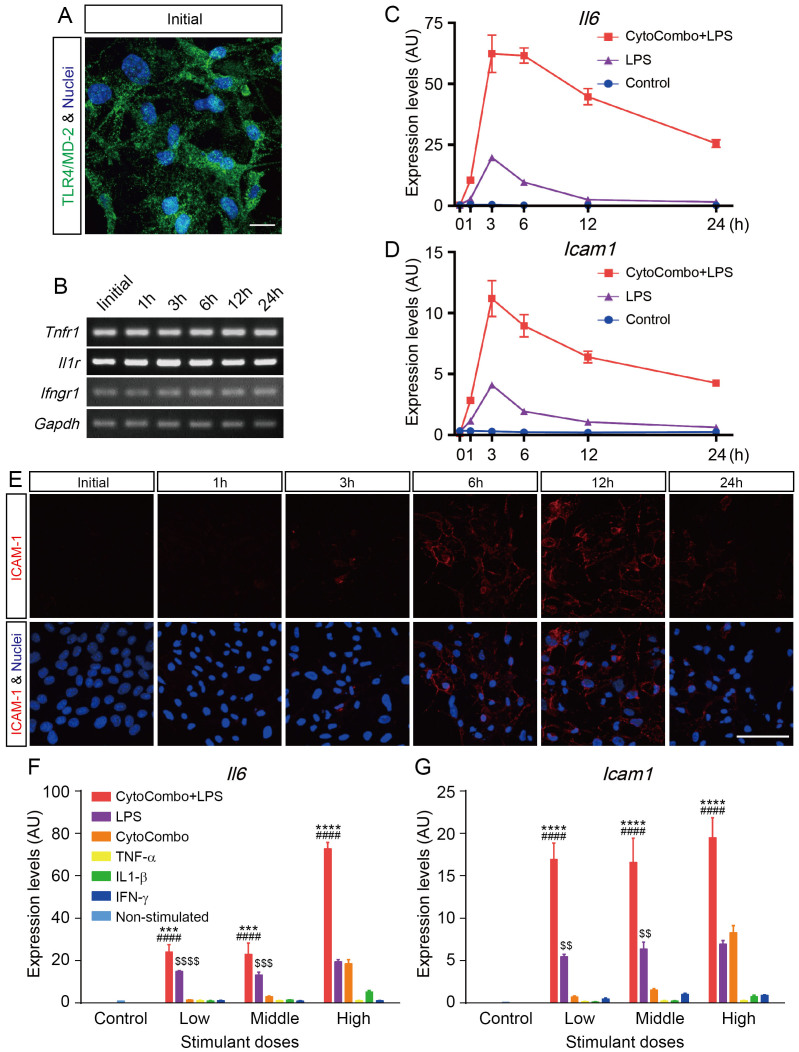
To analyze the pathobiology of E-EVs released in inflammatory conditions, we developed a reproducible in vitro system to induce the production of E-EVs from b.End5 cells, a cerebrovascular endothelial cell line. First, we confirmed that b.End5 cells expressed the LPS receptor TLR4/MD-2 complex under unstimulated conditions by immunocytofluorescence (Fig. 1a). The mRNAs of the inflammatory cytokine receptors Tnfr1 (for TNF-α), Il1r (for IL-1β), and Ifngr1 (for IFN-γ) were detected in unstimulated b.End5 cells by conventional RT-PCR (Fig. 1b). The gene expression levels were consistent up to 24 hours after stimulation with a high dose of CytoCombo + LPS (a mixture of TNF-α, IL-1β, IFN-γ, and LPS; Supplementary Table 1).
Figure 1. Inflammation-related receptor gene and protein expression levels in cerebrovascular endothelial cells.
(a) Expression of the TLR4/MD-2 protein complex was confirmed by immunocytofluorescence (Green). Unstimulated b.End5 cells constitutively express the TLR4/MD-2 protein complex. Nuclei are stained with Hoechst (Blue). Scale bar, 10 μm. (b) Analysis of Tnfr1, Il1r, and Ifngr1 expression by conventional RT-PCR. Unstimulated b.End5 cells constitutively express Tnfr1, Il1r, and Ifngr1, and these genes are continuously expressed after 24 hours of stimulation with CytoCombo + LPS. (c and d) Inflammatory gene expression levels in b.End5 cells. Il6 (c) and Icam1 (d) are markers of inflammatory responsive genes (n = 4 for each time point). Both genes peaked at 3 hours after stimulation. (e) ICAM-1 protein expression levels were confirmed by immunocytofluorescence. ICAM-1 protein expression levels were well correlated with the Icam1 mRNA expression pattern. Scale bars, 50 μm. (f and g) Inflammatory gene expression levels in b.End5 cells incubated with various stimulants. The inflammatory responsive genes Il6 (f) and Icam1 (g) were measured at 3 hours after stimulation (n = 4 for each time point). Cells stimulated with CytoCombo + LPS showed the most intensive inflammation. Non-stimulated control indicates basal expression level of marker genes. **** P < 0.0001, and *** P < 0.001, CytoCombo + LPS vs. LPS; ####P < 0.0001, CytoCombo + LPS vs. CytoCombo; $$$$P < 0.0001, $$$P < 0.001, and $$P < 0.01, LPS vs. CytoCombo.
As inflammatory stimuli have been reported to upregulate IL-6 and ICAM-1 expression levels27,28, we determined the inflammatory responses in b.End5 cells to inflammatory cytokine and endotoxin exposure by measuring Il6 and Icam1 gene expression. In real-time PCR analyses, a high dose of CytoCombo + LPS (Supplementary Table 1) induced Il6 and Icam1 mRNA expression, which peaked after 3 hours of stimulation (Fig. 1c and d). LPS alone also induced Il6 and Icam1 mRNA expression, but at lower levels than CytoCombo + LPS (Fig. 1c and d). In accordance with the Icam1 mRNA expression (Fig. 1d), the immunoreactivity of ICAM-1 in b.End5 cells was detectable at 3 hours and peaked at 6 to 12 hours after CytoCombo + LPS stimulation (Fig. 1e).
