TMET1 is a thylakoid-associated protein that functions in assembly of PSII complexes and interacts with PSII core proteins CP43 and CP47.
Abstract
Photosystem II (PSII) requires constant disassembly and reassembly to accommodate replacement of the D1 protein. Here, we characterize Arabidopsis thaliana MET1, a PSII assembly factor with PDZ and TPR domains. The maize (Zea mays) MET1 homolog is enriched in mesophyll chloroplasts compared with bundle sheath chloroplasts, and MET1 mRNA and protein levels increase during leaf development concomitant with the thylakoid machinery. MET1 is conserved in C3 and C4 plants and green algae but is not found in prokaryotes. Arabidopsis MET1 is a peripheral thylakoid protein enriched in stroma lamellae and is also present in grana. Split-ubiquitin assays and coimmunoprecipitations showed interaction of MET1 with stromal loops of PSII core components CP43 and CP47. From native gels, we inferred that MET1 associates with PSII subcomplexes formed during the PSII repair cycle. When grown under fluctuating light intensities, the Arabidopsis MET1 null mutant (met1) showed conditional reduced growth, near complete blockage in PSII supercomplex formation, and concomitant increase of unassembled CP43. Growth of met1 in high light resulted in loss of PSII supercomplexes and accelerated D1 degradation. We propose that MET1 functions as a CP43/CP47 chaperone on the stromal side of the membrane during PSII assembly and repair. This function is consistent with the observed differential MET1 accumulation across dimorphic maize chloroplasts.
INTRODUCTION
Photosystem II (PSII) is a multi-protein pigment complex that functions as a light-driven water:plastoquinone-oxidoreductase in the thylakoid membranes of cyanobacteria and in higher plant chloroplasts (Kouřil et al., 2012; Pagliano et al., 2013). The organization of active higher plant PSII complexes in the thylakoid grana lamellae is the so-called PSII supercomplex (C2S2M2), consisting of a PSII dimeric core (C2), associated with two pairs of trimers (S2 and M2) of the major light-harvesting complex proteins (LHCII-1,2,3), and two copies of each monomeric minor chlorophyll binding proteins, CP24 (LHCB6), CP26 (LHCB5), and CP29 (LHCB4). The strongly bound S trimers interact mainly with CP26, which, in turn, is associated with the PSII core protein CP43. The more moderately bound M trimers interact with CP24 and CP29 and also require LHCB3, with CP29 interacting with the PSII core protein CP47. Additionally, there are loosely bound major LHCII trimers (L) that can interact peripherally with the C2S2M2 supercomplex to form larger complexes. These L trimers migrate between PSII and photosystem I (PSI) (Caffarri et al., 2009; Ballottari et al., 2012; Kouřil et al., 2012; Pan et al., 2013). Knockout and knockdown mutants in Arabidopsis thaliana have been described for the major and minor LHCII genes and showed that the CP24, CP26, and CP29 play critical roles in association of LHCII trimers to the PSII core (reviewed in Ballottari et al., 2012).
PSII is prone to light-induced damage to the D1 reaction center protein even under nonstress conditions. Consequently, the lifetime of the D1 protein is much shorter than that of other PSII proteins and thylakoid proteins in general (Nath et al., 2013; Nickelsen and Rengstl, 2013). To maintain active PSII, the damaged D1 proteins are continuously removed and replaced by newly synthesized copies. This repair process, coined the PSII repair cycle, requires the partial disassembly of the PSII supercomplex, lateral migration of PSII subcomplexes from the stacked granal regions to the unstacked stroma lamellae, de novo synthesis of chloroplast-encoded D1 protein, reassembly of PSII, and return to the granal region. The basic principle of this stepwise assembly was initially postulated from PSII assembly analysis of high light treatment of Mn-depleted thylakoid membranes by sucrose gradient fractionation, immunoblotting, and light absorption spectroscopy (Barbato et al., 1992). This was followed by pulse-chase labeling studies in isolated thylakoids, chloroplasts, and leaves combined with sucrose gradient fractionation and/or native gels (van Wijk et al., 1995, 1996, 1997; Zhang et al., 1999). Chloroplast protein import assays showed that, in addition, the nuclear-encoded subunits of the water-splitting complex undergo a stepwise maturation and assembly process (Hashimoto et al., 1997). Subsequent analysis of PSII mutants in Arabidopsis that missed one or more PSII subunits refined these initial models (Ohnishi and Takahashi, 2001; Suorsa et al., 2004). Following thylakoid solubilization with nonionic detergents, various partial supercomplexes have been isolated and characterized from Arabidopsis, in particular, the complexes C2S and C2M (∼880 kD), C2S2 (∼1040 kD), S2SM (∼1100 kD), C2S2M (1250 kD), and the full supercomplex C2S2M2 (C2S2M2) (Caffarri et al., 2009). However, the molecular mechanism of the assembly process from a monomer (C) or dimeric PSII core (C2) to full C2S2M2 complex is not well understood; it is not clear to what extent this is a self-assembly process and which steps require auxiliary proteins.
Application of forward and reverse genetics using Arabidopsis over the last 15 years has resulted in the identification of auxiliary proteins aiding in the synthesis and assembly of PSII (reviewed in Mulo et al., 2008; Chi et al., 2012a; Kouřil et al., 2012; Nickelsen and Rengstl, 2013; Pagliano et al., 2013). More than a dozen higher plant PSII-specific biogenesis/repair factors have been reported, including HCF136 (Meurer et al., 1998; Covshoff et al., 2008), LPA1 (Peng et al., 2006), FKBP-2 (Lima et al., 2006), CYP38 (Fu et al., 2007; Sirpiö et al., 2008), TLP18.3 (Sirpiö et al., 2007), LPA2 (Ma et al., 2007), LPA3 (Cai et al., 2010), PAM68 (Armbruster et al., 2010), HCF243 (Zhang et al., 2011), LTO1 (Karamoko et al., 2011), TERC (Schneider et al., 2014), LQY1 (Lu et al., 2011), HHL1 (Jin et al., 2014), and psbN (Torabi et al., 2014). Additionally, the lumenal peptidase CtpA is specifically required for C-terminal processing of the D1 protein (Anbudurai et al., 1994; Oelmüller et al., 1996; Yamamoto et al., 2001); in the absence of this C-terminal processing, no active PSII complex can be formed (Che et al., 2013). Thylakoid bound FtsH and Deg proteases play an important role in degrading damaged D1 (Kapri-Pardes et al., 2007; Sun et al., 2010a, 2010b; Chi et al., 2012b; Kato et al., 2012), even if these proteases are not specific to PSII. Thylakoid protein translocons SecY/E and ALBINO3 (ALB3) have been shown to interact with each other (Klostermann et al., 2002) and are required for co- and posttranslational insertion of plastid- and nuclear-encoded proteins, including components of the PSII reaction center/core complex (Moore et al., 2000; Zhang et al., 2001; Peng et al., 2006; Ma et al., 2007; Cai et al., 2010; Schneider et al., 2014) (reviewed in Richter et al., 2010; Celedon and Cline, 2013). Additional PSII (dis)assembly factors likely remain to be discovered, and much remains to be learned about regulation of the PSII repair cycle, including the handling of pigments and other cofactors.
In the natural environment, light fluctuation during the day occurs frequently at irregular intervals. Plants employ a multilayered strategy to cope with different light conditions to protect photosystems from light damage and optimize the capacities of the light and dark reactions (reviewed in Tikkanen et al., 2012). State transition kinase STN7 and cyclic electron flow component PGR5 play important roles during fluctuating light conditions by protecting PSI (Tikkanen et al., 2010; Grieco et al., 2012; Suorsa et al., 2012). Lumenal protein TLP18.3 protects PSII by regulating the repair cycle under fluctuating light condition (Sirpiö et al., 2007). Under normal growth conditions, mutants for STN7, PGR5, and TLP18.3 (stn7, pgr5, and tlp18.3) did not show altered visible phenotypes, but they showed reduced growth (stn7 and tlp18.3) or died (pgr5) when grown under fluctuating intensities of white light. It is not known whether mutants for all PSII assembly factors show an enhanced phenotype under such fluctuating white light conditions, but this fluctuating light regime provides a tool to generate growth and molecular phenotypes and is therefore quite helpful in elucidating protein functions. A sudden increase in light intensity can result in loss of PSII activity and subsequently requires the PSII repair cycle. It is perhaps not surprising that several of the PSII assembly factor loss-of function mutants are less efficient in this repair process.
Maize (Zea mays) is a NADPH-ME C4-type plant with Kranz anatomy, and photosynthesis is performed via the collaboration of chloroplasts located in two different, but adjacent cell types, the mesophyll (M) cells and bundle sheath (BS) cells (Edwards et al., 2001a; Majeran and van Wijk, 2009). In maize (and other NADPH-ME C4-type plants), active PSII is highly enriched in M chloroplasts compared with BS chloroplasts and BS chloroplasts primarily carry out cyclic electron transport and have low rates of linear electron transport (Woo et al., 1970; Hardt and Kok, 1978; Schuster et al., 1985; Meierhoff and Westhoff, 1993; Majeran et al., 2008). In previous comparative quantitative proteome analyses of isolated M and BS chloroplast fractions of maize leaves, we identified an abundant chloroplast tricopeptide repeat (TPR) protein (GRMZM2G312910) that was 3- to 5-fold enriched in M chloroplasts compared with BS chloroplasts (Majeran et al., 2005; Friso et al., 2010). Consistently, the mRNA ratio between BS and M cells showed a 3-fold enrichment in the M (Li et al., 2010).
Here, we characterized the MET1 homolog in Arabidopsis (AT1G55480) and show that it is a stromal-exposed peripheral thylakoid protein that is enriched in stroma lamellae, but also present at significant levels in the grana stacks. MET1 interacts with PSII core proteins CP43 and CP47, and analysis of a MET1 null mutant shows that it is required for PSII assembly, in particular under fluctuating light conditions. MET1-deficient mutants are also more sensitive to sudden high light treatment, resulting in both loss of PSII supercomplexes and increased damage to PSII. However, they did not show altered phenotypes when grown for longer period at higher light intensities. We therefore propose that this thylakoid TPR protein functions posttranslationally in the assembly of the PSII core, likely both during de novo PSII biogenesis and during the PSII repair cycle. We renamed this protein MET1, for M-enriched thylakoid protein 1.
RESULTS
MET1 Is Conserved across Photosynthetic Eukaryotes
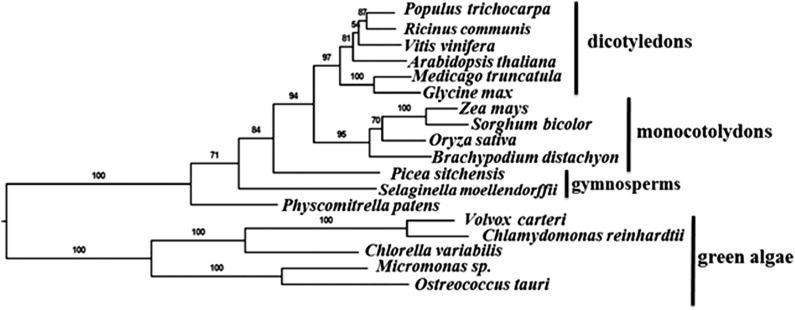
Extensive sequence and phylogenetic analyses show that maize MET1 has a single homolog in each of the green algae, moss, and lower and higher plant species examined (Figure 1) but that homologs are absent in photosynthetic bacteria and nonphotosynthetic organisms. Thus, MET1 is evolutionary conserved in photosynthetic eukaryotes and must have evolved soon after endosymbiosis with cyanobacteria. Consistently, MET1 is also part of a group of proteins that is present only in photosynthetic organisms, the “Greencut” set (Grossman et al., 2010). Sequence alignment of MET1 homologs showed a weak N-terminal PDZ motif and two C-terminal TPR motifs (Supplemental Figure 1A). Proteome analysis of maize, Arabidopsis, and rice (Oryza sativa) chloroplasts identified the respective MET1 homologs (GRMZM2G312910, AT1G55480, and Os07g07540) with high protein sequence coverage (Majeran et al., 2005; Zybailov et al., 2008) and for rice (Supplemental Figure 1B e). Consistently, these three MET1 homologs each has a predicted cTP. After cTP cleavage, mature MET1 has a predicted mass of ∼30 kD protein. None of the homologs has predicted transmembrane domains. A homology model for mature Arabidopsis MET1 shows the PDZ and TPR domains, whereas the highly conserved C terminus is rich in hydrophobic residues and forms a number of short helices (Supplemental Figure 2) (see Discussion). Arabidopsis MET1 was previously assigned as a ZKT protein (for PDZ, K-BOX, and TPR motifs), but neither protein location nor function was determined (Ishikawa et al., 2005), and it does not appear that MET1 contains a genuine K-BOX domain. In the remainder of the article, we determine the function of the Arabidopsis MET1 protein and discuss its significance in chloroplast homeostasis.
Figure 1.
MET1 Proteins Are Conserved across Green Algae and Plants.
Cladogram of MET1 including representatives of green algae, early land plants, monocots, and dicots, based on the alignment in Supplemental Data Set 5. The analysis includes 18 MET1 proteins from different plant species, including 10 angiosperms, 2 gymnosperms, 1 moss, and 5 algae. RAxML bootstrap support values are shown at the nodes of the tree.
Arabidopsis MET1 Accumulates in Photosynthetic Tissue and Is Induced during Greening
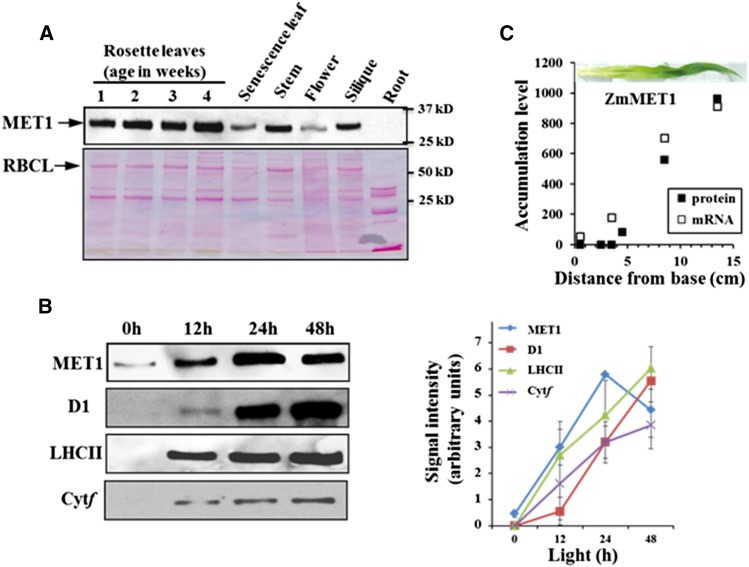
Immunoblot analysis using a specific MET1 antiserum generated against recombinant Arabidopsis MET1 showed that MET1 accumulates as a 30-kD protein in all Arabidopsis tissues except for roots (Figure 2A). The highest levels were observed in young and mature leaves (Supplemental Figure 3A), and MET1 levels strongly decreased during senescence (Figure 2A). mRNA accumulation levels obtained from public data showed comparable accumulation patterns (Supplemental Figure 3B). Low levels of MET1 were present in etiolated leaves and rapidly increased during light-induced greening with comparable kinetics as proteins of the thylakoid photosynthetic apparatus (Figure 2B). Maize MET1 also showed strong induction of mRNA and protein along the developing maize leaf, closely following the buildup of the thylakoid membrane system (Figure 2C).
Figure 2.
