Virus infection altered alternative splicing in >100 Brachypodium distachyon defense-related genes, with little effect on the genome-wide ratios of different splicing types.
Abstract
In eukaryotes, alternative splicing (AS) promotes transcriptome and proteome diversity. The extent of genome-wide AS changes occurring during a plant-microbe interaction is largely unknown. Here, using high-throughput, paired-end RNA sequencing, we generated an isoform-level spliceome map of Brachypodium distachyon infected with Panicum mosaic virus and its satellite virus. Overall, we detected ∼44,443 transcripts in B. distachyon, ∼30% more than those annotated in the reference genome. Expression of ∼28,900 transcripts was ≥2 fragments per kilobase of transcript per million mapped fragments, and ∼42% of multi-exonic genes were alternatively spliced. Comparative analysis of AS patterns in B. distachyon, rice (Oryza sativa), maize (Zea mays), sorghum (Sorghum bicolor), Arabidopsis thaliana, potato (Solanum tuberosum), Medicago truncatula, and poplar (Populus trichocarpa) revealed conserved ratios of the AS types between monocots and dicots. Virus infection quantitatively altered AS events in Brachypodium with little effect on the AS ratios. We discovered AS events for >100 immune-related genes encoding receptor-like kinases, NB-LRR resistance proteins, transcription factors, RNA silencing, and splicing-associated proteins. Cloning and molecular characterization of SCL33, a serine/arginine-rich splicing factor, identified multiple novel intron-retaining splice variants that are developmentally regulated and modulated during virus infection. B. distachyon SCL33 splicing patterns are also strikingly conserved compared with a distant Arabidopsis SCL33 ortholog. This analysis provides new insights into AS landscapes conserved among monocots and dicots and uncovered AS events in plant defense-related genes.
INTRODUCTION
Plant pathogens cause losses of up to 40% in pre- and postharvest agricultural production annually (Flood, 2010). The outcome of plant disease or resistance is predicated on the ability of plants to expediently recognize the invading pathogen and launch robust immune responses. To counter plant defenses, pathogens use a variety of strategies to evade recognition, as well as to perturb host homeostasis. At the cellular level, alternative splicing (AS) represents one means of regulating plant and animal defenses.
AS also has important biological consequences in plant growth and development, flowering, circadian clock function, and stress responses (Howard et al., 2013; Reddy et al., 2013; Staiger and Brown, 2013). In humans, ∼95% of genes are alternatively spliced with exon-skipping events as the predominant AS type (Wang et al., 2008). Recent analysis of Arabidopsis thaliana, soybean (Glycine max), cotton (Gossypium raiimondi), maize (Zea mays), and rice (Oryza sativa) highlighted the prevalence of AS in plants (Filichkin et al., 2010; Zhang et al., 2010; Marquez et al., 2012; Li et al., 2014; Shen et al., 2014; Thatcher et al., 2014). These studies estimate that alternative splicing occurs in ∼60% of Arabidopsis, ∼52% of soybean, ∼40% of cotton, ∼40% of maize, and ∼33% of rice intron-containing genes (Filichkin et al., 2010; Zhang et al., 2010; Marquez et al., 2012; Li et al., 2014; Shen et al., 2014; Thatcher et al., 2014). In contrast to animals, plant AS events predominantly involve intron retention. However, the ratios of the different AS types (intron retention, alternate acceptor, alternate donor, and exon skipping) could vary among monocots and dicots (Shen et al., 2014).
AS arises due to alternate splice-site choices occurring during premature mRNA (pre-mRNA) splicing, resulting in more than one mature mRNA from a single pre-mRNA. AS increases the transcriptome complexity, and thereby the proteome diversity of a cell, and regulates the abundance of functional mRNA (Lewis et al., 2003). AS often incorporates premature stop codons in alternate transcripts that subsequently become targets for nonsense-mediated decay (Kalyna et al., 2012; Drechsel et al., 2013). Furthermore, translation of alternatively spliced transcripts often results in proteins with altered domains, structures, and functions. The mechanism of AS in plants, primarily based on studies of Arabidopsis, involves the concerted action of multiple splicing factors, spliceosomal components, and their interaction with cis-acting regulatory sequences on the pre-mRNA such as exonic and intronic splicing enhancers and suppressors (Hoskins and Moore, 2012; Reddy et al., 2013; Staiger and Brown, 2013). Splicing factors primarily comprise a conserved Ser/Arg-rich (SR) protein family, heterogeneous nuclear ribonucleoproteins, and lysine homology domain-containing proteins. Spatio-temporal changes in the abundance, localization, and activity of these splicing factors in various cells and in different environments could have genome-wide influences on splicing patterns of hundreds of genes (Ali and Reddy, 2008). The identities and the expression dynamics of splicing factors involved in plant defense responses are unknown. Moreover, AS mechanisms and spliceosome functions and their conserved action among monocots and dicots need further investigation.
Compatible and incompatible plant-virus interactions involve the modulation of expression of hundreds of genes affecting metabolism, signal transduction, transcriptional regulation, posttranscriptional gene silencing, protein degradation, and other cellular events (Whitham et al., 2006; Mandadi and Scholthof, 2012; Mandadi and Scholthof, 2013). Yet, the extent of AS occurring among these host genes and specific immune-related gene networks is not known. The significance of AS in plant antiviral responses has been shown for a few individual plant resistance genes (Dinesh-Kumar and Baker, 2000; Zhang and Gassmann, 2003), primarily from studies of dicot-infecting viruses in Arabidopsis, tobacco (Nicotiana species), and tomato (Solanum lycopersicum). For example, alternative splicing of the tobacco N gene is essential for the resistance response against Tobacco mosaic virus (Dinesh-Kumar and Baker, 2000), and alternative splicing of a tomato translation initiation factor 4E is critical for immunity against two potyviruses, Potato virus Y and Pepper mottle virus (Piron et al., 2010).
Despite the significant agricultural relevance of studying grass viral diseases, research on monocot-infecting viruses has generally lagged, compared with studies of dicot-infecting viruses, due to the lack of an amenable plant model similar to Arabidopsis or Nicotiana benthamiana (Mandadi and Scholthof, 2013; Lyons and Scholthof, 2015). Recently, Brachypodium distachyon has gained the status of a model monocot because of its genetic relatedness to key agronomic grasses such as wheat (Triticum aestivum), barley (Hordeum vulgare), maize, and rice (International Brachypodium Initiative, 2010) and because it has a small diploid genome (∼272 Mb), short stature, short generation time, and is amenable to Agrobacterium tumefaciens-mediated transformation (Brkljacic et al., 2011). B. distachyon is currently used to understand diverse aspects of plant biology, including germination, growth and development, biotic and abiotic stress responses, cell wall biology, and vernalization (Brkljacic et al., 2011; Catalan et al., 2014).
We recently established B. distachyon as a host for studying the synergism between Panicum mosaic virus (PMV) and its satellite virus (SPMV) and reported the host transcriptome profiles altered during PMV+SPMV infection (Mandadi and Scholthof, 2012, 2013; Mandadi et al., 2014). PMV and SPMV are both single-stranded, positive-sense RNA viruses. PMV infection alone or coinfection of PMV+SPMV can affect agriculturally important grass species such as switchgrass (Panicum virgatum), maize, and millet (Setaria italica, Pennisetum glaucum, and Panicum miliaceum; Scholthof, 1999). Here, taking advantage of the B. distachyon-PMV pathosystem and next-generation RNA sequencing and genomics tools, we characterized the plant virus-associated spliceome and AS landscape in grasses. In addition to discovering thousands of previously un-annotated transcripts, we identified AS events in key immune-related gene clusters encoding receptor-like kinases, R-proteins, transcription factors, and splicing regulators. Our study extends the utility of B. distachyon for plant-microbe interaction research and provides new insight into AS landscapes and patterns conserved among monocots and dicots.
