Key protein-protein interactions of the plant UV-B photoreceptor UVR8 that regulate UV-B-induced photomorphogenesis and stress acclimation are investigated, focusing on UVR8 interaction with COP1.
Abstract
UV-B photon reception by the Arabidopsis thaliana homodimeric UV RESISTANCE LOCUS8 (UVR8) photoreceptor leads to its monomerization and a crucial interaction with CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1). Relay of the subsequent signal regulates UV-B-induced photomorphogenesis and stress acclimation. Here, we report that two separate domains of UVR8 interact with COP1: the β-propeller domain of UVR8 mediates UV-B-dependent interaction with the WD40 repeats-based predicted β-propeller domain of COP1, whereas COP1 activity is regulated by interaction through the UVR8 C-terminal C27 domain. We show not only that the C27 domain is required for UVR8 activity but also that chemically induced expression of the C27 domain is sufficient to mimic UV-B signaling. We further show, in contrast with COP1, that the WD40 repeat proteins REPRESSOR OF UV-B PHOTOMORPHOGENESIS1 (RUP1) and RUP2 interact only with the UVR8 C27 domain. This coincides with their facilitation of UVR8 reversion to the ground state by redimerization and their potential to interact with UVR8 in a UV-B-independent manner. Collectively, our results provide insight into a key mechanism of photoreceptor-mediated signaling and its negative feedback regulation.
INTRODUCTION
The unavoidable exposure of plants to UV-B radiation (280 to 315 nm) is mitigated by effective toleration mechanisms. UV RESISTANCE LOCUS8 (UVR8) is a unique UV-B photoreceptor that, following the absorption of UV-B photons, initiates changes in gene expression (Heijde and Ulm, 2012; Li et al., 2013; Tilbrook et al., 2013; Jenkins, 2014). Targets include genes involved in phenylpropanoid biosynthesis, resulting in the accumulation of phenolic “sunscreen” metabolites (e.g., flavonols and sinapates) and antioxidants (anthocyanins), as well as genes encoding photolyases, which are involved in DNA repair (Kliebenstein et al., 2002; Brown et al., 2005; Favory et al., 2009; Stracke et al., 2010). The induction of genes associated with UV-B protection and repair highlights the importance of UVR8 for UV-B acclimation (Favory et al., 2009), which is distinct from the UV-B stress pathway involving mitogen-activated protein kinase signaling (González Besteiro et al., 2011). In contrast with a number of UV-B light-induced genes, auxin-responsive genes are widely and rapidly repressed in response to UV-B light, and this response is also dependent on UVR8 (Favory et al., 2009; Hayes et al., 2014; Vandenbussche et al., 2014). This may be the basis of photomorphogenic responses to UV-B such as hypocotyl growth inhibition (Ballare et al., 1995; Kim et al., 1998; Favory et al., 2009; Hayes et al., 2014; Huang et al., 2014; Vandenbussche et al., 2014). In addition to UV-B stress acclimation and hypocotyl growth inhibition, UVR8 also has been implicated in UV-B entrainment of the circadian clock, stomatal closure, phototropic bending, inhibition of shade avoidance, leaf development, and defense responses (Wargent et al., 2009; Fehér et al., 2011; Demkura and Ballaré, 2012; Hayes et al., 2014; Tossi et al., 2014; Vandenbussche et al., 2014). The UVR8 signaling pathway includes CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and ELONGATED HYPOCOTYL5 (HY5) (Ulm et al., 2004; Brown et al., 2005; Oravecz et al., 2006; Stracke et al., 2010; Huang et al., 2012; Binkert et al., 2014) and the negative feedback regulators REPRESSOR OF UV-B PHOTOMORPHOGENESIS1 (RUP1) and RUP2 (Gruber et al., 2010; Heijde and Ulm, 2013).
UVR8 is a β-propeller protein in which intrinsic Trp residues are the basis of UV-B photoreception (Rizzini et al., 2011; Wu et al., 2011, 2012; Christie et al., 2012; Liu et al., 2014). UVR8 exists as a homodimer that readily monomerizes in response to UV-B (Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012). UV-B-activated UVR8 interacts with COP1 (Favory et al., 2009), which is a major factor in the UVR8-mediated signal transduction pathway (Oravecz et al., 2006). The C-terminal C27 domain (UVR8397-423) was found to be necessary and sufficient for UVR8 interaction with COP1, and thus C27 represents the COP1-interaction domain (Cloix et al., 2012). In support of this, UVR8ΔC27 is UV-B-responsive (monomerization, nuclear accumulation) but is impaired in UV-B-dependent COP1 interaction (Cloix et al., 2012). Furthermore, C27 was found to interact constitutively with COP1 in a yeast two-hybrid assay (Cloix et al., 2012). However, it was not known whether the C27 domain is sufficient to activate UV-B-related responses in vivo.
To better understand UVR8-mediated early UV-B signaling, we focused on the β-propeller and the C-terminal regions of UVR8, including the C27 domain, in yeast and plants. We show here that the β-propeller domain of UVR8 interacts with COP1 in a UV-B-dependent manner in the absence of the C-terminal 44 amino acids and, thus, the C27 domain. However, the β-propeller domain alone is not sufficient to activate early UV-B signaling. We further demonstrate that the C-terminal 44 amino acids alone interact constitutively with COP1 and that this depends on a Val-Pro (VP) pair in the C27 domain. Chemically induced expression of the C-terminal 44 amino acids is sufficient to mimic early UVR8 responses. Thus, UVR8 interaction with COP1 is 2-fold: via the β-propeller domain and via the C27 domain, with the latter functionally impinging on COP1. This differs from the mechanism of UVR8-RUP1/RUP2 interaction, coinciding with the differences in activity and dependence on UV-B for interaction with UVR8.