Accumulation of MET1 in Different Organs and during Greening in Leaves.
(A) Accumulation of Arabidopsis MET1 in different organs. Tissues were collected from leaves, stem, flower, siliques, and roots of Arabidopsis plants. Ten micrograms total protein was loaded in each lane. Upper panel shows the MET1 immunoblot and lower panel the Ponceau-stained blot.
(B) Light-induced accumulation of Arabidopsis MET1 during greening. Seeds were grown on agar plates with 0.5× Murashige and Skoog medium in the dark for 5 d and then transferred to light (80 μmol photons m−2 s−1) and sampled after 0, 12, 24, and 48 h. Total leaf proteins were extracted and separated by SDS-PAGE, followed by immunoblotting with specific antisera against MET1, D1 protein, LHCII, and Cyt f. Ten micrograms of protein was loaded in each lane. The graph shows the kinetics of protein accumulation for independent duplicate observations with standard deviations are indicated.
(C) Accumulation of maize MET1 mRNA and protein along the developing maize leaf. mRNA levels (RKPM) were obtained from Li et al. (2010), and protein levels (as normalized adjusted spectral counts [NadjSPC] × 2.106) were from Majeran et al. (2010). The inset shows a representative image of a 9-d-old maize leaf used for the sampling of protein and mRNA.
Identification of Two Independent MET1 Null Alleles
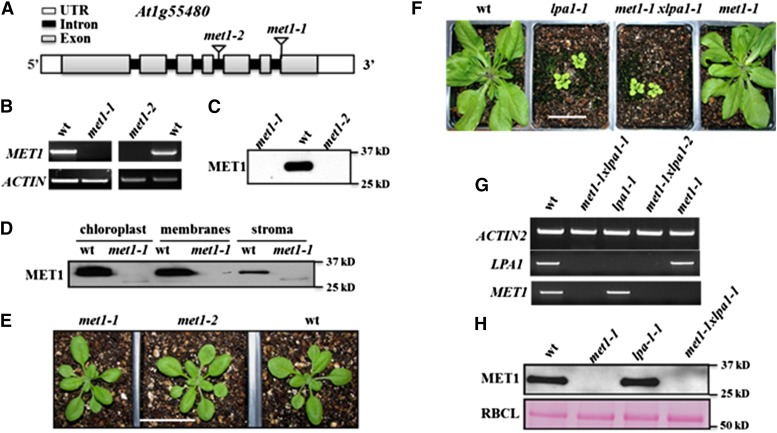
To determine the molecular function of MET1, we identified two T-DNA Arabidopsis mutants in the Columbia-0 (Col-0) background, met1-1 (SAIL_675_E06) and met1-2 (WISCDSLOXHS212_08F) originally obtained from the ABRC Arabidopsis (Figure 3A). RT-PCR analysis showed that MET1 transcript was undetectable in both mutants (Figure 3B). Consistently, immunoblotting of total leaf extracts showed a complete loss of MET1 protein in both lines but, as expected, a 30-kD band in wild-type plants (Figure 3C). Immunoblotting of fractionated chloroplasts of wild-type and met1-1 membranes and stroma showed that MET1 associated primarily with membranes (Figure 3D). Thus, met1-1 and met1-2 are null mutants for chloroplast MET1.
Figure 3.
Reverse Genetic Analysis of Null Mutants in MET1, Chloroplast Localization of MET1, and Effects on Protein and mRNA Levels in Arabidopsis.
(A) Gene model structures and positions of T-DNA insertions in the met1-1 and met1-2 alleles. Exons (gray), introns (black), and 5′ and 3′ untranslated regions (white) are indicated.
(B) met1-1 and met1-2 lines do not accumulate detectable MET1 transcript, as determined by RT-PCR (35 cycles). ACTIN2 was used to normalize MET1 mRNA.
(C) met1-1 and met1-2 do not accumulate detectable MET1 protein. Total protein was extracted from rosette leaves of wild-type and mutant plants. Twenty micrograms of protein was loaded in each lane.
(D) MET1 is primarily located in chloroplast membrane fractions but is absent in met1-1 and met1-2. Total chloroplast, chloroplast membranes, and stroma were used for immunoblotting with MET1 antiserum.
(E) Representative images of 16-d-old wild-type, met1-1, and met1-2 lines grown at 80 μmol photons m−2 s−1 under a 14-h-light/10-h-dark cycle. Older plants (23 d) grown under these conditions and plants (16 and 23 d) grown at similar light intensities at a 18-h-light/6-h-dark cycle are shown in Supplemental Figures 4A and 4B. Bar = 3 cm
(F) Phenotypes of 4-week-old wild-type, lpa1-1, met1-1, and met1-1x lpa-1 grown at 80 μmol photons m−2 s−1 under a 14-h-light/10-h-dark cycle. Additional images for plants grown under an 18-h-light/6-h-dark cycle are shown in Supplemental Figure 5. Bar = 3 cm
(G) mRNA analysis of MET1 and LPA1 in the wild type and single and double mutants of LPA1 and MET1. ACTIN2 is shown as internal reference.
(H) Immunoblot analysis of MET1 in the wild type and single and double mutants of LPA1 and MET1 (upper panel). A Ponceau stain of the RBCL region of the blot is shown as a loading control (lower panel).
met1-1 and met1-2 plants did not show visible phenotypes when grown under 14-h-light/10-h-dark cycle or 18-h-light/6-h-dark cycle at ∼100 μmol photons m−2 s−1 growth light (Figure 3E; Supplemental Figures 4A and 4B). Transfer of these plants to 10-fold higher light intensities did not induce any visible long-term conditional phenotypes as compared with the wild type (Supplemental Figure 4C). No significant differences (P < 0.01) in chlorophyll or carotenoid content (fresh weight basis) between the wild type and met1-1 and met1-2 under these various growth light regimes were observed (Table 1). Thus, under optimal, fully controlled environmental conditions, MET1 does not have an obvious function in plant growth and development, despite MET1 being present and strongly conserved across photosynthetic eukaryotes. This suggests that MET1 is redundant or that MET1 is important under less controlled or different environmental conditions.
Table 1. Chlorophyll and Carotenoid Contents of Leaf Rosettes of the Wild Type, met1-1, and met1-2 Grown under Different Light Regimes.
| Condition | Genotype | Chl a | Chl b | Chl a/b | Carotenoids |
|---|---|---|---|---|---|
| Normal lighta | Wild type | 1.25 ± 0.12 | 0.40 ± 0.02 | 3.1 ± 0.40 | 0.38 ± 0.07 |
| met1-1 | 1.21 ± 0.10 | 0.38 ± 0.03 | 3.1 ± 0.51 | 0.39 ± 0.05 | |
| met1-2 | 1.25 ± 0.11 | 0.39 ± 0.03 | 3.2 ± 0.61 | 0.36 ± 0.02 | |
| 3 d high lightb | Wild type | 0.88 ± 0.02 | 0.33 ± 0.01 | 2.6 ± 0.06 | 0.34 ± 0.01 |
| met1-1 | 0.81 ± 0.02 | 0.29 ± 0.01 | 2.7 ± 0.05 | 0.31 ± 0.02 | |
| met1-2 | 0.83 ± 0.03 | 0.33 ± 0.01 | 2.4 ± 0.05 | 0.31 ± 0.04 | |
| Fluctuating lightc | Wild type | 0.50 ± 0.02 | 0.15 ± 0.01 | 3.4 ± 0.44 | 0.17 ± 0.01 |
| met1-1 | 0.34 ± 0.02** | 0.12 ± 0.03 | 3.0 ± 0.62 | 0.13 ± 0.01* | |
| met1-2 | 0.37 ± 0.03** | 0.12 ± 0.03 | 3.2 ± 0.68 | 0.15 ± 0.01 |
Standard deviations are indicated. Asterisks mark the level of significance using a Student’s t test (*P < 0.01 and **P < 0.001). Chl, chlorophyll.
21-d-old plants grown at 80 μmol photons m−2 s−1 in a 16-h-light/8-h-dark period (n = 3).
Plants grown for 3 weeks at 80 μmol photons m−2 s−1 followed by 3 d at 1000 μmol photons m−2 s−1 in a 16-h-light/8-h-dark period (n = 3).
20-d-old plants grown under fluctuating white light conditions (alternating between 50 and 600 μmol photons m−2 s−1 every 10 min) during the 16-h light period (n = 3).
Genetic Interactions of MET1 with LPA1
BLAST searches of Arabidopsis MET1 identified a single distant homolog based on sequence similarity in the TPR domains (E = 9.10−9). This homolog is the PSII assembly factor LPA1 (Peng et al., 2006). To test a possible genetic interaction between MET1 and LPA1, we generated a met1-1 × lpa1-1 double null mutant and compared this double mutant to both parents and the wild type under different light periods. As expected, lpa1-1 plants showed a clear visible growth and pale leaf phenotype as previously reported (Peng et al., 2006), but the double mutant phenotype was similar to that of lpa1-1, whereas met1-1 had no visible phenotype (Figure 3F; Supplemental Figures 5A and 5B). The steady state MET1 mRNA level in lpa1-1 leaves was similar to that of the wild type; conversely, LPA1 mRNA levels were unaffected in met1-1 compared with the wild type (Figure 3G). Consistently, MET1 protein levels were unchanged in lpa-1 compared with the wild type (Figure 3H). In summary, there is no support for overlapping molecular functions or functional redundancies between LPA1 and MET1, despite their weak homology.
MET1 Is a Peripheral Stromal-Side Thylakoid Protein That Is Enriched in Stroma Lamellae
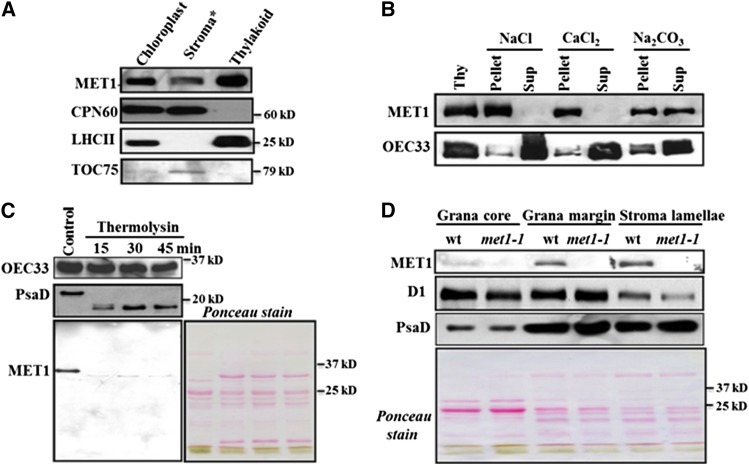
Because MET1 does not have predicted transmembrane domains, we analyzed the intrachloroplast distribution in more detail. Immunoblot analyses of isolated stroma and thylakoid fractions showed that ∼90% of MET1 was located in the thylakoid, with 10% in stroma (Figure 4A). For comparison, CPN60 was nearly exclusively found in stroma, whereas LHCII was 100% located in these thylakoid fractions. Inner envelope protein TOC75 was detected in the stromal fractions, as these envelopes were not pelleted at the g forces used to collect the thylakoid membranes (Figure 4A).
Figure 4.
MET1 Is a Stroma-Exposed Peripheral Thylakoid Protein Enriched in Stroma Lamellae.
(A) Localization of MET1 within the chloroplasts. Total chloroplast, thylakoid membranes, and stroma were used for immunoblotting with MET1 antiserum. Antisera against stromal CPN60, thylakoid LHCII, and outer envelope protein TOC75 were used as markers. Asterisk indicates that the stroma still contains envelopes, as it was collected as the supernatant from broken chloroplasts following a 10-min spin at ∼18,000g.
(B) Salt washing of thylakoid membranes. The membrane was sonicated in the presence of NaCl, CaCl2, and Na2CO3 and incubated on ice for 30 min before centrifugation to separate soluble and membrane fractions. Ten micrograms of proteins from supernatant and pellets was loaded on the SDS-PAGE gel. For control, thylakoids without any treatment of salt or sonication were used.
(C) Thermolysin treatment of thylakoid membranes. Thylakoids isolated from wild-type chloroplasts were treated with thermolysin for 15, 30, and 45 min on ice and then immunoblotted. Proteins were separated by SDS-PAGE and immunoblotted with OEC33 (PsbO), PsaD, and MET1 antisera. The Ponceau-stained image prior to blotting is shown. Ten micrograms of protein was loaded in each lane.
(D) Distribution of MET1 across the different thylakoid membrane regions. Thylakoid proteins were solubilized with digitonin and grana, grana margins, and stroma lamellae were fractionated by ultracentrifugation and analyzed by SDS-PAGE. Ten micrograms of protein was loaded in each lane. Immunoblots for MET1, D1, and PsaD protein are shown. Chlorophyll a/b ratios with standard deviations in parentheses for the wild-type fractions were: 3.4 (±0.41), 2.5 (±0.01), 3.0 (±0.15), and 6.6 (±0.15) for thylakoids, grana core, grana margins, stromal lamellae, respectively. The chlorophyll a/b ratios for the met1 fractions are: 3.6 (±0.5), 2.5 (±0.07), 3.7 (±0.34), and 5.6 (±0.94) for thylakoids, grana core, grana margins, and stromal lamellae, respectively. Ponceau-stained blots are shown for (C) and (D).
To determine the thylakoid association of MET1, isolated thylakoids were treated with the chaotropic reagents NaCl, CaCl2, and Na2CO3. After incubation with these reagents, soluble and membrane fractions were collected by centrifugation and immunoblotted and probed for MET1 and the luminal PSII subunit OEC33. Association of MET1 with the thylakoid was resistant to NaCl and CaCl2, in contrast to lumenal OEC33, which was almost completely released by both NaCl and CaCl2 (Figure 4B). Upon treatment of the thylakoids with Na2CO3, MET1 was partially released from the membrane, suggesting a hydrophobic interaction with the membrane lipids or membrane proteins.
To determine whether MET1 was located on the stromal or lumenal side, thylakoid membranes were treated with the protease thermolysin and incubated for 15, 30, and 45 min, followed by immunoblotting. MET1 protein was completely degraded by thermolysin, whereas lumenal OEC33 was not (Figure 4C). Stromal-exposed thylakoid protein PsaD was partially cleaved by the thermolysin treatment (Figure 4C). Together, these data demonstrate that MET1 was peripherally associated with the stromal side of the thylakoid membrane.
The thylakoid membrane is organized in grana stacks and stroma lamellae. Most functional PSII complexes are located in the grana, whereas PSI and ATP synthase are located predominantly in stromal lamellae. Moreover, most known proteins involved in biogenesis, maintenance, and repair of the photosynthetic apparatus in the thylakoids are located in the stroma lamellae (Chi et al., 2012a; Nath et al., 2013). To determine the distribution of MET1 across these regions, thylakoids were fractionated into grana core, grana margin, and stroma lamellae by detergent solubilization and differential ultracentrifugation. The D1 protein of PSII was used as a marker for grana stacks and PsaD was used as a marker for stroma lamellae. MET1 was clearly enriched in the stroma lamellae and to a lesser degree in the grana margins (Figure 4D).