To determine the optimal stimulatory conditions for inducing inflammatory responses in b.End5 cells, the dose effects of the inflammatory stimuli were determined by real-time PCR analyses of Il6 and Icam1 mRNA at 3 hours after stimulation. The expression of Il6 and Icam1 mRNA was significantly induced by CytoCombo + LPS, CytoCombo (a mixture of TNF-α, IL-1β, and IFN-γ), and LPS (Fig. 1f and g). However, the mRNA levels induced by CytoCombo + LPS were significantly higher than those induced by the other stimuli at all doses examined. The mRNA levels induced by the low and middle doses of LPS were significantly higher than those induced by the low and middle doses of CytoCombo, whereas the high doses of both stimuli induced equivalent mRNA expression levels (Fig. 1f and g). The induction of Il6 and Icam1 mRNA was not significant when b.End5 cells were stimulated with either cytokine alone. As an apoptotic cell death triggered by the inflammatory cytokines and LPS29,30, has been reported to induce the release of apoptotic bodies4, we examined apoptosis in b.End5 cells that were exposed to inflammatory stimuli. As compared with non-stimulated control, 3-hour exposure to any kinds of stimulants did not significantly increase the cleaved caspase 3-positive apoptotic cell. Twenty four-hour exposure to the stimulants except for that of low and middle dose of CytoCombo significantly increased cleaved caspase 3-positive apoptotic cell (Supplementary Figure 1a and b). Based on these data, we exposed b.End5 cells to CytoCombo + LPS, LPS, and CytoCombo for 3 hours when induction of apoptosis is still not evident, and then examined the inflammatory simulation-induced E-EV release from these cells.
Inflammation induced E-EV release from cerebrovascular endothelial cells
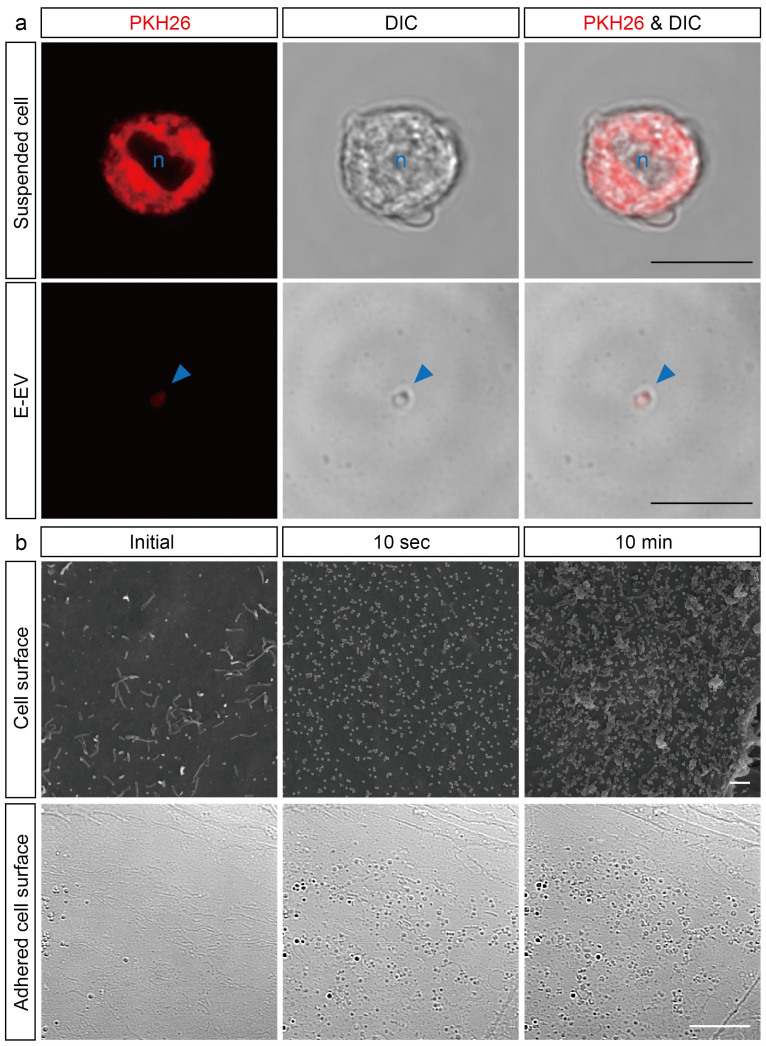
We next examined E-EV release from b.End5 cells that were stimulated by CytoCombo + LPS. To identify the E-EVs released from b.End5 cells, the plasma membranes of these cells were prelabeled with PKH26 and these cells were then exposed to the inflammatory stimuli. Three hours after exposure to a high dose of CytoCombo + LPS, PKH26 fluorescence was detected on the plasma membrane of b.End5 cells (Fig. 2a, upper row). Then, we identified PKH26-positive objects of submicron to micron size within the E-EVs fractionated from the culture medium of PKH26-positive b.End5 cells after a 3-hour stimulation with the high dose of CytoCombo + LPS, and confirmed that these fractionated materials were b.End5-derived globules. We further characterized them using the extracellular vesicle markers, such as vascular endothelial growth factor receptor 2 (VEGFR-2) and CD62E (E-selectin)31. These globular objects in E-EV fraction were immunoreactive for VEGFR-2 and CD62E under confocal microscopy (Supplementary Figure 2). In addition, immuno-reactive bands of VEGFR-2 and CD62E were enriched in the E-EV fraction than in b.End5 cell lysates by western blot analyses (Supplementary Figure 3).
Figure 2. Inflamed cerebrovascular endothelial cells produce shedding-type E-EVs.
(a) Treatment of inflamed cells with PKH26, a plasma membrane-intercalating fluorescent reagent, clearly demonstrates that E-EVs are released from the inflamed endothelial cell membranes. Suspended b.End5 cells stained with PKH26 emit red fluorescence (Upper row). Azure-blue n indicates nucleus. PKH26-positive E-EVs are present in the culture medium (Bottom row). Red fluorescence is colocalized to the micron-sized vesicles as observed by differential interference contrast microscopy (DIC). The azure-blue arrowhead shows an E-EV. Scale bar, 10 μm. (b) Scanning electron microscopy (SEM) images showing that E-EVs are shed from the cell surface immediately after stimulation by CytoCombo + LPS (Upper row). The acute response implies that E-EV production is a non-genomic reaction. DIC image showing E-EVs present on the adhered cell surface (Bottom row), suggesting that E-EVs are bilaterally produced on both the luminal and abluminal surfaces of endothelial cells in the tube formed by the blood vessels in inflammatory conditions in vivo. Scale bar, 10 μm (a and b lower), and 1 μm (b upper).
The surface of the plasma membrane of the inflamed endothelial cells was examined by scanning electron microscopy (SEM). In most of unstimulated b.End5 cells (Initial), we observed few cellular processes like microvilli and limited number of sheddings on the surface of the plasma membrane (Fig. 2b, upper), whereas small fraction of unstimulated cells (2.38 ± 5.83%) showed similar changes as seen in those with stimulation. At 10 seconds after the addition of high dose CytoCombo + LPS to the culture medium, many sheddings were emerged on the surface of plasma membrane, and the microvilli turned to be obscure in all stimulated cells (100%; Fig. 2b, upper row). Higher magnification images further revealed the tiny and dome-shaped swellings (less than 0.1 μm in diameter) on the surface of plasma membrane, which structure may correspond to the early phase of sheddings (Supplementary Figure 4a). The sheddings were further increased at 10 minutes, and most of them ranged between 0.1 μm and 0.4 μm in diameter (Fig. 2b, upper row). At higher magnification, sheddings of larger-size (≥0.3 μm) and those of aggregated form (up to 3 μm) were increased as compared with those at 10-second of stimulation (Supplementary Figure 4b). In addition, similar shedding-like structures were observed on the bottom surface of the cells that were exposed to high-dose CytoCombo + LPS by using the interference contrast microscopy (DIC) mode of a confocal microscope (Fig. 2b, bottom row). These data strongly suggest that E-EV production is mainly membrane-shedding type15,32 and may be partly from multivesicular bodies (MVBs)33, and the plasma membrane shedding of E-EVs induced by inflammatory stimulants is very acute. In addition, our findings may support the possibility that E-EVs could be produced bilaterally, on both the luminal and abluminal surfaces, from inflammation-stimulated endothelial cells in cerebral blood vessels in vivo.