RESULTS
Interaction of the UVR8 β-Propeller Domain with COP1 Is UV-B Dependent
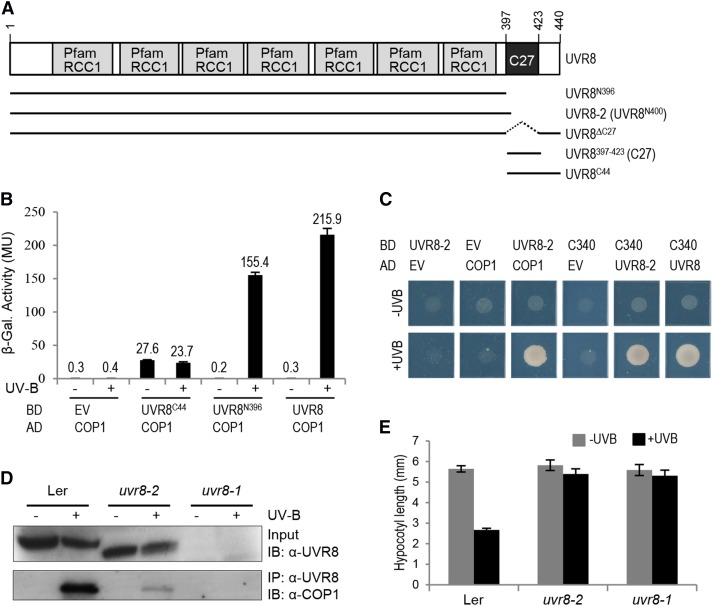
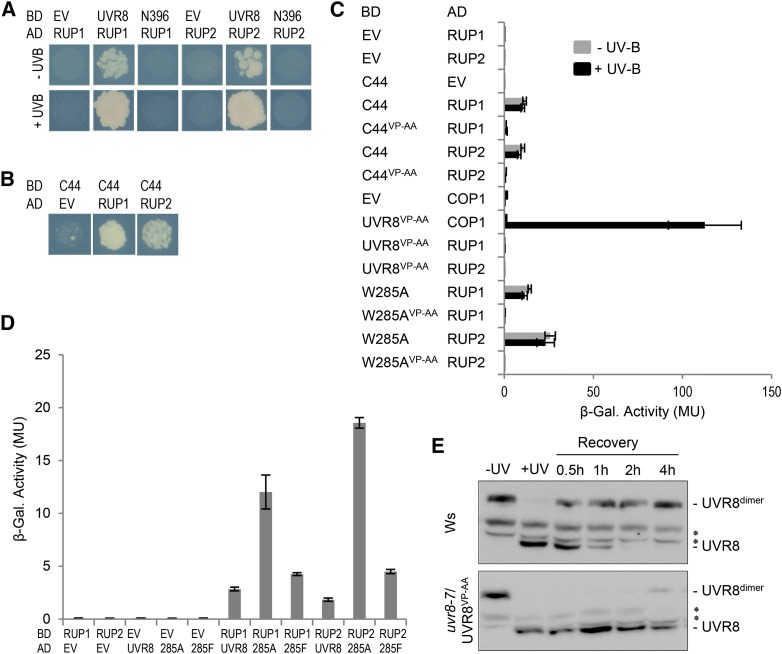
UVR8 is a protein of 440 amino acids (Kliebenstein et al., 2002) containing a seven-bladed β-propeller domain. Previous results suggest that 27 amino acids within the C-terminal 44 amino acids (the C27 domain, amino acids 397 to 423; here given as UVR8397-423) are required for the UV-B-dependent interaction of UVR8 with COP1 (Cloix et al., 2012). Moreover, yeast two-hybrid assays have shown that the C27 domain alone is sufficient for the interaction with COP1, which is UV-B-independent in the absence of the β-propeller core domain (Cloix et al., 2012). To further dissect the function of the UVR8 β-propeller domain and the C-terminal 44 amino acids in UV-B signaling, we divided UVR8 into the two fragments for testing: UVR8N396 (amino acids 1 to 396) and UVR8C44 (amino acids 397 to 440) (Figure 1A). Given that UVR8-COP1 interaction is critical for UV-B signaling, we tested the interaction of UVR8N396 and UVR8C44 with COP1 in a yeast two-hybrid assay. As described before, UVR8 interacts with COP1 in yeast in a UV-B-dependent manner (Figure 1B) (Rizzini et al., 2011; Cloix et al., 2012). UVR8C44 interacted constitutively with COP1 (Figure 1B), similar to UVR8397-423 (C27 domain) (Cloix et al., 2012).
Figure 1.
The UVR8 β-Propeller Core Fragment Lacking the C27 Domain Interacts with COP1 in a UV-B-Dependent Manner, Whereas the C-Terminal C27 Domain Alone Interacts Constitutively with COP1.
(A) Schematic representation of wild-type UVR8 and different variants used in this work. UVR8 contains seven repeats of the RCC1 (Regulator of Chromosome Condensation) Pfam domain (PF00415) (Kliebenstein et al., 2002; Finn et al., 2014) and a C27 interaction domain (Cloix et al., 2012).
(B) Quantitative yeast two-hybrid assays were performed in the presence (+) or absence (−) of UV-B light. Means and se from three biological replicates are shown. AD, activation domain construct; BD, binding domain construct; EV, empty vector; β-Gal., β-galactosidase; MU, Miller units.
(C) Yeast two-hybrid growth assay on selective SD/-Trp/-Leu/-His medium in the presence (+UV-B) or absence (−UV-B) of UV-B irradiation.
(D) Truncated UVR8-2 interacts with COP1 in planta. Coimmunoprecipitation of endogenous COP1 with UVR8 [using anti-UVR8(1–15) antibodies] is shown in extracts from 7-d-old wild-type (Landsberg erecta [Ler]), uvr8-2, and uvr8-1 seedlings. Seedlings were either irradiated with broad-band UV-B light for 15 min (+) or not irradiated (−). IB, immunoblotting; IP, immunoprecipitation.
(E) Hypocotyl lengths of 4-d-old wild-type (Ler), uvr8-2, and uvr8-1 seedlings grown under weak white light with (+UV-B) or without (−UV-B) supplemental narrow-band UV-B. Means and se are shown (n > 20).
However, in contrast with the previous finding that the UVR8 C27 domain is required for COP1 interaction (Cloix et al., 2012), we found clear evidence that UVR8N396 interacts with COP1 in a UV-B-dependent manner, despite the absence of the C27 domain (Figure 1B). We next tested a truncated UVR8 based on the uvr8-2 mutant allele, which has a premature stop codon at Trp-400 (Brown et al., 2005). Indeed, UVR8-2 (UVR8N400) also interacted with COP1 in a UV-B-dependent manner (Figure 1C). Unlike UVR8N396 and UVR8N400, the UVR8ΔC27 version described previously by Cloix et al. (2012) included the C-terminal 17 amino acids, which may explain the difference between our data and theirs. We reproduced the UVR8ΔC27 version and tested it in yeast. As with UVR8N396 and UVR8N400, UVR8ΔC27 also showed UV-B-dependent interaction with COP1 in yeast (Supplemental Figure 1). Independent of this result, we showed previously that UVR8 interacts with the WD40-repeat domain of COP1 (COP1C340) (Favory et al., 2009; Rizzini et al., 2011). In agreement with this, UVR8-2 (UVR8N400) interacted with COP1C340 and thus the WD40 domain of COP1 (Figure 1C).
The yeast two-hybrid data indicate that the truncated UVR8 protein in uvr8-2 mutant plants still interacts with COP1. As our previously generated UVR8 antibodies were specific for epitopes in the C terminus, we developed an antibody against the N-terminal 15 amino acids of UVR8. Using this antibody, we detected the truncated UVR8-2 protein in extracts from uvr8-2 mutant seedlings (Figure 1D) as expected (Cloix et al., 2012). By contrast, UVR8 protein was not detected in uvr8-1, which bears an insertion of five amino acids that is believed to make the protein unstable (Figure 1D) (Kliebenstein et al., 2002; Cloix et al., 2012). In coimmunoprecipitation assays with uvr8-1 as a negative control, COP1 was coimmunoprecipitated with the truncated UVR8 protein from samples of uvr8-2 mutant seedlings (Figure 1D). Similar to wild-type UVR8, UVR8-2 coimmunoprecipitated endogenous COP1 only from UV-B-treated uvr8-2 seedlings, albeit to a lower extent (Figure 1D). By contrast, COP1 was not detected in coimmunoprecipitates from the uvr8-1 negative control (Figure 1D). However, despite the UV-B-dependent interaction of UVR8-2 with COP1, the uvr8-2 mutant displayed no hypocotyl growth inhibition (Figure 1E) or any other UV-B response tested (Brown et al., 2005; Cloix et al., 2012).
We conclude that the seven-bladed β-propeller domain of UVR8 interacts in a UV-B-dependent manner with COP1 and that the interaction of UVR8 with COP1 does not require the C-terminal 44 amino acids that include the C27 domain. Rather, interaction of UVR8 with COP1 via its β-propeller domain after UV-B activation may allow functional interaction of the C27 domain with the COP1 WD40-repeat domain. This interaction of the UVR8 C27 domain and COP1 is required, however, for further stabilization of the interaction and, in particular, for efficient UV-B signaling.
