Abstract
Auxin regulates a vast array of growth and developmental processes throughout the life cycle of plants. Auxin responses are highly context dependent and can involve changes in cell division, cell expansion, and cell fate. The complexity of the auxin response is illustrated by the recent finding that the auxin-responsive gene set differs significantly between different cell types in the root. Auxin regulation of transcription involves a core pathway consisting of the TIR1/AFB F-box proteins, the Aux/IAA transcriptional repressors, and the ARF transcription factors. Auxin is perceived by a transient coreceptor complex consisting of a TIR1/AFB protein and an Aux/IAA protein. Auxin binding to the coreceptor results in degradation of the Aux/IAAs and derepression of ARF-based transcription. Although the basic outlines of this pathway are now well established, it remains unclear how specificity of the pathway is conferred. However, recent results, focusing on the ways that these three families of proteins interact, are starting to provide important clues.
INTRODUCTION
The term auxin is derived from the Greek word “auxein,” which means to grow. Darwin observed the effects of auxin in plants as early as 1880. In his book “The Power of Movement in Plants,” he described how the effects of light on movement of canary grass coleoptiles were mediated by a chemical signal (Darwin and Darwin, 1880). It took another 60 years of research to show that this chemical signal is indole-3-acetic acid, the major naturally occurring auxin in plants (Haagen-Smit et al., 1946; Mauseth, 1991; Raven et al., 1992; Salisbury and Ross, 1992; Arteca, 1996). After this discovery, auxin research advanced rapidly along multiple trajectories. Numerous auxinic compounds were identified, some of which were developed as herbicides and growth regulators (Sterling et al., 1997; Cobb and Reade, 2010). Based on the chemical structures of these compounds, the spatial features of a hypothetical auxin receptor were predicted (Thimman, 1977). This marked the beginning of what turned out to be an extended search for the auxin receptor.
Auxin has been associated with embryogenesis (reviewed in Jürgens, 1995), tropic responses (Firn and Digby, 1980), organogenesis (Li et al., 2005; De Smet et al., 2010), root development (reviewed in Benjamins and Scheres, 2008), shoot development (Vernoux et al., 2011), and plant defense (reviewed in Kazan and Manners, 2009). Understanding how auxin can regulate so many diverse physiological and developmental processes is an active and exciting area of current research.
There are three known classes of auxin receptors: AUXIN BINDING PROTEIN1 (ABP1) (Hertel et al., 1972; Jones et al., 1998; Tromas et al., 2013; Xu et al., 2014), S-PHASE KINASE-ASSOCIATED PROTEIN 2A (SKP2A) (Jurado et al., 2010), and the nuclear SCFTIR1/AFBs-Aux/IAA (SKP-Cullin-F box [SCF], TIR1/AFB [TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX], AUXIN/INDOLE ACETIC ACID) auxin coreceptors (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Calderón-Villalobos et al., 2012). Although there have been some important recent advances in our understanding of ABP1, this review focuses on the SCFTIR1/AFB complexes and their function in auxin perception and the regulation of transcription in Arabidopsis thaliana.
SCFTIR1/AFBs AND AUXIN PERCEPTION
Auxin regulates transcription of auxin-responsive genes through the action of the TIR1/AFB F-box proteins, the Aux/IAA transcriptional repressors, and the auxin response factors (ARFs). The Arabidopsis genome encodes 6 TIR1/AFBs, 29 Aux/IAA proteins, and 23 ARFs. In general, the Aux/IAAs act by directly binding to the ARFs and recruiting the corepressor protein TOPLESS (TPL) to the chromatin (Figure 1; Szemenyei et al., 2008; reviewed in Guilfoyle and Hagen, 2007, 2012; Mockaitis and Estelle, 2008; Chapman and Estelle, 2009; Wang and Estelle, 2014; Guilfoyle, 2015). Degradation of the Aux/IAA repressors is a critical event in auxin signaling and requires a ubiquitin protein ligase E3 called SCFTIR1/AFB (Gray et al., 1999, 2001; Ramos et al., 2001). The substrate recognition subunit of this E3, the F-box protein TIR1 (or related AFB protein), was first identified in a genetic screen for auxin transport inhibitor-response mutants (Ruegger et al., 1998). Since then, a number of elegant studies have shown that auxin promotes degradation of the Aux/IAA proteins through the SCFTIR1/AFB, in an auxin-dependent manner (Gray et al., 2001; Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Tan et al., 2007). The Aux/IAA degron is located in a conserved domain called Domain II (dII). Instead of causing a substrate modification, commonly required for substrate recognition by many other cullin-based E3 ligases, auxin enhances the interaction between SCFTIR1/AFB and the dII by directly binding to TIR1, demonstrating that TIR1 is the long-sought auxin receptor (Dharmasiri et al., 2005a, Kepinski and Leyser 2005; reviewed in Skaar et al., 2013).
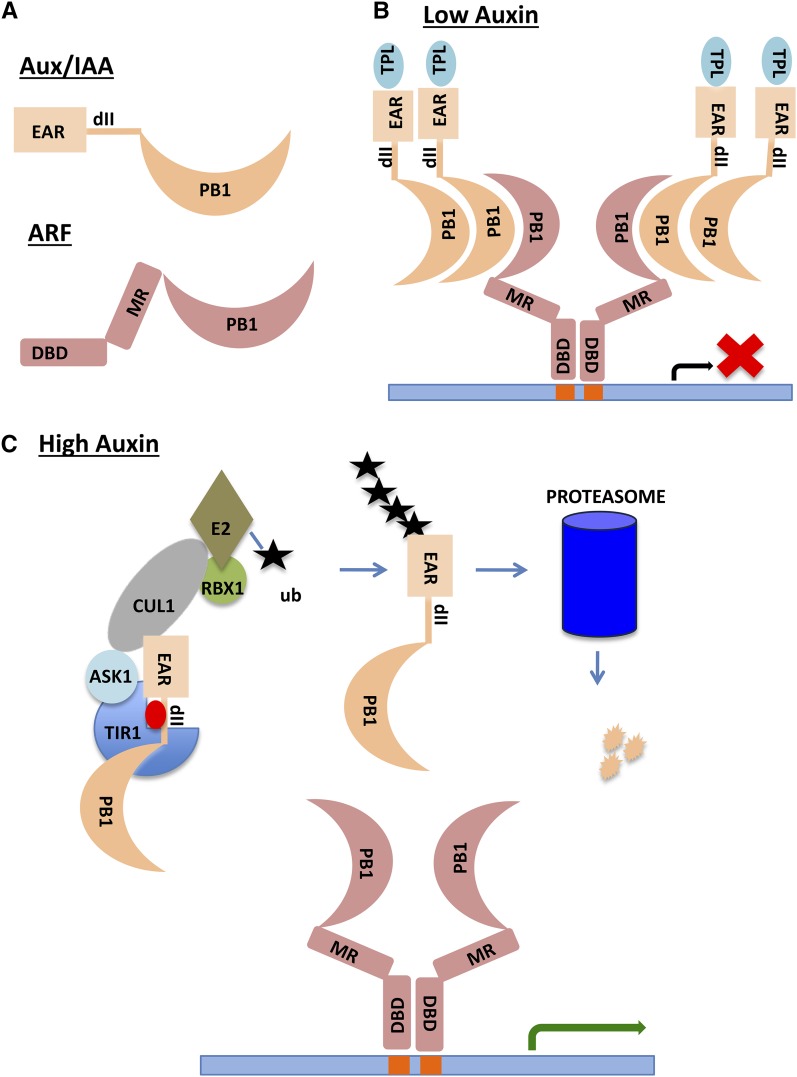
Figure 1.