RESULTS AND DISCUSSION
Mapping of a High-Quality B. distachyon Isoform-Level Transcriptome
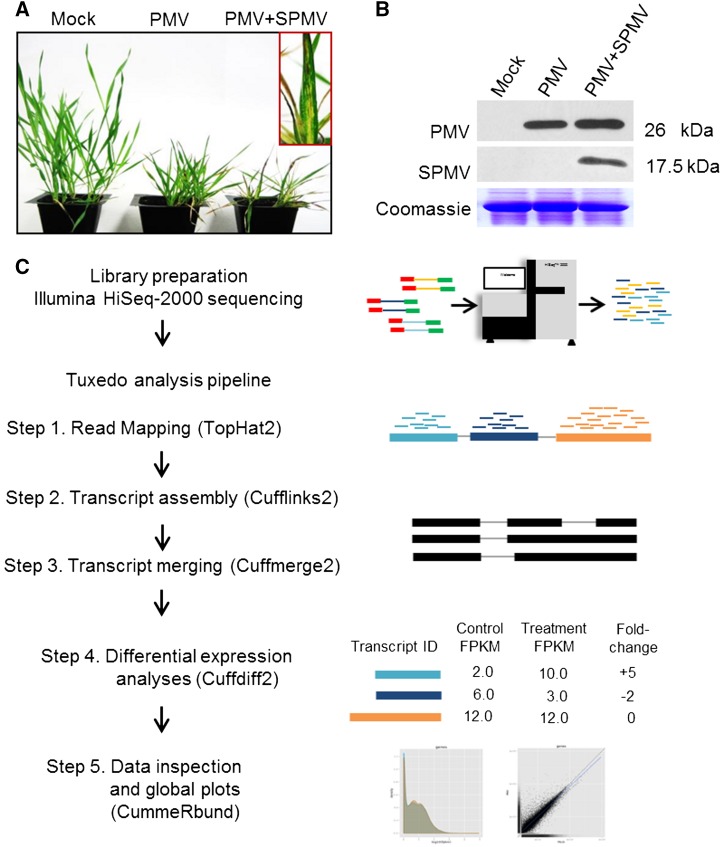
We previously established B. distachyon to study the biology of PMV and its satellite virus, SPMV (Mandadi and Scholthof, 2012; Mandadi et al., 2014). Coinfection of B. distachyon plants with PMV+SPMV exacerbates disease symptoms and additively alters host gene expression compared with PMV alone (Mandadi and Scholthof, 2012). To analyze isoform-level mRNA abundances and AS patterns altered during virus infection, we performed high-throughput RNA sequencing (RNA-seq). B. distachyon plants at the two- to three-leaf stage were mock-inoculated or infected with PMV or PMV+SPMV. Both infections in B. distachyon cause severe chlorosis and necrosis of leaves, reduce plant height, and lower biomass by 21 d postinoculation (dpi) (Figures 1A and 1B). To capture AS changes preceding the disease symptoms, RNA-seq was performed on infected plants at 7 dpi.
Figure 1.
Typical PMV and PMV+SPMV Disease Symptoms in B. distachyon and Workflow of RNA-seq Analysis.
(A) PMV and PMV+SPMV infection of B. distachyon causes severe chlorosis and necrosis of B. distachyon leaves, stunting, and loss of biomass by 21 dpi (Mandadi and Scholthof, 2012; Mandadi et al., 2014).
(B) Immunoblots of mock, PMV-, and PMV+SPMV-infected plants at 7 dpi showing accumulation of viral capsid proteins in the upper, noninoculated leaves.
(C) Workflow of RNA-seq analysis. RNA isolated from mock, PMV-, and PMV+SPMV-infected B. distachyon shoots at 7 dpi were subjected to 100 base paired-end Illumina HiSeq2000 sequencing, followed by quality trimming and filtering. High-quality reads were retained and mapped to the B. distachyon reference genome (Bd21v1.2) using the TopHat2 program, followed by transcript assembly, differential expression, and global analysis using Cufflinks2, Cuffmerge2, Cuffdiff2, and CummeRbund programs (Tuxedo pipeline) (Trapnell et al., 2010). Normalization of expression data was performed using the FPKM mapped fragments metric (Trapnell et al., 2010).
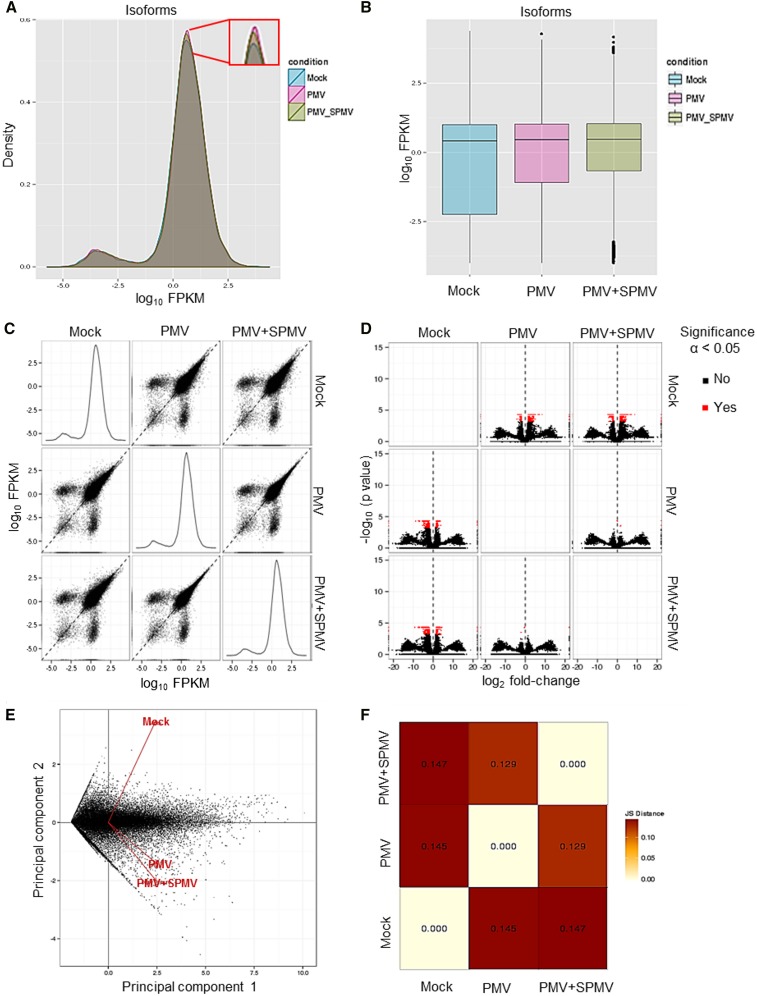
We obtained more than 58 million reads that passed the quality filters (Table 1). These reads were mapped to the B. distachyon reference genome Bd21v1.2 (see Methods), followed by transcript assembly and differential isoform and gene expression analysis using the Tuxedo RNA-seq data analyses pipeline, which employs a robust isoform deconvolution method for estimating gene expression (Trapnell et al., 2010, 2012, 2013) (Figure 1C). The analysis yielded ∼93 to 94% overall alignment rates for mock, PMV, and PMV+SPMV samples (Table 1). Approximately 91% of these reads were unique and mapped only once to a genomic locus, attesting to the high quality of the sequencing reads and of the reference B. distachyon genome. Global inspection of the isoform-level expression (normalized to fragments per kilobase of transcript per million mapped fragments [FPKM]) was further performed using the CummeRbund program (Trapnell et al., 2012). Density and box plots of the isoform-level expression (log10 FPKM) revealed a normal overall distribution of the data with little systematic bias among mock, PMV, and PMV+SPMV gene expression (Figures 2A and 2B) (Dillies et al., 2013). Pairwise scatter matrix plots and volcano matrix plots revealed that large numbers of transcripts were upregulated in the virus-infected samples compared with mock samples, as indicated by positive log10 FPKM and log2 fold changes in the respective plots (Figures 2C and 2D). Multivariate clustering and statistical relationships among the mock, PMV, and PMV+SPMV samples analyzed using principal component analysis and Jensen-Shannon distances further identified significant clustering of PMV and PMV+SPMV data sets when compared with mock (Figures 2E and 2F). Finally, comparison of the expression of ∼11,669 genes from RNA-seq analysis (FPKM ≥ 2) to our previous microarray analysis (Mandadi and Scholthof, 2012) revealed a high degree of correlation as indicated by Pearson product moment correlation coefficient (r) values of >0.54 (Supplemental Figure 1). We also estimated correlation of expression differences of 18 genes using RNA-seq, microarray, and quantitative RT-PCR, and the comparisons showed strong correlation between RNA-seq and quantitative RT-PCR fold changes (r > 0.95) in PMV- and PMV+SPMV-infected samples, followed by RNA-seq and microarray results (r > 0.84) (Supplemental Figure 1). Together, these analyses demonstrate the robustness of the B. distachyon transcriptome data at the isoform-level resolution under both healthy and virus-infected conditions.