The Amino Acids Val-410 and Pro-411 Are Critical for UVR8 Signaling
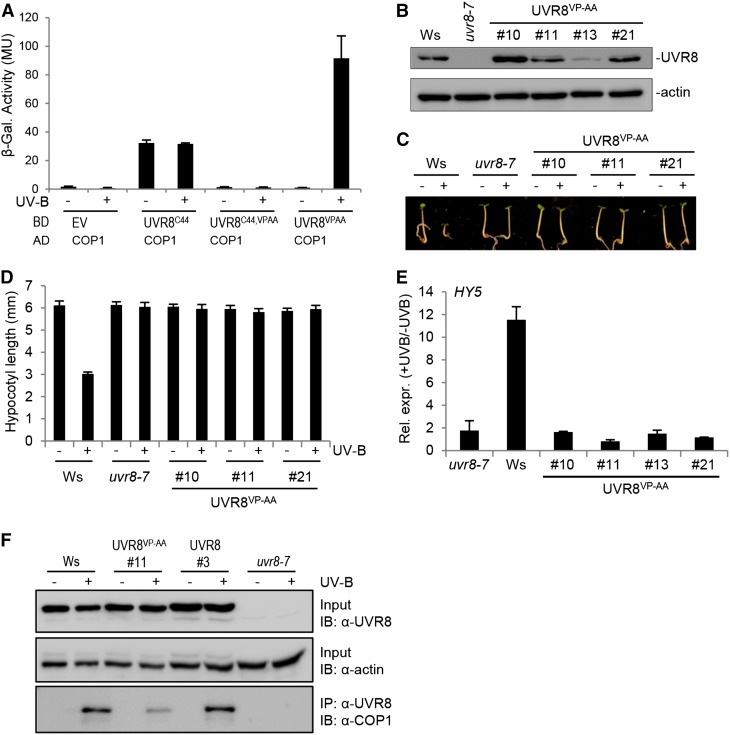
Previous work identified a VP domain with the core sequence V-P-E/D-ϕ-G (where ϕ = a hydrophobic residue) that mediates the interaction of several proteins with the COP1 WD40-repeat domain (Holm et al., 2001; Datta et al., 2006). Indeed, such a potential VP core domain also was identified in the UVR8 C27 domain (Supplemental Figure 2) (Wu et al., 2013). Mutation of the VP core to Ala-Ala (AA) was shown to abrogate COP1 interaction with several target proteins (Holm et al., 2001, 2002; Datta et al., 2006). To analyze the importance of the UVR8 VP pair, both Val-410 and Pro-411 were mutated to Ala (VP-AA). Indeed, UVR8C44/VP-AA did not interact with COP1 (Figure 2A), confirming the importance of these two residues in the UVR8 C27 domain for COP1 interaction. By contrast, a clear interaction of UVR8VP-AA with COP1 was detected in response to UV-B (Figure 2A). These data are consistent with our finding of two COP1 interaction modes in UVR8: via the UVR8 β-propeller core that conserves UV-B-dependent interaction with COP1 in the absence of the C-terminal 44 amino acids and via the C27 domain (including Val-410 and Pro-411) within the C-terminal 44 amino acids that can interact constitutively with COP1 in the absence of the β-propeller core. This explains why the constitutive interaction of UVR8C44 is blocked when mutated (UVR8C44/VP-AA) and why the mutated full-length UVR8 (UVR8VP-AA) still shows UV-B-dependent interaction with COP1.
Figure 2.
The Val-410 and Pro-411 Pair in the C27 Domain Is Critical for UVR8 Signaling.
(A) Quantitative yeast two-hybrid assays were performed in the presence (+) or absence (−) of UV-B. Means and se from three biological replicates are shown. AD, activation domain construct; BD, binding domain construct; EV, empty vector; β-Gal., β-galactosidase; MU, Miller units.
(B) Immunoblot analysis of UVR8 protein levels in wild-type seedlings (Wassilewskija [Ws]), uvr8-7, and four independent uvr8-7/Pro35S:UVR8VP-AA transgenic lines (UVR8VP-AA #10, #11, #13, and #21).
(C) and (D) Images of representative individuals (C) and quantification of hypocotyl lengths (D) of 4-d-old seedlings grown under white light with (+) or without (−) supplementary UV-B light. Means and se are shown (n > 20).
(E) Quantitative RT-PCR analysis of HY5 gene activation in 4-d-old seedlings in response to UV-B irradiation for 1 h. Relative expression is shown as +UV-B/−UV-B (i.e., fold change). Means and se of three biological replicates are shown.
(F) The UVR8VP-AA protein interacts with COP1 in planta. Coimmunoprecipitation of COP1 with UVR8 [using anti-UVR8(1–15) antibodies] is shown from 7-d-old wild-type (Ws), uvr8-7/Pro35S:UVR8VP-AA (line 11), uvr8-7/Pro35S:UVR8 (line 3), and uvr8-7 seedlings. Seedlings were irradiated with broad-band UV-B for 15 min (+) or not (−). IB, immunoblotting; IP, immunoprecipitation.
To investigate the requirement for the UVR8 C-terminal VP pair in planta, we generated transgenic plants expressing UVR8VP-AA in uvr8-7 null mutants. We then selected lines with protein levels similar to those in the wild type (Figure 2B). Relative to the wild type, hypocotyl growth inhibition in response to UV-B was impaired in UVR8VP-AA expression lines, as it was in uvr8-7 (Figures 2C and 2D). Similarly, UVR8VP-AA was impaired in HY5 marker gene activation in response to UV-B (Figure 2E). However, coimmunoprecipitation assays demonstrated that UVR8VP-AA interacted with COP1 in a UV-B-dependent manner in planta, albeit to a lower extent than did endogenous and cauliflower mosaic virus 35S promoter-driven wild-type UVR8 (Figure 2F). This is in agreement with the yeast interaction data (Figure 2D) and the coimmunoprecipitation of endogenous COP1 with truncated UVR8-2 (UVR8N400) protein (Figure 1D). Thus, UVR8VP-AA is blocked specifically in UV-B signaling and cannot complement the uvr8-7 null mutant, despite its photoresponsiveness and UV-B-dependent interaction with COP1.
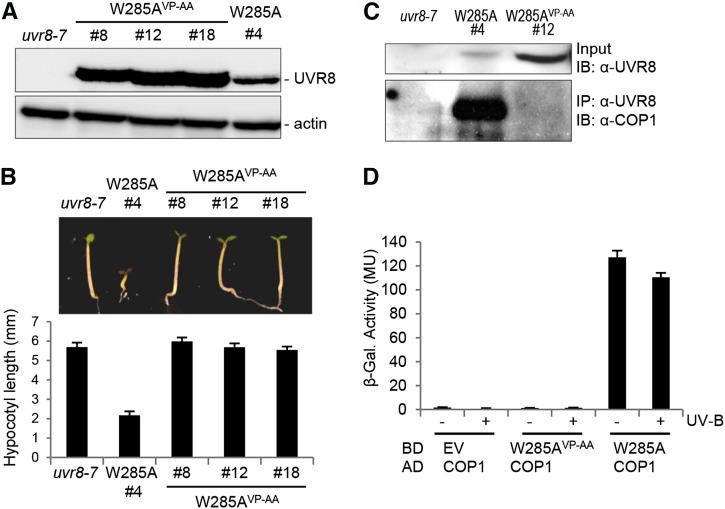
UVR8 Trp-285 is a crucial residue for UV-B photoreception, and its mutation to Phe or Ala rendered UVR8W285F and UVR8W285A UV-B insensitive (Rizzini et al., 2011; Christie et al., 2012; O’Hara and Jenkins, 2012; Wu et al., 2012; Liu et al., 2014). Recently, we and others reported that the single amino acid substitution of Ala in place of Trp in UVR8W285A generates a constitutively active UV-B photoreceptor (Heijde et al., 2013; Huang et al., 2013). To determine whether the activity of UVR8W285A is dependent on the VP pair at the C terminus, we generated transgenic plants expressing UVR8W285A/VP-AA at a level higher than UVR8W285A line 4, all in the uvr8-7 mutant background (Figure 3A). UVR8W285A line 4 developed short hypocotyls in weak white light (Figure 3B), as described before (Heijde et al., 2013). By contrast, this constitutive photomorphogenic phenotype was absent in lines expressing UVR8W285A/VP-AA (Figure 3B), demonstrating that the constitutive activity of UVR8W285A depends on the C-terminal VP pair and its interaction with COP1.
Figure 3.
The Constitutive Interaction of UVR8W285A with COP1 Depends on the C-Terminal VP Pair.