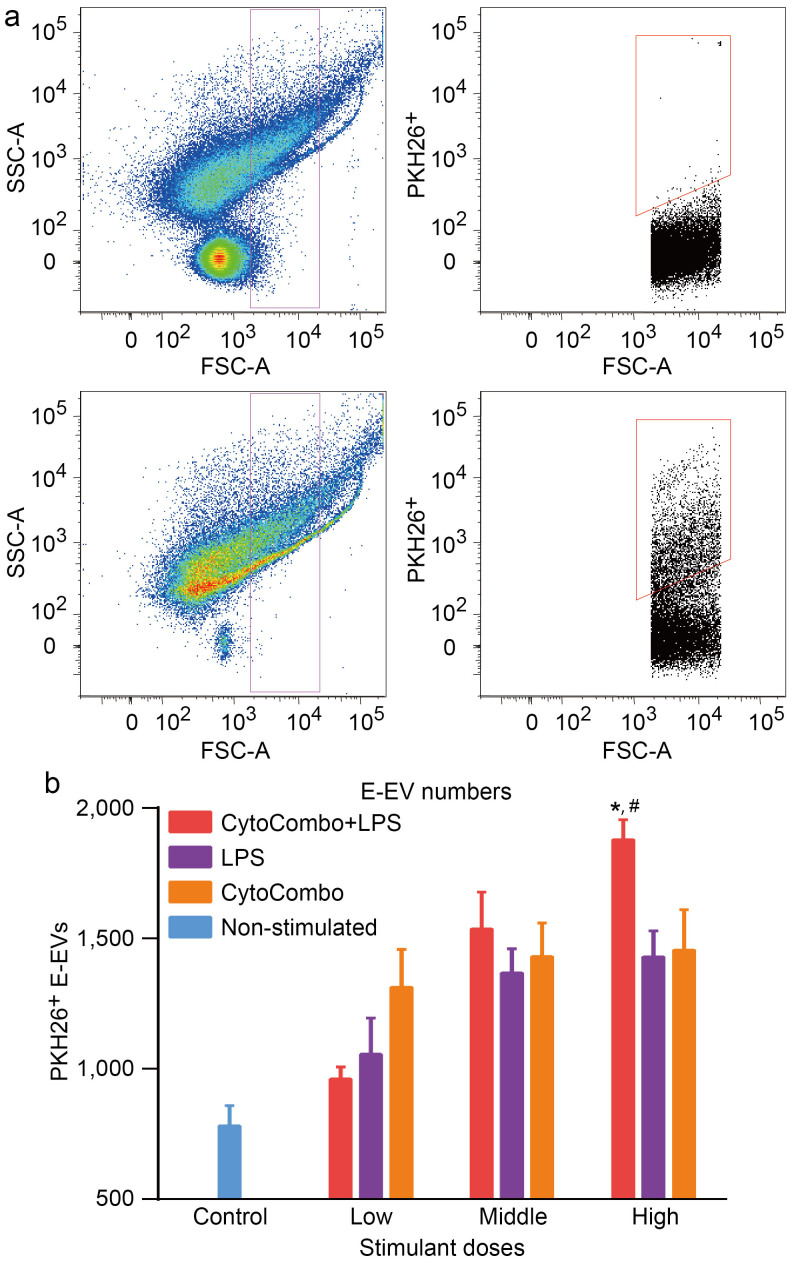
Next, E-EV release from b.End5 cells prelabeled with PKH26 was quantitatively analyzed by flow cytometry. Based on the above mentioned SEM study, we focused on particles between 0.3 μm and 3.0 μm in diameter, which corresponds to the diameter of the E-EVs released in membrane-shedding mode32,34,35. The preculture medium containing CytoCombo + LPS included many particles between 0.3 μm and 3.0 μm in diameter that were negative for PKH26, such as FBS-derived EVs, and EVs from MVBs (Fig. 3a, upper graphs). In contrast, after 3 hours of culture with high dose CytoCombo + LPS, the culture medium contained a significant number of PKH26-positive particles between 0.3 μm and 3.0 μm in diameter (Fig. 3a, bottom graphs). These findings indicated that the plasma membrane-intercalating nature of PKH26, with which the b.End5 cells were prelabeled, allowed us to discriminate foreign substances in the medium from E-EVs and to precisely measure the release of E-EVs. In a comparative assay, the three types of stimulants significantly induced the release of PKH26-positive E-EVs compared to that released from the non-stimulated control (stimulant, P-value vs. the control; CytoCombo + LPS, P < 0.0001; CytoCombo, P < 0.01; LPS, P < 0.05; Fig. 3b). CytoCombo + LPS induced the release of E-EVs in a dose-dependent manner up to the high dose, and the release induced by CytoCombo + LPS was significantly higher than that induced by the other stimuli at the high dose (Fig. 3b). At the high doses, CytoCombo and LPS individually induced the release of E-EVs to similar extents. These changes of E-EV production were well correlated with those of Il6 and Icam1 mRNA induction, both of which were induced by different inflammatory stimuli. Accordingly, these data suggested that E-EV production was correlated with the level of cellular inflammation. At the low and middle doses, CytoCombo induced E-EV release (P < 0.01 and P < 0.001, respectively, vs. the control) but not Il6 and Icam1 mRNA (both n.s. vs. the control; Fig. 1f and g). Thus, E-EV production in cerebrovascular endothelial cells appears to be a more sensitive cellular response than cytokine production based on relative induction by inflammatory stimulants.
Figure 3. Quantitative analysis of E-EVs by flow cytometry.
(a) PKH26-positive E-EVs were detected and measured quantitatively by flow cytometry. The upper graphs show the pre-culture medium with CytoCombo + LPS. The lower graphs show the culture medium from b.End5 cells stained with PKH26 stimulated by CytoCombo + LPS for 3 hours. Vesicles gated from submicron to micron size (0.3 μm to 3.0 μm in diameter, magenta) are shown in the graph on the left. In the graphs on the right, PKH26-positive E-EVs are gated (red). The pre-culture medium contains many FBS-derived vesicles that are negative for PKH26. E-EVs can be observed in the 3-hour culture medium as the PKH26-positive fraction. (b) E-EV production is significantly upregulated after 3 hours of exposure to the high dose of CytoCombo + LPS. In contrast, a lower number of E-EVs are observed in the LPS and CytoCombo groups (reported as the number of PKH26-positive E-EVs in 100 μL of medium, n = 4 for each). Non-stimulated control indicates basal production level of E-EVs. * P < 0.05, CytoCombo + LPS vs. LPS; #P < 0.05, CytoCombo + LPS vs. CytoCombo.