SCFTIR1/AFB-Based Auxin Perception and Response.
(A) Domain structure of the Aux/IAA and ARF proteins. EAR is the ETHYLENE RESPONSE FACTOR-associated amphiphilic repression motif that interacts with the TPL corepressor. The dII domain facilitates interaction with the TIR1/AFB protein in response to auxin. The PB1 domain has both positive and negative electrostatic interfaces for directional protein interaction. DBD is the B3 DNA binding domain, and MR is the middle region that determines the activity of the ARF.
(B) Activating ARFs can form dimers through their DBDs and bind inverted repeat AuxREs (Boer et al., 2014). At low auxin levels, the Aux/IAA proteins form multimers with ARFs and recruit TPL to the chromatin. Note that most AuxREs are not found as inverted repeats in plant genomes, indicating that ARFs bind to DNA in configurations other than shown here.
(C) High levels of auxin promote ubiquitination and degradation of Aux/IAAs through SCFTIR1/AFB and the proteasome. ARFs are free to activate transcription of target genes. The site of Aux/IAA ubiquitination is arbitrary. The actual sites are unknown. Auxin is represented by the red oval.
Single mutants in members of the TIR1/AFB gene family have, at most, a mild auxin-related phenotype. The tir1 mutant is auxin resistant and is slightly shorter than wild-type plants (Ruegger et al., 1998). However, higher order mutants with combinations of afb1, afb2, and afb3 in the tir1 mutant background exhibit severe growth defects and increased auxin resistance. Most of the quadruple tir1 afb1 afb2 afb3 mutants arrest after germination. Occasionally, tir1afb1afb2afb3 plants are able to grow beyond this stage but show defects in multiple auxin responses (Dharmasiri et al., 2005b, Parry et al., 2009). In addition, mutations in other SCF subunits like CUL1, ASK1, and RBX1 cause auxin resistance and stabilize the Aux/IAA proteins (Gray et al., 1999, 2001, 2002; Hellmann et al., 2003; Moon et al., 2007; Gilkerson, et al., 2009). Recently, two new tir1 mutants were identified in a yeast two-hybrid-based screen. The tir1D170E and tir1M473L mutations increase the affinity of TIR1 for the Aux/IAA proteins, whereas plants expressing tir1D170E and tir1M473L transgenes show an auxin hypersensitive phenotype and developmental defects (Yu et al., 2013).
STRUCTURAL INSIGHT INTO AUXIN PERCEPTION BY SCFTIR1/AFBs
All six members of the TIR1/AFB family have been shown to function as auxin receptors (Dharmasiri et al., 2005a, 2005b; Parry et al., 2009; Greenham et al., 2011). Besides the F-box domain, these proteins also contain a leucine-rich repeat (LRR) domain with 18 LRRs. AFB4 and AFB5 proteins are distinct from the other members of this family in that they have an N-terminal extension that is not present in TIR1 and AFB1 to AFB3.
When the TIR1/AFB proteins were first shown to function as auxin receptors, the mechanism of auxin perception was unknown. Later, structural studies revealed the elegant way that auxin acts to facilitate the interaction between TIR1 and the Aux/IAA substrate. The structure of TIR1 was solved in a complex with ASK1, the dII peptide from the Aux/IAA IAA7, and auxin (Tan et al., 2007; reviewed in Calderon-Villalobos et al., 2010) (Figure 2). The TIR1-ASK1 complex is mushroom shaped. The cap of the mushroom, including the auxin binding pocket, is formed by the LRR domain of TIR1. The F-box domain together with ASK1 forms the stem of the mushroom. The LRRs form a slightly twisted, incomplete ring-like solenoid structure of alternating solvent-facing α-helices and core-lining β-strands. The top surface of the LRR domain has a single pocket for auxin binding (Tan et al., 2007; reviewed in Calderon-Villalobos et al., 2010). Strikingly, the structure of the TIR1-ASK1 complex does not change substantially upon auxin binding, indicating that auxin does not induce a conformational change. At the base of the auxin binding pocket lies an inositol hexakisphosphate (InsP6) molecule. Although the biological significance of this InsP6 molecule is not known, it has been suggested that it might act as a structural cofactor (Tan et al., 2007). Structural studies with different auxin compounds revealed that the binding pocket for auxin is somewhat promiscuous. Most importantly, these studies revealed that unlike animal hormones, where the ligand binding site is located distant from the active site of the receptor, auxin acts as a “molecular glue” to stabilize the interaction between TIR1 and the Aux/IAA protein (Tan et al., 2007; reviewed in Calderon-Villalobos et al., 2010; Skaar et al., 2013). So far, the structure of SCFTIR1 has been solved only with the short degron sequence from the Aux/IAA proteins (Tan et al., 2007). It is expected that a complete structure of SCFTIR1 with auxin and a full-length Aux/IAA protein will reveal more structural insights into how auxin triggers ubiquitination of Aux/IAA proteins.
Figure 2.
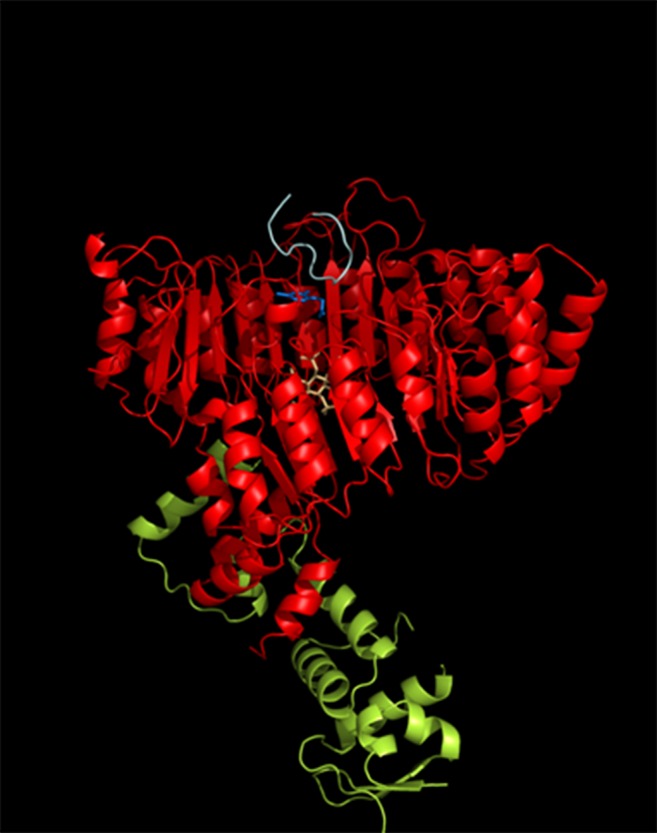
Structure of TIR1-ASK1 in a Complex with IAA and the Degron Peptide from IAA7.