(A) Immunoblot analysis of UVR8 protein expression levels in uvr8-7 and several independent uvr8-7/Pro35S:UVR8W285,VP-AA (W285AVP-AA) and uvr8-7/Pro35S:UVR8W285A (W285A) transgenic lines.
(B) Hypocotyl growth in weak white light. Images of representative individuals (top panel) and quantification of hypocotyl lengths (bottom panel) of 4-d-old seedlings grown under weak white light are shown. Means and se are shown (n > 20).
(C) Coimmunoprecipitation of COP1 in whole cell extracts from seedlings grown for 7 d in white light. IB, immunoblotting; IP, immunoprecipitation.
(D) Quantitative yeast two-hybrid assays were performed in the presence or absence of UV-B. Means and se from three biological replicates are shown. AD, activation domain construct; BD, binding domain construct; EV, empty vector; β-Gal., β-galactosidase; MU, Miller units.
In contrast with the UV-B-dependent UVR8-COP1 interaction (Favory et al., 2009), UVR8W285A coimmunoprecipitated COP1 independently of UV-B, further demonstrating their constitutive interaction in planta (Figure 3C). However, in agreement with the absence of a constitutive photomorphogenic phenotype, UVR8W285A/VP-AA did not coimmunoprecipitate COP1 (Figure 3C). We and others (Rizzini et al., 2011; O’Hara and Jenkins, 2012; Huang et al., 2014) previously showed that UVR8W285A interacts constitutively with COP1 also in yeast (Figure 3D). The absence of an interaction between UVR8W285A/VP-AA and COP1 in yeast further supports the crucial importance of the VP pair in UVR8W285A for an interaction with COP1 (Figure 3D). Thus, rather surprisingly, we conclude that UVR8W285A interacts constitutively with COP1 via its C terminus and not via its β-propeller domain.
Induced Expression of UVR8C44 in uvr8-7 Mimics Early UV-B Signaling
Because the UVR8 C terminus is required for UV-B signaling (Figure 1E) (Cloix et al., 2012), we tested whether the C terminus is sufficient to mimic signaling when released from structural restraints in the UVR8 full-length protein. We generated transgenic Arabidopsis thaliana lines using the estrogen receptor-based XVE transactivator system (Zuo et al., 2000) to induce the expression of UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP by the addition of 17-β-estradiol (NLS, nuclear localization signal; 2A, cotranslational cleavage site; YFP, yellow fluorescent protein). Expression of the 2A peptide should result in cotranslational cleavage of the fusion protein and the release of a triple C44 peptide and YFP (Ryan and Drew, 1994). The cleaved C-terminal fraction of the fusion protein (i.e., YFP plus part of 2A) was detected by immunoblotting with anti-YFP antibody after 5 h of estradiol treatment (Figure 4A), indicating that the fusion protein was properly induced and processed. However, we could not detect the processed triple C44. Estradiol-inducible expression of YFP alone was used as a negative control (Figure 4A). Strikingly, the induced expression of UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP led to HY5 mRNA accumulation (Figure 4B), indicating that the C-terminal 44 amino acids of UVR8 can trigger gene expression even though this C44 peptide does not accumulate to detectable levels, as determined by immunoblot analysis. Because the inducible YFP control did not result in HY5 upregulation despite much higher YFP levels, triggering of gene expression was presumably due to the triple C44 fragment (Figure 4B). Thus, induced expression of UVR8C44 is sufficient to trigger HY5 gene expression in transgenic plants.
Figure 4.
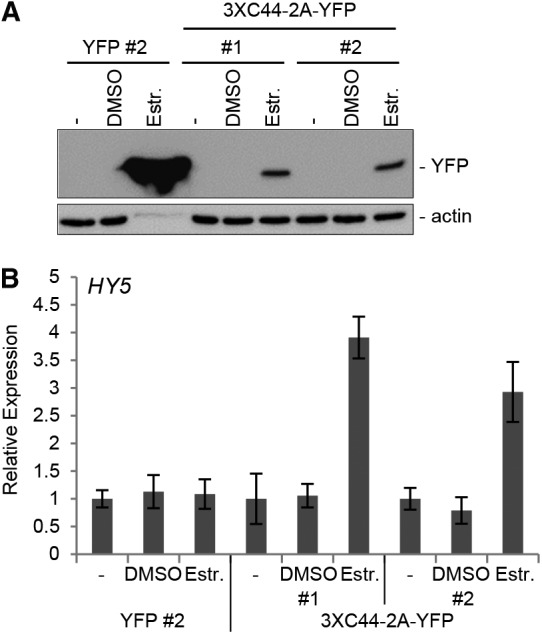
Induced Expression of UVR8C44 Leads to HY5 Transcript Accumulation in Arabidopsis Seedlings in the Absence of UV-B.
(A) Immunoblot analysis of YFP protein accumulation induced by β-estradiol in Arabidopsis seedlings. uvr8-7/XVE:YFP alone (YFP) or uvr8-7/XVE:UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP (3XC44-2A-YFP) was treated for 5 h with 50 μM 17-β-estradiol in DMSO (Estr.) or with DMSO only. Extracts were probed with anti-YFP (top panel) or anti-actin (bottom panel) antibody. Untreated controls (−) were included. 2A encodes a cotranslational cleavage site.
(B) Quantitative real-time PCR analysis of HY5 mRNA accumulation in Arabidopsis seedlings. Seedlings were either treated for 5 h with 50 μM 17-β-estradiol in DMSO or with DMSO only or were left untreated (−) Data were normalized against untreated samples without treatment at time 0. Means and se are shown (n = 3).
Expression of UVR8C44 in uvr8-7 Causes Constitutive UV-B Responses
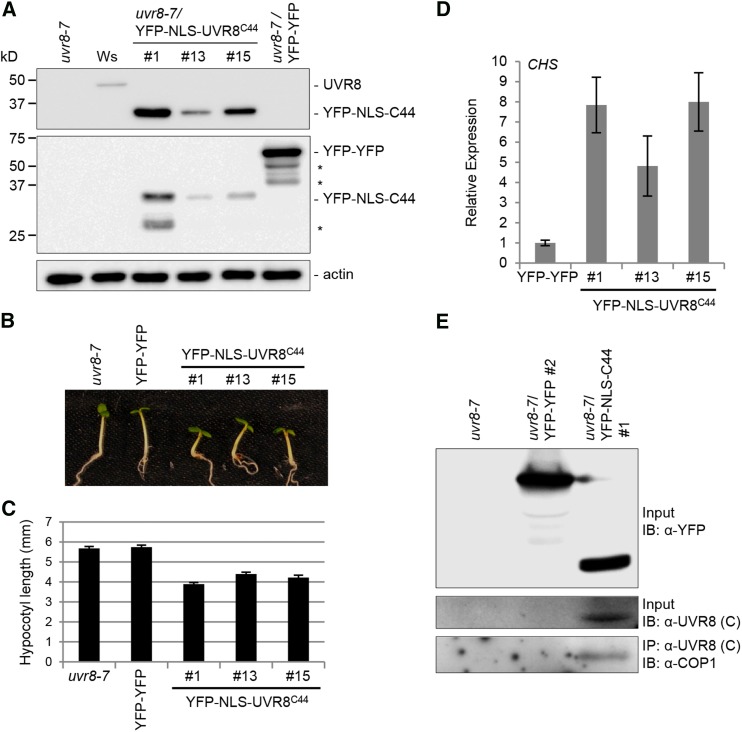
To analyze the effect of stably expressing the UVR8C44 domain in planta, we generated transgenic Arabidopsis lines expressing YFP-NLS-UVR8C44 driven by the 35S promoter in the uvr8-7 null mutant background and selected transgenic lines with different YFP-NLS-UVR8C44 protein levels (Figure 5A). Strikingly, these transgenic lines had shorter hypocotyls than uvr8-7 seedlings when grown in weak light (Figures 5B and 5C). Overexpression of a double YFP fusion (YFP-YFP) in uvr8-7 did not affect hypocotyl length (Figures 5B and 5C), demonstrating that the observed effect is due to NLS-UVR8C44 and not to the YFP tag. In agreement with the short-hypocotyl phenotype in the absence of UV-B, constitutive expression of YFP-NLS-UVR8C44 also resulted in constitutively elevated levels of CHS marker gene expression (Figure 5D). The constitutive responses in YFP-NLS-UVR8C44-expressing transgenic lines (Figures 5B to 5D) together with the constitutive interaction between UVR8C44 and COP1 in yeast (Figure 1A) suggested that YFP-NLS-UVR8C44 interacts constitutively with COP1 in planta. Indeed, endogenous COP1 was coimmunoprecipitated with YFP-NLS-C44 from transgenic seedlings in the absence of UV-B (Figure 5E). Thus, the C44 region in YFP-NLS-UVR8C44-expressing plants interacts constitutively with COP1, leading to elevated marker gene expression and a constitutive photomorphogenic response.
