Maize FASCIATED EAR4 encodes a DNA binding protein that regulates the development of the maize shoot stem cell population, in a manner that may be different from previously known mechanisms.
Abstract
Plant architecture is dictated by precise control of meristematic activity. In the shoot, an imbalance in positive or negative maintenance signals can result in a fasciated or enlarged meristem phenotype. fasciated ear4 (fea4) is a semidwarfed mutant with fasciated ears and tassels as well as greatly enlarged vegetative and inflorescence meristems. We identified FEA4 as a bZIP transcription factor, orthologous to Arabidopsis thaliana PERIANTHIA. FEA4 was expressed in the peripheral zone of the vegetative shoot apical meristem and in the vasculature of immature leaves and conspicuously excluded from the stem cell niche at the tip of the shoot apical meristem and from incipient leaf primordia. Following the transition to reproductive fate, FEA4 was expressed throughout the entire inflorescence and floral meristems. Native expression of a functional YFP:FEA4 fusion recapitulated this pattern of expression. We used chromatin immunoprecipitation-sequencing to identify 4060 genes proximal to FEA4 binding sites, including ones that were potentially bound and modulated by FEA4 based on transcriptional changes in fea4 mutant ears. Our results suggest that FEA4 promotes differentiation in the meristem periphery by regulating auxin-based responses and genes associated with leaf differentiation and polarity, potentially in opposition to factors such as KNOTTED1 and WUSCHEL.
INTRODUCTION
Shoot development is a plastic process that depends on the activity of pluripotent stem cells resident within the shoot apical meristem (SAM). Differences in the initiation, determinacy, and size of different classes of meristems are responsible for variation in vegetative and reproductive architecture within the plant kingdom. Plant architecture, especially inflorescence architecture, is critical for reproductive success and is therefore a primary determinant of crop yield.
The stem cells contained within the SAM have two roles: first, to divide to give rise to daughter cells that supply founder cells for organ initiation; and second, self-replacement. An imbalance in positive or negative signals for stem cell maintenance can result in enlarged or consumed meristem phenotypes; therefore, the stem cell population in the SAM must be precisely regulated. The principal mechanism of stem cell counting in angiosperms is the CLAVATA (CLV)-WUSCHEL (WUS) negative feedback pathway, which was first characterized in Arabidopsis thaliana (Brand et al., 2000; Schoof et al., 2000). WUS is a homeodomain protein expressed in the organizing center beneath the stem cell niche, which acts non-cell-autonomously to promote stem cell fate in the central zone, likely by repressing expression of genes involved in organ differentiation (Mayer et al., 1998; Schoof et al., 2000; Yadav et al., 2013). The stem cells express a gene that encodes a small secreted peptide, CLV3, which is perceived by a battery of receptors, principally CLV1, CLV2, and RECEPTOR-LIKE PROTEIN KINASE2/TOADSTOOL, resulting in the repression of WUS transcription (Clark et al., 1997; Brand et al., 2000; Schoof et al., 2000; Kinoshita et al., 2010).
The framework of this pathway is conserved in the grasses, including the major crop species rice (Oryza sativa) and maize (Zea mays). Two maize fasciated ear mutants, thick tassel dwarf1 and fasciated ear2, have lesions in the maize orthologs of CLV1 and CLV2, respectively, and the rice mutant fon1 harbors a mutation in the CLV1 ortholog (Taguchi-Shiobara et al., 2001; Suzaki et al., 2004; Bommert et al., 2005). The CLV pathway in rice represents an interesting variation on the theme, with three distinct CLV3-like peptides acting to restrict stem cell populations in vegetative, inflorescence, and floral meristems (Suzaki et al., 2009). Aside from the identification of two CLV-like receptors, a number of additional meristem size regulators have been identified in maize. One of these factors, COMPACT PLANT2 (CT2), appears to be directly involved in the CLV pathway, as it encodes the α-subunit of a heterotrimeric GTPase, which physically interacts with FEA2 (Bommert et al., 2013b). Furthermore, double mutants between ct2 and fea2 are not significantly stronger than either single mutant (Bommert et al., 2013b). The association of G-protein signaling with a leucine-rich receptor (LRR)-receptor-like protein provides a plausible mechanism of signaling for FEA2, which lacks a kinase domain (Bommert et al., 2013b). Another mutant, aberrant phyllotaxy1, which encodes an A-type cytokinin response regulator, displays increased SAM size, as well as a switch from alternate to decussate phyllotaxy (Giulini et al., 2004). Molecular cloning of this mutant was the first direct evidence that the cytokinin signaling pathway regulates meristem size in plants (Giulini et al., 2004). Subsequent studies have clarified a role for Arabidopsis response regulators in integrating cytokinin and auxin signals with the CLV-WUS stem cell-counting feedback loop (Lee et al., 2009; Zhao et al., 2010).
At the opposite end of the phenotypic spectrum are mutants that have a reduced meristem size or fail to maintain a productive SAM throughout development. KNOTTED1 (KN1) is a homeodomain protein that is required to maintain meristematic fate, as loss-of-function alleles exhibit smaller meristems and meristem termination in some genetic backgrounds (Kerstetter et al., 1997; Vollbrecht et al., 2000). A recent genome-wide chromatin immunoprecipitation and mRNA sequencing analysis suggested that KN1 promotes meristematic activity by regulating a cascade of transcription factors and hormone biosynthesis/signaling components (Bolduc et al., 2012).
Despite these findings, less is known about the factors controlling meristem size in maize and additional factors are likely to function in this critical developmental process (Pautler et al., 2013). The functional subdomains of the meristem, such as the central zone, organizing center, and peripheral zone, are less well defined in maize due to a paucity of molecular markers. Identification of new regulators through positional cloning of meristem mutants can address these unanswered questions. Here, we describe the phenotypic and molecular characterization of fasciated ear4 (fea4), a maize mutant with enlarged meristems. FEA4 encodes a basic-region leucine zipper (bZIP) transcription factor, orthologous to the Arabidopsis gene PERIANTHIA, a mutant affected in floral organ number, but not meristem size per se (Running and Meyerowitz, 1996; Chuang et al., 1999). Expression analysis by in situ hybridization and fluorescent protein fusions suggest that FEA4 is dynamically and specifically expressed in different meristem types. Genetic analysis of double mutants demonstrates that FEA4 acts in parallel to the canonical CLV-WUS pathway. Chromatin immunoprecipitation-sequencing (ChIP-seq) and expression profiling by RNA sequencing (RNA-seq) suggest that FEA4 is required to regulate the auxin response and leaf differentiation programs in the periphery of the meristem, suggesting a novel mechanism of meristem size regulation that is spatially and mechanistically distinct from the CLV-WUS model. Our results illustrate the power of studying basic plant processes in diverse model systems because novel functions are often revealed. These functions may be masked in specific systems due to genetic or gene network redundancy.
RESULTS
Phenotypic Characterization of the fea4 Mutant
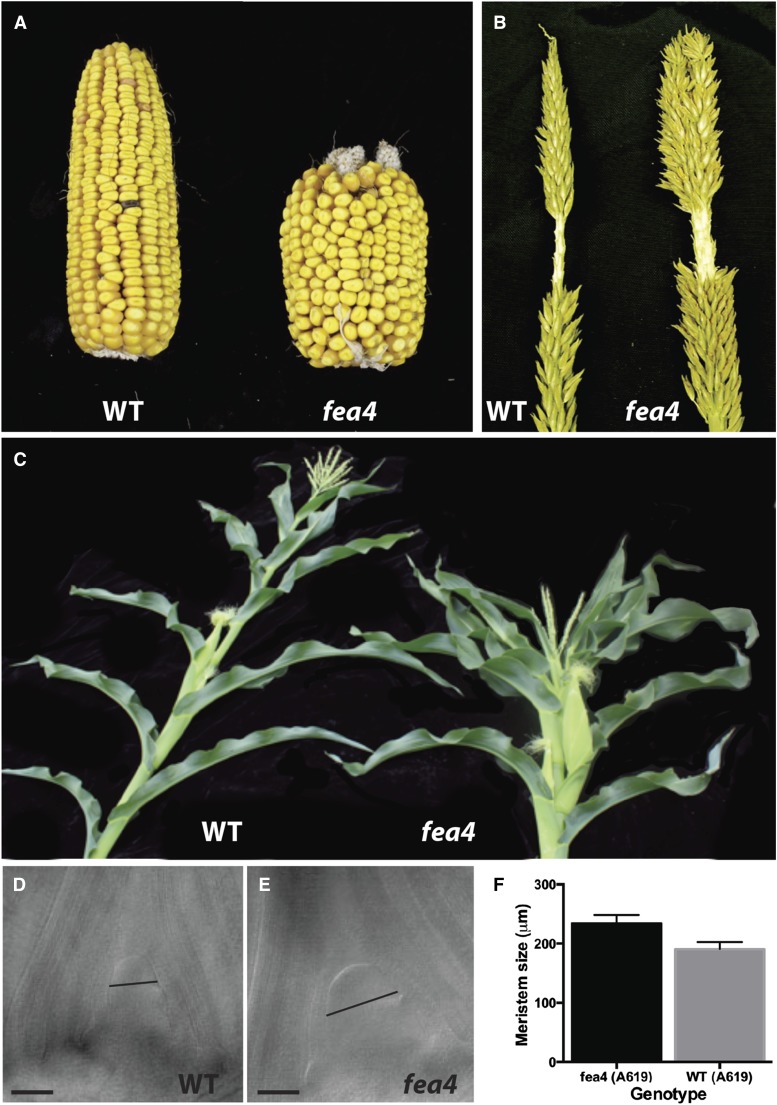
fea4 was originally isolated as a fasciated ear mutant in a screen of ethyl methanesulfonate (EMS)-mutagenized A619 inbred maize. Further phenotypic characterization was performed in the A619, B73, Mo17, and W22 inbred backgrounds. Of these, the phenotype was particularly expressive in A619 and B73. The most dramatic phenotypes in the fea4 mutant were a greatly thickened tassel and massively fasciated ears with disorganized seed rows (Figures 1A and 1B). Mutant tassels had a thicker main rachis and a much higher spikelet density than wild-type siblings (Figure 1B, Table 1). fea4 mutants were also semidwarfed in the A619 inbred background (Figure 1C, Table 1); however, this phenotype was less pronounced in B73 and other inbred lines.