Table 1. Statistical Summary of RNA-seq Mapping and Alignment.
| Total | Mock | PMV | PMV+SPMV | |
|---|---|---|---|---|
| Trimmed reads | ||||
| Left (million) | 29,672,853 | 10,046,371 | 9,422,241 | 10,204,241 |
| Right (million) | 28,867,416 | 9,748,243 | 9,161,016 | 9,958,157 |
| Combined (million) | 58,540,269 | 19,794,614 | 18,583,257 | 20,162,398 |
| Mapped reads | ||||
| Unique left (%) | 26,969,439 (90.8) | 9,197,613 (91.5) | 8,551,336 (90.7) | 9,220,490 (90.3) |
| Nonunique left (%) | 990,103 (3.3) | 314,255 (3.1) | 326,102 (3.4) | 349,746 (3.4) |
| Unique right (%) | 26,180,318 (90.6) | 8,908,250 (91.3) | 8,295,321(90.5) | 8,976,747 (90.1) |
| Nonunique right (%) | 959,212 (3.3) | 302,874 (3.1) | 315,992 (3.4) | 340,346 (3.4) |
| Overall alignment |
94.10% | 94.60% | 94.10% | 93.70% |
| Total aligned pairs | 25,992,902 | 8,864,588 | 8,245,277 | 8,883,037 |
Figure 2.
Global Analysis of Isoform-Level Expression in Mock-Inoculated and Virus-Infected B. distachyon.
(A) to (D) Expression density plots (A), expression box plots (B), expression scatter matrix (C), and expression fold-change volcano matrix (D) show normal distribution of the expression data and significantly greater number of differentially expressed transcripts in virus-infected samples (PMV or PMV+SPMV) compared with mock.
(E) and (F) Principal component analysis (E) and Jensen-Shannon distance matrix (F) shows clustering and positive correlation between PMV and PMV+SPMV infections compared with mock.
The inset in (A) shows the expression density differences among the samples. (C) is a pairwise scatter matrix generated using the csScatterMatrix method of the CummRbund tools (Trapnell et al., 2012) to enable easy visualization of all the possible comparisons. In (D), the significance cutoff used was α < 0.05. The principal component plot in (E) was generated using the PCAplot wrapper of CummeRbund tools (Trapnell et al., 2012) using the log-transformed expression estimates. Together, these analyses attest to the robustness of the B. distachyon transcriptome analysis, revealing little systematic bias among the individual samples (Dillies et al., 2013), as well as enabling identification of the significant differences present among the mock and virus-infected samples at the genomic scale.
The annotated B. distachyon reference genome (Bd21v1.2) comprises ∼31,029 protein-coding transcripts in 26,552 gene loci (International Brachypodium Initiative, 2010). Together with our transcript assemblies, the merged B. distachyon gene annotation has ∼44,443 transcripts in 28,274 gene loci. Of these, ∼2499 (9%) and 10,851 (24%) are previously unannotated gene and transcript models, expressed at FPKM ≥ 2 (Supplemental Data Sets 3 and 4). These transcriptional units can be grouped into two types: (1) transcriptional units mapped to previously unannotated regions of the genome and (2) alternative transcript models of an existing reference gene annotation. In the former category, we found several genes that were induced during virus infection. Representative examples of these genes are shown in Supplemental Figure 2. Each of these transcripts was subjected to BLASTX analysis (E-value cutoff: 1e-10) against the publicly available reference sequence databases. The analysis identified these transcripts as encoding putative proteins with diverse functions such as mitochondrial TIM proteins (XLOC_007079), NADH dehydrogenase (XLOC_020779), F-box protein (XLOC_025395), and DNAJ superfamily (XLOC_026754), as well as a long noncoding RNA (XLOC_014533) (Supplemental Figures 2A to 2E). Expression of these transcripts was further validated by RT-PCR (Supplemental Figure 2F).
A typical example of an alternative transcript annotation identified for an existing reference gene is shown in Supplemental Figure 3. According to the reference annotation, Bradi1g78245 has an assigned transcript length of 114 nucleotides and encodes a 37 amino-acid-long protein, with no known functional annotations (Supplemental Figure 3A). The transcript model we determined here showed that Bradi1g78245 is ∼900 nucleotides long with two exons (Supplemental Figure 3B). BLASTX analysis further identified Bradi1g78245 belonging to the thionin family of pathogenesis-related (PR) proteins (E-value: 1e-10). Expression of Bradi1g78245 was induced by >12-fold in virus-infected plants when compared with mock (Supplemental Figure 3B). This is consistent with activation of PR genes during plant biotic stress. These analyses underscore the sensitivity of our RNA-seq and the transcript assembly pipeline. The ∼30% higher number of additional transcripts and annotations discovered in this study bolsters the utility of RNA-seq approaches for transcript discovery and comprehensive genome annotations.
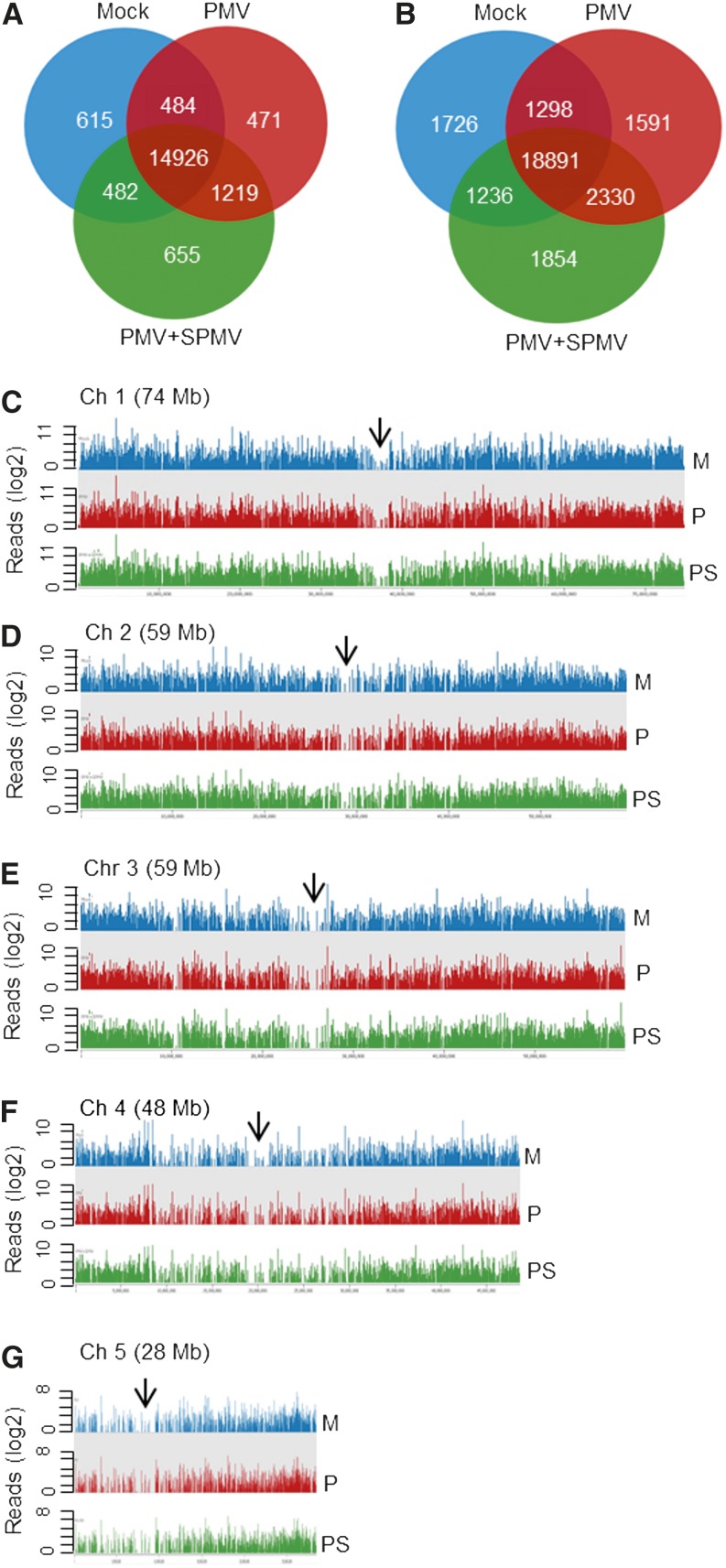
Approximately 18,852 genes and 28,926 transcripts were expressed at >2 FPKM (Figure 3). Plotting the read densities of ∼18,000 genes revealed extensive transcriptional activity on all five B. distachyon chromosomes (Figure 3). The genome-wide distribution of expression densities were largely similar among healthy and PMV and PMV+SPMV-infected samples. Heterochromatic regions of the genome, such as centromeres, showed a similar lack of transcription (Figure 3), suggesting negligible or no chromosome-level biases and/or activation of heterochromatic DNA during PMV and SPMV infection.
Figure 3.
Isoform-Level Transcriptome Maps of Healthy and Virus-Infected B. distachyon.