Figure 5.
Seedlings Stably Expressing UVR8C44 Show a Constitutive Photomorphogenic Phenotype.
(A) UVR8 protein levels in uvr8-7, wild-type (Wassilewskija [Ws]), YFP-NLS-C44-expression line 1, 13, and 15, and YFP-YFP-expression line (far right) seedlings.
(B) and (C) Hypocotyl growth in weak white light. Images of representative individuals (B) and quantification of hypocotyl lengths (C) of 4-d-old seedlings grown under white light are shown. Means and se are shown (n > 20).
(D) Transcript levels of CHS in uvr8-7/Pro35S:YFP-NLS-UVR8C44 transgenic seedlings (lines 1, 13, and 15) compared with a uvr8-7/Pro35S:YFP-YFP line (YFP-YFP) as measured by quantitative RT-PCR analysis. Error bars represent se in triplicate biological measurements.
(E) Coimmunoprecipitation of COP1 in protein extracts from uvr8-7, uvr8-7/Pro35S:YFP-YFP, and uvr8-7/Pro35S:YFP-NLS-UVR8C44 transgenic seedlings grown for 7 d in white light. IB, immunoblotting; IP, immunoprecipitation.
Interaction of UVR8 with RUP1 and RUP2 Is Limited to the C-Terminal C27 Domain
Previous work has shown that UVR8 can interact with RUP1 and RUP2 in the absence of UV-B both in plants and yeast (Gruber et al., 2010; Cloix et al., 2012). This suggests, in contrast with COP1, that RUP1 and RUP2 can interact with the UVR8 homodimer and that the interactions of UVR8 with COP1 and UVR8 with RUP1/RUP2 may differ mechanistically. Interestingly, and in stark contrast with UVR8-COP1, the UVR8-RUP1/RUP2 interaction was dependent on the C-terminal 44 amino acids. Whereas UVR8N396 interacted with COP1 under UV-B light (Figure 1B), interaction of UVR8N396 with RUP1 and RUP2 was not detected (Figure 6A). Thus, in contrast with COP1, UVR8 apparently interacts with RUP1 and RUP2 solely via the C-terminal 44 amino acids and not via the β-propeller domain (Figures 6A and 6B). In agreement with this, UVR8VP-AA (and UVR8C44/VP-AA) also did not interact with RUP1 and RUP2, although UVR8VP-AA was found to interact with COP1 under UV-B (Figure 6C; see also Figure 1B). Consistent with the UV-B-independent interaction of UVR8 with RUP1 and RUP2, the constitutively dimeric UVR8W285F interacted with RUP1 and RUP2 in yeast (Figure 6D), but not with COP1 (Rizzini et al., 2011; O’Hara and Jenkins, 2012). However, compared with UVR8 and UVR8W285F, we found a stronger interaction with the partially active UVR8W285A variant (Figure 6D). This is in agreement with previous data suggesting that the UVR8-RUP1 and UVR8-RUP2 interactions, albeit constitutive, are enhanced by UV-B (Cloix et al., 2012).
Figure 6.
UVR8 Interacts with RUP1 and RUP2 Only via Its C27 Domain.
(A) and (B) Yeast two-hybrid growth assay on selective SD/-Trp/-Leu/-His medium in the presence (+) or absence (−) of UV-B light. AD, activation domain construct; BD, binding domain construct; EV, empty vector.
(C) and (D) Quantitative yeast two-hybrid assays were performed in the presence or absence of UV-B light. Means and se from three biological replicates are shown. β-Gal., β-galactosidase; MU, Miller units.
(E) Dimerization of UVR8 in 7-d-old wild-type (Wassilewskija [Ws]) and uvr8-7/Pro35S:UVR8VP-AA line 11 seedlings. Seedlings were either left unirradiated (−UV) or irradiated with broad-band UV-B light for 15 min (+UV) to induce UVR8 monomerization before being subjected to a recovery period in white light for 0.5 to 4 h. Asterisks indicate nonspecific cross-reacting bands.
We further tested the effect of the VP-AA mutation that impairs UVR8 interaction with RUP1 and RUP2 upon the reversion of UVR8 to its ground state (Heijde and Ulm, 2013). Indeed, redimerization of UVR8VP-AA was strongly impaired compared with wild-type UVR8 (Figure 6E). We conclude that the difference in the UV-B dependence of the UVR8-COP1 and UVR8-RUP1/RUP2 interactions is due to at least two differences in their modes of interaction: (1) the ability of the β-propeller surface exposed in the UVR8 monomer to interact with COP1 but not with RUP1 and RUP2; and (2) the distinct ability of COP1 and RUP1/RUP2 to interact with the UVR8 C-terminal 44 amino acids. In the case of COP1, this interaction occurs only after UV-B activation and monomerization of UVR8, but in the case of RUP1 and RUP2, it occurs also with the nonactivated homodimeric UVR8.
DISCUSSION
UV-B-dependent interaction of the UV-B photoreceptor UVR8 with COP1 is a key event in UV-B signaling that induces photomorphogenic responses and also acclimation to UV-B (Heijde and Ulm, 2012; Li et al., 2013; Jenkins, 2014). Although previous results indicated the importance of a C-terminal 27-amino acid domain (C27), it was not clear how UV-B-mediated UVR8 monomerization results in UVR8-COP1 interaction based solely on C27 as the interaction domain, particularly as C27 alone results in constitutive interaction with COP1 in yeast (Cloix et al., 2012).
It was suggested that C27 is hidden in the homodimeric structure of UVR8 and only exposed upon monomerization after UV-B irradiation (Cloix et al., 2012). However, experimental support was lacking, especially as the C-terminal domain was not included in the UVR8 crystal structure published recently (Christie et al., 2012; Wu et al., 2012). In contrast with the model of C27 exposure after UV-B irradiation, we show here that the seven-bladed β-propeller domain of UVR8 mediates UV-B-dependent interaction of UVR8 and COP1 even in the absence of C27 (Figure 1). This is in agreement with the successful use of the UVR8 core (amino acids 12 to 381) alone with the COP1 WD40 domain to generate a UV-B-responsive split transcription factor based on UV-B-dependent protein-protein interaction as an optogenetic tool in mammalian cells (Müller et al., 2013). Also in planta, it is the WD40-repeat region of COP1 that mediates the interaction with active UVR8 (Favory et al., 2009). Intriguingly, WD40-repeat domains fold into a β-propeller structure similar to RCC1-repeat domain proteins (Xu and Min, 2011), and in fact, a seven-bladed β-propeller structure is predicted for the COP1 C-terminal WD40-repeat domain (Holm et al., 2002; Wu et al., 2013). Our data thus now indicate that the surface of the UVR8 monomer freed by UV-B-dependent monomerization of UVR8 binds to the structurally closely related β-propeller surface of the COP1 WD40-repeat domain.