No Obvious Changes in the Accumulation Levels or Assembly State of the Thylakoid Photosynthetic Machinery Were Observed
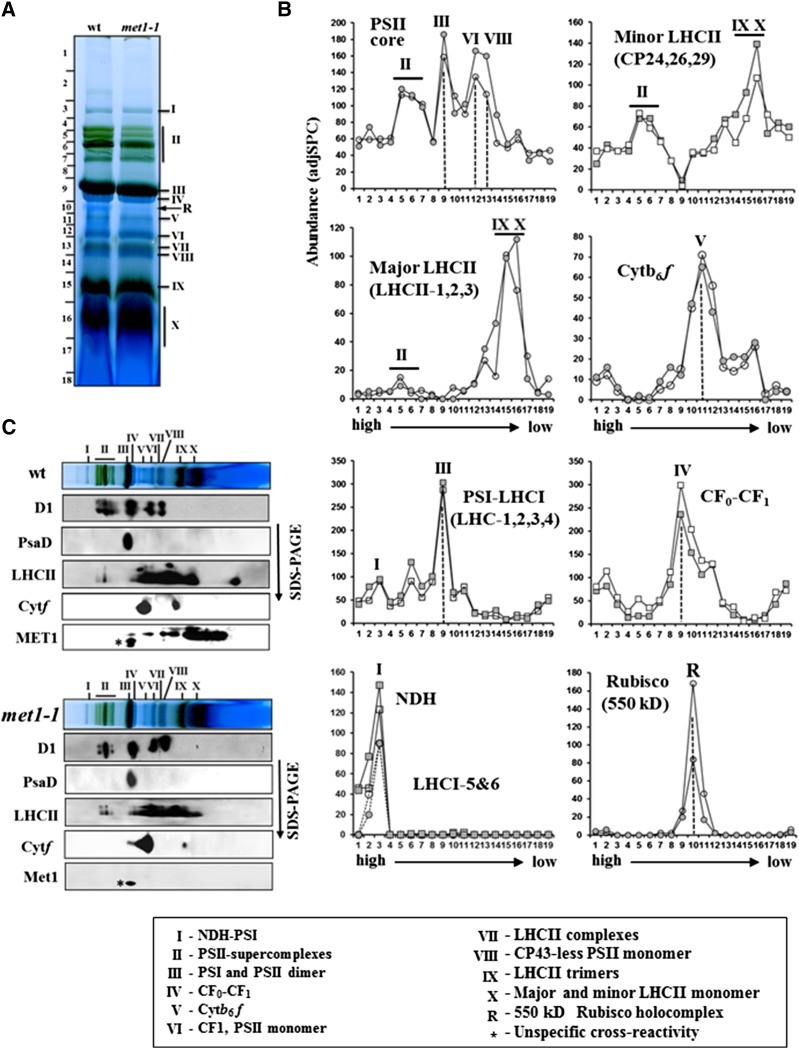
To determine whether loss of MET1 caused a change in thylakoid composition and/or assembly state, thylakoids from the wild type and met1-1 were solubilized with dodecyl maltoside (DM) and analyzed by blue-native PAGE (BN-PAGE) (Figure 5A). Protein accumulation levels and assembly state were then determined by tandem mass spectrometry (MS/MS) with a high-resolution instrument coupled with a nano-HPLC (nanoLC-LTQ-Orbitrap) after in-gel digestion with trypsin using (label-free) spectral counting for quantification (Figure 5B). This identified 250 proteins, of which 94% were plastid localized (Supplemental Data Set 1). Proteins known to be part of one of the five thylakoid-bound photosynthetic complexes were grouped by complex (i.e., PSII, PSI, NDH, Cyt b6f, and ATP synthase) and their assembly state and abundance compared between the wild type and met1-1. Judging from their abundance profiles and staining patterns (Figures 5A and 5B), the complexes were well resolved, including the NDH-PSI supercomplex and its enrichment of LHCI-5,6 (Peng and Shikanai, 2011; Peng et al., 2011; Kouřil et al., 2014), PSII supercomplex, PSII monomers and smaller PSII assemblies, the Cyt b6f complex, and the ATP synthase (both the integral CF0 and peripheral CF1 subcomplexes) (Figure 5B). However, we observed neither genotypic differences in assembly state nor genotypic differences in overall accumulation levels of the five complexes or individual proteins (Figure 6A). Similarly, immunoblot analysis with antisera for PSII (D1 and LHCII), PSI (PsaD), and the Cyt b6f complex (Cyt f) did not reveal differences between the wild type and met1-1 (Figure 5C). Among abundant thylakoid proteins not directly involved in photosynthesis, we identified the thylakoid FtsH complex (FtsH1,2,5,8), STN7, and Ca2+ sensing receptor, but none showed strong genotypic differences between the wild type and met1-1 (Supplemental Data Set 1). In addition, we separated thylakoid proteomes from the wild type and met1-1 by SDS-PAGE (Supplemental Figure 6A) and identified and quantified the proteins by MS/MS using as similar workflow as for the native gels. This identified some 150 thylakoid proteins (Supplemental Data Set 2). The scatterplot in Supplemental Figure 6B shows very similar accumulation profiles for the wild type and met1; no significant differences for individual proteins were observed.
Figure 5.
The Oligomeric State of the Thylakoid Membrane Complexes of 25-d-Old Wild-Type and met1-1 Plants Grown under Constant Light Intensity.
(A) BN-PAGE analysis of thylakoid membrane protein complexes in the wild type and met1-1. DM-solubilized thylakoid membrane proteins were separated by BN-PAGE, followed by staining with Coomassie blue. Proteins in the gel lanes were identified and quantified by MS/MS (Supplemental Data Set 1); together with information from the literature using similar BN-PAGE analysis, this allowed for annotation of the different complexes (Järvi et al., 2011). I, NDH-PSI; II, PSII supercomplexes; III, PSI and PSII dimer; IV, CFo/1 complex; V, Cyt b6f complex; VI, CF1 (peripheral portion of the ATP synthase), PSII monomer; VII, LCHII complexes; VIII, CP43-less PSII monomer; IX, LHCII trimers; X, LHCII monomers and other monomeric proteins. R, the 550-kD Rubisco holocomplex. An equal amount of chlorophyll (12 μg) was loaded for the wild type and met1-1.
(B) Protein abundance accumulation profiles in the BN-PAGE gel of wild type and met1-1 (as in [A]) determined by MS/MS analysis (Supplemental Data Set 1). The y axis shows protein abundance based on the number of matched AdjSPC. Fraction numbers (x axis) correspond to the gel regions (gel slices processed for MS/MS analysis) in (A). Open circle/square represents the wild type, and closed circle/square represents met1-1.
(C) Immunoblotting of two-dimensional BN-PAGE-SDS-PAGE gels of the wild type and met1-1 DM-solubilized thylakoids probed with antisera against MET1, the D1 protein, Cyt f, PsaD, and LHCII-2. Individual lanes from BN-PAGE gels (as in [A]) were excised and solubilized in SDS, and proteins were separated by SDS-PAGE followed by immunoblotting.
Figure 6.
Phenotype and Thylakoid Composition of Wild-Type and met1 Alleles Grown under Fluctuating Light.
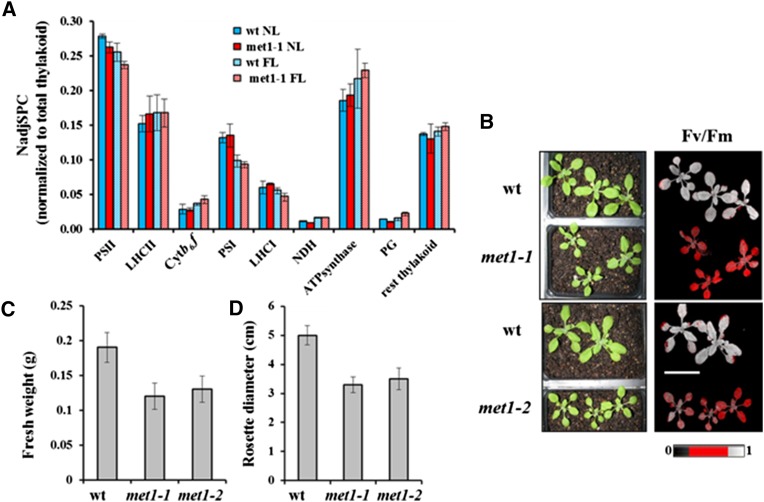
(A) Abundance of thylakoid complexes in met1-1 and wild-type plants under normal growth light (NL) and fluctuating light (FL) based on SDS-PAGE followed by MS/MS using label-free spectral counting for quantification. NadjSPC for each of the protein subunits in each complex was summed and normalized to total thylakoid proteome abundance. Standard deviations (n = 2) are indicated.
(B) Wild-type and met1 alleles mutants grown under fluctuating light for 14 to 16 d. Left panels show plants used for Fv/Fm measurements, and the right panels show false color images representing Fv/Fm. Bar = 3 cm.
(C) and (D) Fresh weight (C) and diameter (D) of rosettes of the wild type, met1-1, and met1-2 grown under fluctuating light for 2 weeks. Standard deviations are indicated (n = 6 [C] and n = 11 [D]).
MET1 Comigrates with PSII Subcomplexes in Arabidopsis
To determine the native mass of MET1 and infer possible interactions between MET1 and the major thylakoid complexes, we analyzed MET1 distribution after BN-PAGE, followed by SDS-PAGE (Figure 5C). MET1 was detected by immunoblotting in several complexes that correlated well with the masses of PSII (sub)complexes, suggesting that a substantial proportion of MET1 protein comigrated with PSII dimers, PSII monomers, CP43-less PSII monomer, PSII reaction centers, and unassembled proteins (Figure 5C).
Loss of MET1 Results in Reduced Growth and Lower PSII Efficiency under Fluctuating White Light Intensities
As mentioned in the Introduction, several mutants for thylakoid proteins (stn7, pgr5, and tlp18.3) did not show any visible phenotypes under normal growth conditions but showed reduced growth or even death when grown under fluctuating white light intensities (Sirpiö et al., 2007; Tikkanen et al., 2010; Grieco et al., 2012; Suorsa et al., 2012). We therefore tested whether such fluctuating light conditions also induced a conditional phenotype in the met1 null alleles. Indeed, when met1-1, met1-2, and the wild type were grown under fluctuating light intensities (alternating between 50 and 600 μmol photons m−2 s−1 every 10 min during the 16-h light period), the met1-1 and met1-2 plants showed significantly reduced growth and biomass compared with the wild type (Figures 6B to 6D). After 12 d of fluctuating light, the fresh weight of the rosette of the mutant was ∼37% (met1-1) and ∼32% (met1-2) lower than that of the wild type (Figure 6C), but the number of leaves in the mutants was the same as in the wild type. The diameter of rosettes was also reduced by ∼34% (met1-1) and ∼30% (met1-2) compared with the wild type (Figure 6D), and the chlorophyll a levels on a fresh weight basis decreased in both null alleles by ∼30% (Table 1). To further analyze this conditional phenotype, the chlorophyll fluorescence of 2-week-old plants was measured. After 20 min of dark adaptation, plants containing either mutant allele showed significantly (P < 0.001) lower maximum quantum efficiency of PSII (Fv/Fm) compared with the wild type (Figure 6B, Table 2). Thus, loss of MET1 results in reduced PSII efficiency and biomass accumulation under fluctuating light conditions but does not affect the rate of leaf initiation.
Table 2. Photosynthetic Electron Transport Parameters from 15- to 18-d-Old Leaf Rosettes of Soil-Grown Wild Type and met1-1 and met1-2 Null Alleles under Fluctuating Light Conditions.
| Parameter | Wild Type | met1-1 | met1-2 |
|---|---|---|---|
| F0 | 0.10 ± 0.01 | 0.12 ± 0.02 | 0.11 ± 0.02 |
| Fm | 0.45 ± 0.04 | 0.39 ± 0.06* | 0.38 ± 0.06* |
| Fv/Fm | 0.77 ± 0.01 | 0.71 ± 0.01** | 0.72 ± 0.02** |
Six plants of each genotype were used and standard deviations are indicated. The asterisk indicates a significant difference between the mutant and the wild type (Student’s t test; *P < 0.01 and **P < 0.001).
To determine the effect of fluctuating light on the relative abundance of thylakoid proteins, we separated thylakoid proteins using SDS-PAGE, with subsequent protein identification and quantification by MS/MS and spectral counting (Supplemental Figures 6A and 6C and Supplemental Data Set 2). We then compared the abundances for each of the five thylakoid complexes (PSI and its LHCI, PSII and its LHCII, ATP-synthase, Cyt b6f, and NDH complexes) in met1-1 and the wild type, based on matched adjusted MS/MS spectra (AdjSPC) and normalization to AdjSPC of the identified proteins (89 in total) in these five complexes. We observed only minor effects on the thylakoid composition (Figure 6A). However, levels of proteins of thylakoid-associated plastoglobules (Lundquist et al., 2012) (24 observed proteins) increased by 48% in met1-1 compared with the wild type, suggesting a stress response in met1-1. Inspection of abundance levels of individual thylakoid proteins did not identify significant differences between individual proteins. Because the effects on the thylakoid proteome of fluctuating growth light are not well understood, we compared this to the thylakoid proteome analysis of the wild type and met1-1 under the normal light regime, as described above. Compared with normal growth light, fluctuating light resulted in increases of Cyt b6f (24 and 54%), ATP synthase (17 and 18%), and NDH (36 and 83%) as well as decreases in PSI core (26 and 31%), LHCI (9 and 29%), and PSII core (8 and 10%) for the wild type and met1-1, respectively, and in the case of met1-1 more than doubling of PG proteome abundance (Figure 6A) for both genotypes.
Loss of MET1 Affects the Oligomeric State of PSII Subcomplexes under Fluctuating Light
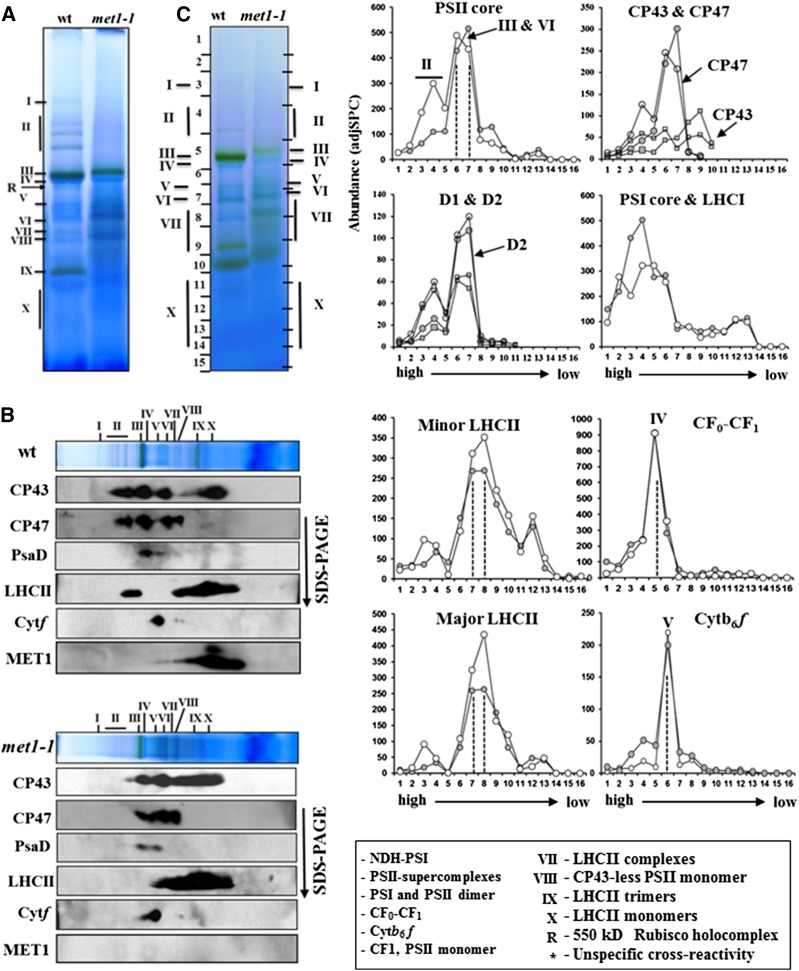
To examine the conditional met1 phenotype in more detail, we analyzed the oligomeric state of thylakoid proteins from the wild type and mutant by BN-PAGE using similar methods as described above for plants grown under normal light conditions. Thylakoid proteins from both genotypes grown under fluctuating light were solubilized by DM, separated by BN-PAGE (Figure 7A), and characterized by either MS/MS (Supplemental Data Set 3) or immunoblotting after separation in the second dimension by SDS-PAGE (Figure 7B). A comparison of BN-PAGE gels from wild-type and met1-1 thylakoids grown under fluctuating light shows that loss of MET1 leads to a loss of PSII supercomplexes proteins under fluctuating light (Figure 7A). Immunodetection of CP43, CP47, and LHCII shows the lack of signal in the region where PSII supercomplexes (II) are detected in the wild type (Figure 7B). By contrast, the oligomeric state of PSI and Cyt b6f complexes was not affected (Figure 7B). Furthermore, the amount of CP43 in CP43 complexes and unassembled CP43 (region IX-X) increased in met1-1. MET1 was detected by immunoblotting in several PSII complexes, including PSII monomers, CP43-less PSII monomer, PSII reaction centers, and unassembled proteins (Figure 7B). However, no signal was detected in the PSII dimer (or supercomplex) and MET1 levels in PSII monomers were also lower than under normal growth conditions (Figure 5C).