Figure 1.
Phenotypes of fea4 Mutants.
fea4 ears were massively fasciated and shorter than wild-type ears and had disorganized seed rows (A). Mutant tassels had a higher spikelet density and thicker rachis diameter than wild-type tassels (B). fea4 mutants were semidwarfed in the A619 inbred background (C), and mutants had a larger vegetative SAM than wild-type siblings 14 d after germination ([D] and [E]), quantified in (F) (significant at P value < 0.05, n = 8 to 9 of each genotype). Black lines in (D) and (E) represent the point of measurement for cleared meristems: the widest point of the meristem above the bulge of the initiating leaf primordium. Error bars in (F) represent sd. Bars = 100 μm.
Table 1. fea4 Mutant Phenotype Quantification in Two Different Inbred Backgrounds, B73 and A619, Grown in Two Locations.
| Trait | n | Wild Type | fea4 |
|---|---|---|---|
| B73, Uplands Farm | |||
| SAM width 14 DAG (μm) | 20 | 179.83 ± 8.38 | 226.16 ± 13.07a |
| Plant height (cm) | 49 | 249.69 ± 11.1 | 213.13 ± 23.94a |
| Spikelet density/cm | 22 | 21 ± 1.97 | 29.8 ± 1.98a |
| Kernel row number | 32 | 17.14 ± 1.51 | 28 ± 2.31a |
| Tassel branch number | 50 | 7.86 ± 1.66 | 6.89 ± 2.21 |
| Stamen number per floret | 50 | 3.0 ± 0 | 3.0 ± 0 |
| B73, Gill Tract | |||
| Plant height (cm) | 11 | 203.20 ± 17.00 | 155.56 ± 33.49a |
| Spikelet density/cm | 11 | 13.55 ± 2.81 | 20.09 ± 6.65a |
| Kernel row number | 11 | 16.91 ± 4.25 | 34.91 ± 12.53a |
| Tassel branch number | 11 | 10.55 ± 3.47 | 7.27 ± 3.77a |
| A619, Uplands Farm | |||
| SAM width 14 DAG (μm) | 20 | 190.67 ± 11.97 | 233.97 ± 14.2a |
| Plant height (cm) | 54 | 204.28 ± 12.19 | 144.61 ± 13.88a |
| Spikelet density/cm | 24 | 18.8 ± 1.39 | 33.5 ± 8.8a |
| Kernel row number | 28 | 15.80 ± 1.48 | 19.67 ± 2.39a |
| Tassel branch number | 33 | 9.4 ± 3.43 | 4.07 ± 1.75a |
| Stamen number per floret | 50 | 3.0 ± 0 | 2.78 ± 0.42a |
Plant phenotypes were quantified at Gill Tract, Albany, CA, and Uplands Farm, Cold Spring Harbor, NY. Plants were grown in the greenhouse for SAM measurements. Measurements represent mean ± sd. Wild-type plants represent true wild-type or heterozygous wild-type phenotype segregants. DAG, days after germination.
Significantly different than wild-type value, Student’s t test, P value < 0.05.
fea4 plants were also characterized by a larger than normal SAM diameter (Figures 1D to 1F). This may explain the decreased plant stature observed in A619 (Figure 1C), as a larger SAM is correlated with decreased plant height in several fasciated mutants (Bommert et al., 2005). fea4 phenotypes were fully recessive and were not observed in heterozygotes (Table 1).
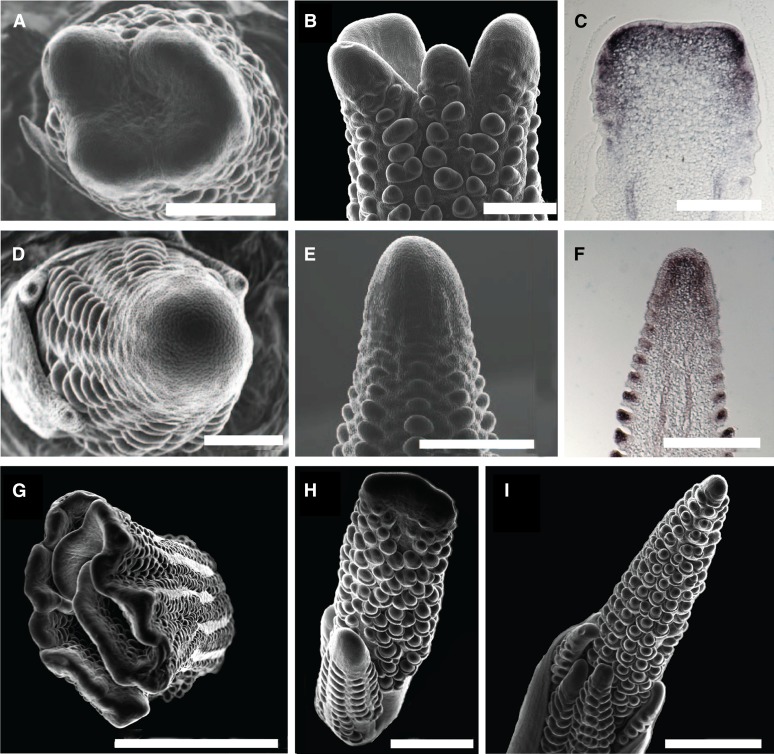
To better understand the ontogeny of the enlarged inflorescence structures, we subjected developing fea4 ears and tassels to scanning electron microscopy. This analysis revealed that the fasciated inflorescences were caused by enlarged tassel and ear inflorescence meristems (IMs) (Figure 2). The large, flattened IMs in the fea4 mutant appeared to function similarly to wild-type IMs, as they initiate rows of axillary meristems and also express the meristem marker kn1 in a pattern resembling the wild type (Figures 2C and 2F). Fasciation became progressively more severe as ear development progressed, culminating in an elongated and folded structure that bore little resemblance to the wild-type ear (Figure 2G). The axillary meristems of the inflorescence, including spikelet pair (SPM), spikelet (SM), and floral meristems (FMs), were not obviously enlarged in the mutant; furthermore, there was no obvious increase in floral organ number, unlike in other fasciated ear mutants, such as fea2 and td1 (Supplemental Figure 1) (Taguchi-Shiobara et al., 2001; Bommert et al., 2005). However, ∼25% of fea4 (A619) florets contained a reduced number of stamens, whereas variation in stamen number was not observed in wild-type florets (Table 1; Supplemental Figure 1). No further abnormalities were observed in floral organ number or patterning, suggesting that FMs were only subtly affected in the balance of organ specification or differentiation.
Figure 2.
Microscopic Phenotypes of fea4 Mutants.
fea4 ear inflorescence meristems were enlarged and flattened ([A] and [B]), whereas wild-type ears had tapered, conical shaped inflorescence meristems ([D] and [E]). In situ hybridization with the KN1 meristem marker showed an expanded domain of expression throughout the enlarged inflorescence meristem in fea4 ears (C) compared with wild-type ears (F). fea4 meristem fasciation became progressively more severe as ear development progressed (G). Tassel inflorescence meristems were similarly affected in fea4 (H) compared with the wild type (I). Bars = 500 μm in (A) and (B), 250 μm in (C) to (F), 2 mm in (G), and 1 mm in (H) and (I).
Cloning of fea4
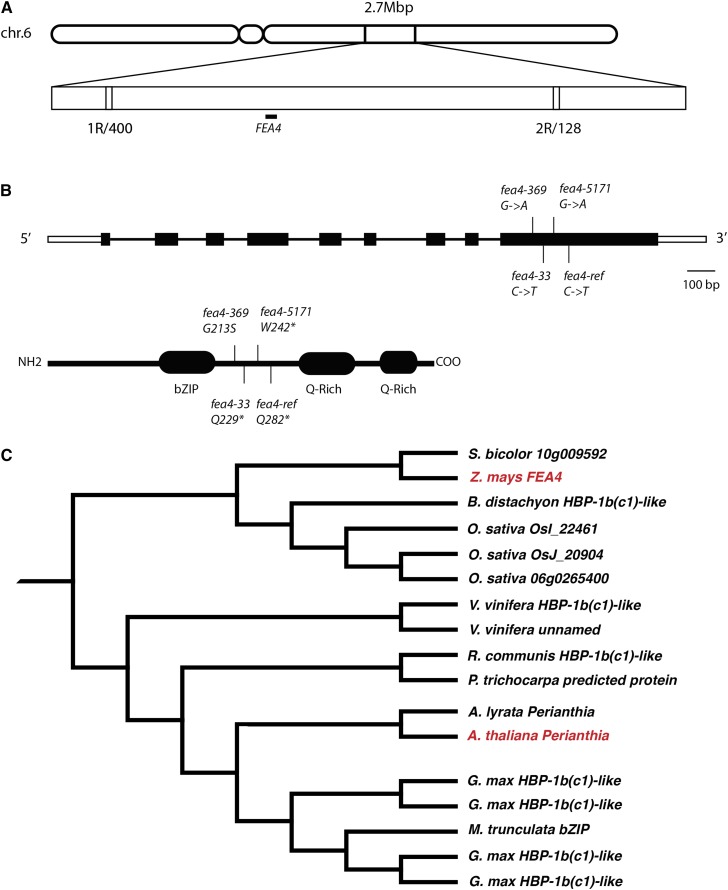
We mapped fea4 to the long arm of chromosome 6 by bulked segregant analysis and performed fine mapping by genotyping 528 mutants from an F2 mapping population (Figure 3A). Two cleaved-amplified polymorphic sequence (CAPS) markers delineated a 2.7-Mb region containing ∼30 predicted genes. We sequenced some of these and found a bZIP transcription factor, in which the reference mutant (fea4-ref) harbored an EMS-induced C-T transition, which caused a premature stop codon. Two additional independently derived alleles (fea4-5171 and fea4-33) also had early stop codons, confirming the identity of the gene underlying the mutant phenotype (Figure 3B). In all three non-sense alleles, the stop codons fall after the bZIP domain, but before two glutamine-rich regions, which are associated with transcriptional activation and may mediate posttranslational regulation of bZIP proteins (Li et al., 2009). Since these alleles remove about half of the FEA4 coding region, they are likely to be null alleles. Subsequently, we isolated four additional alleles from various sources, including EMS and transposon-mutagenized seed stocks, which demonstrated noncomplementation with the fea4 reference allele (Figure 3B; Supplemental Table 1).
