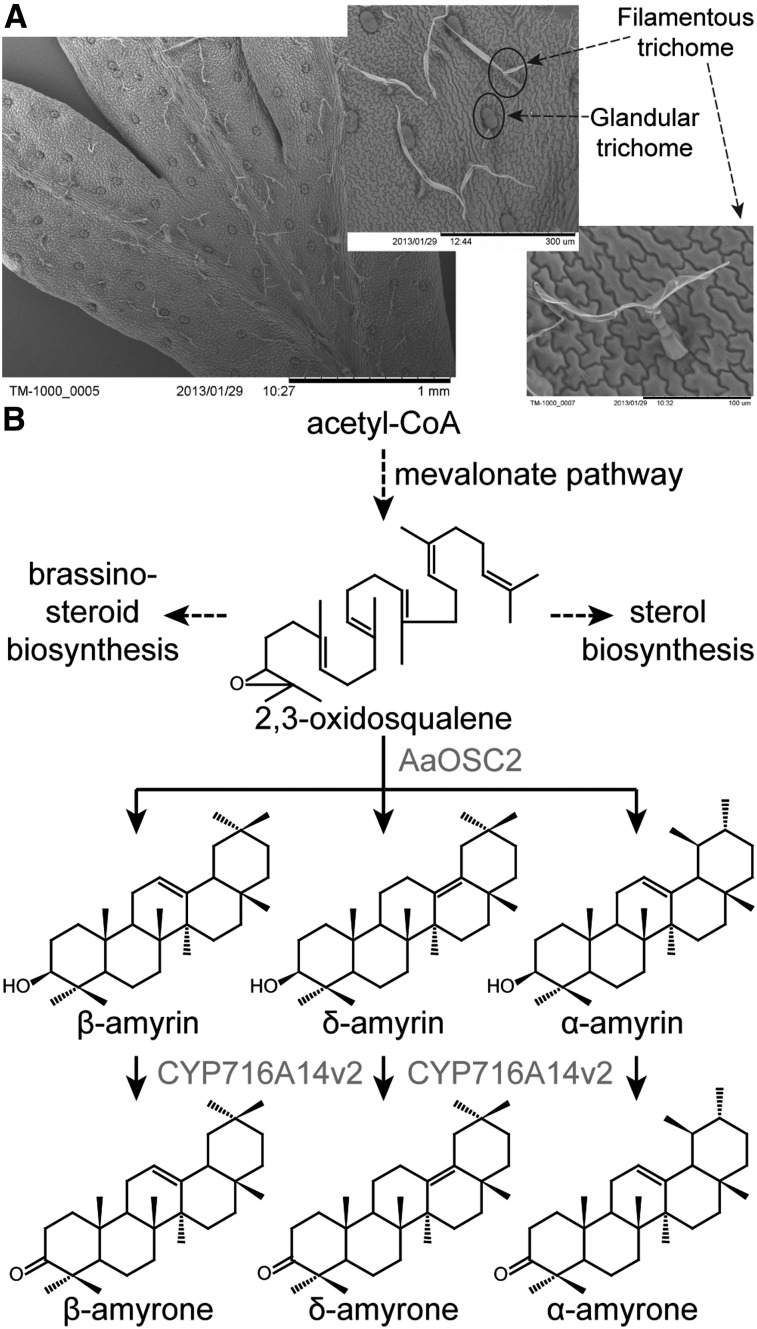
Comparative RNA-seq analysis of glandular and filamentous trichomes from Artemisia annua reveals the biosynthesis pathway to 3-oxo triterpenes that accumulate in the cuticular wax of A. annua’s aerial organs.
Abstract
Artemisia annua is widely studied for its ability to accumulate the antimalarial sesquiterpenoid artemisinin. In addition to producing a variety of sesquiterpenoids, A. annua also accumulates mono-, di-, and triterpenoids, the majority of which are produced in the glandular trichomes. A. annua also has filamentous trichomes on its aerial parts, but little is known of their biosynthesis potential. Here, through a comparative transcriptome analysis between glandular and filamentous trichomes, we identified two genes, OSC2 and CYP716A14v2, encoding enzymes involved in the biosynthesis of specialized triterpenoids in A. annua. By expressing these genes in Saccharomyces cerevisiae and Nicotiana benthamiana, we characterized the catalytic function of these proteins and could reconstitute the specialized triterpenoid spectrum of A. annua in these heterologous hosts. OSC2 is a multifunctional oxidosqualene cyclase that produces α-amyrin, β-amyrin, and δ-amyrin. CYP716A14v2 is a P450 belonging to the functionally diverse CYP716 family and catalyzes the oxidation of pentacyclic triterpenes, leading to triterpenes with a carbonyl group at position C-3, thereby providing an alternative biosynthesis pathway to 3-oxo triterpenes. Together, these enzymes produce specialized triterpenoids that are constituents of the wax layer of the cuticle covering the aerial parts of A. annua and likely function in the protection of the plant against biotic and abiotic stress.