Interestingly, recent Fourier transform infrared spectroscopy difference spectra comparing full-length UVR8 with a C-terminally truncated version could not attribute any structural change to the deleted C-terminal region in response to UV-B (Heilmann et al., 2014). Moreover, the UVR8 C-terminal region is apparently located at the distal ends of the dimer away from the dimer interface (Christie et al., 2012). It is thus not clear how the UVR8 C27 domain is prevented from interacting with COP1 in the homodimeric ground state and what step is required so that this C-terminal region can interact with COP1 after UV-B photoreception by UVR8. However, our data indicate that the UVR8 β-propeller surface is liberated upon UV-B perception, allowing interaction with the COP1 WD40 domain.
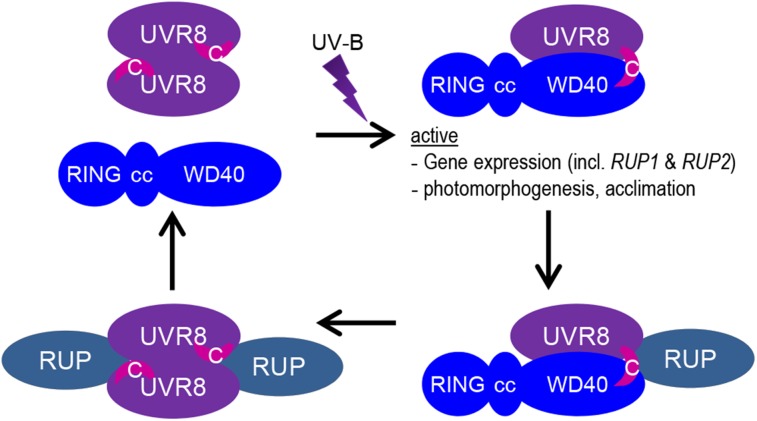
Thus, we propose that interaction of the UV-B-activated monomeric UVR8 β-propeller core with COP1 not only contributes significantly to the crucial UV-B-dependent UVR8-COP1 interaction but also promotes the functional interaction of the UVR8 C terminus with COP1. This indicates the possibility of a sequential two-step interaction mechanism: first, a UV-B-dependent stable interaction of the UVR8 β-propeller core with COP1; next, an interaction of the UVR8 C27 domain affecting COP1 function. However, we cannot exclude a simultaneous interaction of both UVR8 interaction domains with COP1. It seems clear, however, that the UVR8 C27 domain is required for stabilization of the UVR8-COP1 interaction (Figures 1B, 1D, and 2F). Notwithstanding, our data further demonstrate that induced expression of the C-terminal 44 amino acids of UVR8 alone, including the C27 domain, is sufficient to induce HY5 gene expression (Figure 4B) and that sustained expression results in a phenotype resembling continuous UV-B exposure (including hypocotyl growth inhibition) (Figures 5B to 5D; Supplemental Figure 3). Thus, whereas the interaction of the UVR8 β-propeller core with COP1 is central to the UV-B dependence and likely the robustness of the interaction, interaction of the UVR8 C27 domain with COP1 is apparently the key step affecting COP1 function (Figure 7).
Figure 7.
Working Model of the UVR8 Photocycle.
In response to UV-B irradiation, UVR8 homodimers dissociate instantly, which allows the UVR8 seven-bladed β-propeller domain to interact with the COP1 WD40 domain (a structurally related seven-bladed β-propeller). The activated UVR8 monomer then also binds to the COP1 WD40 domain with its C27 domain (indicated by the pink crescent labeled C) and initiates UV-B signaling. Release of the C27 domain from structural constraints in the UV-B light-activated UVR8 is thought to allow its interaction with COP1. The activated UVR8-COP1 signaling pathway induces RUP1 and RUP2 expression, forming a negative feedback loop. RUP1 and RUP2 are WD40-repeat proteins that are phylogenetically and structurally related to COP1. They interact solely with the C27 domain of UVR8 and facilitate UVR8 redimerization and disruption of the UVR8-COP1 interaction. Note that no stable RUP1/RUP2-UVR8-COP1 complex is known, but it is assumed here to occur transiently when RUP1 and RUP2 attach to the C27 domain of UVR8 while UVR8 and COP1 still interact via their β-propeller surfaces. In contrast with COP1, RUP1 and RUP2 proteins are still able to interact with the C27 domain in the inactive homodimeric UVR8. The COP1-interacting SPA proteins as well as the homodimeric constitution of COP1 are omitted from the model. cc, coiled coil; RING, Really Interesting New Gene; WD40, WD40 repeat domain.
A VP pair was identified previously in several unrelated proteins that interact with the COP1 WD-repeat domain (Holm et al., 2001, 2002; Datta et al., 2006). A similar VP pair (Val-410-Pro-411) that is potentially involved in UVR8-COP1 interactions is predicted in the UVR8 C27 domain (Wu et al., 2013; this work). Mutagenesis of this UVR8 VP pair to Ala-Ala impaired the interaction of COP1 with the C-terminal 44 amino acids (UVR8C44,VP-AA) but not with full-length UVR8VP-AA (Figure 2). Despite the UV-B-dependent interaction of UVR8VP-AA, UVR8-2, and UVR8N396 with COP1 that was clearly detectable in yeast and in plants, absence of the C27 domain impaired early UV-B signaling and responses, further supporting the importance of the UVR8 C-terminal region for UV-B signaling.
Interestingly, the previously described constitutive interaction of the singly substituted UVR8W285A with COP1 in yeast and plants (Rizzini et al., 2011; O’Hara and Jenkins, 2012; Huang et al., 2014; this work) was found to depend solely on the C-terminal domain (Figure 3). We showed previously that a weak 4-fold overexpression of UVR8W285A in transgenic Arabidopsis plants (in contrast with 40-fold overexpression of wild-type UVR8) results in a phenotype resembling a constitutive UV-B response (Heijde et al., 2013). By contrast, but in agreement with the impaired COP1 interaction, overexpression of UVR8W285A,VP-AA did not result in a constitutive UV-B response (Figure 3B). Thus, the constitutive activity of UVR8W285A depends on the C27 domain. Moreover, our data indicate that the constitutive interaction of UVR8W285A with COP1 is solely via its C27 domain and not due to a freed β-propeller surface in a potentially monomeric in vivo conformation (Rizzini et al., 2011; Heijde et al., 2013).
Therefore, it is likely that in the UVR8W285A constitutively active photoreceptor variant, the C27 domain is constitutively exposed to interaction with COP1, initiating signaling in the absence of UV-B. Interestingly, although clearly more active than the wild-type UVR8 protein, the UVR8W285A protein is not as active as UV-B-activated UVR8 (otherwise, wild-type or even lower levels of expression would result in a constitutive growth phenotype) (O’Hara and Jenkins, 2012; Heijde et al., 2013; Huang et al., 2013, 2014). This may be explained by the absence of the stabilizing interaction of the UVR8 β-propeller core with COP1 in UVR8W285A. Therefore, we propose that the UV-B-dependent UVR8 β-propeller contributes to target COP1, thereby efficiently directing the C27 domain to the correct protein for signal propagation.
UVR8 inactivation occurs by redimerization of UVR8 monomers to the homodimer (Heijde and Ulm, 2013; Heilmann and Jenkins, 2013). UVR8-COP1 complex formation results in the induction of RUP1 and RUP2 expression, forming a negative feedback loop that facilitates UVR8 redimerization and thus disruption of the UVR8-COP1 interaction (Gruber et al., 2010; Heijde and Ulm, 2013; Ouyang et al., 2014). It was shown previously that RUP1 and RUP2 interact with the C27 domain of UVR8, potentially competing with COP1 (Cloix et al., 2012). We show here, in contrast with COP1, that RUP1 and RUP2 indeed interact solely with the C27 domain and not with the UVR8 core (Figures 6A to 6C). This is despite the fact that RUP1, RUP2, and COP1 are phylogenetically and structurally related: their WD40 domains predict similar seven-bladed β-propeller folds (Gruber et al., 2010). Moreover, in contrast with COP1, the RUP1 and RUP2 proteins can still interact with the C27 domain of inactive homodimeric UVR8 (Figure 6) (Gruber et al., 2010; Cloix et al., 2012). This further indicates that the C27 domain is not simply generally hidden in the UVR8 homodimeric ground state but that interaction with the WD40-repeat proteins RUP1 and RUP2 is possible, and interaction with the WD40-repeat domain of COP1 must be more specifically prevented.