Inflammation-induced E-EVs may target cerebrovascular pericytes
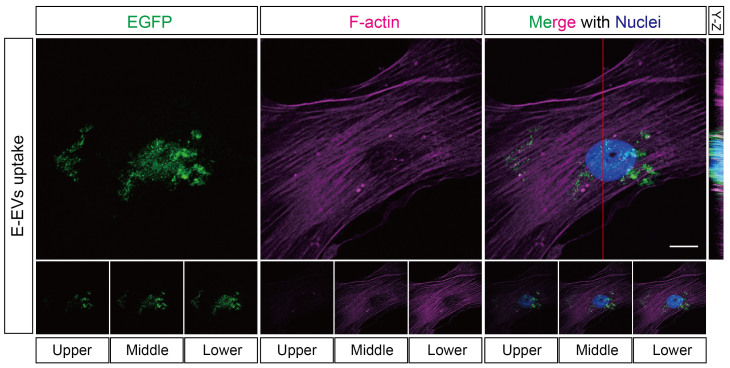
Endothelial cells and pericytes are juxtapositioned and functionally correlated in the vasculature22,36. Next, we determined if the miRNAs in E-EVs could mediate the inflammatory signaling between cerebrovascular endothelial cells and pericytes. To this end, we first assessed whether E-EVs could be incorporated into pericytes. To rule out the effect of modification to the E-EV surface by the plasma membrane-intercalating dye PKH26, we constructed a membrane-localized EGFP (M-EGFP)-coding vector. M-EGFP-positive E-EVs were fractionated from the culture medium of M-EGFP expressing b.End5 cells after a 3-hour stimulation with the high dose of CytoCombo + LPS. The E-EV fractions obtained were washed 3 times with PBS, and then added to the culture medium of human brain pericytes (HBPCs). After a 24-hour incubation, immunocytofluorescence showed that many M-EGFP-positive E-EVs were incorporated into the cytoplasmic region of HBPCs (Fig. 4). Some of the M-EGFP-positive E-EVs were localized near the nucleus, as confirmed by the Y-Z cross-sectional image and single-plane sequential Z images (Fig. 4, upper right and bottom row). These findings suggest that the bioactive molecules encapsulated in the E-EVs were transferred into HBPCs.
Figure 4. E-EVs are incorporated into cerebrovascular pericytes.
Plasma membrane-localized EGFP (M-EGFP)-positive E-EVs were fractionated from M-EGFP-expressing b.End5 cells. When the M-EGFP-positive E-EVs were incubated with cerebrovascular pericytes, the E-EVs were taken up by the cells as confirmed by immunocytofluorescence of EGFP. Incorporation of the E-EVs by the cells was precisely confirmed by a Y-Z cross-sectional image (adjacent to the 3D projection merged image) and by 1-μm, sequential, single Z-plane images (bottom row). Scale bar, 10 μm.
To examine the bioactive molecules encapsulated in the E-EVs, we examined the molecules in the E-EV fractions from b.End5 cells with an miRNA array. Many miRNAs were upregulated in the E-EVs from cells stimulated by a high dose of CytoCombo + LPS compared to those in the E-EVs from non-stimulated control cells (Fig. 5a). Some of them were also upregulated in the E-EVs from cells stimulated with a high dose of CytoCombo, which induced an inflammatory response in b.End5 cells, although to a lesser extent than CytoCombo + LPS (Fig. 5a). These miRNAs sorted into the E-EVs were classified by Ingenuity Pathway Analysis (IPA). A significant proportion of the miRNAs were approved to be related to Inflammatory Disease and Inflammatory Response (bright red) in Disease and Disorders of the IPA (Supplementary Figure 5). Moreover, the miRNAs were abundant in those related to Organismal Injury and Abnormalities, which may associate with inflammation, over-represented (dark red). These data suggest that inflammatory stimuli are able to sort the inflammation-related miRNAs into E-EVs.
Figure 5. MiRNA array data from inflammation-stimulated E-EVs.
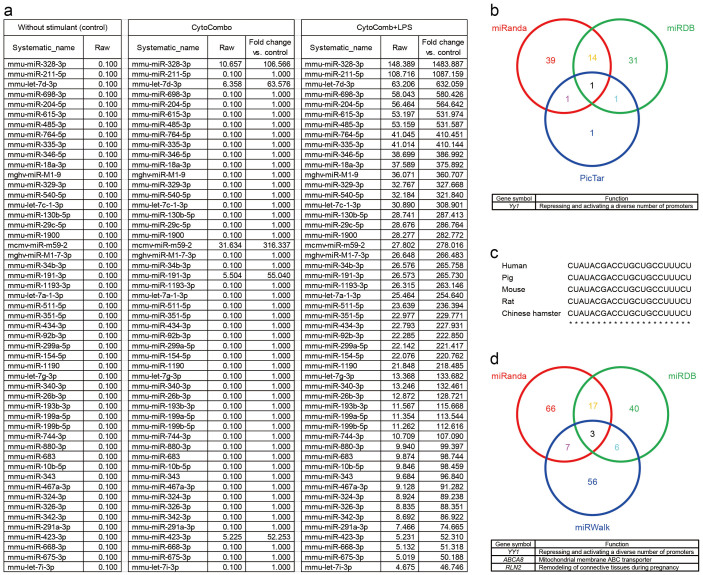
(a) MiRNAs extracted from E-EVs undergo a drastic change following treatment with inflammatory stimulants. Some of the miRNAs are upregulated more than 500 fold by CytoCombo + LPS treatment when compared to the non-stimulated control. (b) The multifunctional transcription factor Yy1 is determined as a target of let-7d-3p in mice that are commonly identified in 3 different public accessible databases. (c) Homology analysis of let-7d-3p. The alignment of let-7d-3p sequences indicates that the mature sequences of five mammalian species are identical. (d) YY1, ABCA8, and RLN2 are determined as a target of let-7d-3p in human that are commonly identified in 3 different public accessible databases.