Figure 7.
The Oligomeric State of the Thylakoid Membrane Complexes of 20-d-Old Wild-Type and met1-1 Plants Grown under Fluctuating Light Intensity.
(A) BN-PAGE of DM-solubilized thylakoid membrane protein complexes in the wild type and met1-1, followed by staining with Coomassie blue. Proteins and protein complexes in the gel lanes were identified and quantified by immunoblotting (B) and MS/MS ([C]; Supplemental Data Set 3). Annotation of the protein complexes (I-X) is as in Figure 5. An equal amount of chlorophyll (10 μg) was loaded in each lane.
(B) Immunoblotting of two-dimensional BN-PAGE-SDS-PAGE gels of the wild type and met1-1 DM-solubilized thylakoids probed with antisera against the D1 protein, CP43, CP47, Cyt f, PsaD, LHCII-1, and MET1. Individual lanes from BN-PAGE gels (as in [A]) were excised and solubilized in SDS, and proteins were separated by SDS-PAGE, blotted, and immunodetected.
(C) Protein abundance accumulation profiles in the BN-PAGE gel of the wild type and met1-1 (as in [A]) determined by MS/MS analysis (see data in Supplemental Data Set 3). The y axis shows protein abundance based on the number of matched AdjSPC. Fraction numbers (x axis) correspond to the gel regions (gel slices processed for MSMS analysis) in (A). Note that the gel slices were cut to accommodate for differential mobility in the wild-type and met1-1 lane as indicated on the sides of the BN-PAGE gel lanes. Open circles represent the wild type, and closed circles represent the mutant.
Quantitative MS/MS analysis also showed the loss of PSII supercomplexes in met1-1 (Figure 7C). Abundance profiles of the complete PSII core (D1, D2, C43, CP47, psbH, PsbTn1,2, and Cyt b559α,β), as well as D1, D2, CP43, and CP47 separately, were plotted in three different panels (Figure 7C). The plot with the complete PSII core shows that the amounts of PSII monomer and dimer (III-IV) were relatively unchanged but that the PSII supercomplex (II) decreased ∼3-fold in met1-1 compared with the wild type. This was also evident from the plots for D1/D2 and CP43/CP47. The amount of CP43 unassociated with the PSII core (fractions 8 and 9) was clearly higher in met1-1, consistent with the immunoblots (Figure 7B). The amount of minor (CP24, 26, and 29) and major LHCII (LCHII-1,2,3) proteins also decreased in met1-1, both in the population associated with the PSII supercomplex (in fraction 3) as well as in other LHCII assemblies (fractions 7 and 8). The amount of LHCI-PSI core in the high mass range (fractions 3 and 4) was increased in met1-1, whereas the typical LHCI-PSI was unaffected (fractions 5 and 6). The assembly state of the ATP synthase complex (CFo-CF1) was not changed in met1, whereas higher amounts of Cyt b6f complex were found in the high mass range (fractions 3 and 4), but the main, typical oligomeric Cyt b6f complex was unaffected (fraction 6). Finally, the amount of monomeric LHCII (IX, X) was strongly decreased in both genotypes when compared with plants grown under normal (constant) light conditions (cf. Figures 5B and 7C).
met1 Alleles Are High Light Sensitive
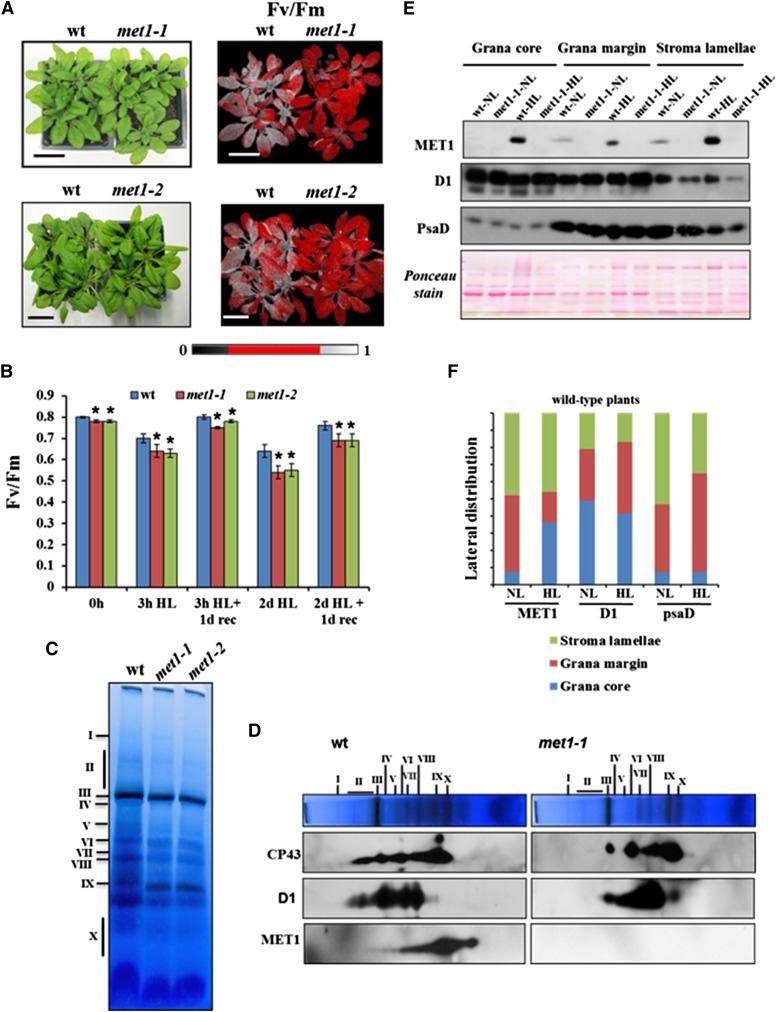
To determine whether loss of MET1 function resulted in enhanced sensitivity to a sudden increase in light intensity, wild-type plants and both met1 mutants were exposed to high light (1200 μmol photons m−2 s−1) for 3 h or 2 d (12-h-light/12-h-dark cycle) (Figure 8A). Subsequently, we measured Fv/Fm as a measure of PSII quantum efficiency by chlorophyll fluorescence imaging 30 min after the light stress or allowing for a 1-d recovery period. All genotypes showed significant loss in Fv/Fm after the high light treatments, with both met1 alleles always more strongly affected than the wild type (Figures 8A and 8B). Even prior to the high light treatment, the Fv/Fm ratio was slightly (3%) but significantly (P < 0.001) reduced in both met1 alleles. To evaluate the impact of the 2-d high light treatment on the PSII assembly state, we performed BN-PAGE of DM solubilized thylakoid membranes followed by either Coomassie blue staining or followed by separation in a second dimension by SDS-PAGE and immunoblotting with antisera against D1, CP43, as well as MET1 (Figures 8C and 8D). As under fluctuating growth light, loss of MET1 resulted in loss of detectible PSII supercomplexes, as evidenced by the lack of D1 and CP43 signals in band II (Figure 8D), whereas PSII supercomplexes were present in the wild type. We also detected significantly less PSII dimer (band III) but an increased level of PSII monomer (VI) in the mutant compared with the wild type (Figure 8D). Moreover, free CP43 was more abundant in the mutant compared with the wild type. We also detected MET1 in PSII dimer, PSII monomer, CP43-less PSII monomer, the PSII reaction center, and unassembled proteins (Figure 8D). To determine whether the light stress affected MET1 lateral thylakoid distribution, we fractionated thylakoids from wild-type plants before and after a 2-d high light treatment; the met1-1 allele was used as negative control (Figure 8E). Immunodetection of D1 and PsaD was used as a marker for PSII and PSI, respectively. Overall MET1 levels increased ∼3-fold after the high light treatment. MET1 distribution shifted from grana margins to grana stacks, but more than half was found in stroma lamellae, as under normal growth light (Figure 8F).
Figure 8.
Response of Wild-Type and met1 Alleles to Short-Term and Long-Term High Light Treatment.
Soil-grown wild type, met1-1, and met1-2 were grown at 100 μmol photons m−2 s−1 (16 h light/8 h dark) followed by 3 h of high light illumination or followed by 2 d of high light. High light was 1200 μmol photons m−2 s−1; 12 h light/12 h dark.
(A) Wild type, met1-1, and met1-2 grown at 100 μmol photons m−2 s−1 (16 h light/8 h dark) exposed for 3 h to high light (1200 μmol photons m−2 s−1). The upper row shows 3-week-old wild-type and met1-1 plants, and the lower row shows 4-week-old wild-type and met1-2 plants after the 3-h light stress. The right panels show false color images representing Fv/Fm. After the 3-h high light treatment, plants were dark adapted for 20 to 30 min prior to fluorescence imaging. Bar = 3 cm.
(B) Fv/Fm values of the wild type, met1-1, and met1-2 grown after 3 h of high light illumination or after 2 d of high light (1200 µmol photons m−2 s−1; 12 h light/12 h dark), in both cases followed by 1 d of recovery at growth light. Plants were dark adapted for 20 to 30 min prior to fluorescence imaging. Standard deviations are indicated (n = 10 to 16). Genotypic differences (wild type and met1) were significant at P < 0.001 using Student's t test as indicated by asterisks.
(C) and (D) BN-PAGE analysis of thylakoid membrane protein complexes in the wild type, met1-1, and met1-2 after 2 d of high light. DM-solubilized thylakoid membrane proteins were separated by BN-PAGE (10 μg chlorophyll per lane), and gel lanes were stained with Coomassie brilliant blue. Individual lanes were run in a second dimension on SDS-PAGE gels after solubilization with SDS, followed by immunoblotting with antisera against D1, CP43, or MET1.
(E) and (F) Distribution of MET1 across the different thylakoid membrane regions before and after high light treatment. Thylakoid proteins were solubilized with digitonin and grana, grana margins, and stroma lamellae were fractionated by ultracentrifugation and analyzed by SDS-PAGE (10 μg protein per lane).
(E) Immunoblots for MET1, D1 (PSII core), and PsaD (PSI core) protein and a representative Ponceau stain of one of the blots are shown.
(F) The quantified distribution for MET1, D1, and PsaD, normalized to the total signal for each protein within each genotype calculated from the blots in (E).
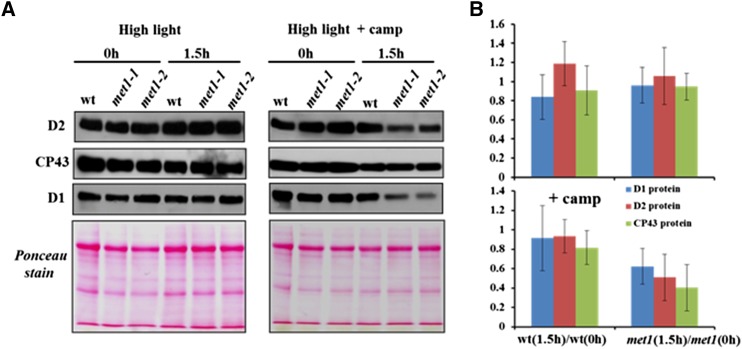
To determine whether PSII degradation rates changed during acute high light stress, detached leaves of the wild type, met1-1, and met1-2 were incubated with the chloroplast translational inhibitor chloramphenicol and subjected to a 1.5-h light stress. Levels of PSII core proteins D1, D2, and CP43 did not significantly change in the wild type and mutants after 1.5 h (Figures 9A and 9B). However, in the presence of chloramphenicol, accumulation of D1 and D2 PSII reaction center proteins decreased strongly in the met1 alleles compared with the wild type (Figures 9A and 9B). Thus, PSII core degradation rates increased in absence of MET1.
Figure 9.
Degradation of PSII Core Proteins after High Light Treatment.
Detached leaves of the wild type, met1-1, and met1-2 grown at 100 μmol photons m−2 s−1 (16 h light/8 h dark) exposed for 1.5 h to high light (1200 μmol photons m−2 s−1) in the absence or presence of the translational inhibitor chloramphenicol. Proteins were extracted from detached leaves and separated by SDS-PAGE gel and subjected to immunoblot with antisera against the PSII core proteins D1, D2, and CP43. Equal amounts of protein (10 μg) were loaded in each lane. (A) shows representative immunoblots and Ponceau stains, whereas (B) shows the average ratios of D1, D2, and CP43 proteins for the wild type and met1 before and after 1.5 h light stress. Data for the met1-1 and met1-2 alleles were averaged within each replicate. Standard deviations are indicated (n = 3). Two-tailed Student’s t tests showed significant differences at P < 0.05 for CP43 and at P < 0.1 for D2 in met1 with chloramphenicol.
Candidates for MET1 Protein Interactors
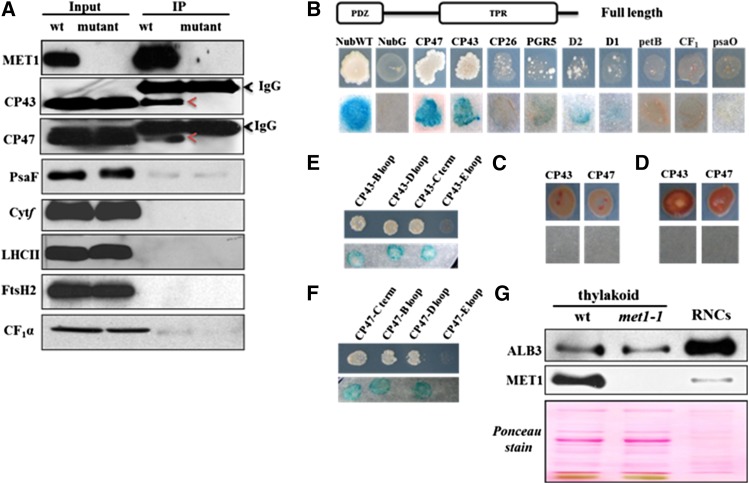
To identify possible proteins interacting with MET1, coimmunoprecipitation (co-IP) experiments were performed with DM-solubilized thylakoid membranes of wild-type and met1-1 chloroplasts using anti-MET1 serum. Immunoblotting and MS/MS analysis showed that the co-IPs were highly enriched for MET1 (Figure 10A; Supplemental Figure 7 and Supplemental Data Set 4). The MS/MS analysis of the co-IPs identified only thylakoid protease FtsH2 (VAR2) as a candidate interactor as it was the only other protein identified in both wild-type replicates with a significant number (26) of matched MS/MS spectra, but never in the met1-1 co-IPs. However, immunoblot analysis of such co-IPs did not confirm the presence of FtsH2 (Figure 10A), suggesting that FtsH2 is less likely to be a functional partner of MET1.