Independent of this, it is not known whether RUP1 and RUP2 proteins must be removed before UVR8 homodimers can be monomerized in response to UV-B. Previous data show that overexpression of RUP2 results in stable UVR8 homodimers (Heijde and Ulm, 2013). But at present, it is not possible to distinguish unequivocally between instantaneous redimerization and blocking of monomerization when RUP2 is overexpressed and interacts constitutively with UVR8. In both cases, however, the generation of signaling-competent UVR8 monomers ultimately requires the dissociation of RUP1 and RUP2 from UVR8 by an as yet unknown mechanism. The complexity of interaction mechanisms, signal propagation, and modulation of the crucial UVR8-COP1-RUP core of the UV-B receptor photocycle demonstrated so far underlines the need for accurate reception and signaling for optimal induction by UV-B of photomorphogenesis and stress acclimation.
METHODS
Plant Material and Growth Conditions
The uvr8-1 and uvr8-2 mutants are in the Arabidopsis thaliana Landsberg erecta background (Kliebenstein et al., 2002; Brown et al., 2005), while uvr8-7 is in the Arabidopsis Wassilewskija background (Favory et al., 2009). The uvr8-7/Pro35S:UVR8W285A line 4 was described previously (Heijde et al., 2013).
The UVR8VP-AA and UVR8W285A,VP-AA mutants were produced by site-directed mutagenesis and were cloned into Gateway vector pB2GW7 (Karimi et al., 2002) to generate uvr8-7/Pro35S:UVR8VP-AA and uvr8-7/Pro35S:UVR8W285A,VP-AA. A flexible peptide linker and a nuclear localization signal were added to UVR8C44 by PCR using primers NLS-C44_Fw (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCTGCAGCCTAAGAAGAAGAGAAAGGTTGGAGGAGGGAAAAGCTGGGTGTCGCCTG-3′) and NLS-C44_Rv (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAAATTCGTACACGCTTGAC-3′). The NLS-UVR8C44 sequence was cloned into the Gateway-compatible vector pB7WGY2 (Karimi et al., 2002), yielding the YFP-NLS-UVR8C44 fusion. The binary vector was transformed into uvr8-7 to generate uvr8-7/Pro35S:YFP-NLS-UVR8C44.
A synthetic nucleotide fragment of UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP (Supplemental Figure 4) was purchased from Biomatik and cloned into the pMDC7 binary vector for estradiol-inducible expression (Curtis and Grossniklaus, 2003). The 2A sequence that provides a cotranslational cleavage site was described previously (Ryan and Drew, 1994). Flexible peptide linkers were added between tandem repeats of UVR8C44. Application of β-estradiol (Sigma-Aldrich) in DMSO (50 μM) was used to induce the expression of this construct in transgenic Arabidopsis seedlings.
Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998). The transgenic lines described in this work were genetically determined to have the transgene integrated at a single locus (∼75% resistant to 25% sensitive in the T2 generation) and were used in the homozygous T3 generation. The growth conditions and UV-B treatments were as described previously (Heijde and Ulm, 2013).
Protein Extraction, Immunoprecipitation, and Immunoblots
Rabbit polyclonal antibodies were generated against a synthetic peptide derived from the UVR8 protein sequence [amino acids 1 to 15 + C: MAEDMAADEVTAPPRC; anti-UVR8(1-15)] and affinity-purified against the peptide (Eurogentec). Protein extracts were incubated with anti-UVR8 antibodies and protein A-agarose (Roche Applied Science) in extraction buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 0.1% Igepal, 2 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 10 mM leupeptin, 10 mM dichloroisocumarin, 1% [v/v] protease inhibitor cocktail for plant extracts [Sigma-Aldrich], and 10 mM each of the proteasome inhibitors MG132, MG115, and ALLN [VWR]) for 2 h at 4°C, and the beads were washed three times in extraction buffer. For immunoblot analysis, immunoprecipitates or total cellular proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes according to the manufacturer’s instructions (Bio-Rad). Polyclonal anti-UVR8(426-440) (Favory et al., 2009), anti-UVR8(1-15) (this work), anti-CHS (sc-12620; Santa Cruz Biotechnology), anti-actin (A0480; Sigma-Aldrich), and monoclonal anti-GFP (632381; Clontech) antibodies were used as primary antibodies, with anti-rabbit, anti-goat, and anti-mouse immunoglobulins (Dako) used as secondary antibodies, as appropriate. Signals were detected using the ECL Western Detection Kit (GE Healthcare).
Yeast Two-Hybrid Interaction Assays
UVR8, UVR8-2 (=UVR8N400), UVR8N396, UVR8C44, UVR8C44,VP-AA, UVR8W285A, UVR8W285A,VP-AA, UVR8VP-AA, and COP1C340 were cloned in-frame to the LexA DNA binding domain in the Gateway-compatible vector pBTM116-D9-GW (Stelzl et al., 2005). Coding regions of COP1, RUP1, RUP2, UVR8, and UVR8-2 were cloned in-frame to the Gal4 activation domain in the Gateway-compatible vector pGADT7-GW (Marrocco et al., 2006). Both vectors were cotransformed into yeast strain L40 using the lithium acetate-based transformation protocol (Gietz and Woods, 2002). Transformed yeast cells were selected on SD/-Trp/-Leu medium (ForMedium). Yeast growth assays to detect colonies with interacting proteins were performed at 30°C on selective SD/-Trp/-Leu/-His medium. The quantitative interaction assays were performed using chlorophenol red-β-d-galactopyranoside (Roche Applied Science) as the substrate as described previously (Rizzini et al., 2011; Crefcoeur et al., 2013). For the yeast two-hybrid assays with UVR8ΔC27 and COP1, UVR8ΔC27 was cloned in-frame to the Gal4 DNA binding domain in pGBKT7-GW, and COP1 was used in the pGADT7-GW vector. Both vectors were cotransformed into yeast strain AH109 using the lithium acetate-based transformation protocol (Gietz and Woods, 2002). For the quantitative yeast two-hybrid assay shown in Figure 6D, UVR8 and its mutant versions were cloned into the pGADT7-GW vector and transformed into yeast strain Y187 (Clontech). RUP1 and RUP2 in the pGBKT7-GW vector were transformed into GoldY2H yeast (Clontech). After mating of appropriate yeast strains, diploids were selected on SD/-Trp/-Leu medium.
If needed, irradiation of yeast cells by narrow-band UV-B (Philips TL20W/01RS; 20 h, 1.5 μmol m−2 s−1) was performed as described previously (Rizzini et al., 2011; Crefcoeur et al., 2013).
Quantitative Real-Time PCR
Arabidopsis total RNA was isolated with the Plant RNeasy Kit (Qiagen) and treated with DNaseI according to the manufacturer’s instructions. cDNA was synthesized using a 1:1 ratio mix of oligo(dT)16 and random hexamer primers using the TaqMan Reverse Transcription Reagents Kit (Life Technologies). PCR was performed and detected using the ABsolute QPCR SYBR Green ROX Mix according to the manufacturer’s instructions (Thermo Scientific). Primers for HY5 were as follows: HY5_Fv (5′-CAAGCAGCGAGAGGTCATCA-3′) and HY5_Rv (5′-CATCGCTTTCAATTCCTTCTTTG-3′). Primers for CHS were as follows: CHS_Fv (5′-CGTGTTGAGCGAGTATGGAAAC-3′) and CHS-Rv (5′-TGACTTCCTCCTCATCTCGTCTAGT-3′). cDNA concentrations were normalized to the 18S rRNA transcript levels using the Eukaryotic 18S rRNA Kit (Applied Biosystems). Quantitative real-time PCR was performed using the 7900HT Real-Time PCR System (Applied Biosystems). Expression was determined in triplicate biological measurements.