Figure 3.
Mapping and Molecular Cloning of fea4.
Two CAPS markers delineated a 2.7-Mb mapping interval on the long arm of chromosome 6, containing ∼30 genes (A). A gene encoding a bZIP transcription factor in this interval (GRMZM2G133331) contained multiple independent lesions (B). fea4 encodes a TGA-class bZIP transcription factor orthologous to the Arabidopsis gene PAN (C). Red font highlights the location of FEA4 and PAN in the tree.
A maximum likelihood phylogenetic tree was constructed from a CLUSTAL alignment of the top 100 FEA4 BLAST hits (Figure 3C; Supplemental Figure 2B and Supplemental Data Set 1). This analysis revealed that FEA4 is more closely related to the Arabidopsis gene PERIANTHIA (PAN) than any other maize gene and therefore likely encodes its ortholog. FEA4 and PAN share ∼59% amino acid identity but have very divergent N-terminal sequences, a common feature of TGA-class bZIP proteins (Supplemental Figure 2) (Jakoby et al., 2002). pan is an Arabidopsis mutant characterized by an increase in floral organ number, without a corresponding increase in FM size (Running and Meyerowitz, 1996). pan mutants also have a mild increase in inflorescence meristem size when the plants are grown in short days (Maier et al., 2011). We also found their IMs to be moderately enlarged (Supplemental Figure 3), but this increase was not anywhere near as severe as the massive fasciation observed in fea4. However, our results suggest that FEA4 and PAN have a conserved function to regulate inflorescence meristem size.
Expression Analysis of fea4
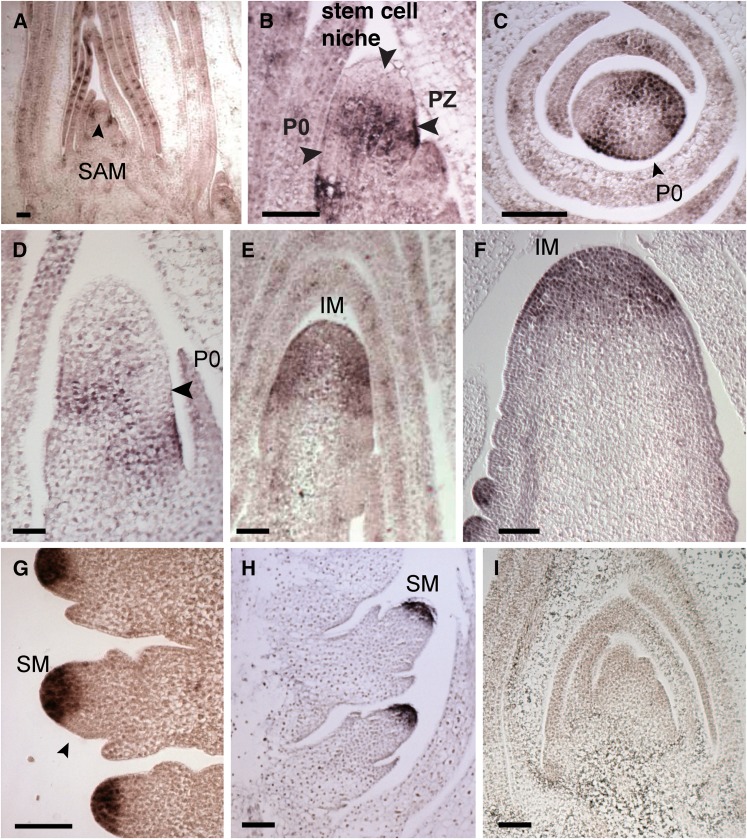
We performed RNA in situ hybridization with a FEA4 antisense probe to determine its expression pattern throughout shoot development. During the vegetative phase, FEA4 was expressed in the peripheral zone of the SAM, but conspicuously excluded from the predicted position of the stem cell niche at the tip of the SAM and from the incipient leaf primordium (P0) (Figures 4A to 4C). FEA4 was also expressed in a domain beneath the P0 and in the vasculature of immature leaves (Figures 4A to 4C). The peripheral zone expression pattern persisted until the inflorescence transition (Figure 4D), but following the transition, FEA4 was expressed throughout the entire IM of the tassel and ear and also throughout the other inflorescence meristem types (SPMs and SMs; Figures 4E to 4G). Similar to the pattern observed in the SAM, FEA4 was downregulated at the site of incipient lateral organ formation (Figure 4G, arrowhead). In control experiments, a shorter antisense probe created from a 5′ portion of the FEA4 cDNA gave rise to a signal identical to the full-length probe (Figure 4H), and a sense probe produced no signal (Figure 4I).
Figure 4.
Expression Analysis of FEA4.
In situ hybridization with a FEA4 antisense probe showing expression in the SAM and vasculature of young leaves surrounding the meristem (A). FEA4 is expressed predominantly in the peripheral zone of the SAM, excluded from the stem cell niche and the incipient leaf P0 (arrowheads, [B]). Transverse SAM section, showing exclusion from the P0 (C). The peripheral zone specific expression pattern persists through the transition stage (D), but FEA4 is expressed throughout the entire inflorescence meristem of the tassel (E) and ear (F) following the floral transition. The arrowhead in (D) indicates lack of expression in the initiating lateral organ primordium. FEA4 is subsequently expressed throughout the entire spikelet pair (F) and spikelet and floral meristems (G) but is again excluded from the site of lateral organ initiation (arrowhead, [G]). A shorter probe from the 5′ half of the cDNA produced the same pattern (H) and a sense FEA4 probe produced no signal after overnight hybridization (I). Bars = 100 μm in all panels.
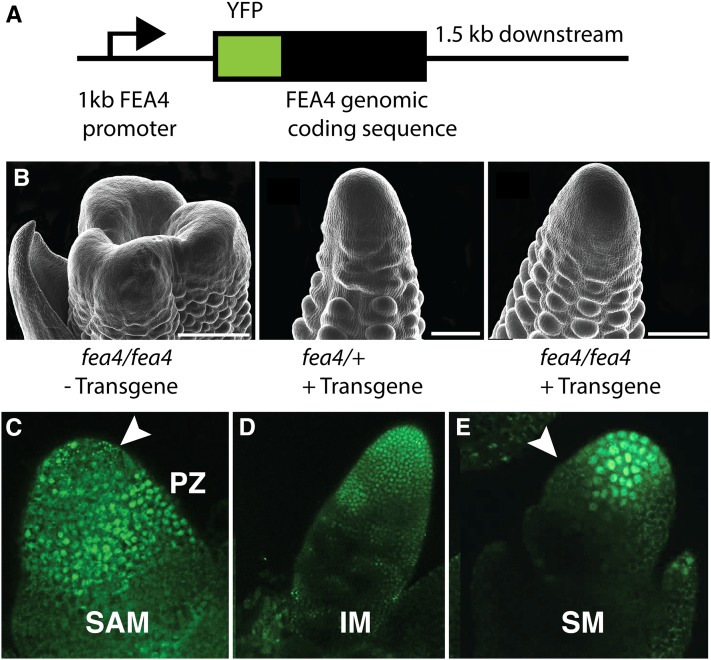
To examine the subcellular and tissue-scale localization of the FEA4 protein, we constructed a translational fusion of the FEA4 coding sequence and yellow fluorescent protein (YFP) under the control of its native promoter (Figure 5A). This construct was transformed into maize and backcrossed twice to the fea4-ref mutant to assess complementation. The presence of the transgene was sufficient to rescue the inflorescence fasciation phenotype in two independent events, indicating that the fusion protein was functional (Figure 5B). Plants with wild-type phenotype from segregating families were genotyped for the fea4-ref mutation to verify the complementation result (n = 10). YFP:FEA4 expression recapitulated the pattern of expression observed by in situ hybridization. Strong nuclear expression was observed in all stages examined, from vegetative to inflorescence, and was also present in young leaves surrounding the SAM (Figures 5C to 5E). The YFP:FEA4 protein was absent from the predicted stem cell niche of the vegetative SAM (Figure 5C, arrowhead) and from sites of lateral organ initiation (Figure 5E, arrow), but it was present throughout the central region of the inflorescence and spikelet meristems (Figures 5D and 5E), similar to the mRNA expression pattern.