Figure 10.
Interaction of MET1 with Thylakoid Proteins Determined by Co-IP and Y2H Analysis.
(A) Co-IP of MET1 with anti-MET1 serum against DM solubilized thylakoids to identify potential protein interactors. Immunoblotting with various specific antisera showed that MET1 was highly enriched in the co-IP and that both CP43 and CP47 interact with MET1. Thylakoids from the met1-1 mutant were used as negative control.
(B) Split-ubiquitin assays for interactions between full-length MET1 and selected thylakoid proteins. Full-length MET1 was used as bait and selected candidate proteins were used as prey. Bait plasmid contains Cub-PLV and prey plasmid contains NubG. NubG moiety was fused to the N terminus of prey proteins. The resulting plasmids were transformed into the yeast bait and prey strains. The transformed yeast strains harboring bait and prey constructs were mated and resulting transformants were analyzed on selective medium lacking Ade, His, Trp, Leu, Ura, and Met (upper lane) and for β-galactosidase (β-Gal) activity (lower lane). Soluble NubG and Nub-WT were used as negative and positive controls, respectively.
(C) Interaction of the TPR domain of MET1 with CP43 and CP47. Diploid cells were analyzed on selective medium lacking Ade, His, Trp, Leu, Ura, and Met (upper lane) and for β-Gal activity (lower lane).
(D) Interaction of PDZ domain of MET1 with CP43 and CP47. Diploid cells were analyzed on selective medium lacking Ade, His, Trp, Leu, Ura, and Met (upper lane) and for β-Gal activity (lower lane).
(E) and (F) Interactions between full-length MET1 and stromal loops (B, D, and C-terminal) and lumenal loop E of CP43 (E) and CP47 (F). Diploid cells were analyzed on selective medium lacking Ade, His, Trp, Leu, Ura, and Met (upper lane) and for β-Gal activity (lower lane).
(G) Enrichment analysis of RNCs extracted from thylakoids of the wild type. ALB3 but not MET1 is highly enriched in such RNC preparations. Solubilized thylakoid proteins of the wild type and met1-1 were used for reference of the immunoblots.
As an alternative approach to finding candidate proteins interacting with MET1, we used the mating-based yeast split ubiquitin system (Grefen et al., 2007), which has been successfully used to identify protein-protein interactions between integral (thylakoid) membrane proteins (Pasch et al., 2005). Full-length MET1 was used as bait and various photosynthetic proteins were selected as prey. Results showed that MET1 interacted with CP43 and CP47 but not with or very weakly with D1, D2, CP26, petB, CF1α, PsaO, or PGR5 (Figure 10B). To determine which domains in MET1 are involved in the interaction with CP43 and CP47, deletion derivatives of MET1 were analyzed for interaction. Constructs with only the TPR domain or only the PDZ domain failed to interact with CP43 and CP47 (Figures 10C and 10D). This suggests that mature MET1 is required for the interaction of MET1 with PSII reaction center proteins CP43 and CP47. To further clarify whether soluble loops of CP43 and CP47 interact with MET1, we tested stromal loops B and D and the C-terminal end using the yeast two-hybrid (Y2H) system (Figures 10D and 10E). We also included here a lumenal loop E from both CP43 and CP47, but we could not find any interaction between MET1 and this lumenal loop (Figures 10D and 10E). Our data suggest that MET1 interacts with the stromal soluble loops of both PSII core proteins. Based on this result, we probed the co-IPs with antisera against CP43 and CP47, confirming the interaction between MET1 and CP43 and CP47 (Figure 10A). By contrast, immunoblotting with specific antisera against D1, D2, PsaF, Cyt f, LHCII, and CF1α did not show interactions between these proteins and MET1.
MET1 Acts Posttranslationally
RT-PCR was used to examine whether transcript accumulation of plastid-encoded thylakoid proteins was effected in the met1-1 plants. The levels of psbA (encoding the D1 subunit of PSII), psbC (encoding the CP43 of PSII), psaD (encoding the PsaD subunit of PSI), and petB (encoding the cytochrome b6 subunit of Cyt b6f complex) were unchanged in the mutant plants (Supplemental Figure 8). This is consistent with the lack of phenotype under normal conditions and also with the observation that loss of MET1, but not accumulation of smaller PSII assembly intermediates, affects supercomplex accumulation. Initiation of translation in the chloroplast takes place on 70S ribosomes, creating so-called ribosome-nascent chain complexes (RNCs). After initiation, elongation proceeds and the nascent chain emerges from the ribosome tunnel. In case of the translating D1 protein, the nascent D1 protein was found to interact directly with SecY (Zhang et al., 2001). Thylakoid ALB3 was shown to interact with SecY (Klostermann et al., 2002), whereas in bacteria, SecY and the ALB3 homolog YidC both function in cotranslational insertion of membrane proteins (reviewed in Richter et al., 2010). In mitochondria (which lack SecY) the ALB3 homolog OXA1 functions in cotranslation insertion of mitochondrial-encoded membranes proteins and OXA1 interacts directly with ribosomes (Richter et al., 2010), but there is no evidence that thylakoid ALB3 directly interacts with 70S ribosomes. However, ALB3 is involved in biogenesis of PSII core complexes, as evidenced by interactions between PSII assembly factors TERC, LPA2, LPA3, and ALB3 (Cai et al., 2010; Schneider et al., 2014). This led us to examine whether MET1 and ALB3 were present in thylakoid RNC fractions. We thus isolated RNCs associated to thylakoid membranes in presence of the translation inhibitor chloramphenicol. As shown in Figure 10F, immunoblotting showed that ALB3 was strongly enriched in RNCs compared with thylakoids, but MET1 was not. This suggests that ALB3, but not MET1, is possibly directly or indirectly involved in cotranslational insertion of chloroplast-encoded membrane proteins.
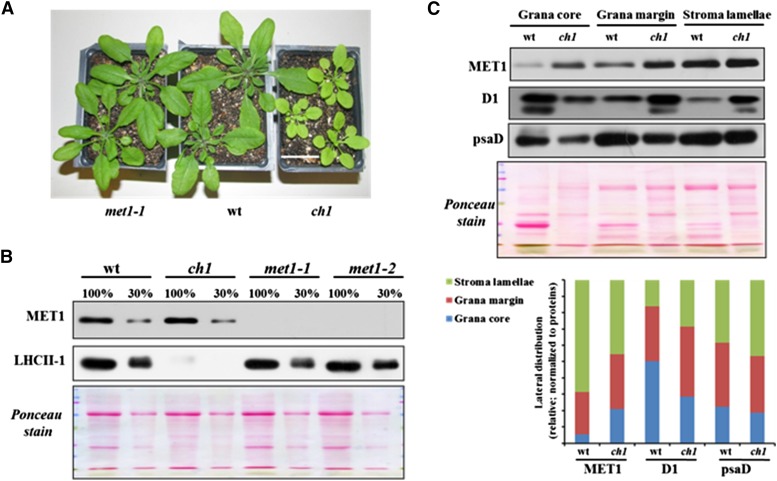
MET1 Accumulation and Lateral Distribution in the Chlorophyll b-Deficient chlorina1 Mutant
Loss of chlorophyll b is known to result in reduced accumulation of LHCII proteins, loss of PSII supercomplex formation, and reduced grana stacking (Murray and Kohorn, 1991; Espineda et al., 1999; Kim et al., 2009). To determine whether MET1 accumulation level and lateral distribution were affected in such chlorophyll b-deficient mutants, we analyzed the chlorina1 (ch1) mutant (Hirono and Redei, 1963), which is a loss-of-function mutant in chlorophyll a oxygenase (Figure 11A). Immunoblot analysis of ch1-1 showed that LHCII-1 (and other LHCII proteins; see Ponceau stain) were strongly reduced to less than ∼10% of wild-type, met1-1, and met1-2 levels (in agreement with previous analysis of ch1; Takabayashi et al., 2011), whereas MET1 levels (normalized to total leaf proteins) were unchanged compared with the wild type (Figure 11B). Compared with the wild type, lateral thylakoid distribution of MET1 in ch1 was slightly shifted toward the grana, when normalized to the distribution of D1 and PsaD distribution (Figure 11C).
Figure 11.
Accumulation of MET1 and LHCII in ch1 and met1 Alleles and the Wild Type.
(A) Plants used for immunoblotting. Bar = 3 cm.
(B) Immunoblot of MET1 and LHCII-1 of thylakoids isolated from ch1, met1-1, met1-2, and the wild type. The gel was loaded based on equal total leaf protein (100% = 10 μg).
(C) Distribution of MET1 across the different thylakoid membrane regions in the wild type and the ch1 mutant. Thylakoid proteins were solubilized with digitonin, and grana, grana margins, and stroma lamellae were fractionated by ultracentrifugation and analyzed by SDS-PAGE (15 μg protein per lane). Immunoblots for MET1, D1 (PSII core), and PsaD (PSI core) protein and the Ponceau stain are shown. The bar diagram shows the relative distribution of MET1, D1, and PsaD across the thylakoid regions in the wild type and ch1 with signals normalized to the total signal for each protein within each genotype.
DISCUSSION
MET1 Is a PSII Assembly Factor in Higher Plants
This study identifies the function of chloroplast MET1, an evolutionary conserved protein in photosynthetic eukaryotes, including algae, but lacking in prokaryotes. Loss of MET1 in Arabidopsis resulted in reduced rosette biomass, reduced PSII efficiency, and photosynthetic electron transport under fluctuating growth light conditions. Furthermore, under fluctuating light intensity, MET1 was essential for the assembly of PSII supercomplexes, the active form of PSII in vivo (see next section). By contrast, under constant daylight intensity, loss of MET1 did not significantly appear to effect plant growth or the PSII assembly state, even if PSII efficiency (Fv/Fm) was slightly (3%) reduced. Also, after sudden high light exposure, PSII supercomplexes were dismantled in the MET1 loss-of-function mutant (met1) but not in the wild type. Furthermore, upon high light exposure, PSII quantum efficiency decreased more in met1 than in the wild type. We showed that MET1 is peripherally attached to the stromal side of thylakoids, in particular stroma lamellae, and that it interacts with various PSII assembly intermediates, including PSII dimers and monomers. Furthermore, both Y2H essays and co-IP showed that MET1 interacts with CP43 and CP47. Loss of MET1 function also resulted in increased levels of unassembled CP43.
The interaction of MET1 with CP43 and CP47 PSII subunits required the TPR and PDZ domains in MET1, and both domains are recognized for their function in establishing protein-protein interactions. TPR domain proteins often mediate specific interactions with partner proteins, acting as cochaperones or providing a scaffold for protein assembly (D’Andrea and Regan, 2003). Examples in chloroplasts include PSII assembly factors LPA1, PSI assembly factors YCF3 and PYG7, as well as many proteins involved in plastid mRNA metabolism. A relatively small number of PDZ domain proteins have been studied in Arabidopsis, most of which are found in the cell wall or are part of the cytoskeleton (Gardiner et al., 2011). Other PDZ domain proteins are various proteases, including chloroplast stromal DEG2 (Sun et al., 2012) and lumenal DEG8 (Sun et al., 2013). PDZ domains typically recognize the C terminus of target proteins, although some also recognize an internal motif, but it does not seem possible to predict the targets. Interestingly, the C termini of both CP43 and CP47 are exposed to the stromal side of the thylakoid membranes, making them a likely target for the PDZ domain of MET1. Indeed, Y2H analysis showed interaction between MET1 and not only these C termini, but also two stromal loops, but not lumenal loops. We speculate that MET1 is involved in folding and/or stabilizing these stromal-exposed CP43/CP47 domains, thereby ensuring the correct configuration for PSII supercomplex formation.
Formation of PSII Supercomplexes in Higher Plants and the Role of MET1
Using cryo-electron microscopy, native gel electrophoresis, and sucrose gradient centrifugation in combination with immunoblotting or mass spectrometry, it is now well established that the active forms of PSII are the so-called supercomplexes (Järvi et al., 2011; Kouřil et al., 2012; Minagawa, 2013; Nickelsen and Rengstl, 2013; Pagliano et al., 2014). After the sequential formation of the monomeric and dimeric PSII core (C2), minor and major LHCII proteins associate to the PSII dimer to form different sized PSII supercomplexes. The minor LHCII proteins (CP24, CP26, and CP29) provide the molecular interface between the PSII core and the LHCII trimers (LHCII-1,2,3) (Ballottari et al., 2012; Kouřil et al., 2012). The PSII supercomplex (C2S2M2) is the major and largest in vivo form of PSII for Arabidopsis.
Numerous reverse genetic studies in Arabidopsis have probed the role of the minor and major LHCII proteins relating to PSII properties and activities, PSII assembly and supercomplex formation, and photosynthetic growth (reviewed in Ballottari et al., 2012; Kouřil et al., 2012; Nickelsen and Rengstl, 2013; Pan et al., 2013). These studies show that there is a surprising robustness in the general light-harvesting function because loss of one gene product can be compensated for by overaccumulation of other gene products. However, loss of specific LHCII proteins can lead to changes in oligomeric organization and light-harvesting and quenching properties. For instance, loss of CP24, CP26, or LHCII-3 resulted in destabilization or loss of PSII supercomplexes (Caffarri et al., 2009; de Bianchi et al., 2011). Also, the various PSII core subunits are important for formation of PSII supercomplexes; e.g., absence of the small PSII core protein PSBW resulted in loss of PSII supercomplexes (García-Cerdán et al., 2011). Finally, environmental conditions appear to influence accumulation of PSII supercomplexes (Kouřil et al., 2013). Interestingly, recent results indicate that state transitions in higher plants thylakoids do not lead to changes in PSII supercomplexes because the migrating LHCII proteins form a separate pool of LHCII and are not trimers within these PSII supercomplexes (Wientjes et al., 2013).
Once assembled and functional, PSII undergoes continuous light-induced damage of the D1 reaction center protein due to oxidative stress. Replacement of damaged D1 with an intact D1 requires partial disassembly of PSII and lateral migration of PSII subcomplexes to the stroma lamellae where cotranslational insertion of D1 protein and reassembly of the PSII core complex take place. This is then followed by PSII dimerization and formation of supercomplexes in the grana regions (Aro et al., 2005; Mulo et al., 2008; Nickelsen and Rengstl, 2013). More than a dozen auxiliary proteins aiding in this PSII repair process have been identified (Mulo et al., 2008; Chi et al., 2012a; Nickelsen and Rengstl, 2013). This underscores the notion that assembly of PSII is a well-organized process involving precise requirements for protein-protein interactions. Nevertheless, the mechanisms driving the sequential formation from PSII monomer to PSII dimer and then PSII supercomplexes are poorly understood.
It is well established that key steps of PSII repair and reassembly take place in stroma lamellae (Mulo et al., 2008), and the enrichment of MET1 in these membrane regions is consistent with its involvement in early steps of PSII core disassembly and/or reassembly. However, MET1 is also found in the intermediate regions and grana, consistent with our observation that MET1 appears associated with PSII monomers and dimers (inferred from the BN-PAGE analysis) and only releases after formation of PSII dimers but prior to PSII supercomplex formation. Presumably, kinetics of the repair/reassembly process requires that most MET1 be localized to the stroma lamellae. In absence of most of the LHCII (through loss of chlorophyll b synthesis in the ch1 mutant), we observed that MET1 partially redistributed to grana regions. This is consistent with our suggestion that MET1 can remain associated with PSII complexes as long as they do not form supercomplexes.