TIR1-ASK1 structure as described by Tan et al. (2007). ASK1 (green) interacts with TIR1 (red) through the F-box domain. IAA (blue) is present in the auxin binding pocket and acts to stabilize the interaction between TIR1 and the degron peptide (pale cyan). A single InsP6 molecule (pale orange) is bound to TIR1 beneath the auxin binding pocket.
The six TIR1/AFB proteins are part of small subclade of F-box proteins with seven members. The seventh protein in the family is CORONATINE INSENSITIVE1 (COI1), known to be essential for the response to jasmonic acid (JA), a hormone that is structurally unrelated to auxin and has a very different role in the plant. Nevertheless, there is a striking similarity between the auxin and JA signaling pathways (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; reviewed in Katsir et al., 2008; Pérez and Goossens, 2013). In the case of JA, degradation of a family of repressors called the JASMONATE ZIM (JAZ) proteins is mediated by an E3 ligase called SCFCOI1. The interaction between the JAZ proteins and COI1 is mediated by direct binding to the JA derivative JA-isoleucine (Thines et al., 2007; Sheard et al., 2010). Thus, plants have evolved a similar mechanism to respond to very different regulatory molecules.
THE AUXIN CORECEPTOR MODEL: A NEW WAY TO THINK ABOUT AUXIN ACTION
Recently, it was shown that efficient binding of auxin to TIR1 requires the assembly of a coreceptor complex consisting of TIR1 and an Aux/IAA protein (Calderón-Villalobos et al., 2012). This may be significant because there are six TIR1/AFB proteins and 29 Aux/IAA proteins in Arabidopsis. Thus, it is possible that different combinations of TIR1/AFB and Aux/IAA will have different biochemical properties (Figure 3). Indeed, auxin binding assays with purified TIR1 and Aux/IAA proteins showed that different coreceptor complexes have different affinities for auxin (Calderón-Villalobos et al., 2012). For example, the TIR1-IAA7 pair has a Kd of 10 to 15 nM for IAA, while TIR1-IAA12 has a Kd of between 250 and 300 nM for IAA. Differences in Kd appear to be determined primarily by the dII sequence of the Aux/IAA proteins, although other sequences may also contribute (Calderón-Villalobos et al., 2012).
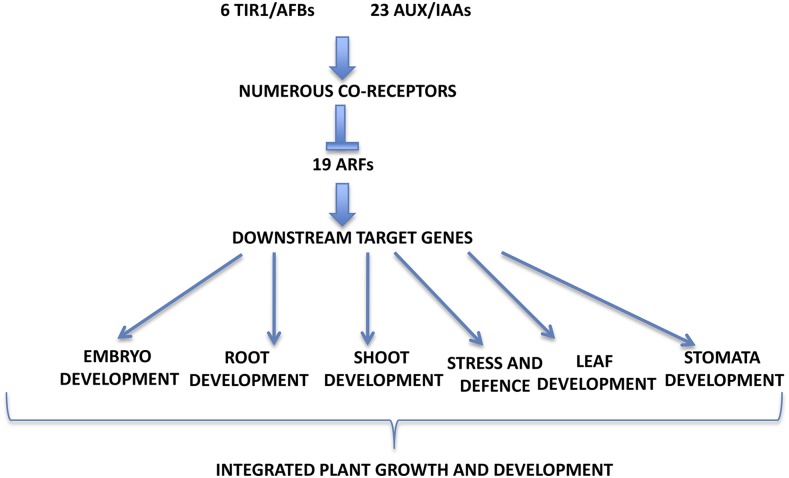
Figure 3.
Different TIR1/AFB-AUX/1AA-ARF Modules May Regulate Different Developmental Processes.
Six TIR1/AFB can interact with the 23 different Aux/IAAs containing the dII to form numerous coreceptor complexes. Each of the Aux/IAA may interact with up to 19 ARFs containing Domains III/IV to regulate distinct sets of target genes that control different physiological processes in the plant.
Localized regulation of auxin levels has a key role in a number of processes including positioning of organ primordia, maintenance of stem cell niches, patterning of the fruit, and ability of auxin to direct cell division, expansion, and differentiation (Jones et al., 1998; Sabatini et al., 1999; Reinhardt et al., 2000; Benková et al., 2003; Li et al., 2005; Sorefan et al., 2009; Jurado et al., 2010; Mähönen et al., 2014). In the root, direct measurement of auxin levels in different cell types, as well as the behavior of auxin reporters, indicate that auxin levels range widely with an auxin maximum around the quiescent center and decreasing auxin levels moving proximally from the quiescent center as well as distally toward the root tip (Petersson et al., 2009; Vernoux et al., 2011; Brunoud et al., 2012; Band et al., 2014). Recently, cell type-specific genome-wide analysis of auxin responses in four different root cell types was reported. One of the highlights of this study was that different cell types have both divergent and parallel transcriptomic response to auxin (Bargmann et al., 2013). These studies highlight the presence of an auxin gradient in the root and the transcriptional complexity of auxin action. It is possible that diverse auxin coreceptors may be necessary to interpret the wide range of auxin levels that exist in the plant. Thus, the coreceptor mechanism could dramatically expand the dynamic range of auxin perception, potentially providing a partial explanation for how auxin controls so many different aspects of plant development (Calderón-Villalobos et al., 2012; Lee et al., 2014).
Aux/IAA AND ARF GENES ACT DOWNSTREAM OF SCFTIR1/AFBs
The Aux/IAA genes were discovered because some members are rapidly induced by auxin. In pea (Pisum sativum) and soybean (Glycine max), the level of several Aux/IAA transcripts increased within a few minutes of auxin treatment (Abel and Theologis, 1996; reviewed in Hagen and Guilfoyle, 2002). It is important to note, however, that some Aux/IAAs, like IAA28 in Arabidopsis, are not auxin induced (Rogg et al., 2001).