INTRODUCTION
The genus Artemisia (Asteraceae) is large and diverse and consists of ∼500 plant species that are a rich source of small molecules that display a wide range of biological activities, including antimalarial, cytotoxic, antihepatotoxic, antibacterial, antifungal, and antioxidant activities (Bora and Sharma, 2011). Artemisia annua (sweet wormwood) is a well-known species of this genus and accumulates several phytochemicals, including flavonoids, coumarins, monoterpenoids, sesquiterpenoids, diterpenoids, triterpenoids, steroids, lipids, and aliphatic compounds (Bhakuni et al., 2001). A. annua is most extensively studied for its inherent ability to accumulate the antimalarial sesquiterpene lactone artemisinin, which is synthesized and stored in epidermal appendages, called glandular trichomes, on the aerial parts of the plant (Olsson et al., 2009).
The leaves, stems, and flowers of A. annua are covered with two kinds of trichomes, glandular and filamentous (Figure 1A), that can be distinguished based on their morphology. The glandular trichomes are secretory in nature and host biosynthetic routes for the synthesis of various terpenoids and flavonoids (Wang et al., 2009). The majority of the monoterpenoids, sesquiterpenoids, and diterpenoids are thought to be biosynthesized in this trichome type. The filamentous trichomes have not hitherto been investigated for their biosynthetic potential. A recent comparative transcriptome analysis has revealed potentially distinct biosynthetic capacities for the glandular and filamentous trichomes of A. annua (Soetaert et al., 2013). Filamentous trichomes were shown to be enriched in transcripts pertaining to triterpenoid and some specific, nonartemisinin, sesquiterpenoid biosynthesis genes. Glandular trichomes were enriched in artemisinin biosynthesis genes, in agreement with the current models, as well as in mono- and diterpenoid genes.
Figure 1.
A. annua Triterpenoid Biosynthesis.
(A) Overview of glandular and filamentous trichomes on the A. annua leaf surface.
(B) OSC2 is an OSC that cyclizes 2,3-oxidosqualene to three products, which are further oxidized by the P450 CYP716A14v2.
Triterpenoids are ubiquitous isoprenoids produced by all eukaryotes. They constitute a structurally diverse class of mostly polycyclic compounds, with different biological functions ranging from primary to secondary metabolism. The vast majority of triterpenoids is derived from the cyclization of 2,3-oxidosqualene, which in turn is derived from isopentenyl pyrophosphate generated through the mevalonate pathway (Figure 1B). The 2,3-oxidosqualene can be cyclized to many different triterpene skeletons, one of which is cycloartenol, the precursor for sterol biosynthesis in plants. Some of the generated phytosterols, like cholesterol, campesterol, and sitosterol, are the precursors for the synthesis of brassinosteroid hormones and steroidal saponins. Other cyclized triterpene skeletons are precursors for the synthesis of specialized triterpenoids and saponins, which are biologically active compounds that help plants survive in their environment and often also display diverse pharmacological activities beneficial to humankind (Moses et al., 2013; Thimmappa et al., 2014).
In addition to functioning as membrane sterols, hormones, and bioactive molecules, the triterpenoids also play a significant role as components of the cuticle, which is the outermost surface of higher plants. The cuticle covers the aerial surfaces of plant organs by forming a hydrophobic layer that acts as a physical barrier to protect the plant from biotic and abiotic stresses. The cuticle is composed of two main components, cutin and wax (Jetter et al., 2000). Triterpenoids are the biologically active constituents of the wax layer (Buschhaus and Jetter, 2011; Szakiel et al., 2012). Pentacyclic oleanane-, ursane-, and lupane-type triterpenes and triterpenoids have been identified in the wax layers of the cuticle covering leaves, stems, flowers, and fruits of multiple plants, such as Madagascar periwinkle (Catharanthus roseus; Murata et al., 2008), tomato (Solanum lycopersicum; Wang et al., 2011), apple (Malus domestica; Bringe et al., 2006), and heather (Calluna vulgaris; Szakiel et al., 2013).
Here, we identified and deciphered the function of two enzymes, OSC2 and CYP716A14v2, that are overrepresented in filamentous trichomes and involved in specialized triterpenoid biosynthesis in A. annua. With these two enzymes we could reconstitute the triterpenoid spectrum of the cuticle of the aerial organs from this medicinal plant in the conventional yeast Saccharomyces cerevisiae and the host plant Nicotiana benthamiana.