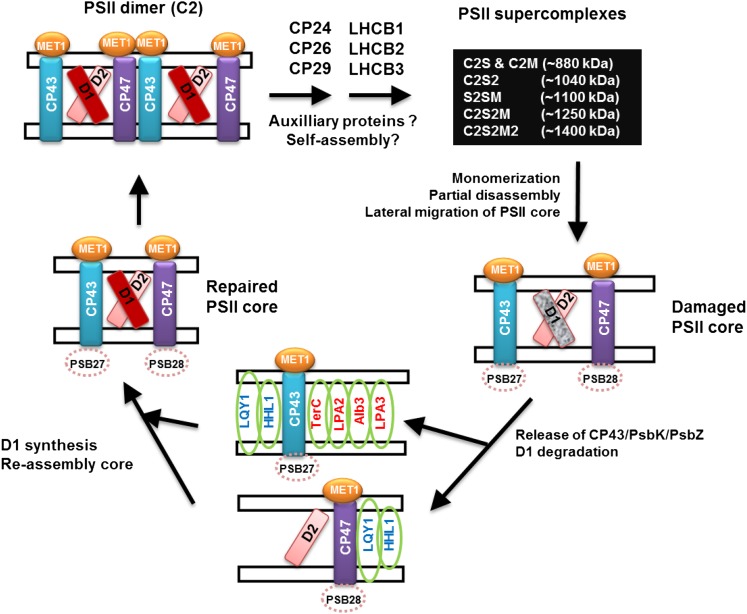
Based on the known mechanisms of PSII repair and reassembly, we summarized the findings for MET1 and five other known auxiliary factors involved with CP43 and/or CP47 in the model shown in Figure 12. Assembly factors involved in D1 synthesis, processing, or reassembly are not shown. MET1 interacts with CP43 and CP47 at every assembly step, except in supercomplexes. We propose that MET1 aids in assembly of the PSII core and dimer, thus helping to prime the PSII dimer for association of minor and major LHCII protein and the formation of PSII supercomplexes. The assembly process from PSII dimer to supercomplex during which minor and major LHCII proteins associate to the core is not understood and could be a self-assembly process perhaps aided by as yet unknown auxiliary factors. The other assembly factors interacting with CP43 are TERC, LPA2, and LPA3 (Pasch et al., 2005; Ma et al., 2007; Schneider et al., 2014), whereas LQY1 and HHL1 interact with both CP43 and CP47 (Lu et al., 2011; Jin et al., 2014). In contrast to MET1, these five factors are all integral membrane proteins. TERC interacts with LPA2, which in turn interacts with LPA3 and each of these three factors interact with the translocon ALB3. This suggests that LPA2, LPA3, and TERC act together in the biogenesis and assembly of CP43. LQY1 also interacts with HHL1, but there is no evidence that they interact with ALB3, LPA2, LPA3, or TERC. This suggests that LQY1 and HHL1 act together in assembly of CP43 and CP47, most likely in a later step than these ALB3 interacting factors. Based on the experimental evidence presented in this study, we suggest that MET1 also acts separate from the ALB3-dependent steps, given its peripheral membrane location (stromal side), and we suggest that it helps folding stromal CP43 loops, once CP43 is fully inserted in the thylakoid membrane. There are two assembly factors, PSB27 and PSB28, in cyanobacteria that have been shown to be involved with CP43 or CP47 assembly. Lumenal PSB27 transiently binds to CP43 to prevent the binding of luminal OEC subunits onto the preassembled monomeric PSII complex and facilitates Mn4CaO5 assembly (Mabbitt et al., 2014). Cyanobacterial PSB28 binds to CP43-less PSII complexes and to unassembled CP47, and analysis of the loss-of-function mutant indicated involvement in chlorophyll synthesis and/or CP47 and PSI reaction center proteins (Dobáková et al., 2009; Mabbitt et al., 2014). The respective homologs in Arabidopsis (AT1G03600 [see Chen et al., 2006] and AT4G28660) likely have similar roles as in cyanobacteria, but this awaits experimental testing. Five of the known Arabidopsis PSII assembly factors (HCF243, LPA2, LPA3, LQY1, and PPL1) and MET1 have no sequence or functional homologs in cyanobacteria but are conserved across higher plants, suggesting that each evolved as an adaption to the higher plant thylakoid system. Out of these six factors, only MET1 and LPA3 are also found in algae such as Chlamydomonas reinhardtii, indicating that MET1 and LPA3 evolved earlier than HCF243, LQY1, LPA2, and PPL1 and therefore are active in a more conserved assembly step than the other factors.
Figure 12.
Model of PSII Assembly Factors Involved with CP43 and/or CP47.
This figure provides a highly simplified outline of the proposed PSII assembly process, emphasizing those assembly factors interacting with CP43 (TERC, LPA2, and LPA3) or both CP43 and CP47 (LQY1 and HHL1) and the role of MET1. MET1 interacts with CP43 and CP47 at every assembly step, except in supercomplexes. We propose that MET1 aids in assembly of the PSII core and dimer, thus helping to prime the PSII dimer for association of minor and major LHCII protein and the formation of PSII supercomplexes,
Why Is MET1 Essential for PSII Supercomplex Formation and Optimal Photosynthetic Rates Only under Strongly Fluctuating Light Conditions and Sudden High Light Exposure?
MET1 plays a key role in PSII assembly and photosynthetic capacity under fluctuating light conditions (changing every 10 min between 50 and 600 μmol photons m−2 s−1) but appears less important under constant light intensity. Similarly, loss-of-function mutants for PSII assembly factor TLP18.3, as well as state transition kinase STN7 and the cyclic electron flow component PGR5, only show conditional growth phenotypes under fluctuating light. TLP18.3 is located on the lumenal side of the thylakoid membrane and is required for the PSII repair cycle under fluctuating growth light. Furthermore, PSII supercomplex formation was reduced but not abolished in the mutant. However, the molecular interactors of TLP18.3 are not yet known (Sirpiö et al., 2007). It is quite likely that both in case of TLP18.3 and MET1, the conditional fluctuating light phenotype indicates that these assembly factors make the process of PSII assembly (and perhaps disassembly) more efficient but are not strictly required. The strong conditional phenotypes of STN7 (but not STN8) and PGR5 under fluctuating light conditions are likely very different and involves the optimization of linear and cyclic electron flow in response of changing light intensities and the changing requirements for ATP and NADPH by chloroplast metabolism. Finally, high light treatment also destabilized PSII supercomplexes in met1-1 but not the wild type, and D1 degradation greatly accelerated, similar as in other PSII assembly mutants (LQY1, HHL1, PPL1, and TLP18.3).
MET1 and the PSII Assembly Process in Dimorphic Chloroplasts of Maize Leaves
We studied MET1 because we initially identified the maize MET1 homolog as a chloroplast protein 3-fold enriched in M cells, compared with BS cells (in the context of understanding regulation of C4 photosynthesis in maize; Majeran and van Wijk, 2009). We correctly hypothesized that MET1 is a PSII assembly factor in Arabidopsis, but it is important to consider how this relates to M and BS chloroplasts in maize.
The photosynthetic capacity of maize dimorphic M and BS chloroplasts has been studied for more than four decades (reviewed in Edwards et al., 2001b, 2004; Majeran and van Wijk, 2009; Sage et al., 2012). Collectively, this has demonstrated that M thylakoids have a complete linear electron transport chain, containing PSII, Cyt b6f, and PSI complexes, similar to C3 plants. By contrast, fully differentiated BS thylakoids have little functional PSII and normal or increased PSI levels, whereas Cyt b6f and ATP synthase complexes are quite evenly distributed between BS and M thylakoids. As a consequence, BS thylakoids carry out mostly cyclic electron flow and have low rates of linear electron flow (Woo et al., 1970; Hardt and Kok, 1978; Schuster et al., 1985). After several conflicting reports on protein levels of PSII and LHCII (Schuster et al., 1985; Bassi and Simpson, 1986; Oswald et al., 1990), a subsequent study clarified both activity and protein accumulation levels of PSI and PSII complexes in BS and M thylakoids (Meierhoff and Westhoff, 1993). This showed that PSII activity in BS was only 10 to 15% of that in M. Immunoblotting showed that BS/M ratios of accumulation levels of oxygen-evolving complex proteins of PSII are ∼0.15, which is lower than the ratios observed for other PSII core subunits (∼0.3) (Meierhoff and Westhoff, 1993). This explained the strongly reduced linear electron transport rates in BS thylakoids, as was shown previously (Schuster et al., 1985). Taking advantage of vast improvements in mass spectrometry, a comprehensive overview of accumulation levels and assembly state of the thylakoid bound photosynthetic apparatus was described (Majeran et al., 2005, 2008, 2010; Friso et al., 2010). This showed that accumulation of the PSII monomer (∼270 kD) and a partially assembled PSII core (∼200 kD) was 3- and 5-fold reduced, respectively, in BS membranes, while the high molecular weight PSII complexes observed in M thylakoids were below detection in BS membranes (Supplemental Figure 9). The accumulation of LHCII trimers (∼140 kD) was only 1.6-fold reduced in the BS membranes (BS/M ∼0.6), suggesting that a subset of LHCII proteins do function in the BS (perhaps by association to PSI, similar as in C3 plants; Galka et al., 2012), consistent with previous findings (Bassi and Simpson, 1986).
The observed MET1 accumulation level was 3- to 5-fold higher in M than in BS chloroplasts (BS/M ratio 0.2 to 0.3), in agreement with the observed enrichment of PSII in M chloroplasts. Also, maize homologs of several other known Arabidopsis PSII assembly factors show similar M enrichment. Examples are LPA3, PSB28, and TLP40 with respective protein BS/M ratios of 0.2, 0.3, and 0.3 (Friso et al., 2010). Thus, PSII assembly factors do not exclusively accumulate in M chloroplasts, and their BS/M ratios are in fact similar to PSII core subunits. BS/M mRNA accumulation ratios based on RNA-seq data are consistent with these protein accumulation ratios (Li et al., 2010; Tausta et al., 2014). More than 10 PSII assembly factors have now been identified in Arabidopsis, and homologs of each are present in maize. So far only PSII assembly factor HCF136 has been studied in maize, and as in Arabidopsis, null mutants (HCF136) did not show accumulation of PSII complexes (Covshoff et al., 2008). Given the sequence homology between maize and Arabidopsis MET1 and the maize BS/M accumulation ratio, it is most likely that maize MET1 functions as a PSII assembly factor also in maize.
Finally, we like to suggest that dimorphic chloroplasts in maize leaves provide a resource for discovery of additional PSII specific assembly factors. Maize BS/M thylakoid protein accumulation ratios below one can potentially identify yet additional PSII assembly factors. A systematic bioinformatics analysis, based on data in the public domain, to identify such factors as well as other factors specifically involved in regulating or accommodating linear and cyclic electron transport is in progress (N.H. Bhuiyan and K.J. van Wijk, unpublished data).
In conclusion, MET1 is a PSII assembly factor required for PSII supercomplex formation, in particular under fluctuating light conditions. Forward genetics analysis of Arabidopsis mutants under such environmental conditions likely will identify additional auxiliary proteins required for PSII biogenesis and homeostasis. More detailed molecular characterization of leaf and chloroplast development of wild-type Arabidopsis grown under fluctuating light is needed to fully understand adaptation and bottlenecks in biogenesis and homeostasis under such environmental conditions. Finally, maize thylakoid proteins with high abundance in M chloroplasts compared with BS chloroplasts provide a resource of potential candidates for PSII assembly factors, state transition, or other regulators of linear electron transport. Exploration of Arabidopsis homologs of such candidates may provide an efficient way to further elucidate this fascinating PSII repair process.
METHODS
Phylogenetic Analysis
To generate a phylogenetic cladogram, 18 MET1 proteins from different plant species including 10 angiosperms, 2 gymnosperms, 1 moss, and 5 algae were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The alignment is available as Supplemental Data Set 5. The aligned sequences were exported in Clustal format and viewed in Jalview (www.jalview.org/). Sequences were then converted in PHYLIP format, and phylogenetic trees were generated (1000 iterations) using the CIPRES Web portal (http://www.phylo.org/) selecting the tool “RAxMLHPC blackbox.” The resulting phylogenetic tree was annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). RAxML bootstrap support values are shown at the nodes of the tree.
mRNA Expression Analysis
In silico mRNA expression data for Arabidopsis thaliana were extracted from the AtGenExpress website (http://www.weigelworld.org/resources/microarray/AtGenExpress/). The Arabidopsis tissue-specific expression profile was derived using the e-FP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi/).
Antibody Generation
The nucleotide sequence encoding amino acids 1 to 233 of MET1 (without TPR domains) were amplified by PCR. The resulting DNA fragment was ligated into BamHI and XhoI sites of the C-terminal His affinity tag of the pET21a expression vector (Novagen). Escherichia coli BL21 cells (New England Biolabs) were transformed with pET21a vector harboring this truncated MET1 gene, and cells were harvested from liquid cultures after addition of 1 mM isopropyl β-d-1-thiogalactopyranoside for 3 h at 37°C. The overexpressed proteins were solubilized in 200 mM NaCl, 50 mM Tris, and 8 M Urea at pH 8.0 and purified on a nickel-nitrilotriacetic acid agarose resin matrix. A polyclonal antibody against this truncated MET1 protein was raised in rabbits by injecting purified antigen (Alfa Diagnostic International). Antisera were affinity purified against the same antigen coupled to a HiTrap N-hydroxysuccinimide (NHS) ester-activated column (GE Healthcare Life Science).
Plant Material, Growth, Light Regimes, Mutants, Genotyping, and RT-PCR Analysis
T-DNA lines (Col-0) MET1-1 (SAIL_675_E06), MET1-2 (WISCDSLOXHS212_08F), and lpa-1 (WiscDsLox506D10) were obtained from the ABRC. T-DNA inserted plant was identified by genotyping and insertion was confirmed by DNA sequencing. The homozygous chlorophyll b-less ch1-1 mutant (Hirono and Redei, 1963) was also obtained from the ABRC (line CS3119); ch1-1 is a loss-of-function mutant in chlorophyll a oxygenase (AT1G44446). A complete list of primers for genotyping can be found in Supplemental Table 1. Seeds were stratified in the dark at 4°C for 2 d and then transferred in a growth chamber with short-day conditions (14 h light/10 h dark) and temperature at 22°C with 80 μmol photons m−2 s−1 light intensity. For high light treatment, 4-week-old soil-grown plants were transferred from 80 μmol photons m−2 s−1 light intensity (normal light condition) to 800 μmol photons m−2 s−1 at 22°C (16 h light/8 h dark). For fluctuating light conditions, soil-grown seedlings (5 to 7 d old) were transferred to an incubator where light conditions were changed every 10 min from 50 μmol photons m−2 s−1 to 600 μmol photons m−2 s−1 at long-day condition (16 h light/8 h dark) and temperature at 22°C.
For transcript analysis, total RNA was extracted from Arabidopsis leaves using the RNeasy plant mini kit (Qiagen). RNA was reverse transcribed with random hexamer primers using Superscript III reverse transcriptase from Invitrogen. mRNA levels were normalized by ACTIN2. PCR conditions were 22 cycles at 94°C for 2 min, 55°C for 30 s, and 72°C for 1 min per cycle. A complete list of primers can be found in Supplemental Table 1.