Figure 5.
Protein Accumulation of YFP:FEA4 Translational Fusion Recapitulates the mRNA Expression Pattern.
YFP was fused in frame to the N terminus of FEA4 and expressed under 1 kb of native upstream sequences and 1.5 kb of downstream sequences (A). Expression of the transgene was sufficient to rescue the mutant phenotype in the inflorescence (B). Strong nuclear fluorescence was observed in the peripheral zone of the SAM (C), and nuclear fluorescence was absent from the stem cell niche (arrowhead, [C]). Note that the small punctate green fluorescence in the stem cell niche is due to autofluorescence (Supplemental Figure 6). Fluorescent signal also accumulated throughout the inflorescence meristem (D). FEA4 was also expressed in the axillary meristems of the ear, such as spikelet meristems (E), and was excluded from the site of lateral organ initiation (arrowhead, [E]). Bars = 500 μm in the leftmost panel in (B) and 250 μm in center and right panels.
Double Mutant Analysis
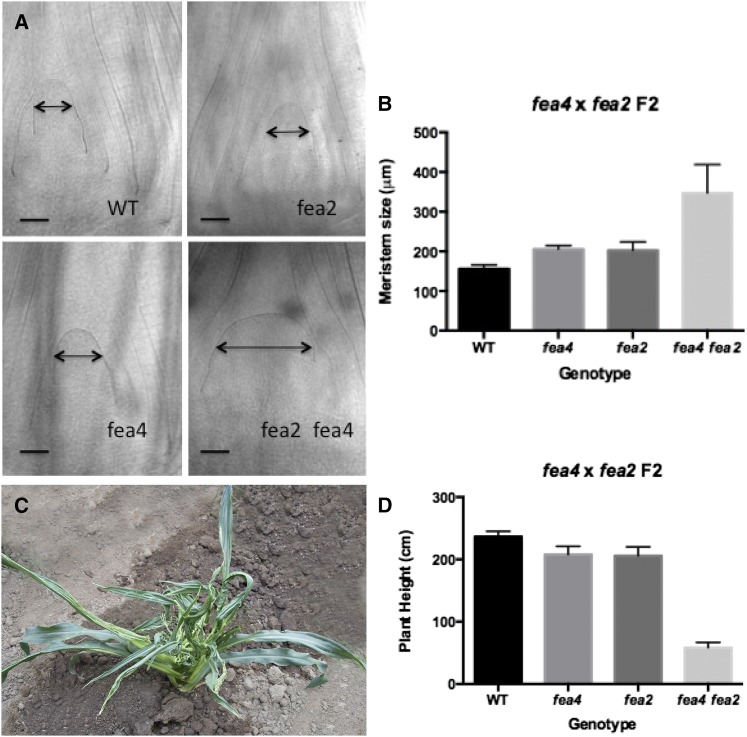
To establish how fea4 interacts with other loci that regulate meristem size, we created F2 populations segregating fea4 and fea2, the maize ortholog of CLV2, in a B73 background. The double mutants displayed a range of highly synergistic phenotypes in vegetative and reproductive structures, including extreme dwarfism and disorganized, split shoots (Figure 6). To acquire a quantitative readout of the genetic interaction, we genotyped plants from a segregating family and measured the size of the SAM at 14 d after planting. SAM diameter was increased by approximately one-third in each of the single mutants relative to the wild type (202.5 ± 21.3 μm for fea2; 205.6 ± 9.7 μm for fea4, compared with 156.1 ± 10.5 μm for the wild type, differences significant at P value < 0.05, Student’s t test). For an additive interaction, we would expect the double mutant to be the wild-type value plus the sum of the mutant differences (156.1 + (202.5 – 156.1) + (205.6 – 156.1) = 252 μm). However, the double mutants were significantly larger, at 347.3 ± 70.9 μm (P value < 0.05, Student’s t test; Figures 6A and 6B). Therefore, we classified this double mutant interaction as a synergistic one and conclude that fea4 acts in parallel to fea2 and is highly unlikely to be involved in transducing CLV-like signals (Laufs et al., 1998; Prigge and Wagner, 2001). In contrast, epistasis between fasciated mutants directly involved in the CLV signaling pathway is readily observed, for example, between fea2 and ct2 (Bommert et al., 2013b).
Figure 6.
SAM Size in F2 Families Segregating fea4 and fea2 Mutants.
Meristems were dissected and cleared with methyl salicylate 14 d after germination (A). fea4 and fea2 meristems were significantly larger than wild-type meristems (P value < 0.05). fea2 fea4 double mutants were significantly larger than both single mutants (P value < 0.05) and also much larger than the predicted additive point (B). n = 4 to 16 of each genotype. Black lines with arrowheads in (A) represent the point of measurement. Mature double mutants ([C] and [D]) showed extreme dwarfism and highly disorganized, split shoots.
Transcriptome Profiling and Genome-Wide Occupancy of FEA4
To gain insight into transcriptional regulation by FEA4, we used RNA-seq to profile genome-wide expression changes in developing ear primordia of fea4 loss-of-function mutants compared with fea4/+ wild-type siblings (Supplemental Figure 4). RNA-seq reads from three biological replicates for the wild type and mutant were mapped to the maize reference genome (AGPv2) and used to quantify expression of 39,656 high-confidence filtered gene set (FGS) gene models (ZmB73v5b.60; gramene.org; Supplemental Table 2 and Supplemental Data Set 2). Using the DESeq software (Anders and Huber, 2010), we identified 422 differentially expressed (DE) genes (P < 0.05); 283 DE genes were upregulated in fea4 mutants, and 172 were downregulated (Supplemental Data Set 3). Downregulated genes were highly enriched for biological processes related to multicellular development (GO:0007275, P = 2.77e-05), transcriptional regulation (GO:0006355; P = 0.002), and response to hormone stimulus (GO:0009725, P = 0.008) (Supplemental Data Set 4). Many downregulated genes in fea4 mutants were key regulators of axillary meristem identity and determinacy in maize, such as BARREN STALK1 (BA1), RAMOSA1 (RA1), and RA3. In addition, homeobox transcription factors (TFs), notably HD-ZIPs, were significantly downregulated in fea4 mutants, as were genes involved in response to auxin stimulus (GO:0009733, P = 0.005), including a number of AUXIN RESPONSE FACTOR (ARF) TFs (Table 2). Upregulated genes in fea4 mutants were enriched for biological processes related to translation (GO:0006412, P = 4.12e-09) as well as cellular biosynthetic (GO:0044249, P = 2.05e-06) and metabolic processes, e.g., sugar metabolism (GO:0005996, P = 1.04e-04; GO:0006006, P = 8.82e-04) (Supplemental Data Sets 3 and 4). Interestingly, a panel of 18 meristem marker genes, comprising CLV and WUS-related genes, as well as additional genes defined by fasciated ear mutants, showed no significant changes in gene expression (Supplemental Table 3), supporting our genetic data that suggest FEA4 acts in parallel to CLV-WUS signaling.
Table 2. Differentially Expressed Genes in fea4 Mutants Involved in Determinacy and Differentiation and/or Auxin-Related Processes.
| Gene ID | Annotationa | Expression in fea4 | Adjusted P Value | FEA4 Boundb | |
|---|---|---|---|---|---|
| Determinacy and differentiation | GRMZM2G397518 | BARREN STALK1 | Down | 0.002 | |
| GRMZM2G003927 | RAMOSA1 | Down | 0.028 | ✓ | |
| GRMZM2G014729 | RAMOSA3 | Down | 1.79e−15 | ||
| GRMZM2G452178 | GNARLEY1 | Down | 0.013 | ✓ | |
| GRMZM2G004957 | HOMEODOMAIN-ZIPPER TF | Down | 0.021 | ||
| GRMZM2G116658 | OUTER CELL LAYER3 | Down | 0.019 | ✓ | |
| GRMZM2G123140 | OUTER CELL LAYER4 | Down | 9.08e−06 | ||
| GRMZM2G001289 | HOMEODOMAIN-ZIPPER TF | Down | 0.0009 | ||
| GRMZM2G346920 | ZINC FINGER-HOMEODOMAIN | Down | 1.54e−06 | ||
| GRMZM2G089619 | ZINC FINGER-HOMEODOMAIN | Down | 0.024 | ||
| GRMZM2G430522 | NAC DOMAIN TF [CUC 3] | Down | 2.07e−05 | ✓ | |
| GRMZM2G393433 | NAC DOMAIN TF [CUC1/2] | Down | 0.004 | ✓ | |
| GRMZM2G144744 | DWARF PLANT8 | Down | 0.0009 | ✓ | |
| GRMZM2G431309 | GRAS TF [SCARECROW-LIKE5] | Down | 0.047 | ||
| GRMZM2G427672 | RNA-BINDING [HUA ENHANCER4] | Down | 0.005 | ||
| GRMZM5G809695 | LRR-KINASE [ERECTA] | Down | 0.025 | ✓ | |
| GRMZM2G003509 | HD-ZIP TF [PHABULOSA/ PHAVOLUTA] | Up | 0.0001 | ✓ | |
| Auxin related | GRMZM2G352159 | AUXIN RESPONSE FACTOR2 [ARF2] | Down | 0.032 | |
| GRMZM2G035405 | AUXIN RESPONSE FACTOR6 [ARF6] | Down | 0.034 | ✓ | |
| GRMZM2G089640 | AUXIN RESPONSE FACTOR6 [ARF6] | Down | 9.17e−05 | ✓ | |
| GRMZM2G475882 | AUXIN RESPONSE FACTOR8 [ARF8] | Down | 0.002 | ||
| GRMZM2G115357 | IAA24 | Down | 0.023 | ✓ | |
| GRMZM2G030125 | AUXIN EFFLUX CARRIER | Down | 0.009 | ||
| GRMZM2G067053 | NPH3 [ENHANCER OF PINOID] | Down | 0.046 | ✓ | |
| GRMZM2G099049 | DORMANCY AND AUXIN ASSOCIATED | Down | 0.004 | ||
| GRMZM5G874163 | AUXIN RESPONSIVE B3 TF [ETTIN] | Up | 0.038 | ✓ |
Annotations based on maize IDs; gene names in brackets are based on closest orthologs in Arabidopsis.