Most of the Aux/IAA proteins have four conserved domains. Domain I has an ETHYLENE RESPONSE FACTOR ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif where the TPL/TOPLESS RELATED corepressor binds (Long et al., 2006; Szemenyei et al., 2008; Causier et al., 2012). Domain II contains the degron sequence, which interacts directly with the TIR1/AFB protein and auxin. Domain III and Domain IV are responsible for dimerization with other Aux/IAA proteins and heterodimerization with ARF proteins (Ulmasov et al., 1997a).
Important insights into the roles of the Aux/IAA genes came from genetic studies. Gain-of-function mutations in several of these genes, including IAA1/AXR5, IAA3/SHY2, IAA7/AXR2, IAA12/BDL, IAA14/SLR, IAA17/AXR3, IAA18/CRANE, IAA19/MSG, and IAA28, lead to stabilization of the respective protein because they are not degraded by SCFTIR1/AFBs (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001; Fukaki et al., 2002; Tatematsu et al., 2004; Yang et al., 2004; Uehara et al., 2008; Ploense et al., 2009). The gain-of-function mutations are all within a stretch of five conserved amino acids in the dII. The mutations prevent SCFTIR1/AFBs binding resulting in stabilization of the protein (Ramos et al., 2001; Dreher et al., 2006). On the other hand, the analysis of loss-of-function mutants has so far failed to reveal robust mutant phenotypes in Arabidopsis, suggesting extensive genetic redundancy among members of the family (Remington et al., 2004; Overvoorde et al., 2005; reviewed in Reed, 2001). This is in contrast to the situation in tomato (Solanum lycopersicum) where several loss-of-function alleles or antisense constructs produce a robust phenotype suggesting that there is less redundancy in this species (Wang et al., 2005; Chaabouni et al., 2009; Bassa et al., 2012; Deng et al., 2012; Su et al., 2014).
The ARF proteins are B3-type transcription factors. Each of the 23 ARFs in Arabidopsis have an N-terminal DNA binding domain (DBD) similar to that found in the transcription factor FUSCA3 (Ulmasov et al., 1995, 1997b; Luerssen et al., 1998; reviewed in Liscum and Reed, 2002). The ARFs bind to auxin response elements (AuxREs), each with the canonical 6-bp TGTCTC sequence in the promoters of auxin-responsive genes. The first four bases in the TGTCTC sequence are absolutely required for ARF binding, while more variation is tolerated in the last two bases (Ulmasov et al., 1997b, 1999a; Boer et al., 2014; reviewed in Guilfoyle and Hagen, 2007).
Based on activity in a protoplast assay the ARFs are divided into activators and repressors (reviewed in Guilfoyle and Hagen, 2012). ARF5, 6, 7, 8, and 19 proteins have a middle region that is Gln (Q) rich and function as activators. All the rest, except for ARF23, have a middle region rich in serine, proline, or leucine/glycine and are thought to act as repressors, although this has not been experimentally tested for every member of this group. In addition, ARF3, 13, and 17 lack Domains III/IV. ARF23 consists of a truncated DBD only. Although the ARFs have been classified as either activators or repressors, it is important to note that their behavior in the plant may be much more complex.
For the activating ARFs, a working model for ARF regulation is now well established (Figure 1; reviewed in Guilfoyle and Hagen, 2007, 2012). At low auxin levels, these ARFs are bound to the Aux/IAA proteins, which recruit the TPL corepressor and other associated chromatin modifying proteins via the EAR motif in Domain I, resulting in the repression of auxin-responsive genes (Tiwari et al., 2001; Szemenyei et al., 2008). At higher auxin levels (Figure 1), Aux/IAAs are ubiquitinated and degraded via the 26S proteasome machinery, thus freeing ARFs to activate expression of auxin responsive genes (Figure 1). Since the phenotype of gain-of-function aux/iaa mutants is caused by stabilization of the respective Aux/IAA proteins and constitutive repression of ARF proteins, loss-of-function ARF activator mutants should have a similar phenotype to Aux/IAA gain-of-function mutants. This is the case for several mutants, such as iaa12/bdl and arf5/mp, both of which have a rootless phenotype (Hardtke and Berleth, 1998; Hamann et al., 1999; Weijers et al., 2006).
Recently, a large-scale analysis of Aux/IAA and ARFs interactions was done using systemic large-scale yeast two-hybrid assays and bimolecular fluorescence complementation assays. The major conclusion of this study was that Aux/IAA-Aux/IAA and Aux/IAA-activator ARF interactions are common, whereas interactions between ARFs or between Aux/IAAs and repressor ARFs were less common (Vernoux et al., 2011). However, a recent study provides genetic evidence for an interaction between ARF9, characterized as a repressor, and IAA10, suggesting either the function of the ARFs is more complex or that the Aux/IAAs can interact with repressor ARFs in vivo (Rademacher et al., 2012).
Recent structural studies of ARFs have led to exciting new insight into the molecular function of the ARF and Aux/IAA proteins (Boer et al., 2014; Korasick et al., 2014; Nanao et al., 2014). Because the ARF proteins readily form homodimers through Domains III/IV, this became the focus of studies on ARF interaction. However Domain III/IV-independent ARF dimerization was reported as long ago as 1999 (Ulmasov et al., 1999b). More recently, Boer et al. (2014) solved the structure of the DNA binding domains from ARF5 and its distant paralog ARF1 in complex with a generic AuxRE element and showed that the DNA binding domains homodimerize to generate cooperative DNA binding (Boer et al., 2014). Furthermore, this study proposed that ARF1 and ARF5 differ in the spacing between adjacent binding sites, potentially contributing to ARF specificity.
Further insight was gained by structural studies of the C-terminal domain of ARF5 (Nanao et al., 2014) and ARF7 (Korasick et al., 2014). This work revealed that Domains III and IV, present in most of the Aux/IAA and ARFs, form a Phox and Bem1p (PB1) domain as first proposed by Guilfoyle and Hagen (2012). The PB1 domains provide both positive and negative electrostatic surfaces for directional protein interaction (reviewed in Guilfoyle, 2015). Biochemical analysis confirmed that a mutation that affects one or the other of the surfaces in the ARF protein still permits dimerization with itself or an Aux/IAA protein, whereas an ARF protein with substitutions in both faces is unable to form a dimer (Korasick et al., 2014; Nanao et al., 2014). Additional insight was gained by studies of the Aux/IAA proteins. Expression of stabilized forms of these proteins results in a strong auxin response defect. However, if one of the two PB1 faces is mutated, this defect is strongly ameliorated, implying that the formation of Aux/IAA multimers is required for efficient repression. So far, this effect has only been demonstrated for IAA16, but seems likely to be general. These discoveries constitute a major refinement of the auxin-signaling model (Figure 1; Korasick et al., 2014; Nanao et al., 2014).