(A) and (B) Venn diagrams of expressed genes (A) and their corresponding transcripts (B) with FPKM ≥ 2 among mock, PMV-, and PMV+SPMV-infected plants.
(C) to (G) Read densities mapped along the five B. distachyon chromosomes in mock (M), PMV- (P), and PMV+SPMV-infected (PS) plants show extensive transcriptional activity throughout the genome. The arrowheads indicate centromeric regions. Read densities (log2 RPM) were quantitated and plotted against reference chromosome models (Bd21v1.2) using Seqmonk tools.
Virus-Modulated Alternative Splicing Landscapes
Although AS occurs in plants and animals, they differ in the relative proportion and frequency of AS types (Reddy et al., 2013). Intron retention (IR) events are the most predominant AS events in plants. In animals, exon skipping (ES) events are the predominant AS type. Stress can induce the number and type of AS events. For example, heat stress decreased the frequency of IR events, while increasing the alternate acceptor (AA) and alternate donor (AD) events in Physcomitrella patens (Chang et al., 2014).
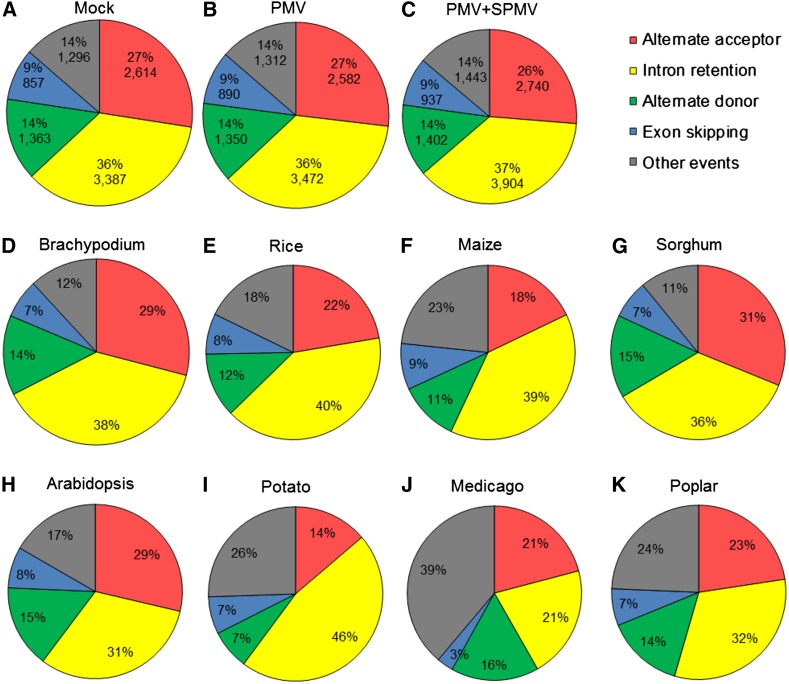
To analyze the AS landscapes of B. distachyon and determine their patterns during virus infection, we used the Alternative Splicing Transcriptional Landscape Visualization (ASTALAVISTA) tool and categorized the AS events (Foissac and Sammeth, 2007). Of ∼21,490 intron-containing, multiexonic genes detected in our study, ∼8911 genes (42%) were alternatively spliced. The splice junctions of these genes were used to identify, extract, and classify the AS events into: (1) IR, (2) ES, (3) AA, (4) AD, and (5) other events. The “other events” are complex AS events comprising duplicated IR, AA, AD, and ES events or a combination thereof. In total, we identified ∼9517 AS events in mock-inoculated B. distachyon plants (Supplemental Data Set 5). Among the different AS types, IR events predominated (∼3387, 36%), followed by AA (∼2614, 27%), AD (∼1296, 14%), and ES (∼857, 9%) events. Approximately 1296 events (14%) were classified as complex AS events (Figure 4A; Supplemental Data Set 5). Virus infection increased the number of AS events in most AS types in B. distachyon (Figures 4A to 4C). We observed little change in the overall ratios of the different AS types in virus-stressed B. distachyon, in contrast to the effect of heat stress on AS patterns in P. patens (Chang et al., 2014). An increased number of AS events during PMV+SPMV infections compared with PMV alone (Figure 4C; Supplemental Data Set 5) supports our previous report of additive effects of SPMV on altered host gene expression profiles (Mandadi and Scholthof, 2012).
Figure 4.
AS Landscapes of Healthy and Virus-Infected B. distachyon.
Frequency of AS types in mock (A), PMV- (B), and PMV+SPMV-infected (C) B. distachyon. Reference AS landscapes of four monocots, B. distachyon (Bd21v1.2) (D), rice (MSU6.0) (E), maize (AGPv3) (F), and sorghum (Sorbi1) (G), and four dicots, Arabidopsis (TAIR10) (H), potato (SolTub3.0) (I), M. truncatula (MedtrA17_3.5) (J), and poplar (JGI2.0) (K). Reference gene annotations were obtained from Ensemble. AS events were extracted and classified using the ASTALAVISTA tool (Foissac and Sammeth, 2007).
In a recent analysis of AS in soybean, Shen et al. (2014) suggested that the ratio of different AS types could vary between monocots and dicots (Shen et al., 2014). To test this, we analyzed the AS landscapes and the relative proportion of AS types among four reference monocots, including B. distachyon (Bd21v1.2), rice (MSU6.0), maize (AGPv3), and sorghum (Sorghum bicolor; Sorbi1), as well as four reference dicots, Arabidopsis (TAIR10), potato (Solanum tuberosum; SolTub3.0), Medicago truncatula (MedtrA17_3.5), and poplar (Populus trichocarpa; JGI2.0) using the ASTALAVISTA tool (Supplemental Data Set 5). The B. distachyon reference genome (Bd21v1.2) had fewer AS events (∼3975) compared with those identified in this study (∼9517; Figure 4A). This result shows that many AS transcripts remain unannotated in the B. distachyon reference genome (Figure 4D). Nevertheless, the ratios of the different AS types and landscapes of B. distachyon were strikingly similar to those of rice, maize, and sorghum (Figures 4D to 4G). This result also reflects the high degree of collinearity of the B. distachyon genome with the genomes of agronomic grasses (International Brachypodium Initiative, 2010) and supports the utility of B. distachyon as a robust laboratory model for economically important grains. We found little difference in the ratios of the different AS types among monocots and dicots (Figures 4D to 4K). IR events were the predominant AS type among all dicots and monocots analyzed, followed by AA and AD types. ES events were the least abundant AS type among all plants (Figures 4D to 4K). Together, these results show that the intron-processing mechanisms and AS processes are broadly conserved among monocots and dicots and support the view that IR events predominate in plants compared with animals (Reddy et al., 2013).