DE gene is located within 10 kb of a high-confidence FEA4 binding site.
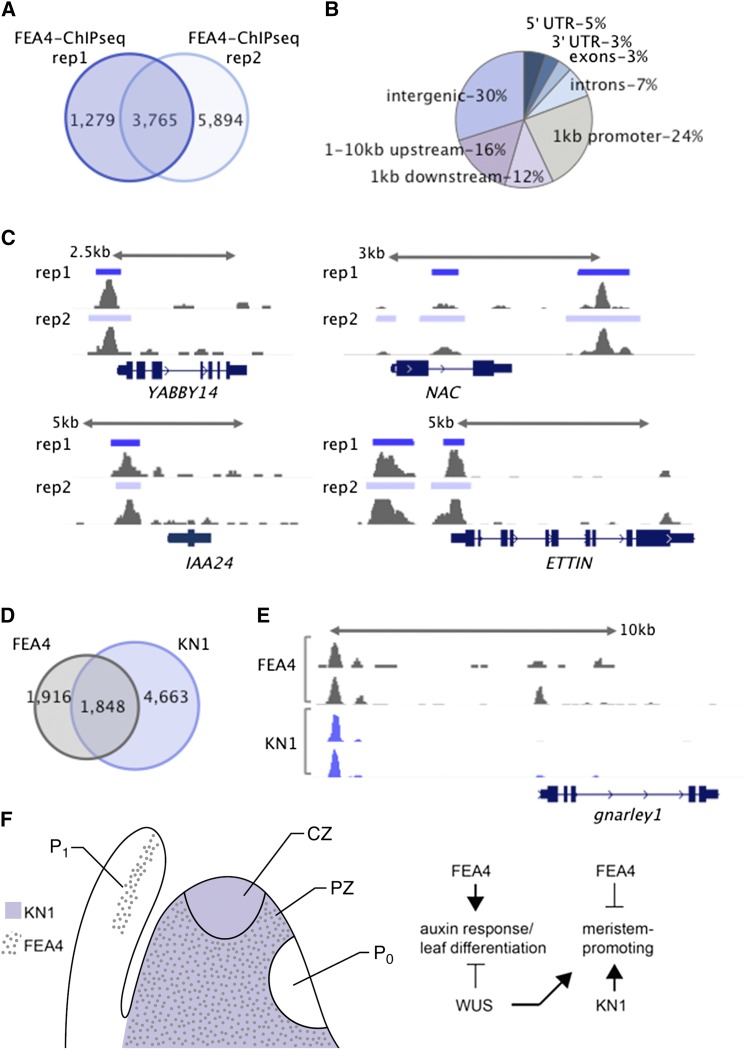
To map genome-wide occupancy of FEA4 and define its putative transcriptional targets, we performed ChIP-seq using the ProFEA4-YFP:FEA4 transgenic lines. Based on analysis of two biological replicates, we identified 3765 high-confidence peaks, i.e., where significantly enriched peaks of FEA4 binding compared with input control (P < 1e-05) overlapped in the two replicates (Figure 7A; Supplemental Table 4 and Supplemental Data Set 5). A subset of these binding regions was validated by ChIP-qPCR (Supplemental Figure 5). FEA4 bound in various genomic contexts, with 18% of binding associated with gene bodies and 24% occurring within 1-kb promoter regions upstream of transcriptional start sites (Figure 7B). We considered FGS genes within 10 kb of high-confidence peaks as putative targets of FEA4 (Supplemental Data Set 6). Among these 4065 putative target genes, overrepresented biological processes were related to auxin response (P = 7.39e-06), leaf vascular tissue patterning (GO:0010305, P = 2.05e-05), primary SAM specification (GO:0010072, P = 8.85e-04), and regionalization (GO:0003002, P = 2.50e-05), i.e., a pattern specification process defining cell differentiation (Supplemental Data Set 7). Putative FEA4 targets included a number of known regulators of lateral organ differentiation, including ROUGH SHEATH2 and ROUGH SHEATH1, as well as KANADI and YABBY family members (Figure 7C; Supplemental Data Set 6).
Figure 7.
Genome-Wide Binding Profiles and Model for FEA4 Function.
ChIP-seq using two biological replicates revealed 3765 high-confidence FEA4 binding peaks, which were shared between replicates (A). Distribution of high-confidence peaks shows FEA4 binds in various genomic contexts, with 24% of peaks located within 1-kb promoter regions (B). FEA4-bound genes involved in organ differentiation; shown are examples of FEA4 bound to the promoter of YABBY14 (GRMZM2G005353) and to the intron of a NAC TF (GRMZM2G393433) orthologous to CUC1/2 from Arabidopsis as well as ∼2 kb downstream of the gene. FEA4 also targeted genes involved in auxin response; shown are examples of FEA4 bound to the promoter of IAA24 (GRMZM2G115357) and to both the promoter and 5′ untranslated region of a putative ortholog of ETTIN (GRMZM5G874163) (C). Approximately half of FEA4 high-confidence peaks overlapped directly with high-confidence peaks bound by the KN1 TF (D). GNARLEY1 (GRMZM2G452178) is cobound by FEA4 and KN1 and oppositely modulated, activated, and repressed, respectively (E). FEA4 potentially acts as an antagonist to meristem maintaining factors, such as KN1 and WUS, to promote differentiation in the peripheral zone (F). CZ, central zone; PZ, peripheral zone; P0, P1, plastochron zero and plastochron one leaf primordia.
Approximately 25% (102) of genes DE in fea4 mutants were also bound by FEA4, including 49 downregulated and 53 upregulated genes, suggesting that FEA4 can act as an activator or repressor of gene expression. In general, FEA4 tended to activate genes involved in organ differentiation, e.g., NAC TFs similar to CUP-SHAPED COTYLEDON (CUC) genes from Arabidopsis, as well as genes related to auxin response including IAA and ARF genes (Figure 7C, Table 2). Interestingly, FEA4 repressed an ortholog of ETTIN/ARF3, which in Arabidopsis was shown to act redundantly with PAN in floral development (Sessions et al., 1997).
We next tested the extent to which FEA4 bound to genomic regions also bound by the KN1 TF, a master regulator of shoot stem cell maintenance, using published ChIP-seq data (Bolduc et al., 2012). Approximately half of the FEA4 high-confidence peaks overlapped with high-confidence KN1 peaks, which were also identified based on two biological replicates (Figure 7D). Within 10 kb of these cobound regions were 2080 putative target genes, of which 12% were differentially expressed either in fea4 and/or kn1 loss-of-function mutant ears. These 245 cobound and modulated target genes suggested differential modes of regulation by FEA4 and/or KN1 (Supplemental Data Set 8). For example, the TF, GNARLEY1, which affects vegetative and floral development in maize (Foster et al., 1999), is positively modulated by FEA4 and repressed by KN1 (Figure 7E). Additional genes identified as being oppositely modulated by FEA4 and KN1 included the ETTIN/ARF3 putative ortholog (GRMZM5G874163), GENERAL REGULATORY FACTOR1, which encodes a 14-3-3 protein, an LRR kinase (GRMZM2G166413) similar to RECEPTOR LIKE PROTEIN29 from Arabidopsis, and an HD-ZIP TF (GRMZM2G003509) with sequence similarity to PHAVOLUTA/PHABULOSA. In addition, FEA4 and KN1 bound and positively modulated a CUC-like NAC TF (GRMZM2G430522), consistent with positive regulation of CUC genes by SHOOT MERISTEMLESS in Arabidopsis (Spinelli et al., 2011), an LRR kinase (GRMZM5G809695) with sequence similarity to ERECTA and implicated in specification of organs originating at the SAM (Torii et al., 1996; Mandel et al., 2014), and a NPH3 domain protein (GRMZM2G067053) similar to ENHANCER OF PINOID. The latter two have been implicated in regulation of auxin transport (Treml et al., 2005; Chen et al., 2013). While FEA4 and KN1 share largely overlapping expression patterns with both excluded from the P0 (Jackson et al., 1994), KN1 is expressed in the central zone, whereas FEA4 is not. These common yet slightly distinct spatiotemporal expression patterns may reflect both opposing and synergistic modes of gene regulation by FEA4 and KN1. Consistent with the suite of FEA4 bound and modulated genes, our model suggests that FEA4 promotes organ differentiation in opposition to the meristematic fate-promoting role of KN1 (Figure 7F).
DISCUSSION
FEA4 Controls Meristem Size by a Novel Mechanism, in Parallel to the CLV-WUS Pathway
In this study, we isolated a fasciated ear mutant, fea4. Most of the previously identified maize fasciated ear genes, such as fea2, td1, and ct2, are directly involved in perceiving or transducing signals in the CLV pathway (Taguchi-Shiobara et al., 2001; Bommert et al., 2005, 2013b). The synergistic phenotypes of fea4 fea2 double mutants suggest that these genes operate in separate (parallel) pathways that converge on the common process of shoot meristem size control. However, we did not detect significant differences in expression of WUS or CLV pathway genes, so we cannot predict where the pathways converge. We also believe that FEA4 defines a novel way in which meristem size can be perturbed because of its distinct expression pattern in the peripheral zone. In summary, we propose that FEA4 acts in parallel to the CLV pathway to regulate meristem size homeostasis (Laufs et al., 1998; Prigge and Wagner, 2001). Interestingly, fea4 is the most severe fasciated ear mutant isolated to date, highlighting a critical requirement for this parallel function in maize.