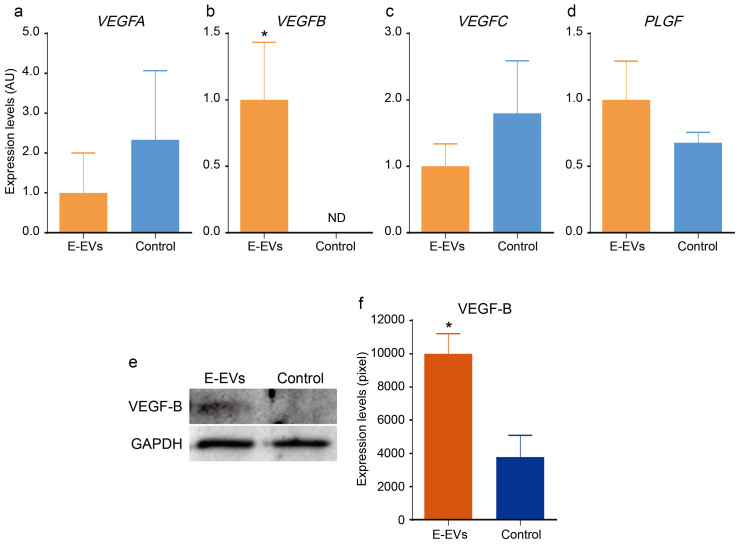
Among the various inflammation-related miRNAs detected (Fig. 5a), we focused on two, miR-328-3p and let-7d-3p, because these miRNAs were extensively induced after stimulation with CytoCombo + LPS, and were significantly induced, but to a lesser extent, after CytoCombo stimulation. Real-time PCR showed that the cultured HBPCs expressed Yin and yang 1 (YY1), which is a target mRNA of let-7d-3p and is an orthologue of mouse Yy1 that was identified through in silico analysis (Figure 5b), but not the target mRNAs of miR-328-3p including Proviral integration site for Moloney murine leukemia virus 1 (PIM1), Testis-specific kinase 2 (TESK2), and Ataxin-2 binding protein 1 (A2BP1) (data not shown). The well-conserved, mature let-7d-3p sequences in mice and humans are identical (Fig. 5c) and were shown to target YY1 mRNA through in silico analyses (Fig. 5d). VEGF family proteins are synthesized in pericytes/vSMCs37,38 and are believed to be the downstream targets of YY1, since YY1 knockdown upregulates their expression levels, in which the upregulation of VEGF-B was most striking among VEGF family proteins39. Therefore, we investigated whether inflammation-induced E-EVs modulated the expression of VEGF family mRNAs and proteins. We observed that VEGF-B mRNA and protein levels were low in the controls, but were extensively induced in the E-EV-supplemented group (Fig. 6b, e, f). In contrast, E-EV supplementation did not affect VEGFA, VEGFC, or PLGF mRNA levels (Fig. 6a, c, d). The miRNAs carried by E-EVs may mediate the inflammatory responses of endothelial cells by regulating gene expression profiles in pericytes/vSMCs, especially in cerebrovascular pericytes.
Figure 6. Effects of E-EVs in cerebrovascular pericytes.
(a–d) The mRNA levels of the VEGF family members, VEGFA, VEGFC, and PLGF in the E-EV-supplemented and non-stimulated control groups did not differ significantly (a, c, and d). In contrast, VEGFB was significantly upregulated in the E-EV-supplemented group compared to the non-stimulated control group (b) (n = 3, P < 0.05). (e and f) VEGF-B protein levels were measured by western blotting (e), using GAPDH as an internal control. VEGF-B protein was significantly upregulated in the E-EV-supplemented group compared to the non-stimulated control group (f) (n = 3, P < 0.05).
Discussion
Emerging evidence has suggested the importance of EVs in cell-to-cell communication as carriers of small molecules such as peptides, proteins, lipids, mRNAs, and miRNAs40,41. The bioactive molecules encapsulated in EVs can be transferred to and function in target recipient cells6,42,43,44. In addition, miRNAs act as regulatory molecules of diverse biological phenomena, such as immune responses, inflammation, and tumor growth18,19. However, our knowledge of EV- and encapsulated miRNA-mediated cell-to-cell communication is mainly limited to tumor cell biology. Although it is known that the proinflammatory agent TNF-α induces the release of E-EVs45, the significance of these E-EVs remains to be elucidated. To the best of our knowledge, we demonstrated here, for the first time, that the inflammation-related miRNAs carried by the E-EVs from endothelial cells mediated inflammatory responses in pericytes. The induction of E-EV shedding was very acute and sensitive compared to previously established inflammation markers like IL-6 and ICAM-127,28, and it may be useful as a very sensitive marker for inflammatory diseases.
In recent years, a variety of pathogens and disease states have been shown to affect the composition and function of these EVs. In vitro studies demonstrated that environmental stresses such as heat, hypoxia, irradiation, and changing of conditioned medium, differentially modified the composition of EVs46,47,48,49. Specific coding and non-coding RNAs, retrotransposon RNAs, Alu transposable elements, and chromosomal and mitochondrial DNA have been reported to be enriched in tumor cell-derived EVs50,51,52,53,54. The results of our present study showed that when exposed to inflammatory stimuli, cerebrovascular endothelial cells could sort many inflammation-related miRNAs into E-EVs ad initium. Moreover, the type and amount of miRNAs that were sorted into the E-EVs was strongly correlated with the severity of endothelial inflammation.
EVs from tumor cells could educate the bone marrow progenitor cells to mobilize out from their niche55. Vascular endothelial cells in tumors could be “educated” by EVs derived from tumor cells to exhibit a highly proliferative phenotype and karyotype abnormalities7,56. Along these same lines, our results show that the molecules in the inflammation-induced E-EVs function in target pericytes to increase VEGF-B mRNA and protein expression. Furthermore, this “education” was suggested to be mediated by the miRNA encapsulated in the E-EVs since VEGFB is thought to be a downstream target of the highly-accumulated miRNA in the inflammation-induced E-EVs. Similarly, various organ-derived EVs have been shown to be incorporated into bone marrow cells in vitro57. As a result, the mRNAs were delivered and the expression of organ-specific mRNAs was induced in the bone marrow cells. E-EVs and encapsulated miRNA were shown to be involved in the cascade of cellular events in the inflammatory vascular response.
VEGF-B is a specific ligand of VEGFR-1 that is expressed in endothelial cells and pericytes/vSMCs58,59. VEGFR-1 mediates pathological angiogenesis by targeting both endothelial cells and pericytes in the pathological neovascularization of choroidal and retinal tissues60. Similarly, VEGFR-1 stimulates the endothelial differentiation and the formation of blood vessels in infantile hemangioma61 and vascular formation in implanted tumors62. Furthermore, VEGFR-1 ablated vascular pericytes and aggravated vascular leakage to induce cancer-associated retinopathy63. Accordingly, inflammation-induced E-EVs enhanced VEGF-B expression in pericytes, which may mediate pathological neovascularization and/or vascular leakage in response to inflammation of endothelial cells. Furthermore, since E-EVs are carried in the blood stream in many pathological conditions, E-EVs may spread the local inflammatory responses of endothelial cells to the systemic vasculature. Inhibition of E-EVs may limit the extent of inflammation-induced cellular responses from endothelial cells to the surrounding cells, and E-EVs may be a new therapeutic target for inflammatory diseases.