In addition to interactions between themselves, the Aux/IAAs and ARFs have also been reported to regulate and be regulated by other transcription factors. A MYB transcription factor, MYB77, was shown to interact with the ARF7 protein and contribute to auxin-regulated transcription (Shin et al., 2007). In sunflower (Helianthus annuus), HaIAA27 was shown to bind to the heat shock transcription factor HaHSFA9 and repress its activity during seed development. As in the case of the ARFs, auxin acted to relieve repression of the HaHSFA9 protein (Carranco et al., 2010). In another recent report, phosphorylation of ARF7 and ARF19 by BRASSINOSTEROID INSENSITIVE2 (BIN2) was shown to suppress their interaction with Aux/IAAs and this in turn enhanced transcription of LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16) and LBD29 during lateral root initiation, independent of auxin perception (Cho et al., 2014).
Despite these recent advances, our understanding of how the ARFs work remains quite superficial compared with fungal and animal systems. For example, we are just beginning to learn about the coactivators and corepressors that collaborate with the ARFs to regulate transcription. Similarly, the chromatin states associated with ARF function are unknown. Finally, the function and activity of the repressor ARFs is poorly understood.
DEGRADATION OF Aux/IAA IS CRUCIAL FOR AUXIN ACTION
Understanding how Aux/IAA proteins are degraded is a crucial step in unraveling how auxin triggers diverse developmental responses. Domain II of the Aux/IAAs is thought to be the primary determinant for degradation by SCFTIR1/AFB (Gray et al., 2001; Ramos et al., 2001; Dreher et al., 2006). However, in addition to the Domain II degron motif, a conserved lysine between Domain I and Domain II contributes to Aux/IAA degradation (Ouellet et al., 2001; Dreher et al., 2006). It is interesting to note that the half-life of the Aux/IAAs varies widely. The half-life of IAA7 is ∼10 min, while the half-life of IAA28 is 80 min, despite the fact that these two proteins have an identical degron sequence. These results indicate that determinants outside of Domain II also contribute to degradation rate. On the other hand, IAA31, which has a degenerate Domain II, without the conserved lysine, has a half-life of >20 h, although this drops to ∼4 h after auxin treatment. A small group of Aux/IAAs, namely, IAA20, IAA30, and IAA32-34, do not have the classical Domain II, but overexpression of IAA20 and IAA30 show strong auxin-related defects implying that these proteins repress auxin regulated transcription (Sato and Yamamoto, 2008).
Recently, a synthetic biology approach has been applied to the study of auxin signaling (Havens et al., 2012; Pierre-Jerome et al., 2014). By engineering the core auxin-signaling pathway into budding yeast, these workers developed a novel and powerful platform for studies of the pathway. Using this system, they confirmed that the Aux/IAA proteins are degraded at very different rates, but in addition, the rate is dependent on the TIR1/AFB protein (Havens et al., 2012). More importantly, the system enabled them to define a minimal auxin response circuit sufficient to recapitulate auxin-induced transcription in yeast. By building and testing circuits composed of different Aux/IAA and ARF proteins, they were able to show that the behavior of the circuit varied significantly depending on the circuit components. Furthermore, circuits with multiple coexpressed Aux/IAAs displayed unique behaviors that may be relevant during plant development. This work provides a new approach for dissecting auxin signaling and demonstrates the key role of Aux/IAAs in tuning the dynamic pattern of auxin response (Pierre-Jerome et al., 2014).
In a related study, Shimizu-Mitao and Kakimoto (2014) tested the auxin-dependent degradation of all Arabidopsis Aux/IAAs in combination with TIR1 or AFB in yeast. They found that TIR1 and AFB2, but not AFB1, or AFB3-5 were effective in Aux/IAA degradation in the yeast system. All Aux/IAAs, except those lacking Domain II (IAA20, IAA30, IAA32, and IAA34), were degraded in an auxin-dependent manner. As in earlier studies (Calderón-Villalobos et al., 2012), the effective auxin concentration for Aux/IAA degradation depended on the identity of both the Aux/IAA and TIR1/AFB2 protein (Shimizu-Mitao and Kakimoto, 2014).
REGULATORY LOOPS IN AUXIN SIGNALING
Regulatory complexity is a recurring theme in plant development, so it is not surprising that feedback and regulatory loops exist in the auxin-signaling pathway (Figure 4). The most striking of these is the negative feedback loop generated by auxin-induced transcription of the Aux/IAA genes. Clearly this feedback loop will result in rapid dampening of auxin response upon auxin treatment. However, given that the kinetics of auxin regulation of Aux/IAAs is complex, a complete understanding of this regulatory system will require additional experiments in conjunction with a modeling approach.
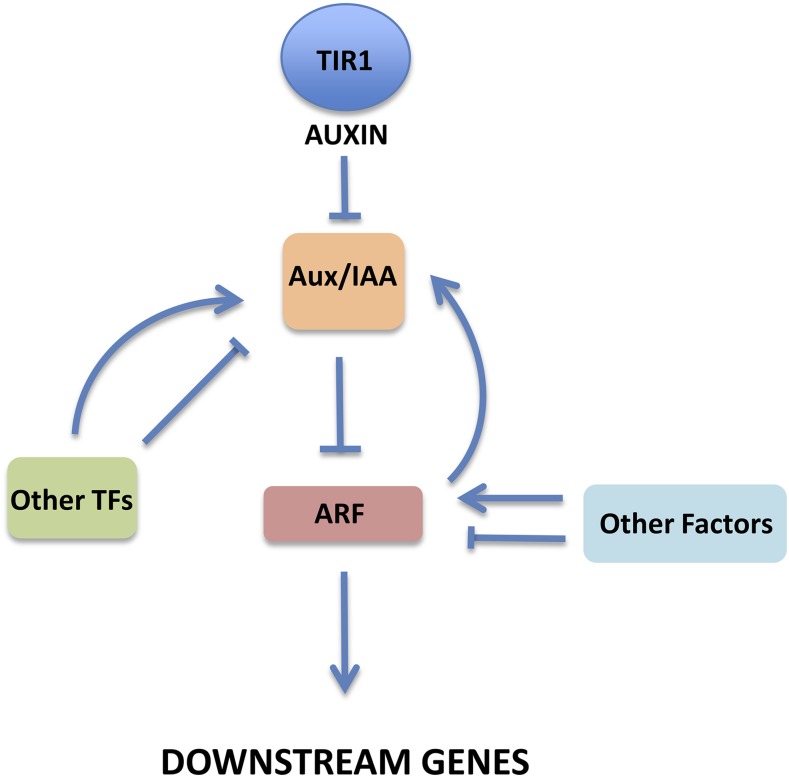
Figure 4.
Regulation of the TIR1/AFB Pathway.