FEA4 Encodes the Ortholog of the Arabidopsis PAN Gene
Phylogenetic analysis revealed that although FEA4 belongs to a large gene family, it is more closely related to PAN than any other maize gene and therefore encodes the ortholog of PAN. pan mutants have increased floral organ numbers in the petal and sepal whorls as well as reduced numbers of stamens (Running and Meyerowitz, 1996). Careful analysis of floral meristems (Running and Meyerowitz, 1996) revealed no change in meristem size, cell size, or cell number in pan mutants. The authors hypothesized that rather than having extra perianth organs due to an increase in meristem size, pan mutants were compromised in their ability to specify the identity of floral organs in response to positional cues (Running and Meyerowitz, 1996; Chuang et al., 1999). Interestingly, fea4 mutants also displayed reduced stamen number without extreme FM fasciation or disorganization (Supplemental Figure 1), suggesting a similar function during floral development. Changes in the pan phenotype in different environmental conditions provide further insight regarding PAN function, as it was noted that pan mutants had larger inflorescence meristems under short-day conditions (Maier et al., 2011), which we also confirmed. However, the magnitude of change in IM size is much less in pan mutants than in fea4.
fea4 loss of function differentially affects different classes of meristems; for example, the IM is severely affected, while the axillary meristems of the inflorescence are not. The IM remains indeterminate for an extended period of time, and this could explain why it is more sensitive to loss of FEA4 than short-lived, determinate meristems, such as axillary SPMs or SMs. However, the indeterminate vegetative SAM is relatively mildly affected in fea4, compared with the ear IM. One possible explanation for this difference is that the ear inflorescence meristem is sensitized to genetic perturbation due to selection for increased meristem size during domestication and subsequent improvement of maize (Taguchi-Shiobara et al., 2001; Brown et al., 2011; Bommert et al., 2013a). Alternatively, the absence of, or weak phenotypes in certain classes of shoot meristems in Arabidopsis and maize, may reflect the redundant activities of other bZIP or unrelated genes. Both maize and Arabidopsis lineages have a complex pattern of genome duplications, followed by gene loss or subfunctionalization in the different lineages (Bennetzen, 2007). Therefore, each species has a unique set of redundancies, arising either from individual gene function, or emergent network-level properties.
There are also both striking similarities and differences between the FEA4 and PAN expression patterns. Both genes have a peripheral zone-specific expression pattern: FEA4 in the vegetative SAM and PAN in the IM. In contrast, FEA4 is expressed throughout the central and peripheral zones of reproductive meristems, whereas pan is expressed throughout vegetative meristems. Therefore, the expression patterns for the two genes are somewhat conserved, but shifted heterochronically. Changes in the expression pattern of conserved genes could underlie morphological differences between the different lineages (Carroll, 2008). It is also interesting to consider whether differences in phenotype between maize and Arabidopsis can be explained by protein coding changes. FEA4 and PAN are very different in their N-terminal regions (Supplemental Figure 2), which could translate into different DNA binding activities or protein-protein interactions.
In Arabidopsis, in situ hybridization with a pan antisense probe revealed an expression pattern strongly enriched in the peripheral zone of the IM (Maier et al., 2011). However, immunolocalization of PAN protein using a peptide antibody showed accumulation throughout all cell layers of the IM (Chuang et al., 1999); this discrepancy suggests that PAN might move from cell to cell, similar to other plant transcription factors (Lucas et al., 1995; Gallagher et al., 2004; Maier et al., 2011). We did not observe any evidence of FEA4 protein movement, as the expression pattern of the YFP:FEA4 translational fusion line closely matched the in situ mRNA pattern. Furthermore, in situ hybridization of YFP:FEA4 transgenic plants using a YFP antisense probe demonstrated that the mRNA and protein expression patterns were in concordance (Supplemental Figure 6).
Significance of the Peripheral Zone
The peripheral zone (PZ) is defined as the population of cells that has been displaced to the flanks of the meristem by cell division of stem cells and their daughters in the central zone (Reinhardt et al., 2003). Studies involving inducible WUS expression suggest that the stem cell promoting function of WUS can be attributed in part to its role in repressing the expression of organ differentiation genes, which start to be expressed in the organ initiation sites in the PZ (Yadav et al., 2013). Therefore, the cells in the PZ may be more simply defined as the population of cells that are relieved of this repression and are on their way toward a determinate fate. Meristem size phenotypes of both the pan and fea4 mutants indicate that disruptions in the PZ can have dramatic developmental consequences. Similar effects are seen for the Arabidopsis LOST MERISTEMS genes, which encode GRAS domain transcription factors, as mutations in these genes prevent differentiation of cells in the PZ and cause an overaccumulation of meristematic cells (Schulze et al., 2010). Beyond this, the contribution of the PZ to meristem size control is not well established. It would be of interest to develop molecular markers to better define the different meristem domains in maize.
Potential Targets and Effectors of the fea4 Phenotype
Our RNA-seq analyses revealed a number of key developmental regulators misexpressed in fea4 mutants, including several homeobox-containing TFs and genes implicated in auxin response pathways. Furthermore, genome-wide analysis of FEA4 occupancy using ChIP-seq indicated overrepresentation of functional processes, including auxin response and leaf and vascular differentiation among target genes of FEA4. Many of these target genes implicated in leaf initiation and differentiation appear to be co-opted constituents of gene modules that also function in lateral organ differentiation in inflorescences (Eveland et al., 2014). Based on expression profiles from fea4 mutants, genes in these categories were largely activated by FEA4, suggesting that FEA4 functions in promoting this lateral organ differentiation program in both vegetative and reproductive development. Therefore, FEA4 may act as a counterweight to the meristem promoting factors WUS and KN1, by accelerating the differentiation of cells in the PZ. It may accomplish this by activating genes that control differentiation and possibly antagonizing repression of WUS target genes such as KANADI, ASYMMETRIC LEAVES2, and YABBY3, as described in Arabidopsis (Yadav et al., 2013).
Similar to the recently described basic helix-loop-helix factor HECATE in Arabidopsis, FEA4 provides another example of a TF adding a layer of complexity to meristem regulation by integrating CLV-WUS and hormone signaling (Schuster et al., 2014). However, we note that FEA4 does not appear to directly target any of the core CLV-WUS machinery and that CLV- and WUS-related genes did not show statistically significant changes in expression in fea4 mutant inflorescences (Supplemental Table 6). In Arabidopsis floral meristems, PAN promotes WUS repression indirectly, by activating the floral homeotic gene, AGAMOUS (AG), which PAN binds in the regulatory second intron (Das et al., 2009). Interestingly, FEA4 bound a maize ortholog of AG, Zea AGAMOUS1 (ZAG1) (Schmidt et al., 1993), but the binding appeared to be in the third intron (Supplemental Data Sets 5 and 6). This differential binding of PAN and FEA4 to AG and ZAG1, respectively, could reveal a different evolution of regulatory mechanisms between Arabidopsis and maize, and it would be interesting to test whether the PAN-AG-WUS module functions in maize. ZAG1 was not identified as differentially expressed in our RNA-seq analysis of fea4 mutants, and there are several possible explanations for this. For example, as demonstrated for KN1, redundant binding of multiple bZIP TFs could mask the modulation of target genes (Bolduc et al., 2014). Alternatively, modulation of a target may require a cofactor that is present in a restricted spatiotemporal pattern, or binding may regulate a different gene at long range (Farnham, 2009).
We also found that FEA4 positively modulates, either directly or indirectly, key genes controlling axillary meristem identity and determinacy in maize (Table 2). Positive regulation of these genes is consistent with a role for FEA4 in promoting differentiation in lateral organs. For example, FEA4 binds and activates RA1, which encodes a C2H2 TF that imposes determinacy on axillary branches in maize inflorescences, and ra1 loss-of-function mutants display increased branching in tassels and aberrant branching in ears (Vollbrecht et al., 2005). RNA-seq analyses in ra1 mutants showed that FEA4 expression was also significantly downregulated during ear primordium development, suggesting that FEA4 and RA1 promote and reinforce expression of each other by way of a feed-forward mechanism (Eveland et al., 2014).
FEA4 and PAN as Transcription Factors
Many developmentally relevant transcription factors were downregulated in fea4 inflorescences, relative to the wild type, suggesting that FEA4 could be an important activator of these genes. In support of this, PAN was shown in a yeast transactivation assay to activate transcription of a reporter gene, but only in the presence of coactivators, such as GARP-domain proteins (Maier et al., 2009). Furthermore, PAN has also been shown to physically interact with the BTB-POZ domain transcriptional coactivators BLADE ON PETIOLE1 (BOP1) and BOP2 (Hepworth et al., 2005). ChIP experiments indicate that BOP1/BOP2 colocalize with TGA-class bZIP binding sites in the genome (Xu et al., 2010). FEA4 has two glutamine-rich domains that are traditionally associated with transcriptional activators (Xiao and Jeang, 1998). These same domains may be necessary for the posttranslational redox regulation of PAN by the glutaredoxin proteins ROXY1 and ROXY2, but this hypothesis remains to be tested (Li et al., 2009). Interestingly, a maize ROXY-related mutant also affects meristem size (Yang et al., 2015), suggesting this regulation might be relevant in the context of meristem size regulation.