In conclusion, we developed a reproducible in vitro E-EV production system and a quantitative E-EV measurement method. Using these tools, we demonstrated that inflammation-induced E-EVs could “educate” target cells to modulate growth factor production. We also showed evidence to suggest that part of this cell education signal might be an miRNA encapsulated in the E-EVs. Our findings offer a new clinical approach, targeting E-EVs, for the prevention and treatment of acute and chronic inflammatory diseases. Further experiments are needed to elucidate the molecular mechanisms by which E-EVs transfer their “educating” messages.
Methods
Cell lines and culture conditions
The mouse brain endothelial cell line, b.End5, and human brain pericytes (HBPCs) were purchased from DS Pharma Biomedical (Osaka, Japan), and from Applied Cell Biology Research Institute (Kirkland, WA), respectively. The cells were cultured using the EBM-2 bullet kit (Takara, Kyoto, Japan) at 37°C in a humidified atmosphere containing 5% CO2. Membrane-localized EGFP (M-EGFP)-expressing b.End5 cells were generated by transfection of the GAP43 membrane targeting sequence-Egfp fusion gene, which was constructed as follows. Briefly, the human GAP43 membrane targeting sequence was cloned from the plasmid (Clone ID No. IRAL020F10, RIKEN BRC, Tsukuba, Japan) and inserted into the multi-cloning site of the pEGFP-N1 vector (Takara). The vector containing the resultant fusion gene, consisting of the GAP43 membrane targeting sequence and Egfp, was transfected into b.End5 cells using X-tremeGENE transfection reagent (Roche, Basel, Switzerland) according to the manufacturer's protocol.
Reagents
The inflammatory cytokines TNF-α, IL-1β, and IFN-γ were purchased from PeproTech (Rocky Hill, NJ). Lipopolysaccharide (LPS) was obtained from List Biological Laboratories (Campbell, CA). TNF-α, IL-1β, and IFN-γ were used separately, as a mixture (referred to as CytoCombo), or as a mixture of CytoCombo and LPS (CytoCombo + LPS) (Supplementary Table 1). The plasma membrane intercalating fluorescent dye PKH26 was purchased from Sigma-Aldrich (St. Louis, MO) and was used according to the manufacturer's protocol. The cell tracer fluorescence dye 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CSFE) was purchased from eBioscience (San Diego, CA) and was used according to the manufacturer's protocol.
Fractionation of endothelial cell-derived extracellular vesicles (E-EVs)
Ten-centimeter dishes coated with type I collagen (BD Biosciences, San Jose, CA) of confluent b.End5 cells were rinsed with PBS, and stimulated with medium containing CytoCombo + LPS, CytoCombo, or LPS alone to induce inflammation. Culture medium without any stimulant was used as a control. After a 3-hour incubation, the conditioned culture medium was centrifuged at 1,000 rpm for 10 minutes at 4°C to eliminate the cellular debris. The supernatant was then ultracentrifuged at 100,000 × g for 90 minutes at 4°C, and the pellets were used as the E-EV fraction in the following analyses.
Immunocytofluorescence
For immunocytofluorescence, cells were seeded into 8-well culture slides (BD Biosciences). After treatment with various stimuli, the slides were fixed in 4% paraformaldehyde for 15 minutes on ice, and then rinsed with PBS. All specimens were permeabilized in a 0.3% Triton X-100/PBS solution. Permeabilized specimens were incubated at 4°C overnight with primary antibodies diluted in 0.03% Triton X-100/PBS with 10% normal goat serum (VECTOR LABORATORIES, Burlingame, CA), which was used for blocking. The following primary antibodies were used: rat anti-TLR4/MD-2 complex (1:200; eBioscience), mouse anti-ICAM-1 (1:200; Abcam, Cambridge, UK), rabbit anti-cleaved caspase 3 (1:200; Cell Signaling Technology, Danvers, MA), rabbit anti-GFP (1:100; Frontier Institute, Hokkaido, Japan). The Alexa-Fluor488, Alexa-Fluor568, or Alexa-Fluor633-conjugated secondary antibodies (Life Technologies Corporation, Carlsbad, CA) were used at 1:500 dilutions. For F-actin staining, Falloidin-Alexa-Fluor647 (1:500; Cytoskeleton, Denver, CO) was used. Nuclei were stained with Hoechst 33258 (Nacalai Tesque, Koto, Japan). For immunofluorescence of E-EVs, fractionated E-EVs from culture media of b.End5 cells pre-stained by PKH26 or CSFE were incubated with 3% normal mouse serum (VECTOR LABORATORIES) at 4°C for blocking. The following primary antibodies were used: rat anti-Flk-1-FITC (1:20; BD Biosciences, San Jose, CA), rat anti-CD62E-PE (1:20; BD Biosciences). The imaging system was a Leica TCS SP5 confocal system (Leica Microsystems, Wetzlar, Germany).
FACS analysis
E-EVs released into culture media were analyzed by flow cytometry. E-EVs in the media were gated as events of 0.3 to 3.0 μm in diameter, corresponding to the size of the membrane shed EVs32,33,34. E-EVs were further gated as PKH26-positive events, by using b.End5 cells pre-labeled with PKH26, a membrane intercalating dye. This gating allowed us to discriminate E-EVs from the other microparticles of similar diameters in the medium. We used 3.0- to 3.4-μm rainbow calibration particles (BD Biosciences) as benchmarks. Flow cytometry was performed with a FACSCanto II (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR) was used for data analysis.
Scanning electron microscopy
Endothelial b.End5 cells were seeded on cover slips and cultured until confluent. Confluent cells were then exposed to inflammatory stimuli (CytoCombo + LPS) for 10 seconds or 10 minutes. After stimulation, the cells were fixed with 2% formaldehyde (Wako Pure Chemical Industries, Osaka, Japan) and 2.5% glutaraldehyde (Wako Pure Chemical Industries) prepared in 0.1 M sodium phosphate buffer (pH 7.4) for 2 hours at room temperature, and then postfixed in 1% osmium tetroxide in the same buffer for 2 hours on ice. Cells were then dehydrated in a graded series of ethanol, substituted with t-butyl alcohol and freeze-dried. Cells were sputter-coated with osmium and then observed in a S4200 scanning electron microscope (Hitachi).
