Abstract
NADH is an essential redox cofactor in numerous metabolic reactions, and the cytosolic NADH-NAD+ redox state is a key parameter in glycolysis. Conventional NADH measurements rely on chemical determination or autofluorescence imaging, which cannot assess NADH specifically in the cytosol of individual live cells. By combining a bacterial NADH-binding protein and a fluorescent protein variant, we have created a genetically encoded fluorescent biosensor of the cytosolic NADH-NAD+ redox state, named Peredox (Hung et al., Cell Metab 14:545–554, 2011). Here, we elaborate on imaging methods and technical considerations of using Peredox to measure cytosolic NADH:NAD+ ratios in individual live cells.
Keywords: NADH, glycolysis, lactate dehydrogenase, sensor calibration, single cell imaging
1. Introduction
Nicotinamide adenine dinucleotide (oxidized: NAD+; reduced, NADH) is an essential redox cofactor in numerous metabolic reactions. In mammalian cells, the cytosolic NADH-NAD+ redox state is a key parameter in glucose metabolism by glycolysis (1). Conventional NADH measurements rely on chemical determination or autofluorescence imaging; however, these approaches cannot assess NADH specifically in the cytosol of individual live cells: Chemical methods cannot measure NADH in different cell types within a population or in single cells, while autofluorescence reflects the combined signal of both NADH and NADPH, thus not specific for NADH alone (2, 3).
We have therefore created a genetically encoded fluorescent biosensor to report on the cytosolic NADH-NAD+ redox state (4). This biosensor, named Peredox, is constructed by linking a green fluorescent protein (GFP) variant, circularly permuted T-Sapphire (5), to a bacterial NADH-binding protein Rex (6–8). Upon binding NADH, Rex undergoes a conformational change that is coupled to an increase in the GFP fluorescence. Through competition between NADH and NAD+ for binding, Peredox reports the cytosolic NADH:NAD+ ratios in mammalian cells (see Fig. 1). Peredox has been optimized to be pH-resistant to facilitate imaging experiments. We calibrate the sensor response by varying the amounts of exogenous lactate and pyruvate, and we have monitored the cytosolic NADH-NAD+ redox dynamics in various cultured and primary cell types during energetic challenges. Together with other fluorescent biosensors (9–11), Peredox can reveal how glucose metabolism and the cytosolic NADH-NAD+ redox state are regulated in the context of intact cells. Here, we elaborate on imaging methods, biosensor calibration using lactate and pyruvate, and other technical considerations of using Peredox to measure cytosolic NADH:NAD+ ratios in individual live cells.
Fig. 1.
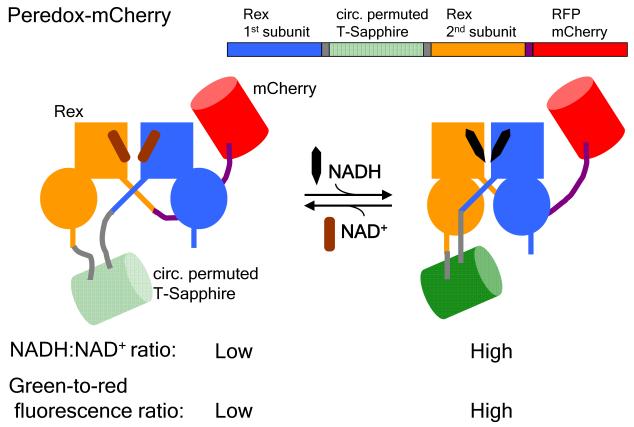
Schematic showing the design of the fluorescent sensor of the cytosolic NADH-NAD+ redox state, Peredox-mCherry: A circularly permuted GFP T-Sapphire (green) is interposed between the two Rex subunits (blue and orange), with a C-terminal RFP mCherry to normalize for the green fluorescence. Upon binding to NADH (black) but not NAD+ (brown), Rex undergoes a conformational change that is coupled to an increase in the GFP fluorescence. Thus, the green-to-red fluorescence ratio increases with NADH:NAD+ ratio. Adapted from Hung et al., 2011.
2. Materials
2.1 Cell Culture
Plasmid DNA encoding Peredox-mCherry or other versions of this reporter in a mammalian expression vector, such as pcDNA3.1 or GW1 (see Notes 1–4; available from www.addgene.org).
Mouse neuroblastoma cell line Neuro-2a or other adherent mammalian cell lines (American Type Culture Collection; see Note 5).