Meristem Size, Kernel Row Number, and Crop Yield
This study suggests a mechanism by which meristem size could be regulated, through FEA4’s function acting in parallel to the CLV-WUS pathway. As such, FEA4 provides another potential target for manipulation of inflorescence meristem size and kernel row number, as has been recently demonstrated for fea2. Hypomorphic fea2 alleles do not produce obviously fasciated ears, but significantly increase kernel row number around the diameter of the ear and kernels per ear (Bommert et al., 2013a). It would be useful to further study if natural variation in FEA4 expression or activity could contribute to variation in meristem size, in order to deepen understanding of meristem biology and to facilitate crop improvement.
METHODS
Plant Stocks and Growth Conditions
fea4-ref was isolated from an M2 screen of EMS-mutagenized A619 inbred maize (Zea mays) and was deposited in the Maize Genetics Co-Op Stock Center as fea179. The mutation was introgressed four to five times into various inbred lines for phenotypic analysis in segregating families. fea4-rel*09-5171 and fea4-rel*07-167 were found in a screen for enhancers of ra2 in the A619 inbred background. Trait Utility System in Corn alleles were identified from a PCR-based screen of Mutator element-mutagenized populations, using FEA4 and Mu-specific primer sets. Plants were grown in field locations in Cold Spring Harbor, NY; Berkeley, CA, Davis, CA; Valle de Banderas, Mexico; or under standard greenhouse conditions.
Scanning Electron Microscopy Analysis
Fresh tissues were dissected, mounted on stubs with silver conductive paint, and the stubs were kept on ice before imaging on a Hitachi S-3500N variable pressure scanning electron microscope.
Mapping and Molecular Cloning
fea4-ref (A619) was crossed to the B73 and W23 inbred lines, and the F1 plants were self-pollinated to produce F2 mapping populations. F2 individuals with the mutant phenotype were selected and subjected to bulked segregant analysis and further genotyping with CAPS markers. CAPS markers based on single nucleotide polymorphisms in the introns of GRMZM2G166366 and GRMZM2G042889 were used to screen recombinants and establish an interval of 2.7 Mb. The fea4 coding sequence was amplified in 1-kb fragments from genomic DNA extracted from a pool of F2 mutants and sequenced by Sanger sequencing.
Phylogenetic Analysis
A phylogenetic tree was constructed with the PHYML program (http://www.atgc-montpellier.fr/phyml/) (Guindon et al., 2005) using a CLUSTAL alignment of the top 100 BLAST hits with FEA4 protein sequence as input. Branch support was determined by the aLRT SH-like fast likelihood-based method.
In Situ Hybridization
The coding sequence of FEA4 was amplified from cDNA using primers MP815 and MP816 (Supplemental Table 5) and TOPO cloned into pCR2.1 (Invitrogen). Digoxigenin-labeled riboprobes in the sense and antisense orientation were synthesized by in vitro transcription from the T7 promoter. Hybridization was performed according to Jackson (1991), with the addition of 8% polyvinyl alcohol to the detection buffer to minimize diffusion of reaction products. Slides were exposed for ∼12 to 15 h before mounting and imaging. The short FEA4 probe specific to the 5′ region of the gene was synthesized as above from a cloned fragment amplified with primers MP833 and MP834 (Supplemental Table 5). KN1 in situ hybridizations were performed using a mix of three probes, according to Jackson et al., (1994).
Fluorescent Protein Fusions and Confocal Microscopy
A translational fusion of the coding sequence of FEA4 and YFP driven by the FEA4 native promoter was created with the Multisite Gateway Pro kit (Invitrogen). Approximately 1 kb of upstream promoter sequence, the entire coding region plus introns, and 1.5 kb of downstream genomic sequence were included. Primers used are listed in Supplemental Table 5. The fragments were cloned into a Gateway-compatible version of PTF101 (pAM1006-RL), and this binary vector was transformed into the maize HiII line at the Iowa State Plant Transformation Facility (Ames, IA) (Mohanty et al., 2009). Plant apices and inflorescences were dissected, mounted on glass slides in water, and imaged on a Zeiss 710 confocal microscope.
Double Mutant Analysis
fea4-ref mutants introgressed four times into B73 were crossed to fea2-0 in B73, and F1 plants were self-pollinated to create F2 mapping populations segregating both single and double mutants. Seedlings were genotyped for fea2 using gene-specific (fea2-D and fea2-ASA) and Mu-element specific (Mu58) primers (Supplemental Table 5). fea4 genotype was determined using primers MP900 and MP901, followed by Cac8I digestion (Supplemental Table 5). Shoot apices were dissected from seedlings after 14 d of growth. Meristems, with surrounding leaves still attached, were fixed in FAA (10% formalin, 5% acetic acid, and 45% ethanol), dehydrated in an ethanol series, and cleared with methyl salicylate (Jackson and Hake, 1999). Apices were mounted on glass slides and imaged on a light microscope with attached camera. Meristem width was measured just above the bulge of the P1 leaf primordium, using ImageJ. For mature plant analysis, F2 families were grown in the field in Newark, DE, or Valle de Banderas, Mexico.
RNA-seq Library Preparation and Analysis
Triplicate pools of more than 10 1-mm ears were harvested from homozygous fea4 mutants and heterozygous wild-type sibling plants. Freshly dissected ear tissue was fixed in ice-cold acetone, followed by vacuum infiltration for 20 min, and three acetone changes of 1 h each (Park et al., 2012). Total RNA was extracted from pools of ear tissue using the PicoPure RNA Isolation kit (Life Technologies), according to manufacturer instructions. mRNA was enriched by two successive purifications with oligo(dT)-coupled Dynabeads (Invitrogen). Approximately 50 ng of mRNA was used as input for the ScriptSeq v2 RNaseq system (Epicentre). This kit allowed the addition of barcoded adapters to enable multiplexed sequencing in a single lane of an Illumina HiSeq2000. Prior to sequencing, the average size distribution of the libraries was verified on a high sensitivity Bioanalyzer chip. The libraries were diluted to 10 nM, and concentration was verified by quantitative PCR using standards (KAPA Biosystems).
The Tuxedo suite (Trapnell et al., 2010) was used for mapping and analysis of the RNA-seq data. We used Tophat (version 2.0.9) to align the 101-bp paired-end reads to the maize reference genome (AGPv2) based on a set of 110,028 predicted maize gene models (Working Gene Set v5b.60; maizesequence.org). We then quantified read counts per gene and normalized fragments per kilobase exon per million reads mapped values using Cufflinks (version 2.0.2) and a high-confidence subset of 39,656 maize gene models, the FGS (v5b.60; www.maizesequence.org/). Biological replicates showed strong correlations (r > 0.96) in gene expression. Differential expression analysis was performed using the R package DESeq (v.1.14.0; Anders and Huber, 2010). An adjusted P value of < 0.05 was used to call differentially expressed genes after multiple testing correction.
Functional annotations for maize gene IDs were obtained via xref in Ensembl, incorporating Refseq, Entrez, and InterPro entries, accessible through GrameneMart v.36 (www.gramene.org; Spooner et al., 2012). Closest putative orthologs of maize genes in Arabidopsis thaliana were identified through EnsemblCompara (Vilella et al., 2009), and functional descriptions for Arabidopsis orthologs were based on TAIR10. Gene Ontology (GO) enrichment analyses were performed using the R package GOStats (Falcon and Gentleman, 2007). GO classifications for maize FGS gene IDs were obtained via GrameneMart v.36 and v.36b. Overrepresentation of GO biological processes was determined based on a hypergeometric test and a corrected P value of < 0.05.
ChIP-seq and Analysis
Developing ear primordia, ∼2 to 5 mm in size, were harvested from YFP:FEA4 plants grown under greenhouse conditions. These experiments used families containing a transgene integration event that complemented the mutant (A399S1-7), and expression of the transgene was verified by fluorescence microscopy. Following dissection, inflorescences were immediately cross-linked in buffer containing 1% formaldehyde for 15 min under vacuum, followed by addition of glycine to a concentration of 0.1 M, which was infiltrated for 5 min. Following three washes with distilled water, the cross-linked tissues were dried with paper towels and flash frozen in liquid nitrogen.
Chromatin extracts from YFP:FEA4 plants were immunoprecipitated with anti-green fluorescent protein (ab290; Abcam) antibody. Protein A Dynabeads (Invitrogen) blocked with yeast tRNA and BSA were used to capture antibody-chromatin complexes (Morohashi et al., 2012). Following purification of immunoprecipitated DNA, libraries were constructed using the Ovation Low Input DR kit (Nugen) and barcoded to allow multiplexed sequencing. Two input and two IP libraries were sequenced on one run of an Illumina MiSeq machine.
ChIP-seq reads were aligned to the maize reference genome (AGPv2) using Bowtie (v.0.12.7; Langmead et al., 2009), and peak calling was performed with MACS version 1.4.0rc2 using only uniquely mapped reads (Zhang et al., 2008). Peaks were identified as significantly enriched (P < 1e-05) in each of the ChIP-seq libraries compared with input DNA. Peaks were determined as high confidence if significant peaks from individual biological replicates were overlapping (i.e., their summits were positioned within 300 bp of each other). Coordinates for KN1 peaks were extracted from the published data set (Bolduc et al., 2012; GEO [GSE39161]). FGS gene models within 10 kb of high-confidence peak summits were considered putative targets of FEA4.