RESULTS
Identification of OSC2
A comparative transcriptome analysis of the glandular and filamentous trichomes from the flower buds of the A. annua Anamed A3 cultivar revealed differential expression of terpene biosynthesis genes with triterpene-related contigs being overrepresented in the filamentous trichomes (Soetaert et al., 2013). Comp7642 is one such higher expressed contig that corresponds to an uncharacterized oxidosqualene cyclase (OSC) that we annotated as OSC2. Compared with the glandular trichomes, OSC2 expression levels were significantly higher (41-fold, P = 0.000109) in the filamentous trichomes (Supplemental Table 1).
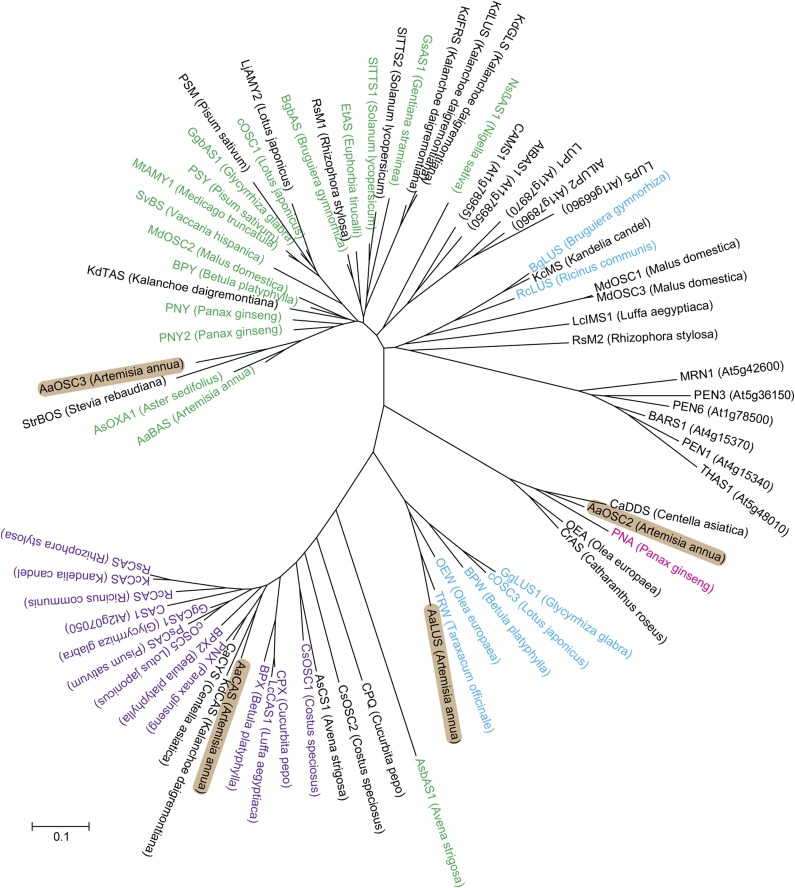
The full-length coding region of OSC2 corresponds to an open reading frame of 760 amino acids and was isolated from cDNA prepared from leaves of the A. annua Anamed A3 cultivar. OSC2 shows 83% sequence similarity to the multifunctional Centella asiatica dammarenediol synthase (DDS; Kim et al., 2009) and 81% sequence similarity to the Panax ginseng dammarenediol synthase (Tansakul et al., 2006). A phylogenetic analysis with characterized OSCs involved in triterpene biosynthesis grouped OSC2 with the multifunctional OSCs that cyclize 2,3-oxidosqualene to more than one product (Figure 2; Supplemental Data Set 1), such as CaDDS (Kim et al., 2009) and CrAS (Huang et al., 2012; Yu et al., 2013).
Figure 2.
Phylogenetic Analysis of A. annua Triterpenoid Biosynthesis Enzymes.
A. annua OSC2, CAS, LUS, and OSC3 are highlighted in brown. Specific β-amyrin synthases are in green, lupeol synthases in blue, dammarenediol synthase in pink, and cycloartenol synthases in purple. All mixed OSCs that cyclize 2,3-oxidosqualene to multiple products are in black. The amino acid sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The scale bar represents the number of amino acid substitutions per site.
OSC2 showed 61% sequence similarity to the previously characterized A. annua β-amyrin synthase (BAS) that cyclizes 2,3-oxidosqualene to β-amyrin (Kirby et al., 2008) and that groups together with other OSCs that only generate one product from the cyclization reaction (Figure 2). In our trichome-specific transcriptome data set reported by Soetaert et al. (2013), A. annua BAS was represented by the contig comp33386, which was expressed 72-fold higher (though not significantly, P = 0.192) in the filamentous trichomes compared with the glandular trichomes.