Virus-Modulated Alternatively Spliced Gene Clusters
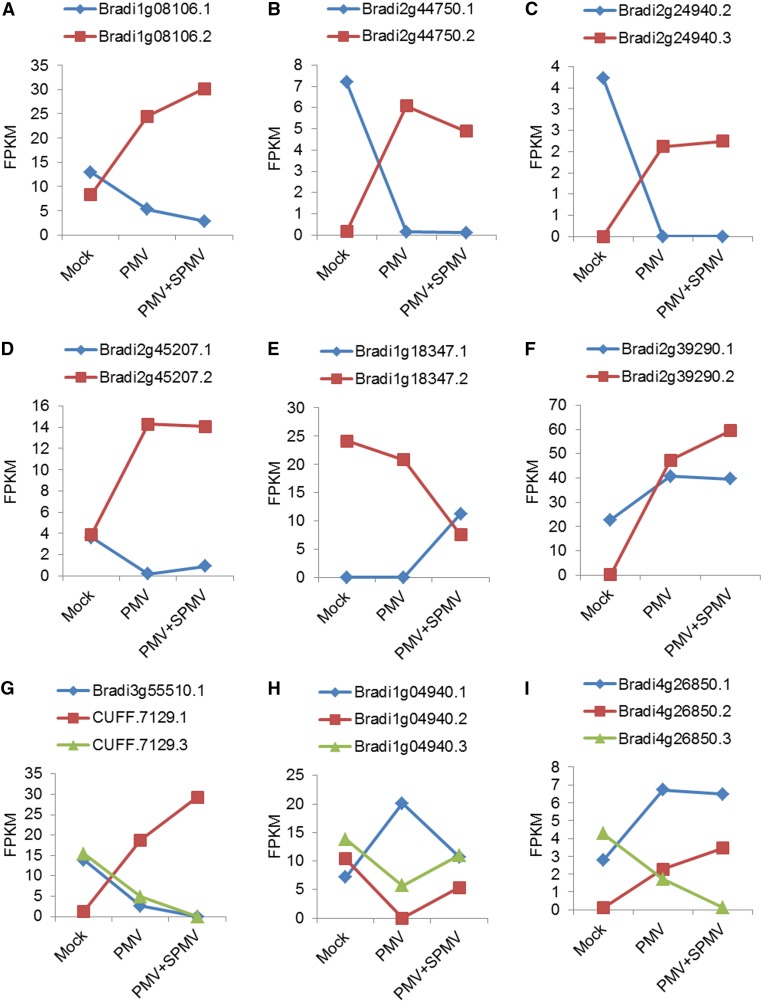
To identify alternatively spliced gene clusters that were differentially regulated by virus infection, we performed differential splicing analysis using the Cuffdiff program (Trapnell et al., 2010, 2013). After statistical cutoffs (false discovery rate [FDR] < 0.05), we found posttranscriptional splicing overload, a measure of differential splicing (Trapnell et al., 2010, 2013), in ∼669 genes in PMV- and PMV+SPMV-infected plants compared with mock. These genes encompass ∼4403 transcripts. Of these, ∼1970 (44.7%) were known reference annotated transcripts, while the remaining, ∼2433 (55.3%) are novel transcripts or annotations different from the B. distachyon reference annotations. Comparison of the isoform-level expression of the differentially spliced genes between mock- and virus-infected plants revealed varying expression dynamics ranging from simple isoform-switching to complex splicing patterns (Trapnell et al., 2010) (Figure 5). Instances of simple isoform switching (Figures 5A to 5F) often showed little change in total gene expression, despite substantial differences among the individual transcript-level abundance. For example, gene-level expression change of Bradi1g08106, a WRKY transcription factor with two annotated splice variants (Bradi1g08106.1 and Bradi1g08106.2), was induced only 1.3- and 1.5-fold in PMV and PMV+SPMV infection, respectively (Figure 5A). However, measurement of isoform-level expression revealed that the primary transcript, Bradi1g08106.1, was downregulated by 2.5- and 4.6-fold, while Bradi1g08106.2 expression was upregulated by 2.9- and 3.6-fold in PMV- and PMV+SPMV-infected plants, respectively. Complex splicing overloads often resulted in a net change in gene expression in the direction of the predominant isoform fold-change differences (Figures 5G to 5I). Together, these findings highlight the strengths of RNA-seq and isoform deconvolution methods to accurately determine transcript-level dynamics and robust gene annotations over conventional microarray-based or count-based gene expression analyses (Trapnell et al., 2010, 2013).
Figure 5.
Splicing Overload and Transcript-Level Dynamics Modulated during Virus Infection.
Levels of expression (FPKM) of individual isoforms within a splicing group that are significantly differentially expressed (FDR < 0.05) among mock, PMV-, and PMV+SPMV-inoculated samples are plotted to reveal simple isoform switching ([A] to [F]) and complex splicing overloads ([G] to [I]).
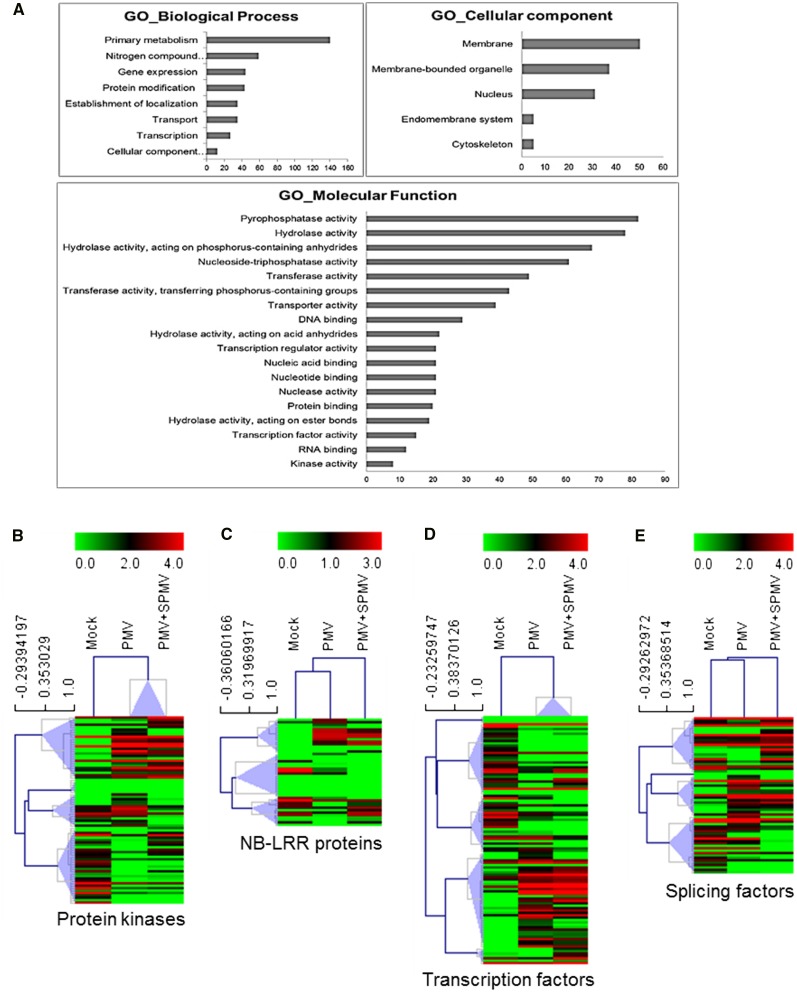
We next determined putative functions enriched among the ∼600 genes differentially spliced during virus infection using Gene Ontology (GO) terms (Du et al., 2010). Gene set enrichment analysis revealed significantly enriched (FDR < 0.05) terms in metabolism and transport, gene expression and transcription, and protein modification (Figure 6A). Through manual curation of GO, PANTHER, PFAM, and KEGG functional annotations (Kanehisa and Goto, 2000; Thomas et al., 2003; Du et al., 2010; Goodstein et al., 2012; Punta et al., 2012), we identified ∼100 defense-related AS gene clusters that are differentially spliced. These genes encode ∼20 protein kinases, nine nucleotide-binding site leucine-rich repeat (NB-LRR) family resistance (R) proteins, 25 transcription factors, 15 RNA silencing/methylation-related proteins, 16 splicing factors, and 10 proteasomal degradation-related proteins (Supplemental Data Set 1). We found novel splice variants among key defense genes such as WALL-ASSOCIATED KINASE25 (Bradi1g69340), CTR1/MAPKKK7-like (Bradi1g28110), RGA4 (Bradi2g60230), RPP8-like (BRADI1G29427), NICOTIANA LESION INDUCING-like (Bradi5g00790), AUTOPHAGY PROTEIN12 (Bradi3g57280) WRKY81 (Bradi1g16120), DICER2-like (Bradi1g15440), and RNA-DEPENDENT RNA POLYMERASE6 (Bradi1g55350). The novel splice variants comprised AA-, AD-, IR-, and ES-type AS events and often encoded putative proteins with truncated and/or altered sequences (Supplemental Data Set 5). Further hierarchical cluster analysis of the select defense-related gene clusters showed discernable patterns of coregulation among the splice variants during PMV and PMV+SPMV infection compared with mock plants (Figures 6B to 6E). Together, these analyses identified AS events and splice variants in several defense-related genes and greatly expand the AS gene repertoire involved in plant disease beyond a handful of known R genes (Staiger and Brown, 2013).
Figure 6.
Virus-Modulated Differentially Spliced Gene Clusters in B. distachyon.
(A) GO biological processes, cellular components, and molecular functions enriched (FDR < 0.05) among differentially spliced genes in PMV and PMV+SPMV infection compared with mock. The x axis indicates the number of genes corresponding to the enriched GO term.
(B) to (E) Hierarchical clustering of expression profiles of the AS transcripts encoding putative protein kinases (B), NB-LRR proteins (C), transcription factors (D), and splicing factors (E). Average linkage method and Pearson correlation metric was used for the clustering. Node heights of the gene clusters are displayed above the gene trees. Colors represent log2 FPKM expression values of the corresponding alternatively spliced transcripts.
Virus-Modulated Alternative Splicing of Splicing Factors
The mechanism of pathogen stress-induced AS in plants is not known. It likely involves spliceosomal proteins and splicing factors that are themselves alternatively spliced, perhaps to maintain cellular splicing homeostasis (Reddy et al., 2013; Staiger and Brown, 2013). We identified multiple B. distachyon SR proteins, an U2 snRNP auxiliary factor, and a polypyrimidine tract binding protein that are differentially spliced during virus infection (Supplemental Data Set 1; Figure 6E). These splicing factors could mediate AS homeostasis during plant-microbe interactions.