RNA extraction, cDNA synthesis, conventional PCR, and real-time PCR
Total RNA was isolated from b.End5 cells or HBPCs using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA (100 ng) was reverse-transcribed as described in the PrimeScript RT reagent Kit protocol (Takara). Conventional PCR was performed with a Bio-Rad C100 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA). The cDNAs were diluted 1:20 with the reaction mixture from the Ex Taq PCR kit (Takara), and the PCR was performed by standard 3 step amplification protocol consisting of hot start enzyme activation at 95°C for 10 minutes followed by 35 cycles of amplification at 95°C for 30 seconds, 60°C for 60 seconds, and 72°C for 60 seconds. For real-time PCR, which was performed with a Takara Thermal Cycler Dice Real Time System TP800 (Takara), the cDNAs were diluted 1:25 in the reaction mixture containing SYBR Premix EX Taq II (Takara). The real-time PCR program consisted of hot start enzyme activation at 95°C for 10 seconds followed by 45 cycles of amplification at 95°C for 10 seconds and 60°C for 40 seconds. Finally, to obtain a dissociation curve, a final cycle was performed at 95°C for 1 minute, 60°C for 30 seconds, and 95°C for 10 seconds. For data analysis, the mouse or human glyceraldehyde-3-phosphate dehydrogenase (Gapdh or GAPDH) housekeeping gene was used as an internal control. Induction values were calculated using analysis software (Takara). Primer sequences are available upon request from the TAKARA BIO INC. website (http://www.takara-bio.co.jp/).
MiRNA array
The miRNA expression profile was obtained by using a Mouse miRNA microarray 8 × 15 K (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. Total RNA was prepared from fractionated E-EVs using the miRNA easy Mini Kit (Qiagen). RNA quality was measured with an Agilent 2100 Bioanalyzer (Agilent Technologies). Synthesis of probes, and hybridization to the Mouse miRNA microarray 8 × 15 K were performed by using the miRNA Complete Labeling and Hyb Kit (Agilent Technologies). Hybridized samples were analyzed using a Microarray Scanner (G2505B, Agilent Technologies) and GeneSpring GX software (Agilent Technologies) according to standard protocols. The miRNA array data were analyzed using Ingenuity Pathway Analysis (Ingenuity Systems, http://www.ingenuity.com/).
Western blotting
For E-EV marker analyses, E-EVs that were fractionated from the culture media of b.End5 cells after CytoCombo + LPS stimulation were used as sample. For the control, b.End5 cells were used. For VEGF-B analyses, E-EVs that were fractionated from the culture media of b.End5 cells after CytoCombo stimulation were added (100 μg) to HBPCs cultured in a collagen type I-coated 24-well plate (BD Bioscience), and the plate was incubated at 37°C for 48 hours. Sample preparations and all other procedures for western blotting have been described elsewhere64. Briefly, cells were lysed using RIPA buffer (Thermo Fisher, Waltham, MA) on ice. Whole cell lysates were separated by SDS-PAGE and then electrophoretically transferred to polyvinylidene difluoride membranes. The membranes were incubated for 1 hour at room temperature in a 50 mM Tris buffer containing 150 mM NaCl, 0.05% Tween 20, and 5% BSA for blocking. The membranes were then probed with rabbit anti-VEGFR-2 (1:500; Cell Signaling Technology), rabbit anti-CD62E (1:100; Abcam), rabbit anti-VEGF-B (1:100; Abcam), and mouse anti-GAPDH (1:5,000; Merck Millipore, Billerica, MA) antibodies. The membranes were washed in a 50 mM Tris buffer containing 150 mM NaCl, and 0.05% Tween 20, and were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies. Immunoreactive bands were detected using enhanced chemiluminescence reagents (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer's instructions.
Statistical analysis
Statistical significance was determined using Student's t-test, or one- or two-way analysis of variance (ANOVA) with Tukey's or Newman-Keuls multiple comparisons as a post hoc analysis for ANOVA. P values less than 0.05 were considered statistically significant. Graphs were drawn using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). Quantified data are presented as mean ± SEM.
Author Contributions
S.Y. and S.N. conceived and designed the project. S.Y., E.A., T.Y., M.M., T.-T.H., H.S., S.H., M.I., Y.N., K.M. and H.M. performed experiments. S.Y. wrote the manuscript with significant input from K.T., T.H., Y.I., H.M., N.M., S.N. and M.S. All authors reviewed the manuscript.
Supplementary Material
Supplementary data
Acknowledgments
We thank the members of the Department of Pathology at the University of Toyama for helpful discussions. We also thank M. Satake, A. Watanabe, D. Inoue, M. Dohmoto for technical assistance and discussions, and N. Yamamoto, A. Yamamoto, R. Yamamoto for broad-based support. This work was supported by a Grant-in-Aid for Scientific Research (21590589 and 24659112; to S.Y.) from The Ministry of Education, Culture, Sports, Science, and Technology and by The Research Funding for Longevity Sciences (23-9; to S.N.) from the National Center for Geriatrics and Gerontology, and The Advanced Research for Products Mining Programme (10–43; to S.N.) from the National Institute of Biomedical Innovation.
References
- Raposo G. & Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J. & Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2, 20389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H. & Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101, 2087–2092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer C. & Pisetsky D. S. The role of microparticles in the pathogenesis of rheumatic diseases. Nat Rev Rheumatol 6, 21–29 (2010). [DOI] [PubMed] [Google Scholar]
- Antonyak M. A. et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A 108, 4852–4857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J., Wysoczynski M., Hayek F., Janowska-Wieczorek A. & Ratajczak M. Z. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 20, 1487–1495 (2006). [DOI] [PubMed] [Google Scholar]
- Kawamoto T. et al. Tumor-derived microvesicles induce proangiogenic phenotype in endothelial cells via endocytosis. PLoS One 7, e34045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3, 15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostefai H. A. et al. Circulating microparticles from patients with septic shock exert protective role in vascular function. Am J Respir Crit Care Med 178, 1148–1155 (2008). [DOI] [PubMed] [Google Scholar]
- Simak J., Gelderman M. P., Yu H., Wright V. & Baird A. E. Circulating endothelial microparticles in acute ischemic stroke: a link to severity, lesion volume and outcome. J Thromb Haemost 4, 1296–1302 (2006). [DOI] [PubMed] [Google Scholar]
- Petrozella L. et al. Endothelial microparticles and the antiangiogenic state in preeclampsia and the postpartum period. Am J Obstet Gynecol 207, 140 e120–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger C. M., Amabile N. & Tedgui A. Circulating microparticles: a potential prognostic marker for atherosclerotic vascular disease. Hypertension 48, 180–186 (2006). [DOI] [PubMed] [Google Scholar]
- Koga H. et al. Elevated levels of VE-cadherin-positive endothelial microparticles in patients with type 2 diabetes mellitus and coronary artery disease. J Am Coll Cardiol 45, 1622–1630 (2005). [DOI] [PubMed] [Google Scholar]
- Agouni A. et al. Endothelial dysfunction caused by circulating microparticles from patients with metabolic syndrome. Am J Pathol 173, 1210–1219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M. C., Tesse A., Zobairi F. & Andriantsitohaina R. Shed membrane microparticles from circulating and vascular cells in regulating vascular function. Am J Physiol Heart Circ Physiol 288, H1004–1009 (2005). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R. L. & van Rooij E. MicroRNA regulation as a therapeutic strategy for cardiovascular disease. Curr Drug Targets 11, 936–942 (2010). [DOI] [PubMed] [Google Scholar]