ARF-mediated regulation of the Aux/IAA genes constitutes a robust negative feedback loop. Other pathways may regulate transcription of auxin response genes in both a positive and negative manner. For example, the cytokinin responsive transcription factor ARR1 promotes transcription of IAA3 in the root, resulting in downregulation of the ARF target PIN1. This results in a change in auxin distribution that affects cell differentiation (Dello Ioio et al., 2008). In addition, other pathways may act directly on the ARFs. For example, the BIN2 kinase regulates the interaction between ARF7 and Aux/IAA by directly phosphorylating the ARF (Cho et al., 2014).
Apart from the negative regulatory loop involving the Aux/IAAs, members of the auxin efflux carrier PIN-FORMED (PIN) family were also shown to be under control of the Aux/IAAs and ARFs (Vieten et al., 2005). As cellular auxin levels rise, PIN gene expression increases, resulting in more auxin efflux and a reduction in auxin levels (reviewed in Adamowski and Friml, 2015). Thus, this regulatory circuit contributes to auxin homeostasis. Among the features of this regulation is a striking compensatory mechanism that may act to stabilize auxin gradients. In this system, the loss of a PIN protein results in an increase in cellular auxin levels. This in turn causes the ectopic expression of other PIN proteins, thus compensating for the original PIN deficiency (Vieten et al., 2005). In addition, accumulation of auxin during de novo organ formation leads to rearrangements in the subcellular polar localization of PIN auxin transporters. This effect is cell specific, independent of PIN transcription, and involves the Aux/IAA-ARF signaling pathway (Sauer et al., 2006).
The PINs also factor into another auxin-dependent regulatory loop that affects behavior of cells in the root meristem. Dello Ioio et al. (2008) showed that the cytokinin response factor ARR1 activates transcription of the Aux/IAA gene SHY2/IAA3. The IAA3 protein in turn represses transcription of PIN1 resulting in a change in auxin distribution that promotes cell differentiation. It is likely that many additional regulatory nodes that involve the Aux/IAAs and ARF will be identified going forward (Figure 4).
THE EVOLUTIONARY HISTORY OF AUXIN SIGNALING
Colonization of land by plants was a major event in evolution. However, the time at which auxin signaling emerged is not clear (reviewed in De Smet and Beeckman, 2011). The auxin-signaling pathway is conserved in land plants. Genes encoding Aux/IAA, ARF, and TIR1 homologs are present within the genomes of the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii (Lau et al., 2009; Paponov et al., 2009; reviewed in De Smet and Beeckman, 2011; Finet and Jaillais, 2012). In the case of P. patens, genetic studies have shown that the mechanism of auxin signaling is very similar to that of angiosperms (Prigge et al., 2010; Lavy et al., 2012). The presence of auxin in algal species has been reported, but the physiological significance of this is not clear. In the case of Chlorophyta, a division of the green algae, no orthologs of TIR1/AFB, Aux/IAA, and ARFs were found (Paponov et al., 2009; reviewed in Lau et al., 2009; De Smet and Beeckman, 2011; Finet and Jaillais, 2012). A recent report of a draft genome sequence of the filamentous terrestrial alga Klebsormidium flaccidum indicates that this species lacks a TIR1-like auxin receptor but does have other auxin-related proteins such as ABP1, AUXIN RESISTANT1, and PIN (Hori et al., 2014). It is also interesting to note that most of the SCF-dependent plant hormone signaling components, such as TIR1, COI1, and GA INSENSITIVE DWARF1, are missing in K. flaccidum genome (Hori et al., 2014).
USE OF AUXIN-INDUCIBLE DEGRONS AS A TOOL IN ANIMAL SYSTEMS
In the last several years, SCFTIR1/AFB and the Aux/IAA proteins have provided the basis for a novel method of regulating protein levels in non-plant species. This system is called the auxin-inducible degron system (Nishimura et al., 2009; Holland et al., 2012; Kanke et al., 2012; Farr et al., 2014; Nishimura and Kanemaki, 2014; Samejima et al., 2014). All eukaryotes possess SCF ubiquitin ligases, and the architecture of Arabidopsis TIR1, including the F-box domain, is sufficiently conserved to allow assembly into an SCFTIR1 complex in yeast and animals. When a protein of interest is fused to the Aux/IAA degron, called the auxin-induced degron in this context, and introduced into yeast cells expressing TIR1, the tagged protein will be degraded in an auxin-dependent manner (Nishimura et al., 2009). The system provides a rapid and, more importantly, reversible way to regulate protein levels. The auxin-inducible degron system has been adapted for a number of vertebrate cell types and is proving to be a useful tool for a wide range of studies
NEW TECHNOLOGIES TO DISSECT THE AUXIN-SIGNALING PATHWAY
As mentioned above, there appears to be extensive redundancy in both the ARF and Aux/IAA families of proteins. Consequently, the role of each Aux/IAA and ARF protein has not been defined (Okushima et al., 2005; Overvoorde et al., 2005). Because the creation of higher order mutants by genetic crossing is a time-consuming process, the emergence of precise genome editing tools like CLUSTERED REGULARLY INTERSPACED SHORT PALINDROMIC REPEAT (CRISPR)-CRISPR ASSOCIATED SYSTEM (Cas9) is a welcome development (Cong et al., 2013; Mali et al., 2013). The CRISPR-Cas9 system has been successfully used to create multiple mutants in a mouse model in a short time (Wang et al., 2013). Several reports of successful precise genome editing in Arabidopsis and other plants using CRISPR-Cas9 are very promising (Li et al., 2013; Feng et al., 2014; Jiang et al., 2014; Schiml et al., 2014; reviewed in Lozano-Juste and Cutler, 2014; Hyun et al., 2015). The CRISPR-Cas9 system should decrease the amount of time it takes to generate the higher order mutants required for analysis of Aux/IAA and ARF gene families.
Over the last three decades genetics, biochemistry and molecular approaches have provided an explanation for how auxin controls many aspects of plant growth. However, partly because of the complexity of auxin biology, our view of this regulatory system remains incomplete. A more complete understanding will certainly require the application of systems level and computational approaches. Several groups have developed instructive mathematical models that help to explain several auxin-related events like phyllotaxy, lateral branching, and root growth (Reinhardt et al., 2003; Jönsson et al., 2006; Shinohara et al., 2013; Band et al., 2014; Mähönen et al., 2014). This insightful approach will become even more powerful as the models become increasingly parameterized by experimental data.