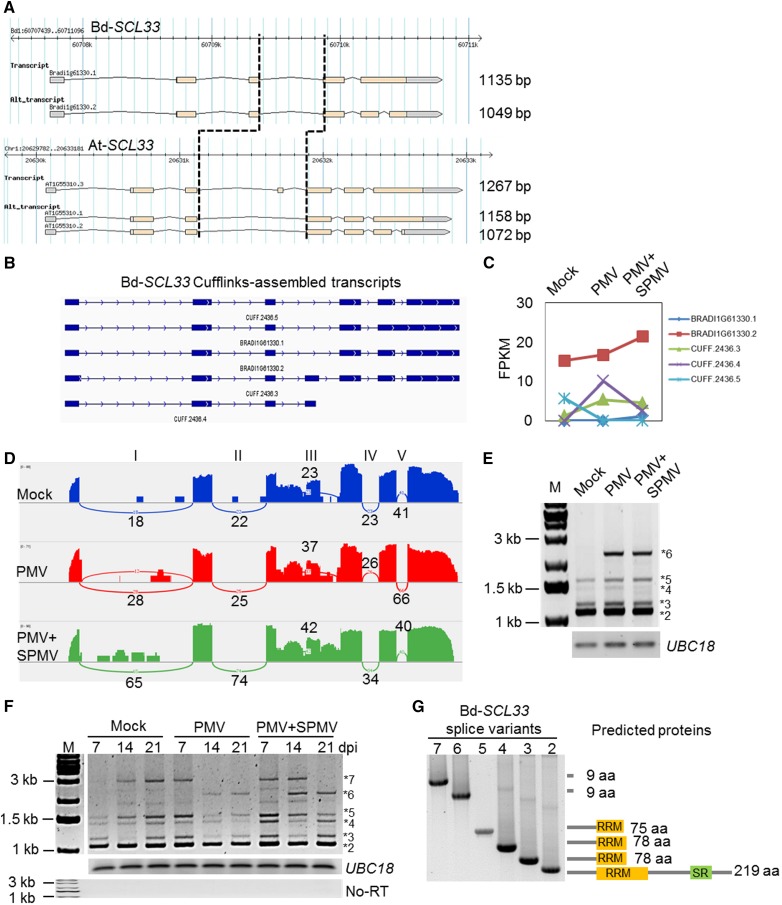
In Arabidopsis, a splicing factor, SCL33 (At1g55310), functions in flower development and autoregulates its alternative splicing (Thomas, 2012; Thomas et al., 2012). Expression of Arabidopsis SCL33 splice variants are also modulated by stresses such as heat and cold (Palusa et al., 2007). We serendipitously discovered an IR AS event in a B. distachyon SR protein (Bradi1g61330) that was reminiscent of the Arabidopsis SCL33 IR AS pattern (Palusa et al., 2007; Thomas et al., 2012). Through further comparisons of splice junctions of Bradi1g61330, we validated it as the B. distachyon ortholog of At-SCL33 (Figures 7A and 7B). The B. distachyon reference genome (Bd21v1.2) has two transcript annotations for Bd-SCL33. The primary transcript is Bradi1g61330.1 (designated #1) and Bradi1g61330.2 is the splice variant (designated #2), encoding 246 and 219 amino-acid-long proteins, respectively (Figure 7A). Both predicted SCL33 proteins contain an N-terminal RNA recognition motif (RRM) and an SR-rich domain in the C terminus. Expression of Bradi1g61330.1 was below detectable levels in our analysis, while Bradi1g61330.2 was the predominant splice variant with FPKM = 15.3 (Figure 7C). Expression of Bradi1g61330.2 was not significantly altered during virus infection (Figure 7C). However, we identified multiple SCL33 splice variants in the Cufflinks transcript assembly that displayed varying expression patterns during virus infection (Figures 7B and 7C). A Sashimi plot visualization of the SCL33 splice variants revealed a substantial number of RNA-seq reads mapping to SCL33 intron I and intron III (Figure 7D). Subsequent RT-PCR analysis using the same RNA used for RNA-seq confirmed the presence of multiple SCL33 splice variants (Figure 7E).
Figure 7.
Virus-Modulated Alternative Splicing Patterns of Bd-SCL33.
(A) Reference gene annotations of Bd-SCL33 and At-SCL33 and their splice variants. The intron III of SCL33 is indicated by the vertical black dotted lines.
(B) Cufflinks-assembled transcript models of Bd-SCL33 showing intron III-retaining splice variants.
(C) Expression dynamics of SCL33 splice variants analyzed by RNA-seq in mock, PMV-, and PMV+SPMV-infected plants.
(D) Sashimi plots showing RNA-seq reads mapping to Bd-SCL33 locus in mock, PMV, and PMV+SPMV samples. Heights of the bars represent overall read coverage. Splice junctions supported by ≥10 split reads are displayed as loops. Intron numbers I through V are indicated on the top of the panel. Note the read coverage on introns I and III.
(E) RT-PCR validation of novel SCL33 splice variants in mock, PMV-, and PMV+SPMV-infected samples at 7 dpi used for RNA-seq.
(F) Temporal expression analysis of SCL33 splice variants in mock, PMV-, and PMV+SPMV-infected plants at 7, 14, and 21 dpi in an independent experiment using different biological samples than those used for RNA-seq analysis. UBC18 was used as a control for approximately equal cDNA amounts in the different samples. No RT control was performed to evaluate genomic DNA contamination in the cDNA.
(G) Cloning and sequencing of the SCL33 splice variants confirmed intron retention events in the longer splice variants. Note that the longer splice variants contain premature termination codons in their open reading frame resulting in truncated SCL33 proteins devoid of an RRM and SR domain.
As demonstration of the robustness of the bioinformatics approach to empirically predict splice variants, we further determined temporal expression patterns of SCL33 in virus-infected B. distachyon plants during disease progression, using RT-PCR, followed by cloning and sequencing the various splice variants. We found dynamic expression patterns of the putative intron-retained splice variants (Figure 7F). For instance, in healthy mock-inoculated plants, the shortest SCL33 splice variant (#2) was the most abundant transcript and is expressed constitutively. The longer SCL33 splice variants (#4, #5, and #7) were predominant in mature plants (14 and 21 dpi). Splice variant #3 showed a unique pattern of downregulation around 14 dpi. Together, the dynamic temporal expression patterns of SCL33 splice variants suggest a role for SCL33 in plant development and/or senescence (Figure 7F). Strikingly, virus infection prematurely induced the abundance of several splice variants (#4, #5, and #7) as early as 7 dpi, as well as uniquely triggered the production of another splice variant #6 (Figure 7F). By cloning and sequencing the SCL33 splice variants, we confirmed splice variant #2 as the mature SCL33 transcript and splice variants #3 through #6 as intron-retaining splice variants with either partial or complete segments of intron I and intron III (Supplemental Table 1). The longest splice variant #7 is the full-length SCL33 pre-mRNA (Supplemental Table 1).
How the altered splicing patterns and expression dynamics of SCL33 splice variants and perhaps of the remaining splicing regulators contribute to AS homeostasis during virus infection remains to be investigated. However, analysis of the open reading frames of SCL33 splice variants (#3 through #7) revealed multiple premature termination codons, which could result in truncated proteins lacking a complete C-terminal SR domain and/or partial segments of the RRM domain (Figure 7G; Supplemental Table 1). These truncated SCL33 proteins could compete with the full-length SCL33 for binding to its targets, acting as dominant-negative mutants. Alternatively, the splice variants containing premature stop codons could become targets of nonsense-mediated decay machinery and contribute to altering the levels of functional SCL33 protein through regulated unproductive splicing and translation mechanisms (Filichkin et al., 2010; Kalyna et al., 2012).
In conclusion, here, we have shown the extent of genome-wide changes in alternative splicing occurring during plant-virus infections using the B. distachyon-PMV pathosystem. Our RNA-seq analysis identified ∼30% more splice variants than those annotated in the B. distachyon reference genome and discovered key immune-related gene clusters encoding R proteins, protein kinases, and transcription and splicing factors that are differentially spliced during virus infection. These splice variants potentially expand the B. distachyon proteome diversity and function by encoding truncated proteins and/or altered protein sequences that could influence plant immune signaling and defense homeostasis. As an example of the importance of the genome-wide AS dynamics revealed from our analysis, we highlighted one virus-modulated alternative splicing event in a B. distachyon splicing factor, SCL33. Alternative splicing of SCL33 resulted in production of at least six SCL33 splice variants whose expression patterns are dynamically regulated during plant development and during viral disease progression. Furthermore, splicing patterns of B. distachyon SCL33 appears conserved with a distant Arabidopsis SCL33 ortholog (Palusa et al., 2007; Thomas et al., 2012) that diverged ∼150 million years ago (Chaw et al., 2004) and suggests a conserved role for SCL33 splice variants in plants. Our analysis of B. distachyon fills a knowledge gap that exists in understanding the extent of genome-wide alternative splicing dynamics occurring during plant-virus infection and provides the impetus for further exploration of AS in plant defense signaling and homeostasis.