Data Access
All RNA-seq and ChIP-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under accession number GSE61954 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=atqhmuyevbozdmjandacc=GSE61954).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MAIZE actin, J01238.1; PAN, NM_105536.3; ROXY1, AY910752.1; KN1, AY260164.1; RA1, GQ891946.1; BOP1, NM_115572.1; and Zea AGAMOUS1, NM_001111851.1.
Supplemental Data
Supplemental Figure 1. fea4 Floral Phenotypes.
Supplemental Figure 2. A CLUSTAL Alignment of FEA4 and PERIANTHIA, and Extended Phylogeny.
Supplemental Figure 3. Reproductive Phenotypes of the perianthia (pan) Mutant of Arabidopsis.
Supplemental Figure 4. Samples Collected for RNA-seq and Correlations in Expression.
Supplemental Figure 5. ChIP-qPCR Validations.
Supplemental Figure 6. YFP Antisense in Situ Hybridizations.
Supplemental Table 1. fea4 Allele Table.
Supplemental Table 2. RNA-seq Summary Statistics.
Supplemental Table 3. Meristem Marker Genes.
Supplemental Table 4. ChIP-seq Summary Statistics.
Supplemental Table 5. Primers Used in This Study.
Supplemental Data Set 1. CLUSTAL Alignment of the Top 100 FEA4 BLAST Hits.
Supplemental Data Set 2. Expression Information for All Filtered Gene Set Genes Based on Analysis with Cufflinks v.2.0.2 and Differential Expression Analysis with DESeq.
Supplemental Data Set 3. Differentially Expressed Genes in fea4 Mutants Compared with the Wild Type.
Supplemental Data Set 4. Gene Ontology Enrichment Analysis for Differentially Expressed Genes.
Supplemental Data Set 5. All Significant Peaks Called from Combined FEA4 ChIP-seq Analyses and Distance to Nearest Gene Models.
Supplemental Data Set 6. Putative FEA4 Target Genes (FEA4 High-Confidence Peak within 10 kb).
Supplemental Data Set 7. Gene Ontology Enrichment Analysis for Putative FEA4 Target Genes.
Supplemental Data Set 8. Genes Cobound by FEA4 and KN1 and Differentially Expressed in fea4 and/or kn1 Mutant Ear Primordia.
Supplementary Material
Acknowledgments
We thank Anand Narayanan for help with initial mapping of fea4 and phenotyping and Tim Mulligan for plant care. We thank Gerry Neuffer for generating the EMS stocks that produced the original allele of FEA4. This work was funded by the National Science Foundation Plant Genome Program (Grant IOS-1238202), the Agriculture and Food Research Initiative Competitive Grants Program Grant 2010-04112 from the USDA National Institute of Food and Agriculture, a Pioneer Hi-Bred International Research Collaboration Program Agreement, and NSERC PGS fellowship to M.P.
AUTHOR CONTRIBUTIONS
M.P. conducted phenotypic analysis, mapped and cloned fea4, conducted RNA and protein expression analysis, RNA-seq, and ChIP-seq experiments, and wrote the article. A.L.E. performed all bioinformatics analysis for RNA-seq and ChIP-seq and helped write the article. T.L. assisted with ChIP-seq and validated ChIP-seq peaks by quantitative PCR. F.Y. performed in situ hybridizations. R.W. and E.V. helped map fea4 and provided alleles that confirmed molecular cloning. C.L. performed phenotyping of fea4 mutants. B.I.J. and R.M. provided alleles that confirmed molecular cloning. M.K. and H.S. participated in mapping of fea4. D.J. contributed to genetics experiments, designed the research, and edited the article.
Glossary
- SAM
shoot apical meristem
- ChIP-seq
chromatin immunoprecipitation-sequencing
- RNA-seq
RNA sequencing
- EMS
ethyl methanesulfonate
- IM
inflorescence meristem
- SPM
spikelet pair
- SM
spikelet
- FM
floral meristem
- CAPS
cleaved-amplified polymorphic sequence
- FGS
filtered gene set
- DE
differentially expressed
- TF
transcription factor
- PZ
peripheral zone
- GO
Gene Ontology
Footnotes
Articles can be viewed online without a subscription.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10: 176–181. [DOI] [PubMed] [Google Scholar]
- Bolduc N., Tyers R.G., Freeling M., Hake S. (2014). Unequal redundancy in maize knotted1 homeobox genes. Plant Physiol. 164: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P., Nagasawa N.S., Jackson D. (2013a). Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 45: 334–337. [DOI] [PubMed] [Google Scholar]
- Bommert P., Je B.I., Goldshmidt A., Jackson D. (2013b). The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558. [DOI] [PubMed] [Google Scholar]
- Bommert P., Lunde C., Nardmann J., Vollbrecht E., Running M., Jackson D., Hake S., Werr W. (2005). thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245. [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Brown P.J., Upadyayula N., Mahone G.S., Tian F., Bradbury P.J., Myles S., Holland J.B., Flint-Garcia S., McMullen M.D., Buckler E.S., Rocheford T.R. (2011). Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 7: e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Chen M.K., Wilson R.L., Palme K., Ditengou F.A., Shpak E.D. (2013). ERECTA family genes regulate auxin transport in the shoot apical meristem and forming leaf primordia. Plant Physiol. 162: 1978–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.F., Running M.P., Williams R.W., Meyerowitz E.M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13: 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Das P., Ito T., Wellmer F., Vernoux T., Dedieu A., Traas J., Meyerowitz E.M. (2009). Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136: 1605–1611. [DOI] [PubMed] [Google Scholar]
- Eveland A.L., et al. (2014). Regulatory modules controlling maize inflorescence architecture. Genome Res. 24: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S., Gentleman R. (2007). Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258. [DOI] [PubMed] [Google Scholar]
- Farnham P.J. (2009). Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 10: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T., Yamaguchi J., Wong B.C., Veit B., Hake S. (1999). Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11: 1239–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette A.J., Nakajima K., Benfey P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14: 1847–1851. [DOI] [PubMed] [Google Scholar]
- Giulini A., Wang J., Jackson D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Guindon S., Lethiec F., Duroux P., Gascuel O. (2005). PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33: W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Zhang Y., McKim S., Li X., Haughn G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. (1991). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and P. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174. [Google Scholar]
- Jackson D., Hake S. (1999). Control of phyllotaxy in maize by the abphyl1 gene. Development 126: 315–323. [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413. [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F., bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111. [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Johnston R., Yang Y., Gallavotti A., Kojima M., Travencolo B.A., Costa L.F., Sakakibara H., Jackson D. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P., Grandjean O., Jonak C., Kiêu K., Traas J. (1998). Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10: 1375–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lauri A., Ziemann M., Busch A., Bhave M., Zachgo S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983. [DOI] [PubMed] [Google Scholar]
- Maier A.T., Stehling-Sun S., Offenburger S.L., Lohmann J.U. (2011). The bZIP transcription factor PERIANTHIA: A multifunctional hub for meristem control. Front. Plant Sci. 2: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A.T., Stehling-Sun S., Wollmann H., Demar M., Hong R.L., Haubeiss S., Weigel D., Lohmann J.U. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136: 1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel T., Moreau F., Kutsher Y., Fletcher J.C., Carles C.C., Eshed Williams L. (2014). The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development 141: 830–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- Mohanty A., Yang Y., Luo A., Sylvester A.W., Jackson D. (2009). Methods for generation and analysis of fluorescent protein-tagged maize lines. Methods Mol. Biol. 526: 71–89. [DOI] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Schatz M.C., Lippman Z.B. (2012). Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler M., Tanaka W., Hirano H.Y., Jackson D. (2013). Grass meristems I: shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 54: 302–312. [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Wagner D.R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2003). Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083. [DOI] [PubMed] [Google Scholar]
- Running M.P., Meyerowitz E.M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122: 1261–1269. [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Veit B., Mandel M.A., Mena M., Hake S., Yanofsky M.F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5: 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Schulze S., Schäfer B.N., Parizotto E.A., Voinnet O., Theres K. (2010). LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 64: 668–678. [DOI] [PubMed] [Google Scholar]
- Schuster C., Gaillochet C., Medzihradszky A., Busch W., Daum G., Krebs M., Kehle A., Lohmann J.U. (2014). A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28: 438–449. [DOI] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Spinelli S.V., Martin A.P., Viola I.L., Gonzalez D.H., Palatnik J.F. (2011). A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 156: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner W., Youens-Clark K., Staines D., Ware D. (2012). GrameneMart: the BioMart data portal for the Gramene project. Database (Oxford) 2012: bar056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ohneda M., Toriba T., Yoshida A., Hirano H.Y. (2009). FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 5: e1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Sato M., Ashikari M., Miyoshi M., Nagato Y., Hirano H.-Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F., Yuan Z., Hake S., Jackson D. (2001). The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R.F., Komeda Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treml B.S., Winderl S., Radykewicz R., Herz M., Schweizer G., Hutzler P., Glawischnig E., Ruiz R.A. (2005). The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132: 4063–4074. [DOI] [PubMed] [Google Scholar]
- Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. (2009). EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Springer P.S., Goh L., Buckler E.S. IV, Martienssen R. (2005). Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126. [DOI] [PubMed] [Google Scholar]
- Xiao H., Jeang K.T. (1998). Glutamine-rich domains activate transcription in yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 22873–22876. [DOI] [PubMed] [Google Scholar]
- Xu M., Hu T., McKim S.M., Murmu J., Haughn G.W., Hepworth S.R. (2010). Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 63: 974–989. [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G.V. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Bui H.T., Pautler M., Llaca V., Johnston R., Lee B., Kolbe A., Sakai H., Jackson D. (2015). A maize glutaredoxin gene, Abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 27: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.