Culture medium, serum, and supplements appropriate for the chosen cell line (see Note 5).
Phosphate-buffered saline (PBS) and trypsin/ethylenediamine tetraacetic acid (EDTA).
Standard 10-cm tissue culture dishes and multi-well tissue culture plates
Transfection reagents (for instance, QIAGEN Effectene) or viral transduction reagents (see Notes 6 and 10).
Flame-sterilized glass coverslips coated with protamine (Sigma) at a concentration 1 mg/ml in water (see Note 7).
2.2 Cell microscopy
Extracellular solution: 121.5 mM NaCl, 25 mM NaHCO3, 2.5 mM KCl, 2 mM CaCl2, 1.25 mM NaH2PO4, and 1 mM MgCl2. (see Note 8).
Sodium lactate (Sigma) at a stock concentration of 500 mM in water (see Note 9).
Sodium pyruvate (Sigma) at a stock concentration of 500 mM in water (see Note 9).
Widefield fluorescence microscope, equipped with appropriate optical filters, and environmental control (see Notes 11–13).
3. Methods
3.1 Cell Culture
Neuro-2a or other adherent cell lines are passaged before confluence and plated for transient transfection: Cells are rinsed with PBS, dissociated using trypsin/EDTA, and plated sparsely onto sterilized protamine-coated coverslips in a 24-well plate.
After several hours or the next day, the adherent cells are transfected with plasmid DNA encoding Peredox-mCherry (or other reporters of interest) using the transfection reagent according to the manufacturer’s protocol. The following day, Neuro-2a cells are maintained in low-serum culture medium supplemented with 10–20 μM retinoic acid for differentiation.
Cells are imaged 1–5 days after transient transfection.
3.2 Imaging Peredox in various lactate:pyruvate ratios
Beyond qualitative assessment, Peredox can report quantitatively the cytosolic NADH:NAD+ ratios in individual live cells (see Note 14). To do so, we first need to calibrate the biosensor: to establish the relationship between its fluorescence response and the cytosolic NADH:NAD+ ratio. The quantitative details of the biosensor response can vary depending on the fluorescence optics and the light source, as well as the intracellular environment, making it important to calibrate the sensor in the target cells. As detailed in this section, we accomplish this by varying the extracellular concentrations of lactate and pyruvate, in order to poise the cytosolic NADH-NAD+ redox state in individual cells (12). Presumably, lactate and pyruvate enter cells via monocarboxylate transporters, equilibrate readily between extracellular and intracellular compartments, and lead to exchange between NADH and NAD+ catalyzed by endogenous lactate dehydrogenases (LDH):
By setting the extracellular lactate:pyruvate ratios, we poise the cytosolic NADH:NAD+ ratios and measure the corresponding biosensor readouts (see Notes 15 and 16).
Prior to imaging, prepare the following lactate and pyruvate solutions by diluting the lactate and/or pyruvate stocks in the extracellular solution (see Table 1 and Materials 2.2.1).
All solutions are maintained at a near physiological temperature of ~35°C and bubbled with 95% air and 5% CO2. Continuous perfusion of solutions to the imaging chamber is performed using a peristaltic pump at a flow rate of ~3 ml/min. We begin with Solution A (the extracellular solution containing 10 mM lactate).
Place the coverslip containing transfected Neuro-2a cells onto the imaging chamber. Immobilize the coverslip using wax or other adhesives.
To monitor Peredox-mCherry, acquire green (T-Sapphire) and red fluorescence images at constant time intervals, for instance, 20 s. The exposure time depends on the optics (magnification and numerical aperture of the objective, density filters), pixel binning, and biosensor expression. To reduce sensor photobleaching and cell phototoxicity, choose the minimum exposure time that yields a good signal-to-noise ratio for fluorescence detection. For both green and red images, we typically used a 20× objective and an exposure time of 20 ms.
Measure a baseline sensor response in Solution A for ~15 min. Switch sequentially to Solutions B, C, D, E, F and G (the order can be varied), and measure the sensor response in each condition for ~15 min. In the case of real-time monitoring by image acquisition software, one can select a region of interest per cell and monitor its green-to-red fluorescence ratio throughout the experiment; one can switch the solutions once the steady state is achieved for the previous solution condition.
6. To verify the lactate:pyruvate ratio sensing of Peredox, repeat the experiment with an extracellular solution containing different total concentrations of lactate and pyruvate. For instance, double the lactate and pyruvate concentrations in Methods 3.2.1 while keeping the indicated lactate:pyruvate ratios constant.