Accession Numbers
Sequence data from this work can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G13930 (CHS), AT2G32950 (COP1), AT5G11260 (HY5), AT5G52250 (RUP1), AT5G23730 (RUP2), and AT5G63860 (UVR8).
Supplemental Data
Supplemental Figure 1. UVR8ΔC27 Interacts with COP1 in Yeast in a UV-B-Dependent Manner.
Supplemental Figure 2. Sequence Alignment between UVR8C44 and Partial Sequences of Several Known COP1-Interacting Proteins.
Supplemental Figure 3. Sustained Expression of UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP Leads to Hypocotyl Growth Inhibition in the Absence of UV-B.
Supplemental Figure 4. Sequence of the Synthesized UVR8C44-UVR8C44-NLS-UVR8C44-2A-YFP Fragment.
Supplementary Material
Acknowledgments
We thank Daniel Kliebenstein and Gareth Jenkins for providing the uvr8-1 and uvr8-2 mutants, respectively, and Florence Ares-Orpel and Joël Nicolet for excellent technical assistance. This study was supported by the State of Geneva, by European Research Council Grant 310539 under the European Union’s Seventh Framework Programme, and by Swiss National Science Foundation Grants SNF 31003A-132902 and SNF 31003A-153475.
AUTHOR CONTRIBUTIONS
R.Y. performed all experiments, except for that shown in Figure 6D, which was done by A.B.A. M.B. contributed new tools. R.Y. and R.U. designed the experiments, analyzed the data, and wrote the article.
Glossary
- VP
Val-Pro
Footnotes
Articles can be viewed online without a subscription.
References
- Ballare C.L., Barnes P.W., Flint S.D. (1995). Inhibition of hypocotyl elongation by ultraviolet-B radiation in de-etiolating tomato seedlings. I. The photoreceptor. Physiol. Plant. 93: 584–592. [Google Scholar]
- Binkert M., Kozma-Bognár L., Terecskei K., De Veylder L., Nagy F., Ulm R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102: 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M., Arvai A.S., Baxter K.J., Heilmann M., Pratt A.J., O’Hara A., Kelly S.M., Hothorn M., Smith B.O., Hitomi K., Jenkins G.I., Getzoff E.D. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C., Kaiserli E., Heilmann M., Baxter K.J., Brown B.A., O’Hara A., Smith B.O., Christie J.M., Jenkins G.I. (2012). C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl. Acad. Sci. USA 109: 16366–16370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crefcoeur R.P., Yin R., Ulm R., Halazonetis T.D. (2013). Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat. Commun. 4: 1779. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkura P.V., Ballaré C.L. (2012). UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 5: 642–652. [DOI] [PubMed] [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér B., Kozma-Bognár L., Kevei E., Hajdu A., Binkert M., Davis S.J., Schäfer E., Ulm R., Nagy F. (2011). Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67: 37–48. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- González Besteiro M.A., Bartels S., Albert A., Ulm R. (2011). Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 68: 727–737. [DOI] [PubMed] [Google Scholar]
- Gruber H., Heijde M., Heller W., Albert A., Seidlitz H.K., Ulm R. (2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 20132–20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S., Velanis C.N., Jenkins G.I., Franklin K.A. (2014). UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. USA 111: 11894–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237. [DOI] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2013). Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. USA 110: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Binkert M., Yin R., Ares-Orpel F., Rizzini L., Van De Slijke E., Persiau G., Nolf J., Gevaert K., De Jaeger G., Ulm R. (2013). Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 20326–20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann M., Jenkins G.I. (2013). Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 161: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann M., Christie J.M., Kennis J.T., Jenkins G.I., Mathes T. (2014). Photoinduced transformation of UVR8 monitored by vibrational and fluorescence spectroscopy. Photochem. Photobiol. Sci., http://dx.doi.org/10.1039/C4PP00246F. [DOI] [PubMed] [Google Scholar]
- Holm M., Hardtke C.S., Gaudet R., Deng X.W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Chen L., Wei N., Deng X.W. (2013). Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 110: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Li G., Li J., Chen H., Deng X.W. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Yang P., Ouyang X., Chen L., Deng X.W. (2014). Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 10: e1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.I. (2014). The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 26: 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kim B.C., Tennessen D.J., Last R.L. (1998). UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J. 15: 667–674. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Lim J.E., Landry L.G., Last R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang L., Jin D., Nezames C.D., Terzaghi W., Deng X.W. (2013). UV-B-induced photomorphogenesis in Arabidopsis. Protein Cell 4: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Li X., Zhong F.W., Li J., Wang L., Shi Y., Zhong D. (2014). Quenching dynamics of ultraviolet-light perception by UVR8 photoreceptor. J. Phys. Chem. Lett. 5: 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K., Zhou Y., Bury E., Dieterle M., Funk M., Genschik P., Krenz M., Stolpe T., Kretsch T. (2006). Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45: 423–438. [DOI] [PubMed] [Google Scholar]
- Müller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C., Ulm R., Timmer J., Zurbriggen M.D., Weber W. (2013). Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 41: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara A., Jenkins G.I. (2012). In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell 24: 3755–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám E., Schäfer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., Huang X., Jin X., Chen Z., Yang P., Ge H., Li S., Deng X.W. (2014). Coordinated photomorphogenic UV-B signaling network captured by mathematical modeling. Proc. Natl. Acad. Sci. USA 111: 11539–11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106. [DOI] [PubMed] [Google Scholar]
- Ryan M.D., Drew J. (1994). Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 13: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U., et al. (2005). A human protein-protein interaction network: A resource for annotating the proteome. Cell 122: 957–968. [DOI] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103. [DOI] [PubMed] [Google Scholar]
- Tilbrook K., Arongaus A.B., Binkert M., Heijde M., Yin R., Ulm R. (2013). The UVR8 UV-B photoreceptor: Perception, signaling and response. The Arabidopsis Book 11: e0164, doi/10.1199/tab.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi V., Lamattina L., Jenkins G.I., Cassia R.O. (2014). Ultraviolet-B-induced stomatal closure in Arabidopsis is regulated by the UV RESISTANCE LOCUS8 photoreceptor in a nitric oxide-dependent mechanism. Plant Physiol. 164: 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Máté Z., Adám E., Oakeley E.J., Schäfer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Tilbrook K., Fierro A.C., Marchal K., Poelman D., Van Der Straeten D., Ulm R. (2014). Photoreceptor-mediated bending towards UV-B in Arabidopsis. Mol. Plant 7: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Wargent J.J., Gegas V.C., Jenkins G.I., Doonan J.H., Paul N.D. (2009). UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 183: 315–326. [DOI] [PubMed] [Google Scholar]
- Wu D., Hu Q., Yan Z., Chen W., Yan C., Huang X., Zhang J., Yang P., Deng H., Wang J., Deng X., Shi Y. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219. [DOI] [PubMed] [Google Scholar]
- Wu M., Eriksson L.A., Strid A. (2013). Theoretical prediction of the protein-protein interaction between Arabidopsis thaliana COP1 and UVR8. Theor. Chem. Acc. 132: 1371. [Google Scholar]
- Wu M., Grahn E., Eriksson L.A., Strid A. (2011). Computational evidence for the role of Arabidopsis thaliana UVR8 as UV-B photoreceptor and identification of its chromophore amino acids. J. Chem. Inf. Model. 51: 1287–1295. [DOI] [PubMed] [Google Scholar]
- Xu C., Min J. (2011). Structure and function of WD40 domain proteins. Protein Cell 2: 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.