METHODS
Plant Growth Conditions and Virus Inoculations
Brachypodium distachyon (Bd21-3) seeds were stratified at 4°C for 3 to 4 d. Plants were grown in a chamber with 14 h light (21°C)/10 h dark (18°C) with a light intensity of ∼250 to 280 μmol/m2s and 60% relative humidity (Mandadi and Scholthof, 2012; Mandadi et al., 2014). Virus infections were performed on B. distachyon plants at the two- to three-leaf stage (Mandadi and Scholthof, 2012; Mandadi et al., 2014). Briefly, full-length cDNA clones of PMV and SPMV in plasmid vectors were linearized using EcoICR1 and BglII restriction endonucleases, respectively (Turina et al., 1998). The linearized plasmid (∼500 ng) was subsequently used for in vitro transcription reactions using T7 RNA polymerase (Fermentas) following the manufacturer’s instructions. The integrity of the transcripts was verified in agarose gels. Equal amounts of PMV and SPMV transcripts (∼100 ng) were resuspended in RNA inoculation buffer (50 mM KH2PO4, 50 mM glycine, pH 9.0, 1% bentonite, and 1% Celite) and rub-inoculated on B. distachyon leaves (Mandadi and Scholthof, 2012; Mandadi et al., 2014). Inoculated plants were kept in the dark for 24 h under humid conditions and were subsequently transferred to the growth chamber.
Plant Sampling, Protein and RNA Extraction, and Immunoblot Assays
Three independent biological replicate samples of aboveground shoots were collected from infected B. distachyon plants before the onset of chlorosis symptoms (7 dpi), along with mock-inoculated plants. The presence of PMV and SPMV in the infected samples was determined by immunoblot analysis using antibodies against PMV and SPMV capsid proteins, as described previously (Mandadi and Scholthof, 2012; Mandadi et al., 2014). A primary antibody concentration of 1:5000 and 1:1000 was used for PMV and SPMV detection, respectively. A 1:10,000 dilution was used for the secondary peroxidase-conjugated goat anti-rabbit IgG secondary antibodies (Rockland), and the visualization was performed using SuperSignal West Pico (Pierce) chemiluminescence detection reagents and x-ray film. Infected plant samples identified as positive by immunoblots (Figure 1B) were further subjected to spin column-based RNA purification and DNase treatment using the Direct-zol RNA technique (Zymo Research), following the manufacturer’s instructions.
RNA-seq and Bioinformatics Analyses
Poly(A) RNA enrichment and RNA-seq library preparation was performed by pooling the RNA of three biological replicates, followed by multiplex adapter ligation, and 100 base paired-end sequencing on the Illumina Hiseq-2000 platform. The sequencing yielded a sum total of ∼59 million reads. The libraries are publicly available through NCBI under the following accession numbers: BioProject-PRJNA259564; Experiments-SRX746906, SRX747740 and SRX747746. Successfully demultiplexed and prefiltered reads were further quality-trimmed and quality-filtered using FASTQC tools to remove poor-quality, adapter-containing, and ambiguous nucleotide-containing sequences. Subsequent mapping of the RNA-seq reads to the B. distachyon reference genome (Bd21v1.2) was performed using the Tuxedo RNA-seq analysis pipeline (Trapnell et al., 2010, 2012, 2013) and at the iPlant Discovery environment (Goff et al., 2011). The Tuxedo pipeline comprised TopHat2 (mapping), Cufflinks2 (transcript assembly), Cuffmerge2 (transcript merging), and Cuffdiff2 (differential gene and transcript expression, and differential splicing analysis) (Figure 1C). Normalization of RNA-seq expression was performed using the FPKM mapped fragments metric (Trapnell et al., 2010). The Perl scripts used to implement these programs are publicly available through the iPlant’s GitHub repository https://github.com/iplant/de-wrapper-scripts. The following parameters were used for each program with minor modifications according to the specifications of the iPlant RNA-seq analysis workflow: TopHat2, –align_reads ‘serial’ –mate-inner-dist ‘50’ –library-type ‘fr-unstranded’ –min-anchor-length ‘8’ –splice-mismatches ‘0’ –min-intron-length ‘70’ –max-intron-length ‘50000’ –min-isoform-fraction ‘0.15’ –max-multihits ‘20’ –min-segment-intron ‘50’ –max-segment-intron ‘500000’ –segment-mismatches ‘2’ –segment-length ‘20’ –num-threads ‘6’ –b2-sensitive–tophat_version ‘2.0.9’ –bowtie_version ‘2.1.0’; Cufflinks2, –junc-alpha ‘0.0010’ –max-bundle-length ‘3500000’ –max-intron-length ‘300000’ –max-mle-iterations ‘5000’ –min-frags-per-transfrag ‘10’ –min-intron-length ‘50’ –min-isoform-fraction ‘0.1’ –num-importance-samples ‘1000’ –num-threads ‘4’ –overhang-tolerance ‘8’ –pre-mrna-fraction ‘0.15’ –small-anchor-fraction ‘0.09’ –trim-3-avgcov-thresh ‘10’ –trim-3-dropoff-frac ‘0.1’ –version ‘2.1.1’; Cuffmerge2, -p ‘6’ –min-isoform-fraction ‘0.1’ –version ‘2.1.1’; and Cuffdiff2, –min-alignment-count ‘10’ ‘–compatible-hits-norm’–FDR ‘0.05’ –min-reps-for-js-test ‘1’ –frag-bias-correct–version ‘2.1.1’.
The intermediate and final computational output files from these analyses including the transcript assemblies have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.jc68d. Initial data inspection and comparisons of the Cufflinks assembled transcript qualities between mock, PMV, and PMV+SPMV samples with the reference B. distachyon assemblies was performed using Cuffcompare (Trapnell et al., 2010) on the Galaxy platform (Goecks et al., 2010). The locus-level sensitivity (Sn) and specificity (Sp) scores for the Cufflinks assembled transcripts that matched to the reference annotations were ∼97 and 90%, respectively, attesting to the robustness of the transcript assembly (Supplemental Table 2). Chromosome-level visualization of read densities was performed using Seqmonk tools (Babraham Bioinformatics) and the Integrated Genome Viewer (Broad Institute) (Robinson et al., 2011), according to the recommended program documentation. Sashimi plots were constructed using the MISO tools framework (Katz et al., 2010, 2015). RNA-seq read densities were plotted along the splice junctions to visualize gene structure and read coverage. A split-read cutoff ≥10 was used to indicate evidence for splice junction reads and is displayed by loops.
During the preparation of this article, the Bd21v2.1 genome assembly was released by the DOE-Joint Genome Initiative (JGI). The Bd21 v2.1 annotation consisted of ∼31,694 genes and ∼42,868 transcripts. To determine the AS events in the Bd21v2.1 genome, we performed ASTALAVISTA analysis and found ∼9541 AS events. Overall, the number of gene, transcript, and AS events in the reference Bd21v2.1 genome are comparable to those we discovered here and supports our transcript assembly and AS analysis pipeline. However, transcript assemblies and AS events from this study could vary qualitatively compared with the Bd21v2.1 reference gene annotations, as they include virus stress-associated transcriptional and splicing events. For example, in the Bd21v2.1 genome, SCL33-locus (Bradi1g61330) still contains only two annotated reference transcripts (Bradi1g61330.1 and Bradi1g61330.2) and lacks the intron-retained splice variants we identified here. In order to include additional annotations into the reference genome, we will communicate with the curators of Brachypodium.org and Phytozome (DOE-JGI) for incorporation into the next reference genome revision.
Alternative Splicing Landscapes and Differential Splicing Analysis
To analyze the various types of AS events prevalent in B. distachyon, and other plants, we used the ASTALAVISTA tool (Foissac and Sammeth, 2007). ASTALAVISTA extracts and classifies the different splicing events based on the transcript model and splice junctions. The final output files from ASTALAVISTA analyses have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.jc68d. For differential splicing analysis, we used Cuffdiff2 (Trapnell et al., 2012, 2013), which categorizes individual transcripts based on their transcription start site, thus grouping together all the transcripts with a common pre-mRNA molecule. Cuffdiff then estimates significant differences in expression of each transcript in a group relative to one another using the Jensen-Shannon divergence metric across the different samples. Significantly divergent splicing groups or “splicing overloads” among the samples was further corrected for Type-I error using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995; Trapnell et al., 2012, 2013).