Table 1.
Extracellular solution with various lactate:pyruvate ratios
| [Lactate] (mM) | [Pyruvate] (mM) | Lac:Pyr ratio | Solution |
|---|---|---|---|
| 10 | 0.00 | ∞ | A |
| 10 | 0.02 | 500 | B |
| 10 | 0.07 | 160 | C |
| 10 | 0.20 | 50 | D |
| 10 | 0.50 | 20 | E |
| 10 | 1.67 | 6 | F |
| 0 | 10 | 0 | G |
3.3 Data analysis and sensor calibration
Using image analysis software such as ImageJ (http://rsb.info.nih.gov/ij/) or TILLvisION 4.0.1 (TILL photonics), subtract background from fluorescence images.
For each time point, divide the green image by the red image to generate a pixel-by-pixel green-to-red fluorescence ratio image. Set a signal threshold for the red image to avoid ratioing artifacts.
For each cell, select a region of interest, and plot the time course of its green-to-red fluorescence ratio using Excel (Microsoft), MATLAB (MathWorks), Origin 6.0 (MicroCal), or other graphic programs.
To normalize the data in each cell, divide its fluorescence ratio time course by the minimum ratio, which is achieved by perfusion with pyruvate alone. The normalized response of Peredox-mCherry should range from 1 to ~2.5 (see Fig. 2; Notes 10 and 17).
From the normalized fluorescence ratios of multiple cells, plot the steady-state response against the corresponding lactate:pyruvate ratio in the extracellular solution. The sensor response versus the lactate:pyruvate ratio is fitted using a logistic function (see Note 18).
By assuming a constant physiological pH of 7.4 and that the LDH reaction is at equilibrium, we convert the lactate:pyruvate ratios into predicted NADH:NAD+ ratios. Therefore, by calibrating the biosensor using lactate and pyruvate, we can determine from its response the cytosolic NADH:NAD+ ratio (see Fig. 3; Notes 16 and 19).
Fig. 2.
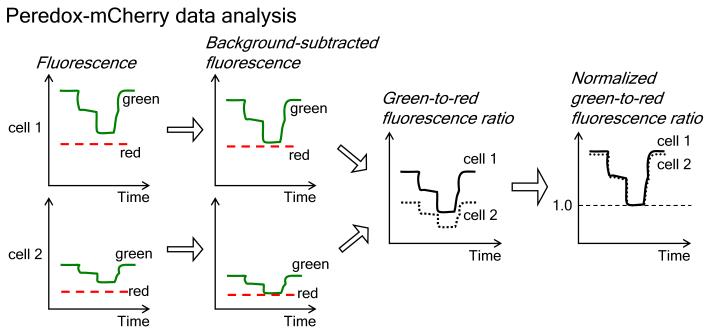
Flow chart depicting data analysis of Peredox-mCherry: After plotting the time course of both green and red fluorescence for each cell, we first subtract the background. We then generate the green-to-red fluorescence ratio at each time point for each cell. Finally, we normalize the data by the minimal green-to-red ratio signal in pyruvate alone (or by the maximal ratio signal in lactate); the normalized green-to-red ratio data for each cell facilitates comparison of biosensor responses among cells. Data analysis is described in Subheading 3.3.
Fig. 3.
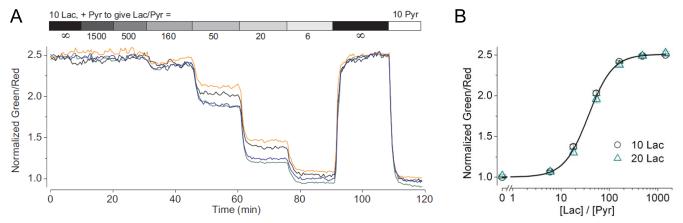
Example of Peredox-mCherry calibration data by varying lactate:pyruvate ratios in the extracellular solution. (A) Time course of normalized green-to-red fluorescence ratios of four individual cultured mouse neuroblastoma Neuro-2a cells transfected with Peredox-mCherry; cells were placed in extracellular solutions with indicated lactate:pyruvate ratios. (B) Steady-state green-to-red fluorescence ratios of Neuro-2a cells plotted against extracellular lactate:pyruvate ratios, with lactate of 10 mM or 20 mM (mean ± SEM, n = 12–15 cells). Adapted from Hung et al., 2011.