- Small E. M. & Olson E. N. Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan F. et al. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One 7, e50837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S. et al. Functional role of NF-kappaB in expression of human endothelial nitric oxide synthase. Biochem Biophys Res Commun 448, 101–107 (2014). [DOI] [PubMed] [Google Scholar]
- Armulik A., Genove G. & Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21, 193–215 (2011). [DOI] [PubMed] [Google Scholar]
- Armulik A. et al. Pericytes regulate the blood-brain barrier. Nature 468, 557–561 (2010). [DOI] [PubMed] [Google Scholar]
- Daneman R., Zhou L., Kebede A. A. & Barres B. A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. D. et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 68, 409–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E. A., Bell R. D. & Zlokovic B. V. Central nervous system pericytes in health and disease. Nat Neurosci 14, 1398–1405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M. Interleukin-6: from an inflammatory marker to a target for inflammatory diseases. Trends Immunol 33, 571–577 (2012). [DOI] [PubMed] [Google Scholar]
- Hua S. Targeting sites of inflammation: intercellular adhesion molecule-1 as a target for novel inflammatory therapies. Front Pharmacol 4, 127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague A. H. & Khalil R. A. Inflammatory cytokines in vascular dysfunction and vascular disease. Biochem Pharmacol 78, 539–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerman D. D. & Goldblum S. E. Mechanisms of bacterial lipopolysaccharide-induced endothelial apoptosis. Am J Physiol Lung Cell Mol Physiol 284, L899–914 (2003). [DOI] [PubMed] [Google Scholar]
- Dignat-George F. & Boulanger C. M. The many faces of endothelial microparticles. Arterioscler Thromb Vasc Biol 31, 27–33 (2011). [DOI] [PubMed] [Google Scholar]
- Shai E. & Varon D. Development, cell differentiation, angiogenesis—microparticles and their roles in angiogenesis. Arterioscler Thromb Vasc Biol 31, 10–14 (2011). [DOI] [PubMed] [Google Scholar]
- Fujita Y., Kuwano K., Ochiya T. & Takeshita F. The impact of extracellular vesicle-encapsulated circulating microRNAs in lung cancer research. Biomed Res Int 2014, 486413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese J. & Dainiak N. Modulation of intercellular communication mediated at the cell surface and on extracellular, plasma membrane-derived vesicles by ionizing radiation. Exp Hematol 31, 455–464 (2003). [DOI] [PubMed] [Google Scholar]
- Delabranche X., Berger A., Boisrame-Helms J. & Meziani F. Microparticles and infectious diseases. Med Mal Infect 42, 335–343 (2012). [DOI] [PubMed] [Google Scholar]
- Armulik A., Abramsson A. & Betsholtz C. Endothelial/pericyte interactions. Circ Res 97, 512–523 (2005). [DOI] [PubMed] [Google Scholar]
- Darland D. C. et al. Pericyte production of cell-associated VEGF is differentiation-dependent and is associated with endothelial survival. Dev Biol 264, 275–288 (2003). [DOI] [PubMed] [Google Scholar]
- Yonekura H. et al. Placenta growth factor and vascular endothelial growth factor B and C expression in microvascular endothelial cells and pericytes. Implication in autocrine and paracrine regulation of angiogenesis. J Biol Chem 274, 35172–35178 (1999). [DOI] [PubMed] [Google Scholar]
- de Nigris F. et al. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc Natl Acad Sci U S A 107, 14484–14489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barteneva N. S., Maltsev N. & Vorobjev I. A. Microvesicles and intercellular communication in the context of parasitism. Front Cell Infect Microbiol 3, 49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S. & Losordo D. W. Exosomes and cardiac repair after myocardial infarction. Circ Res 114, 333–344 (2014). [DOI] [PubMed] [Google Scholar]
- Valadi H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- Skog J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias D. M. et al. Stem cell microvesicles transfer cystinosin to human cystinotic cells and reduce cystine accumulation in vitro. PLoS One 7, e42840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis A. M. et al. p38 mitogen-activated protein kinase targets the production of proinflammatory endothelial microparticles. J Thromb Haemost 7, 701–709 (2009). [DOI] [PubMed] [Google Scholar]
- Chen T., Guo J., Yang M., Zhu X. & Cao X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J Immunol 186, 2219–2228 (2011). [DOI] [PubMed] [Google Scholar]
- Tetta C., Bruno S., Fonsato V., Deregibus M. C. & Camussi G. The role of microvesicles in tissue repair. Organogenesis 7, 105–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J. et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856 (2006). [DOI] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G. & Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3 22 (2006). [DOI] [PubMed] [Google Scholar]
- Ronquist G. K., Larsson A., Stavreus-Evers A. & Ronquist G. Prostasomes are heterogeneous regarding size and appearance but affiliated to one DNA-containing exosome family. Prostate 72, 1736–1745 (2012). [DOI] [PubMed] [Google Scholar]
- Guescini M., Genedani S., Stocchi V. & Agnati L. F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm 117, 1–4 (2010). [DOI] [PubMed] [Google Scholar]
- Balaj L. et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2, 180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak J. & Guha A. Extracellular vesicles--vehicles that spread cancer genes. Bioessays 34, 489–497 (2012). [DOI] [PubMed] [Google Scholar]
- Waldenstrom A., Genneback N., Hellman U. & Ronquist G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One 7, e34653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18, 883–891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K., Meehan B., Kerbel R. S., Allison A. C. & Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A 106, 3794–3799 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliotta J. M. et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol 38, 233–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H. & Shibuya M. The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin Sci (Lond) 109, 227–241 (2005). [DOI] [PubMed] [Google Scholar]
- Cao Y. Positive and negative modulation of angiogenesis by VEGFR1 ligands. Sci Signal 2, re1 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang F. et al. VEGF-B is dispensable for blood vessel growth but critical for their survival, and VEGF-B targeting inhibits pathological angiogenesis. Proc Natl Acad Sci U S A 106, 6152–6157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscolo E., Mulliken J. B. & Bischoff J. VEGFR-1 mediates endothelial differentiation and formation of blood vessels in a murine model of infantile hemangioma. Am J Pathol 179, 2266–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Yamamoto S., Osawa T. & Shibuya M. Vascular endothelial growth factor receptor-1 signaling promotes mobilization of macrophage lineage cells from bone marrow and stimulates solid tumor growth. Cancer Res 70, 8211–8221 (2010). [DOI] [PubMed] [Google Scholar]
- Cao R. et al. VEGFR1-mediated pericyte ablation links VEGF and PlGF to cancer-associated retinopathy. Proc Natl Acad Sci U S A 107, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S. et al. Essential role of Shp2-binding sites on FRS2alpha for corticogenesis and for FGF2-dependent proliferation of neural progenitor cells. Proc Natl Acad Sci U S A 102, 15983–15988 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data