High light treatments on detached leaves were conducted using detached leaves as described by Ishihara et al. (2007). Detached leaves were placed (adaxial side up) on filter paper soaked with sodium phosphate/NaOH buffer, pH 7.0. Leaves were illuminated with 1200 μmol photons m−2 s−1 at 22°C and sampled at different time points as described in the text. For chloramphenicol treatment, detached leaves were incubated with the same buffer with 200 μg mL−1 chloramphenicol under reduced pressure for 15 min prior to light illumination. Leaves were collected at different time points and ground immediately in liquid nitrogen. To analyze PSII core proteins, total proteins were extracted from ground leaves and analyzed by SDS-PAGE and immunoblotted with respective antibodies.
Light Sources and Details
The following light sources were used: incandescent lamp 40 W (GE), Ceramalux high-pressure sodium noncycling 400 W ED-18 (clear) lamp (Phillips-C400S51), and metal halide 400 W M59/E (Sylvania BT37 Metalarc). Light sources for normal growth light were as follows: 80 μmol photons m−2 s−1 light intensity (in this case, the distance between plant and light was ∼38 inches), 2× metal halide and 2× incandescent lamp. Light sources for light fluctuation experiments were: 50 μmol photons m−2 s−1 light intensity (in this case, the distance between plant and light was ∼38 inches), 1× metal halide and 2× incandescent lamp; 600 μmol photons m−2 s−1 light intensity (in this case, the distance between plant and light was ∼38 inches), 4× metal halide, 2× sodium lamp, and 4× incandescent lamp. For high light experiments, the sources were: 800 μmol photons m−2 s−1 light intensity (in this case, the distance between plant and light was ∼38 inches), 4× metal halide, 3× sodium lamp, and 4× incandescent lamp; 1200 μmol photons m−2 s−1 light intensity (in this case, the distance between plant and light was ∼16 inches), 4× metal halide, 4× sodium lamp, and 4× incandescent lamp.
Chlorophyll Measurement and Chlorophyll Fluorescence
Chlorophyll concentrations were determined by absorbance spectrometry after extraction of chlorophyll in 80% acetone (Porra et al., 1989). Chlorophyll fluorescence parameters Fo and Fm were measured for 14- to 16-d-old wild-type and met1-1 plants after 20 min dark adaption using the MAXI version of the Imagine-PAM M-series chlorophyll fluorometer (Heinz-Walz Instruments).
Extraction and Analysis of Total Cellular Proteome
To quantitatively extract total cellular proteins from leaves, stems, roots, inflorescences, and siliques, the respective organs were ground in liquid nitrogen, followed by extraction, filtering, and centrifugation as described (Friso et al., 2011).
Chloroplast and Thylakoid Isolation
Chloroplasts were isolated on Percoll step gradients from mature rosettes of 6-week-old plants as described (Olinares et al., 2010), Leaves were briefly homogenized in grinding medium (50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol, 2 mM EDTA, 5 mM ascorbic acid, 5 mM cysteine, and 0.03% BSA) and filtered through a nylon mesh. Crude plastids were then collected by a 5-min spin at 2000g and further purified on 40 to 85% Percoll cushions (Percoll in 0.6% Ficoll and 1.8% polyethylene glycol) by a 20-min spin at 4000g and one additional wash in the grinding medium without ascorbic acid, cysteine, and BSA. Chloroplasts were subsequently lysed in lysis buffer (10 mM HEPES-KOH, pH 8.0, and 5 mM MgCl2) containing a mixture of protease inhibitors that we assembled from individual stocks (antipain, bestatin, chymostatin, pefabloc, and aprotinin) (Friso et al., 2011) with mild mechanical disruption using a glass homogenizer with a tight-fitting pestle. The lysate was then subjected to centrifugation at 18,000g (Beckman coulter centrifuge 22R) for 10 min at 4°C to pellet the membranes. Protein amounts were determined using the BCA protein assay kit (Thermo Scientific). Alternatively, crude thylakoids were isolated without Percoll step gradients. Leaves were briefly homogenized in grinding medium and filtered through a nylon mesh. The crude plastid extract was collected by a 5-min spin at 2000g and the pellet was washed in the grinding buffer without ascorbic acid, cysteine, and BSA. The pellet was then lysed in lysis medium and plastids were mechanically disrupted using a glass homogenizer with a tight-fitting pestle. Thylakoid membranes were pelleted by centrifugation at 18,000g (Beckman coulter centrifuge 22R) for 10 min at 4°C.
Subfractionation of Thylakoid Membranes
Thylakoid membranes were subfractionated into grana and stromal lamellae using the method described by Lu et al. (2011). Thylakoids were diluted to 1 mg/mL chlorophyll by adding resuspension buffer (25 mM Tricine-NaOH, pH 7.8, 150 mM sorbitol, 10 mM NaCl, and 5 mM MgCl2). Membranes (at 0.5 mg/mL chlorophyll concentration) were solubilized for 15 min on ice in the same solution in presence of 1% digitonin. The reaction was stopped by adding a 10-fold volume of ice-cold resuspension buffer. After centrifugation at 1000g for 3 min at 4°C, supernatant was collected and grana lamellae were collected by centrifugation at 10,000g for 30 min at 4°C. The supernatant was centrifuged at 40,000g for 30min at 4°C to collect the grana margins. Finally, to pellet stroma lamellae enriched membrane, the supernatant was centrifuged at 145,000g for 1 h at 4°C.
Thermolysin Treatment
Thylakoids were resuspended in lysis medium and adjusted to 0.3 mg of chlorophyll/mL and treated with thermolysin (0.1 mg/mL final concentration) for 15, 30, and 45 min on ice. Protease digestion was stopped by addition of EGTA (50 mM final concentration).
Two-Dimensional BN-PAGE
BN-PAGE was performed essentially as described (Järvi et al., 2011) with the following modifications. Thylakoid membranes were resuspended in the BN-PAGE medium (10 mM HEPES, pH 8.2, 5 mM MgCl2, 50 mM aminocaproic acid, and 20% glycerol) to a final chlorophyll concentration of 0.5 mg/mL. Thylakoid membranes were solubilized with 1% DM on ice for 5 min, and unsolubilized material was removed by centrifugation at 25,000g at 4°C for 10 min. Coomassie Brilliant Blue G 250 was added to the solubilized materials (0.25% w/v). Proteins were loaded on native page gradient gels (4 to 16% acrylamide, Novex Bis-Tris gel system; Invitrogen). For 2D analysis, individual lanes from the BN-PAGE gel were sliced and equilibrated in equilibration buffer (50 mM Tris, pH 6.8, 6 M urea, 20% glycerol, 2% SDS, and 5 mM TBP) for 1 h at room temperature and embedded on the top of a 2D gel using 1% agarose. After separation of protein complexes, the gel was subjected to immunoblotting with relevant antibodies.
Isolation of Ribosome Nascent Chain Complexes
RNCs were isolated as described (Houben et al., 1999; Nilsson et al., 1999) with some modifications. Thylakoid proteins from young seedling were solubilized with 1% DM in RNCs buffer (250 mg/mL chloramphenicol, 50 mM HEPES, pH 8.0, 5 mM Mg acetate, 50 mM K acetate, 2 mM DTT, and protease cocktail; described above). Solubilization was performed at 0.5 μg/mL chlorophyll on ice for 60 min and then briefly spun at 1000g for 1 min to remove unsolubilized material. The supernatant was loaded onto 33% (w/v) sucrose cushions containing RNC buffer. Sucrose cushions were spun at 200,000g for 90 min. The pellet was dissolved in 2% SDS and analyzed by immunoblotting with Alb3 and MET1 antibodies.
Co-IP
For co-IP, purified MET1 antiserum was incubated with Dynabeads protein A (Invitrogen) for 1 to 2 h at 4°C with PBS solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 8.0) and 0.1% (v/v) Igepal CA-630 (Sigma-Aldrich). Then beads were washed with same buffer (900 μL each time) three times to remove excess antiserum. Both wild-type and met1-1 thylakoids were solubilized using 1% DM at 1 μg chlorophyll/mL for 30 min on ice, followed by centrifuged at 12,000g for 20 min. The resulting supernatant (50 to 100 μg solubilized membrane proteins) was incubated with antiserum-Dynabead complex for 2 to 3 h in co-IP buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1% Igepal CA-630 [Sigma-Aldrich], and 5 μg mL−1 aprotinin) at 4°C. The beads were washed seven times with co-IP buffer. Bound proteins were eluted with 50 μL of 1.5× Laemmli buffer (90 mM Tris-HCl, pH 6.8, 3% SDS, 10% glycerol, and a few grains of bromophenol blue) at 75°C.
Yeast Split Ubiquitin Assay
Fragments containing the coding sequence of mature MET1 (amino acids 74 to 335) or constructs TPR (amino acid 217 to 335) and PDZ (amino acid 74 to 216) were cloned in pMetYC-DEST vector (Grefen et al., 2007) and used as bait for interaction studies. Different membrane proteins were cloned in to pXN22-DEST or pNX32-DEST (Grefen et al., 2007) and used as prey for the interaction with bait proteins. Haploid yeast strains THY.AP4 and THY.AP5 (ABRC) were transformed by bait constructs and prey constructs, respectively. The bait plasmid contains Cub-PLV and the prey plasmid contains NubG. NubG is a mutated form of N-terminal ubiquitin domain (mutated at Ile-13 to glycine) that has reduced affinity for Cub (C-terminal ubiquitin); functional ubiquitin can only be reconstituted when NubG and Cub are in close vicinity by fusion via interacting proteins (Grefen et al., 2007). The NubG moiety was fused to the N terminus of prey proteins. After mating THY.AP4 with THY.AP5, diploid cells were selected on synthetic medium lacking Trp and Leu. Positive colonies were used for further testing of interaction. Interactions between bait and prey proteins were performed according to the protocol described (Grefen et al., 2007). Interactions were verified by growing yeast colonies on synthetic minimal medium lacking Ade, His, Trp, Leu, Ura, and Met and also by β-galactosidase activity (Grefen et al., 2007). Soluble NubG and Nub-WT were used as negative and positive controls, respectively. Interactions between MET1 and stromal soluble loops of CP43 and CP47, loops B (amino acids 129 to 173 for CP43 and amino acids 111 to 150 for CP47) and D (amino acids 254 to 271 for CP43 and amino acids 220 to 236 for CP47), and C-terminal loop (amino acids 438 to 473 for CP43 and amino acids 471 to 508 for CP47), as well as lumenal loops E (amino acids 294 to 425 for CP43 and amino acids 258 to 449 for CP47), were cloned and tested by the Y2H system using the same method as for intact CP43 and CP47.
Proteomics, Mass Spectrometry, and Display in PPDB
For protein identification and quantification, each gel lane was cut in consecutive gel slices, followed by in-gel digestion using trypsin and subsequent peptide extraction as described previously (Friso et al., 2011). Peptide extracts for each gel band were then analyzed by online nano-liquid chromatography-MS/MS using an LTQ-Orbitrap (Thermo). Resulting spectral data were searched against the predicted Arabidopsis proteome (TAIR10), including a small set of typical contaminants and the decoy, as described (Nishimura et al., 2013). Only proteins with two or more matched spectra were considered. Protein abundances were quantified according to the number of matched AdjSPC as explained (Friso et al., 2011). MS-derived information, as well as annotation of protein name, location, and function for the identified proteins, can be found in the Plant Proteome Database (http://ppdb.tc.cornell.edu).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: maize MET1, GRMZM2G312910; Arabidopsis MET1, AT1G55480; rice MET1, Os07g07540; Arabidopsis LPA1, AT1G02910; Arabidopsis LPA1-like, AT4G28740; and ACTIN2, AT3G918780. Germplasm used included Arabidopsis T-DNA mutants in the Col-0 background from ABRC: met1-1 (SAIL_675_E06); met1-2 (WISCDSLOXHS212_08F); and ch1-1 (ABRC line CS3119, deficient in AT1G44446).
Supplemental Data
Supplemental Figure 1. Sequence Alignment of the MET1 Homologs in Selected Monocots and Dicots and Experimental Sequence Coverage of Rice MET1.
Supplemental Figure 2. Structural Model for Mature MET1 Generated by i-TASSER.
Supplemental Figure 3. Protein and mRNA Accumulation Patterns during Arabidopsis Development in Leaves and Other Organs.
Supplemental Figure 4. Testing the Effect of Different Light Regimes on Growth and Development of met1-1 and met1-2.
Supplemental Figure 5. Growth and Developmental Phenotypes of Single and Double Mutants in LPA1 and MET1.
Supplemental Figure 6. Scatterplots and Coomassie-Stained SDS-PAGE Gel with 60 μg Proteins of Isolated Thylakoid Proteins of Wild-Type and met1-1 Grown on Soil under Normal Light or Fluctuating Growth Light Used for MS/MS Analysis and Quantification by Label-Free Spectral Counting.
Supplemental Figure 7. Identification of MET1-1 Candidate Protein Interactors.
Supplemental Figure 8. Accumulation of Selected Plastid Gene Transcripts in the Wild Type and met1-1 Mutant as Determined by RT-PCR.
Supplemental Figure 9. MS/MS Analysis of BN-PAGE Gel Lanes of Maize M and BS Thylakoids.
Supplemental Table 1. Primers Used for Genotyping, RT-PCR Analysis and Cloning of the MET1 Antigen.
Supplemental Data Set 1. MS/MS Analysis of BN-PAGE Gel Lanes of Wild-Type and met1-1 Plants Grown under Normal Light Conditions.
Supplemental Data Set 2. MS/MS Analysis of SDS-PAGE Gel Lanes of Wild-Type and met1-1 Plants Grown under Fluctuating Light or Normal Light Conditions.
Supplemental Data Set 3. MS/MS Analysis of BN-PAGE Gel Lanes of Wild-Type and met1-1 Plants Grown under Fluctuating Light Conditions.
Supplemental Data Set 4. Proteins Detected by MS/MS in THE Wild Type and met1-1 (Negative Control) from co-IP with MET1 Antiserum.
Supplemental Data Set 5. Sequence Alignment of the MET1 Proteins in Arabidopsis and in Other Plant and Algal Species.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grants IOS-0922560 and IOS-1127017 to K.J.v.W. We thank Eva Mari-Aro for providing D1 and D2 antisera, Alice Barkan for CP43, OEC33, and PsaD antisera, Zach Adam for FtsH2 antiserum, Danny Schnell for TOC75 antiserum, and Hendrick Scheller for PsaF antiserum. CP47 and LHCII antisera were purchased from Agrisera. We thank Jian Hua for providing yeast two-hybrid vectors and Tom Owens for assistance with chlorophyll fluorescence imaging.