3.4 Measuring cytosolic NADH:NAD+ ratios in individual live cells
Acquire the fluorescence responses of cells expressing Peredox-mCherry in the condition of interest (see Methods 3.2.2 and 3.2.3; Notes 19–21).
Perfuse cells with the extracellular solution containing lactate (10 mM), and measure the maximum green-to-red ratio signal. Then, perfuse cells with the extracellular solution containing pyruvate (10 mM), and measure the minimum ratio signal (see Notes 15 and 16).
On the acquired imaging data, perform background subtraction, ratio determination, and data normalization (see Methods 3.3.1 to 3.3.4).
Using the calibration curve determined in Methods 3.3.6, we convert the normalized fluorescence ratio response into measurements of cytosolic NADH:NAD+ ratios.
4. Notes
Both pcDNA3.1 (Invitrogen) and GW1 (from the former British Biotech) are mammalian expression vectors that are driven by the cytomegalovirus (CMV) promoter. For most cell lines, the pcDNA3.1 expression vector has been widely used, whereas the GW1 expression vector has been used in neuronal cell lines.
Based on the GFP T-Sapphire, Peredox is not spectrally ratiometric, as its fluorescence excitation or emission spectrum contains only a single major peak. To normalize its readouts for sensor concentration, Peredox is attached in tandem to the RFP mCherry (or the YFP mCitrine); the green-to-red (or green-to-yellow) fluorescence ratio then indicates the sensor readout. As the two fluorescent proteins in Peredox-mCherry (or Peredox-mCitrine) differ in photostability, the fluorescence ratio may drift over time; however, depending on the illumination condition, such drifts appear minor in most imaging setups.
Many RFPs including mCherry can aggregate in lysosomes (15). Indeed, we observe bright red puncta in certain cell types expressing Peredox-mCherry. These fluorescent puncta render the green-to-red fluorescence ratios heterogeneous within a cell and difficult to compare among distinct cells. Nonetheless, we minimize puncta by using stable low biosensor expression rather than transient transfection with high expression. We can also preclude these puncta by using Peredox-mCitrine or nuclear-targeted Peredox-mCherry.
Peredox-mCitrine can be used together with a red biosensor (such as pHRed (13)) or a red small-molecule indicator (such as tetramethylrhodamine methyl ester TMRM) for simultaneous imaging of two metabolic readouts.
Consult the American Type Culture Collection (http://www.atcc.org) for protocols on cell culture. We purchase all cell culture media, serum, and supplements from Invitrogen.
Consult (14) for protocols on generation of stable cell lines using retroviral transduction.
For coating the growth surface for cell adhesion, other cell types may prefer poly-D-lysine or collagen.
When bubbled with 95% air and 5% CO2, this extracellular solution should yield an osmolarity of ~280 Osm and a pH of 7.4. For other cell types, the salt concentration can be varied to optimize the solution osmolarity.
Once prepared, stock solutions can be stored at −20°C for several months. Repeated freezing and thawing are not recommended (16).
The NADH biosensor can be expressed in mammalian cells transiently via transfection or stably via viral transduction of the encoding plasmid DNA. To decide between transient and stable expression, the two main considerations are time and uniformity of sensor expression. On one hand, the major advantage of transient over stable expression is time: Given an optimized protocol, cells can be imaged as soon as 1–2 days after transient transfection, whereas it takes several weeks to generate and expand individual stable cell clones. On the other hand, the major advantage of stable expression is that both the sensor concentration and the fluorescence ratio response appear more uniform, rendering signal comparison among individual cells more reliable. In contrast, early on after transient transfection, the baseline green-to-red (or green-to-yellow) fluorescence ratios can vary widely across cells, as the two fluorescent proteins in Peredox-mCherry (or Peredox-mCitrine) differ in their maturation periods, and it could take days for their fluorescence in cells to arrive at steady states. Here, while the baseline green-to-red fluorescence ratios can be nonuniform, fluorescence ratios from each cell can be normalized after sensor calibration. We acquire the sensor signal of each cell in well-defined conditions in terms of cytosolic NADH-NAD+ redox state: We obtain the minimum green-to-red fluorescence ratio in each cell by perfusion with pyruvate alone, and we use that signal to normalize the data series of that cell accordingly. Once normalized, fluorescence responses can be compared among cells (see Fig. 2).