Functional Enrichment, Clustering, and Isoform-Level Differential Expression Analysis
To identify functional terms enriched among the differentially spliced genes, we performed set enrichment analysis using the agriGO resource (Du et al., 2010). The GO enrichment analysis was performed against a precalculated B. distachyon reference genome, employing a χ2 test for significance, followed by FDR correction using the Hochberg method (FDR < 0.05) to enrich for statistically significant functional terms. Hierarchical clustering of the differentially spliced gene clusters encoding protein kinases, NB-LRR proteins, transcription factors, and splicing regulators was performed using the multiexperiment viewer application (Saeed et al., 2003), using the average linkage method and the Pearson correlation metric. The detailed isoform-level expression dynamics are presented in Supplemental Data Set 1. Differential expression of select gene loci (XLOC_007079, XLOC_014533, XLOC_020779, XLOC_025395, and XLOC_026754) and SCL33 (Bradi1g61330) in mock-, PMV-, and PMV+SPMV-infected samples was validated by RT-PCR using the same RNA used for the RNA-seq analysis. Expression of UBIQUITIN18 (UBC18; Bradi4g00660) was used to indicate the amount of cDNA used as template for the RT-PCR. Temporal expression analysis of SCL33 was performed in an independent experiment using mock-, PMV-, and PMV+SPMV-infected plants at 7, 14, and 21 dpi. Cloning and characterization of the different SCL33 splice variants was performed using the pGEM-T Easy Vector System (Promega) and their sequences determined by the chain-termination method on an ABI 3100 automated sequencer. Primers used in this study are listed in Supplemental Table 3.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g55310 (At-SCL33), Bradi1g61330 (Bd-SCL33), Bradi4g00660 (Bd-UBC18), Bradi1g78245, PRJNA259564, SRX746906, SRX747740, and SRX747746.
Supplemental Data
Supplemental Figure 1. Validation of RNA-seq Expression Data.
Supplemental Figure 2. Select Examples of Gene Loci Activated in Virus-Infected B. distachyon.
Supplemental Figure 3. Select Example of a Splice Junctions Identified in the RNA-seq Analysis.
Supplemental Table 1. Cloning and Sequencing of Bd-SCL33 Splice Variants.
Supplemental Table 2. Cuffcompare Summary of Locus-Level Sensitivity and Specificity Match Scores of the Cufflinks Transcript Assemblies to Reference B. distachyon (Bd21 v1.2) Assembly.
Supplemental Table 3. Primers Used in This Study.
Supplemental Data Set 1. Virus-Modulated Alternatively Spliced Gene Clusters.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.jc68d.
Supplemental Data Set 2. TopHat Output.
Supplemental Data Set 3. Cufflinks Output.
Supplemental Data Set 4. Cuffmerge Output.
Supplemental Data Set 5. ASTALAVISTA Output.
Supplementary Material
Acknowledgments
We thank iPlant and Galaxy cyberinfrastructure platforms for the bioinformatics tools and resources. We thank Herman Scholthof, Jesse Pyle, Will Cody, and Regina Mendoza (Texas A&M University) for the critical reading and comments during the preparation of this article. This study was supported by funds from The Norman Hackerman Advanced Research Program (000517-0002-2009) awarded to K.-B.G.S.
AUTHOR CONTRIBUTIONS
K.K.M. and K.-B.G.S. designed the research. K.K.M. performed the experiments and analyzed the data. K.K.M. and K.-B.G.S. wrote the article.
Glossary
- AS
alternative splicing
- SPMV
Panicum mosaic virus satellite virus
- PMV
Panicum mosaic virus
- RNA-seq
RNA sequencing
- dpi
days postinoculation
- FPKM
fragments per kilobase of transcript per million mapped fragments
- IR
intron retention
- ES
exon skipping
- AA
alternate acceptor
- AD
alternate donor
- FDR
false discovery rate
- GO
Gene Ontology
- NB-LRR
nucleotide-binding site leucine-rich repeat
References
- Ali G.S., Reddy A.S.N. (2008). Spatiotemporal organization of pre-mRNA splicing proteins in plants. In Nuclear pre-mRNA Processing in Plants, Reddy A.N. and Golovkin M., eds (Berlin, Heidelberg:Springer; ), pp. 103–118. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Brkljacic J., et al. (2011). Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan P., Chalhoub B., Chochois V., Garvin D.F., Hasterok R., Manzaneda A.J., Mur L.A.J., Pecchioni N., Rasmussen S.K., Vogel J.P., Voxeur A., International Brachypodium Initiative (2014). Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends Plant Sci. 19: 414–418. [DOI] [PubMed] [Google Scholar]
- Chang C.-Y., Lin W.-D., Tu S.-L. (2014). Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165: 826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S.-M., Chang C.-C., Chen H.-L., Li W.-H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58: 424–441. [DOI] [PubMed] [Google Scholar]
- Dillies M.-A., et al. ; French StatOmique Consortium (2013). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 14: 671–683. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Baker B.J. (2000). Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97: 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A.K., Stauffer E., Behr J., Drewe P., Rätsch G., Wachter A. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.-K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood J. (2010). The importance of plant health to food security. Food Security 2: 215–231. [Google Scholar]
- Foissac S., Sammeth M. (2007). ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35: W297–W299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J., Galaxy Team (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2011). The iPlant Collaborative: Cyberinfrastructure for plant biology. Front. Plant Sci. 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins A.A., Moore M.J. (2012). The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 37: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B.E., Hu Q., Babaoglu A.C., Chandra M., Borghi M., Tan X., He L., Winter-Sederoff H., Gassmann W., Veronese P., Heber S. (2013). High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS ONE 8: e74183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, Y., Wang, E.T., Stilterra, J., Schwartz, S., Wong, B., Thorvaldsdóttir, H., Robinson, J.T., Mesirov, J.P., Airoldi, E.M., and Burge, C.B. (2015). Quantitative visualization of alternative isoform expression from RNA-seq data. Bioinformatics pii: btv034. [DOI] [PMC free article] [PubMed]
- Lewis B.P., Green R.E., Brenner S.E. (2003). Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 100: 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Xiao G., Zhu Y.-X. (2014). Single-nucleotide resolution mapping of the Gossypium raimondii transcriptome reveals a new mechanism for alternative splicing of introns. Mol. Plant 7: 829–840. [DOI] [PubMed] [Google Scholar]
- Lyons C.W., Scholthof K.-B.G. (2015). Watching grass grow: The emergence of Brachypodium distachyon as a model for the Poaceae. Archimedes 40:in press. [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2012). Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 160: 1432–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2013). Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25: 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Pyle J.D., Scholthof K.-B.G. (2014). Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveal conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant Microbe Interact. 27: 1277–1290. [DOI] [PubMed] [Google Scholar]
- Marquez Y., Brown J.W.S., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S.N. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49: 1091–1107. [DOI] [PubMed] [Google Scholar]
- Piron F., Nicolaï M., Minoïa S., Piednoir E., Moretti A., Salgues A., Zamir D., Caranta C., Bendahmane A. (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40: D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S.N., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Scholthof K.-B.G. (1999). A synergism induced by satellite panicum mosaic virus. Mol. Plant Microbe Interact. 12: 163–166. [Google Scholar]
- Shen Y., Zhou Z., Wang Z., Li W., Fang C., Wu M., Ma Y., Liu T., Kong L.-A., Peng D.-L., Tian Z. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26: 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J.W.S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher S.R., Zhou W., Leonard A., Wang B.-B., Beatty M., Zastrow-Hayes G., Zhao X., Baumgarten A., Li B. (2014). Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell 26: 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. (2012). Functional Analysis of Three Arabidopsis SR Proteins (SCL33, SC35, SCL30A) in Plant Development and Splicing. PhD dissertation (Fort Collins, CO: Colorado State University). [Google Scholar]
- Thomas J., Palusa S.G., Prasad K.V.S.K., Ali G.S., Surabhi G.-K., Ben-Hur A., Abdel-Ghany S.E., Reddy A.S.N. (2012). Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J. 72: 935–946. [DOI] [PubMed] [Google Scholar]
- Thomas P.D., Kejariwal A., Campbell M.J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J.A., Doremieux O. (2003). PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turina M., Maruoka M., Monis J., Jackson A.O., Scholthof K.-B.G. (1998). Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology 241: 141–155. [DOI] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S.A., Yang C., Goodin M.M. (2006). Global impact: elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 19: 1207–1215. [DOI] [PubMed] [Google Scholar]
- Zhang G., et al. (2010). Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 20: 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-C., Gassmann W. (2003). RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 15: 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.