FUTURE DIRECTIONS
Auxin plays a role almost every aspect of plant development. Although the general framework of auxin action has been established, the specific elements involved in each developmental signal remain to be discovered. Because the Aux/IAA proteins are central and dynamic regulators of auxin signaling, further studies of their role in auxin perception, their interactions with the ARF proteins, and their ultimate effect on the transcriptional output will be an important way forward. The ability of the Aux/IAAs to form auxin coreceptors with TIR1/AFBs further expands the dynamic range of auxin perception. In addition, recent exciting studies show that ABP1 functions as a cell surface-based auxin receptor (Chen et al., 2001; Chen et al., 2012; Xu et al., 2014). How auxin perception at the cell surface and in the nucleus are coordinated is an important outstanding question (Tromas et al., 2013; Paque et al., 2014). Finally, the effects of auxin on cell cycle regulation may be mediated in part by SCFSKP2A, which binds to auxin in a cell-free assay (Jurado et al., 2010). Discovering how information from these different perception mechanisms is integrated during plant development will be an exciting challenge for the future.
Acknowledgments
M.S. and R.B. thank Rebecca Dickstein for critically reading the article and for helpful suggestions. This work was supported by grants from the Howard Hughes Medical Institute, the Gordon and Betty Moore Foundation, and the National Institutes of Health (Grant GM43644).
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
References
- Abel S., Theologis A. (1996). Early genes and auxin action. Plant Physiol. 111: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteca R. (1996). Plant Growth Substances: Principles and Applications. (New York: Chapman & Hall; ). [Google Scholar]
- Band L.R., et al. (2014). Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann B.O.R., Vanneste S., Krouk G., Nawy T., Efroni I., Shani E., Choe G., Friml J., Bergmann D.C., Estelle M., Birnbaum K.D. (2013). A map of cell type-specific auxin responses. Mol. Syst. Biol. 9: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassa C., Mila I., Bouzayen M., Audran-Delalande C. (2012). Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol. 53: 1583–1595. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Boer D.R., Freire-Rios A., van den Berg W.A.M., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., Weijers D., Coll M. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L.I., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos L.I., Tan X., Zheng N., Estelle M. (2010). Auxin perception—structural insights. Cold Spring Harb. Perspect. Biol. 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranco R., Espinosa J.M., Prieto-Dapena P., Almoguera C., Jordano J. (2010). Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc. Natl. Acad. Sci. USA 107: 21908–21913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S., Jones B., Delalande C., Wang H., Li Z., Mila I., Frasse P., Latché A., Pech J.-C., Bouzayen M. (2009). Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 60: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Chen J.-G., Ullah H., Young J.C., Sussman M.R., Jones A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Naramoto S., Robert S., Tejos R., Löfke C., Lin D., Yang Z., Friml J. (2012). ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr. Biol. 22: 1326–1332. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Cho H., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16: 66–76. [DOI] [PubMed] [Google Scholar]
- Cobb, A.H., and Reade, J.P.H. (2010). Herbicides and Plant Physiology. (Hoboken, NJ: Wiley-Blackwell). [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C., Darwin F.E. (1880). The Power of Movement in Plants. (New York: D Appleton and Co.). [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- Deng W., Yang Y., Ren Z., Audran-Delalande C., Mila I., Wang X., Song H., Hu Y., Bouzayen M., Li Z. (2012). The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 194: 379–390. [DOI] [PubMed] [Google Scholar]
- De Smet I., Beeckman T. (2011). Asymmetric cell division in land plants and algae: the driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12: 177–188. [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr C.J., Antoniou-Kourounioti M., Mimmack M.L., Volkov A., Porter A.C. (2014). The α isoform of topoisomerase II is required for hypercompaction of mitotic chromosomes in human cells. Nucleic Acids Res. 42: 4414–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C., Jaillais Y. (2012). Auxology: when auxin meets plant evo-devo. Dev. Biol. 369: 19–31. [DOI] [PubMed] [Google Scholar]
- Firn R.D., Digby J. (1980). The establishment of tropic curvatures in plants. Annu. Rev. Plant Physiol. 31: 131–148. [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29: 153–168. [DOI] [PubMed] [Google Scholar]
- Gilkerson J., Hu J., Brown J., Jones A., Sun T.P., Callis J. (2009). Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 181: 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., del Pozo J.C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W.L., Yang M., Ma H., Estelle M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13: 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Hellmann H., Dharmasiri S., Estelle M. (2002). Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14: 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- Greenham K., Santner A., Castillejo C., Mooney S., Sairanen I., Ljung K., Estelle M. (2011). The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr. Biol. 21: 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guilfoyle T.J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2012). Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 190: 82–88. [DOI] [PubMed] [Google Scholar]
- Haagen-Smit A.J., Dandliker W.B., Wittwer S.H., Murneek A.E. (1946). Isolation of 3-indoleacetic acid from immature corn kernels. Am. J. Bot. 33: 118–120. [Google Scholar]
- Hagen G., Guilfoyle T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49: 373–385. [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jürgens G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens K.A., Guseman J.M., Jang S.S., Pierre-Jerome E., Bolten N., Klavins E., Nemhauser J.L. (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H., Hobbie L., Chapman A., Dharmasiri S., Dharmasiri N., del Pozo C., Reinhardt D., Estelle M. (2003). Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 22: 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R., Thomson K.-S., Russo V.E.A. (1972). In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107: 325–340. [DOI] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Han J.S., Cleveland D.W. (2012). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 109: E3350–E3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Kim J., Cho S.W., Choi Y., Kim J.S., Coupland G. (2015). Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241: 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Yang B., Weeks D.P. (2014). Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9: e99225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Im K.H., Savka M.A., Wu M.J., DeWitt N.G., Shillito R., Binns A.N. (1998). Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282: 1114–1117. [DOI] [PubMed] [Google Scholar]
- Jönsson H., Heisler M.G., Shapiro B.E., Meyerowitz E.M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S., Abraham Z., Manzano C., López-Torrejón G., Pacios L.F., Del Pozo J.C. (2010). The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. (1995). Axis formation in plant embryogenesis: cues and clues. Cell 81: 467–470. [DOI] [PubMed] [Google Scholar]
- Kanke M., Kodama Y., Takahashi T.S., Nakagawa T., Masukata H. (2012). Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 31: 2182–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J.K., Howe G.A. (2008). Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14: 373–382. [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Westfall C.S., Lee S.G., Nanao M.H., Dumas R., Hagen G., Guilfoyle T.J., Jez J.M., Strader L.C. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Shao N., Bock R., Jürgens G., De Smet I. (2009). Auxin signaling in algal lineages: fact or myth? Trends Plant Sci. 14: 182–188. [DOI] [PubMed] [Google Scholar]