The basic requirements for live-cell fluorescence imaging include a fluorescence microscope with appropriate filters, image acquisition software, and environmental control. For long-term imaging, we recommend a widefield microscope over a confocal microscope. Despite a lack of optical sectioning, widefield microscopy requires lower illumination intensity, thus reducing sensor photobleaching and cell phototoxicity. We use either an upright microscope (such as Olympus BX51) or an inverted microscope (such as Nikon Eclipse Ti), although the use of a dipping objective will slow solution exchange.
Sensor fluorescence can be acquired using appropriate filter sets controlled by automated filter wheels (see Table 2). When using a light source with an integrated monochromator (such as TILL Photonics Polychrome IV), a filter set can be used with selective excitation to acquire both T-Sapphire and mCherry fluorescence (see Table 3) or both T-Sapphire and mCitrine fluorescence (see Table 4).
Proper environmental control is a prerequisite for cell viability and long-term imaging. Cells can be continuously perfused with defined extracellular solutions (see Materials 2.2.2), which have been bubbled with 95% air and 5% CO2 at a near physiological temperature of ~35°C. To avoid outgassing, solutions are first warmed to physiological temperature before bubbling. Temperature can be maintained using an in-line heater or chamber heater, and perfusion rate can be regulated by a peristaltic pump or gravity flow. As an alternative to continuous perfusion, cells can be placed within an environmental chamber with temperature and CO2 regulation but no solution exchange, for instance, in a multi-well microplate format.
In qualitative assessment, we monitor the relative changes in the cytosolic NADH:NAD+ ratio, whether it increases, decreases, or remains unchanged. In contrast, with quantitative measurements, we determine the actual cytosolic NADH:NAD+ ratio. Unlike qualitative assessment, quantitative measurements require proper calibration of the biosensor and are thus experimentally more demanding.
We calibrate the biosensor response by setting cytosolic NADH:NAD+ ratios in individual live cells using exogenous lactate and pyruvate without cell permeabilization. As this calibration method relies on the equilibration between intracellular and extracellular lactate and pyruvate concentrations, cells lacking monocarboxylate transporters or lactate dehydrogenases may necessitate other calibration method: For instance, one can permeabilize cells using α-toxin or saponin (17) and perfuse solutions containing NADH and NAD+ at various NADH:NAD+ ratios.
For effective live-cell calibration using lactate and pyruvate, we need to establish the following two conditions: First, we eliminate the effect of glycolysis on the cytosolic NADH-NAD+ redox state; otherwise, lactate and pyruvate alone would not be sufficient to set the NADH-NAD+ redox state, and the calibration curve would be shifted upward (4). Thus, in the calibration procedure, we do not supply cells with glucose. Alternatively, in conditions where glucose cannot be easily removed, cells can be calibrated in the presence of a glycolytic inhibitor, such as iodoacetate; after application of 10 mM pyruvate with 0.5 mM iodoacetate, we can obtain the minimal ratio response of Peredox (4). Second, we effectively control the extracellular concentrations of lactate and pyruvate by using continuous perfusion of fresh solutions; only then can we set intracellular lactate:pyruvate ratios and cytosolic NADH:NAD+ ratios. Notably, for multi-well microplates and other formats that are incompatible with continuous perfusion, extracellular concentrations of lactate and pyruvate can vary dramatically due to cell metabolism. To ensure constant extracellular concentrations of lactate and pyruvate for calibration, one can plate small number of cells in a large volume of solution, for instance, 1,000 cells in 3 ml of solution in the well of a 24-well plate.
With optimal optics, Peredox-mCherry has a dynamic range of roughly 2.5-fold, whereas Peredox-mCitrine has a smaller dynamic range of about 2-fold, due to the greater spectral overlap between the containing fluorescent proteins. The dynamic range may appear smaller with nonoptimal optics.
-
We find that a lactate:pyruvate ratio of 37 corresponds to a half-maximal response in Peredox-mCherry. For data fitting by a logistic function, we use a Hill coefficient of 1.7. From the calibration data, we derive the following equation:
where F is the normalized fluorescence response, and X is the extracellular lactate:pyruvate ratio.Assuming a constant physiological pH of 7.4 and that the LDH reaction is at equilibrium,We derive the following equation:Combining the equations, we get the following equation:
orWhere F is the normalized fluorescence response, and R is the cytosolic NADH:NAD+ ratio.