AUTHOR CONTRIBUTIONS
N.H.B. and K.J.v.W. designed the experiments, analyzed the data, and wrote the article. G.F. and A.P. carried out the mass spectrometry analysis. N.H.B. carried out all other experiments described in this article. L.P. carried out statistical analysis of SPC data sets and other bioinformatics analysis of SPC. K.J.v.W. supervised the project.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- M
mesophyll
- BS
bundle sheath
- Col-0
Columbia-0
- DM
dodecyl maltoside
- BN-PAGE
blue-native PAGE
- MS/MS
tandem mass spectrometry
- co-IP
coimmunoprecipitation
- Y2H
yeast two-hybrid
- RNC
ribosome-nascent chain complex
- AdjSPC
adjusted MS/MS spectra
References
- Anbudurai P.R., Mor T.S., Ohad I., Shestakov S.V., Pakrasi H.B. (1994). The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. USA 91: 8082–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E.M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005). Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56: 347–356. [DOI] [PubMed] [Google Scholar]
- Ballottari M., Girardon J., Dall’osto L., Bassi R. (2012). Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817: 143–157. [DOI] [PubMed] [Google Scholar]
- Barbato R., Friso G., Rigoni F., Dalla Vecchia F., Giacometti G.M. (1992). Structural changes and lateral redistribution of photosystem II during donor side photoinhibition of thylakoids. J. Cell Biol. 119: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., Simpson D.J. (1986). Differential expression of LHCII genes in mesophyll and bundle sheath cells of maize. Carlsberg Res. Commun. 51: 363–370. [Google Scholar]
- Caffarri S., Kouril R., Kereïche S., Boekema E.J., Croce R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28: 3052–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Ma J., Chi W., Zou M., Guo J., Lu C., Zhang L. (2010). Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol. 154: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Celedon J.M., Cline K. (2013). Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y., Fu A., Hou X., McDonald K., Buchanan B.B., Huang W., Luan S. (2013). C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 16247–16252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., Lu C., Zhang L. (2006). A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61: 567–575. [DOI] [PubMed] [Google Scholar]
- Chi W., Ma J., Zhang L. (2012a). Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3420–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Sun X., Zhang L. (2012b). The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim. Biophys. Acta 1817: 239–246. [DOI] [PubMed] [Google Scholar]
- Covshoff S., Majeran W., Liu P., Kolkman J.M., van Wijk K.J., Brutnell T.P. (2008). Deregulation of maize C4 photosynthetic development in a mesophyll cell-defective mutant. Plant Physiol. 146: 1469–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea L.D., Regan L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28: 655–662. [DOI] [PubMed] [Google Scholar]
- de Bianchi S., Betterle N., Kouril R., Cazzaniga S., Boekema E., Bassi R., Dall’Osto L. (2011). Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23: 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M., Sobotka R., Tichý M., Komenda J. (2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G.E., Franceschi V.R., Ku M.S., Voznesenskaya E.V., Pyankov V.I., Andreo C.S. (2001b). Compartmentation of photosynthesis in cells and tissues of C(4) plants. J. Exp. Bot. 52: 577–590. [PubMed] [Google Scholar]
- Edwards G.E., Franceschi V.R., Voznesenskaya E.V. (2004). Single-cell C(4) photosynthesis versus the dual-cell (Kranz) paradigm. Annu. Rev. Plant Biol. 55: 173–196. [DOI] [PubMed] [Google Scholar]
- Edwards G.E., Furbank R.T., Hatch M.D., Osmond C.B. (2001a). What does it take to be C4? Lessons from the evolution of C4 photosynthesis. Plant Physiol. 125: 46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espineda C.E., Linford A.S., Devine D., Brusslan J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., Majeran W., Huang M., Sun Q., van Wijk K.J. (2010). Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 152: 1219–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., Olinares P.D.B., van Wijk K.J. (2011). The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. In Chloroplast Research in Arabidopsis, Jarvis R.P., ed (New York: Humana Press; ), pp. 265–282. [DOI] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H.S., Lima A., Buchanan B.B., Luan S. (2007). A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15947–15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka P., Santabarbara S., Khuong T.T., Degand H., Morsomme P., Jennings R.C., Boekema E.J., Caffarri S. (2012). Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24: 2963–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cerdán J.G., Kovács L., Tóth T., Kereïche S., Aseeva E., Boekema E.J., Mamedov F., Funk C., Schröder W.P. (2011). The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65: 368–381. [DOI] [PubMed] [Google Scholar]
- Gardiner J., Overall R., Marc J. (2011). PDZ domain proteins: ‘dark matter’ of the plant proteome? Mol. Plant 4: 933–937. [DOI] [PubMed] [Google Scholar]
- Grefen, C., Lalonde, S., and Obrdlik, P. (2007). Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. 5: 5.27. [DOI] [PubMed] [Google Scholar]
- Grieco M., Tikkanen M., Paakkarinen V., Kangasjärvi S., Aro E.M. (2012). Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 160: 1896–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Karpowicz S.J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K.K., Johnson X., Alric J., Wollman F.A., Li H., Merchant S.S. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106: 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt H., Kok B. (1978). Comparison of photosynthetic activities of spinach chloroplasts with those of corn mesophyll and corn bundle sheath tissue. Plant Physiol. 62: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A., Ettinger W.F., Yamamoto Y., Theg S.M. (1997). Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: Sites of assembly and mechanism of binding. Plant Cell 9: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono Y., Redei G.P. (1963). Multiple allelic control of chlorophyll b level in Arabidopsis thaliana. Nature 4674: 1324–1325. [Google Scholar]
- Houben E., de Gier J.W., van Wijk K.J. (1999). Insertion of leader peptidase into the thylakoid membrane during synthesis in a chloroplast translation system. Plant Cell 11: 1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A., Tanaka H., Kato C., Iwasaki Y., Asahi T. (2005). Molecular characterization of the ZKT gene encoding a protein with PDZ, K-Box, and TPR motifs in Arabidopsis. Biosci. Biotechnol. Biochem. 69: 972–978. [DOI] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Paakkarinen V., Aro E.M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439: 207–214. [DOI] [PubMed] [Google Scholar]
- Jin H., Liu B., Luo L., Feng D., Wang P., Liu J., Da Q., He Y., Qi K., Wang J., Wang H.B. (2014). HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26: 1213–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. (2007). The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M., Cline S., Redding K., Ruiz N., Hamel P.P. (2011). Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23: 4462–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sun X., Zhang L., Sakamoto W. (2012). Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 159: 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.H., Li X.P., Razeghifard R., Anderson J.M., Niyogi K.K., Pogson B.J., Chow W.S. (2009). The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: a study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 1787: 973–984. [DOI] [PubMed] [Google Scholar]
- Klostermann E., Droste Gen Helling I., Carde J.P., Schünemann D. (2002). The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem. J. 368: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouřil R., Dekker J.P., Boekema E.J. (2012). Supramolecular organization of photosystem II in green plants. Biochim. Biophys. Acta 1817: 2–12. [DOI] [PubMed] [Google Scholar]
- Kouřil R., Strouhal O., Nosek L., Lenobel R., Chamrád I., Boekema E.J., Šebela M., Ilík P. (2014). Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J. 77: 568–576. [DOI] [PubMed] [Google Scholar]
- Kouřil R., Wientjes E., Bultema J.B., Croce R., Boekema E.J. (2013). High-light vs. low-light: effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana. Biochim. Biophys. Acta 1827: 411–419. [DOI] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lima A., Lima S., Wong J.H., Phillips R.S., Buchanan B.B., Luan S. (2006). A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 103: 12631–12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hall D.A., Last R.L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Poliakov A., Bhuiyan N.H., Zybailov B., Sun Q., van Wijk K.J. (2012). The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 158: 1172–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Peng L., Guo J., Lu Q., Lu C., Zhang L. (2007). LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mabbitt P.D., Wilbanks S.M., Eaton-Rye J.J. (2014). Structure and function of the hydrophilic Photosystem II assembly proteins: Psb27, Psb28 and Ycf48. Plant Physiol. Biochem. 81: 96–107. [DOI] [PubMed] [Google Scholar]
- Majeran W., Cai Y., Sun Q., van Wijk K.J. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., Ponnala L., Connolly B., Huang M., Reidel E., Zhang C., Asakura Y., Bhuiyan N.H., Sun Q., Turgeon R., van Wijk K.J. (2010). Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Zybailov B., Ytterberg A.J., Dunsmore J., Sun Q., van Wijk K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7: 1609–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., van Wijk K.J. (2009). Cell-type-specific differentiation of chloroplasts in C4 plants. Trends Plant Sci. 14: 100–109. [DOI] [PubMed] [Google Scholar]
- Meierhoff K., Westhoff P. (1993). Differential biogenesis of photosytem II in mesophyll and bundle sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: the non-stoichiometric abundance of the subunits of photosystem II in the bundle sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191: 23–33. [Google Scholar]
- Meurer J., Plücken H., Kowallik K.V., Westhoff P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17: 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J. (2013). Dynamic reorganization of photosynthetic supercomplexes during environmental acclimation of photosynthesis. Front. Plant Sci. 4: 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M., Harrison M.S., Peterson E.C., Henry R. (2000). Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275: 1529–1532. [DOI] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501. [DOI] [PubMed] [Google Scholar]
- Murray D.L., Kohorn B.D. (1991). Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol. Biol. 16: 71–79. [DOI] [PubMed] [Google Scholar]
- Nath K., Jajoo A., Poudyal R.S., Timilsina R., Park Y.S., Aro E.M., Nam H.G., Lee C.H. (2013). Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Lett. 587: 3372–3381. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64: 609–635. [DOI] [PubMed] [Google Scholar]
- Nilsson R., Brunner J., Hoffman N.E., van Wijk K.J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18: 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Asakura Y., Friso G., Kim J., Oh S.H., Rutschow H., Ponnala L., van Wijk K.J. (2013). ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R., Herrmann R.G., Pakrasi H.B. (1996). Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 271: 21848–21852. [DOI] [PubMed] [Google Scholar]
- Ohnishi N., Takahashi Y. (2001). PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. J. Biol. Chem. 276: 33798–33804. [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald A., Streubel M., Ljungberg U., Hermans J., Eskins K., Westhoff P. (1990). Differential biogenesis of photosystem-II in mesophyll and bundle-sheath cells of ‘malic’ enzyme NADP(+)-type C4 plants. A comparative protein and RNA analysis. Eur. J. Biochem. 190: 185–194. [DOI] [PubMed] [Google Scholar]
- Pagliano C., Saracco G., Barber J. (2013). Structural, functional and auxiliary proteins of photosystem II. Photosynth. Res. 116: 167–188. [DOI] [PubMed] [Google Scholar]
- Pagliano C., Nield J., Marsano F., Pape T., Barera S., Saracco G., Barber J. (2014). Proteomic characterization and three-dimensional electron microscopy study of PSII-LHCII supercomplexes from higher plants. Biochim. Biophys. Acta 1837: 1454–1462. [DOI] [PubMed] [Google Scholar]
- Pan X., Liu Z., Li M., Chang W. (2013). Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 23: 515–525. [DOI] [PubMed] [Google Scholar]
- Pasch J.C., Nickelsen J., Schünemann D. (2005). The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl. Microbiol. Biotechnol. 69: 440–447. [DOI] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shikanai T. (2011). Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 155: 1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yamamoto H., Shikanai T. (2011). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807: 945–953. [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Richter C.V., Bals T., Schünemann D. (2010). Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur. J. Cell Biol. 89: 965–973. [DOI] [PubMed] [Google Scholar]
- Sage R.F., Sage T.L., Kocacinar F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63: 19–47. [DOI] [PubMed] [Google Scholar]
- Schneider A., Steinberger I., Strissel H., Kunz H.H., Manavski N., Meurer J., Burkhard G., Jarzombski S., Schünemann D., Geimer S., Flügge U.I., Leister D. (2014). The Arabidopsis Tellurite resistance C protein together with ALB3 is involved in photosystem II protein synthesis. Plant J. 78: 344–356. [DOI] [PubMed] [Google Scholar]
- Schuster G., Ohad I., Martineau B., Taylor W.C. (1985). Differentiation and development of bundle sheath and mesophyll thylakoids in maize. Thylakoid polypeptide composition, phosphorylation, and organization of photosystem II. J. Biol. Chem. 260: 11866–11873. [PubMed] [Google Scholar]
- Sirpiö S., Allahverdiyeva Y., Suorsa M., Paakkarinen V., Vainonen J., Battchikova N., Aro E.M. (2007). TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem. J. 406: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S., Khrouchtchova A., Allahverdiyeva Y., Hansson M., Fristedt R., Vener A.V., Scheller H.V., Jensen P.E., Haldrup A., Aro E.M. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55: 639–651. [DOI] [PubMed] [Google Scholar]
- Sun R., Fan H., Gao F., Lin Y., Zhang L., Gong W., Liu L. (2012). Crystal structure of Arabidopsis Deg2 protein reveals an internal PDZ ligand locking the hexameric resting state. J. Biol. Chem. 287: 37564–37569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Gao F., Fan H., Shan X., Sun R., Liu L., Gong W. (2013). The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases. Acta Crystallogr. D Biol. Crystallogr. 69: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. (2010b). The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 152: 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ouyang M., Guo J., Ma J., Lu C., Adam Z., Zhang L. (2010a). The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J. 62: 240–249. [DOI] [PubMed] [Google Scholar]
- Suorsa M., Järvi S., Grieco M., Nurmi M., Pietrzykowska M., Rantala M., Kangasjärvi S., Paakkarinen V., Tikkanen M., Jansson S., Aro E.M. (2012). PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24: 2934–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Regel R.E., Paakkarinen V., Battchikova N., Herrmann R.G., Aro E.M. (2004). Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur. J. Biochem. 271: 96–107. [DOI] [PubMed] [Google Scholar]
- Takabayashi A., Kurihara K., Kuwano M., Kasahara Y., Tanaka R., Tanaka A. (2011). The oligomeric states of the photosystems and the light-harvesting complexes in the Chl b-less mutant. Plant Cell Physiol. 52: 2103–2114. [DOI] [PubMed] [Google Scholar]
- Tausta S.L., Li P., Si Y., Gandotra N., Liu P., Sun Q., Brutnell T.P., Nelson T. (2014). Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. J. Exp. Bot. 65: 3543–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Grieco M., Kangasjärvi S., Aro E.M. (2010). Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 152: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Grieco M., Nurmi M., Rantala M., Suorsa M., Aro E.M. (2012). Regulation of the photosynthetic apparatus under fluctuating growth light. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S., Umate P., Manavski N., Plöchinger M., Kleinknecht L., Bogireddi H., Herrmann R.G., Wanner G., Schröder W.P., Meurer J. (2014). PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J., Andersson B., Aro E.M. (1996). Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 271: 9627–9636. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Bingsmark S., Aro E.M., Andersson B. (1995). In vitro synthesis and assembly of photosystem II core proteins. The D1 protein can be incorporated into photosystem II in isolated chloroplasts and thylakoids. J. Biol. Chem. 270: 25685–25695. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Roobol-Boza M., Kettunen R., Andersson B., Aro E.M. (1997). Synthesis and assembly of the D1 protein into photosystem II: processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36: 6178–6186. [DOI] [PubMed] [Google Scholar]
- Wientjes E., Drop B., Kouřil R., Boekema E.J., Croce R. (2013). During state 1 to state 2 transition in Arabidopsis thaliana, the photosystem II supercomplex gets phosphorylated but does not disassemble. J. Biol. Chem. 288: 32821–32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K.C., Anderson J.M., Boardman N.K., Downton W.J., Osmond C.B., Thorne S.W. (1970). Deficient photosystem II in agranal bundle sheath chloroplasts of C(4) plants. Proc. Natl. Acad. Sci. USA 67: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Inagaki N., Satoh K. (2001). Overexpression and characterization of carboxyl-terminal processing protease for precursor D1 protein: regulation of enzyme-substrate interaction by molecular environments. J. Biol. Chem. 276: 7518–7525. [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhou G., Liu B., Kong Y., Chen N., Qiu Q., Yin H., An J., Zhang F., Chen F. (2011). HCF243 encodes a chloroplast-localized protein involved in the D1 protein stability of the Arabidopsis photosystem II complex. Plant Physiol. 157: 608–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K.J., Aro E.M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274: 16062–16067. [DOI] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., Suorsa M., Aro E.M. (2001). A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J. Biol. Chem. 276: 37809–37814. [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.