- Lavy M., Prigge M.J., Tigyi K., Estelle M. (2012). The cyclophilin DIAGEOTROPICA has a conserved role in auxin signaling. Development 139: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Sundaram S., Armitage L., Evans J.P., Hawkes T., Kepinski S., Ferro N., Napier R.M. (2014). Defining binding efficiency and specificity of auxins for SCF(TIR1/AFB)-Aux/IAA co-receptor complex formation. ACS Chem. Biol. 9: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125. [DOI] [PubMed] [Google Scholar]
- Li J.F., Norville J.E., Aach J., McCormack M., Zhang D., Bush J., Church G.M., Sheen J. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Reed J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49: 387–400. [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J., Cutler S.R. (2014). Plant genome engineering in full bloom. Trends Plant Sci. 19: 284–287. [DOI] [PubMed] [Google Scholar]
- Luerssen H., Kirik V., Herrmann P., Miséra S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15: 755–764. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., ten Tusscher K., Siligato R., Smetana O., Díaz-Triviño S., Salojärvi J., Wachsman G., Prasad K., Heidstra R., Scheres B. (2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauseth J.D. (1991). Botany: An Introduction to Plant Biology. (Philadelphia: Saunders; ). [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80. [DOI] [PubMed] [Google Scholar]
- Moon J., Zhao Y., Dai X., Zhang W., Gray W.M., Huq E., Estelle M. (2007). A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 143: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Walker L.M., Young J.C., Sonawala A., Timpte C., Estelle M., Reed J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao M.H., et al. (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5: 3617. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6: 917–922. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kanemaki M. (2014). Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID). Curr. Protoc. Cell Biol. 64: 20.9.1–20.9.16. [DOI] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F., Overvoorde P.J., Theologis A. (2001). IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P.J., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov I.A., Teale W., Lang D., Paponov M., Reski R., Rensing S.A., Palme K. (2009). The evolution of nuclear auxin signalling. BMC Evol. Biol. 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S., Mouille G., Grandont L., Alabadí D., Gaertner C., Goyallon A., Muller P., Primard-Brisset C., Sormani R., Blázquez M.A., Perrot-Rechenmann C. (2014). AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis. Plant Cell 26: 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L.I., Prigge M., Peret B., Dharmasiri S., Itoh H., Lechner E., Gray W.M., Bennett M., Estelle M. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 106: 22540–22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez A.C., Goossens A. (2013). Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ. 36: 2071–2084. [DOI] [PubMed] [Google Scholar]
- Petersson S.V., Johansson A.I., Kowalczyk M., Makoveychuk A., Wang J.Y., Moritz T., Grebe M., Benfey P.N., Sandberg G., Ljung K. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E., Jang S.S., Havens K.A., Nemhauser J.L., Klavins E. (2014). Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl. Acad. Sci. USA 111: 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense S.E., Wu M.F., Nagpal P., Reed J.W. (2009). A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Lavy M., Ashton N.W., Estelle M. (2010). Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 20: 1907–1912. [DOI] [PubMed] [Google Scholar]
- Rademacher E.H., Lokerse A.S., Schlereth A., Llavata-Peris C.I., Bayer M., Kientz M., Freire Rios A., Borst J.W., Lukowitz W., Jürgens G., Weijers D. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 17: 211–222. [DOI] [PubMed] [Google Scholar]
- Ramos J.A., Zenser N., Leyser O., Callis J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven P.H., Evert R.F., Eichhorn S.E. (1992). Biology of Plants, 5th ed. (New York: Worth Publishers). [Google Scholar]
- Reed J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6: 420–425. [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Remington D.L., Vision T.J., Guilfoyle T.J., Reed J.W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135: 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg L.E., Lasswell J., Bartel B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D., Mackay P., Stirnberg P., Estelle M., Leyser O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W.M., Hobbie L., Turner J., Estelle M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Samejima K., Ogawa H., Ageichik A.V., Peterson K.L., Kaufmann S.H., Kanemaki M.T., Earnshaw W.C. (2014). Auxin-induced rapid degradation of Inhibitor of Caspase Activated DNase (ICAD) induces apoptotic DNA fragmentation, caspase activation and cell death. J. Biol. Chem. 289: 31617–31623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury F.B., Ross C.W. (1992). Plant Physiology. (Belmont, CA: Wadsworth; ). [Google Scholar]
- Sato A., Yamamoto K.T. (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 133: 397–405. [DOI] [PubMed] [Google Scholar]
- Sauer M., Balla J., Luschnig C., Wisniewska J., Reinöhl V., Friml J., Benková E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20: 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80: 1139–1150. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Mitao Y., Kakimoto T. (2014). Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol. 55: 1450–1459. [DOI] [PubMed] [Google Scholar]
- Shin R., Burch A.Y., Huppert K.A., Tiwari S.B., Murphy A.S., Guilfoyle T.J., Schachtman D.P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N., Taylor C., Leyser O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar J.R., Pagan J.K., Pagano M. (2013). Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S.J., Ljung K., Robles P., Galván-Ampudia C.S., Offringa R., Friml J., Yanofsky M.F., Østergaard L. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586. [DOI] [PubMed] [Google Scholar]
- Sterling, T.M., and Hall, J.C. (1997). Mechanism of action of natural auxins and the auxinic herbicides. In Herbicide Activity: Toxicology, Biochemistry, and Molecular Biology, R.M. Roe, J.D. Burton, and R.J. Kuhr, eds (Amsterdam: IOS Press), pp. 111–141. [Google Scholar]
- Su L., Bassa C., Audran C., Mila I., Cheniclet C., Chevalier C., Bouzayen M., Roustan J.P., Chervin C. (2014). The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 55: 1969–1976. [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M.K., Harper R.M., Liscum E., Yamamoto K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimman K.V. (1977). Hormone Action in the Whole Life of Plants. (Amherst, MA: University of Massachusetts Press; ). [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Tian Q., Reed J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Wang X.J., Hagen G., Guilfoyle T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A., Paque S., Stierlé V., Quettier A.-L., Muller P., Lechner E., Genschik P., Perrot-Rechenmann C. (2013). Auxin-binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway. Nat. Commun. 4: 2496. [DOI] [PubMed] [Google Scholar]
- Uehara T., Okushima Y., Mimura T., Tasaka M., Fukaki H. (2008). Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 49: 1025–1038. [DOI] [PubMed] [Google Scholar]
- Ulmasov T. Hagen G., Guilfoyle T.J. (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999a). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999b). Dimerization and DNA binding of auxin response factors. Plant J. 19: 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z.B., Hagen G., Guilfoyle T.J. (1995). Composite structure of auxin response elements. Plant Cell 7: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wisniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., Chaabouni S., Latché A., Pech J.C., Bouzayen M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Estelle M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21C: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Schlereth A., Ehrismann J.S., Schwank G., Kientz M., Jürgens G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10: 265–270. [DOI] [PubMed] [Google Scholar]
- Xu T., et al. (2014). Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Lee S., So J.H., Dharmasiri S., Dharmasiri N., Ge L., Jensen C., Hangarter R., Hobbie L., Estelle M. (2004). The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J. 40: 772–782. [DOI] [PubMed] [Google Scholar]
- Yu H., Moss B.L., Jang S.S., Prigge M., Klavins E., Nemhauser J.L., Estelle M. (2013). Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 162: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]