Common cell culture medium such as Dulbecco’s Modified Eagle’s Medium (DMEM) contains millimolar pyruvate, which can be catalyzed by lactate dehydrogenase to lower the cytosolic NADH:NAD+ ratios in cells. To elevate the baseline response of Peredox, cells can be placed in a pyruvate-deficient culture medium such as Roswell Park Memorial Institute (RPMI)-1640.
While tuned to sensing the cytosolic NADH:NAD+ ratio, Peredox cannot report physiological changes in the mitochondrial NADH:NAD+ ratio, which has been estimated to be 100- to 1000-fold higher than the cytosolic NADH:NAD+ ratio (18). Although a fluorescent sensor has been engineered to monitor the mitochondrial NADH pool (19), it does not report the mitochondrial NADH:NAD+ ratio.
Beyond live-cell microscopy, Peredox may be monitored by fluorescence-activated cell sorting (FACS) or a fluorescence platereader. For adherent cells, we do not recommend using Peredox with FACS, as metabolic state may be altered by cell detachment required by this technique (20).
Table 2.
Chroma filters for imaging Peredox-mCherry or -mCitrine without a monochromator
| FP | Filter set | Excitation filter | Dichroic | Emission filter |
|---|---|---|---|---|
| T-Sapphire | - | ET405/20x | T425LPXR | ET525/50m |
| RFP mCherry | 41043 | HQ575/50x | Q610LP | HQ640/50m |
| YFP mCitrine | 41028 | HQ500/20x | Q515LP | HQ535/30m |
Table 3.
Filters for imaging Peredox-mCherry with a monochromator
| FP | Excitation | Excitation filter | Dichroic | Emission filter |
|---|---|---|---|---|
| T-Sapphire RFP mCherry |
405 nm 575 nm |
Chroma 69002x | Chroma 69002bs | Semrock FF01- 524/628-25 |
Table 4.
Chroma filters for imaging Peredox-mCitrine with a monochromator
| FP | Excitation | Excitation filter | Dichroic | Emission filter |
|---|---|---|---|---|
| T-Sapphire | 405 nm | - | 515DCXR | D535/25m |
| YFP mCitrine | 485 nm |
Acknowledgments
We thank Mathew Tantama for careful reading of this manuscript. This work was supported by the Albert J. Ryan fellowship, the Stuart H.Q. and Victoria Quan predoctoral fellowship in neurobiology (both to Y.P.H.), and the U.S. National Institutes of Health (R01 NS055031 to G.Y.).
References
- 1.Nicholls DG, Ferguson SJ. Bioenergetics. Third Edition Academic Press; London: 2002. [Google Scholar]
- 2.Avi-Dor Y, Olson JM, Doherty MD, Kaplan NO. Fluorescence of pyridine nucleotides in mitochondria. J Biol Chem. 1962;237:2377–2383. [Google Scholar]
- 3.Rocheleau JV, Head WS, Piston DW. Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J Biol Chem. 2004;279:31780–31787. doi: 10.1074/jbc.M314005200. [DOI] [PubMed] [Google Scholar]
- 4.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sickmier EA, Brekasis D, Paranawithana S, Bonanno JB, Paget MSB, Burley SK, Kielkopf CL. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, von Wachenfeldt C. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol. 2008;69:466–478. doi: 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin KJ, Strain-Damerell CM, Xie K, Brekasis D, Soares AS, Paget MS, Kielkopf CL. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol Cell. 2010;38:563–575. doi: 10.1016/j.molcel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev. 2009;38:2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tantama M, Hung YP, Yellen G. Optogenetic reporters: Fluorescent protein-based genetically encoded indicators of signaling and metabolism in the brain. Prog Brain Res. 2012;196:235–263. doi: 10.1016/B978-0-444-59426-6.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bücher T, Brauser B, Conze A, Klein F, Langguth O, Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem. 1972;27:301–317. doi: 10.1111/j.1432-1033.1972.tb01840.x. [DOI] [PubMed] [Google Scholar]
- 13.Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 15.Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A. GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct. 2008;33:1–12. doi: 10.1247/csf.07011. [DOI] [PubMed] [Google Scholar]
- 16.Passonneau JV, Lowry OH. Enzymatic analysis: a practical guide. Humana Press; Totowa: 1993. [Google Scholar]
- 17.Schulz I. Permeabilizing cells: some methods and applications for the study of intracellular processes. Methods Enzymol. 1990;192:280–300. doi: 10.1016/0076-6879(90)92077-q. [DOI] [PubMed] [Google Scholar]
- 18.Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Jin J, Hu Q, Zhou H-M, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]