In Vivo Enzymatic Activity of OSC2
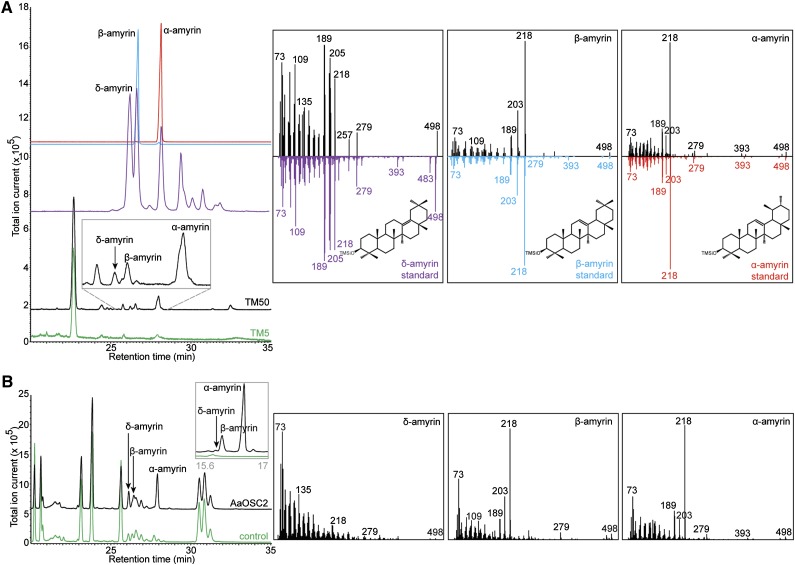
To determine the cyclization specificity of OSC2, we expressed its gene in a sterol engineered S. cerevisiae strain TM1 (Moses et al., 2014). Strain TM50, expressing OSC2 from a galactose-inducible promoter, was generated from strain TM1 and compared with a control strain (TM5). The genotypes of all yeast strains generated and used in this study are provided in Supplemental Table 2. Strains TM50 and TM5 were cultured in medium containing galactose and methyl-β-cyclodextrin (MβCD), which sequesters triterpenes from yeast cells and transports them to the growth medium (Moses et al., 2014). Organic extracts of their spent media were compared by gas chromatography-mass spectrometry (GC-MS) analysis.
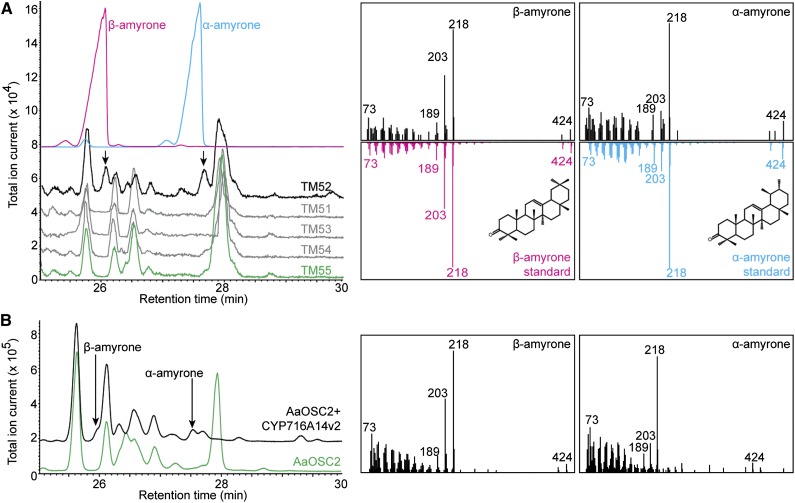
Three distinct peaks were unique in the GC-MS chromatogram of strain TM50 (Figure 3A). The GC retention time and electron ionization-mass spectrum (EI-MS) of two of the three peaks were identical to authentic α-amyrin and β-amyrin standards, and the third peak matched δ-amyrin prepared from tomato wax extracts, which accumulate high amounts of this compound (Wang et al., 2011) (Figure 3A). Additionally, the relative intensities of the three peaks in the GC-MS chromatogram of strain TM50 suggest a higher accumulation of α-amyrin, compared with β-amyrin and δ-amyrin. Relative quantification of the peak intensities showed accumulation of α-amyrin:β-amyrin:δ-amyrin to be in the ratio 5.5:2.7:1.8. Hence, OSC2 is a multifunctional OSC that cyclizes 2,3-oxidosqualene to α-amyrin, β-amyrin, and δ-amyrin (Figure 1B), with a preference for the first product when exogenously expressed in S. cerevisiae.
Figure 3.
OSC2 Is a Multifunctional Oxidosqualene Cyclase.
(A) Overlay of GC-MS chromatograms showing the cyclization of 2,3-oxidosqualene to δ-amyrin, β-amyrin, and α-amyrin by OSC2 in the sterol engineered yeast strain TM50 (black) but not the control yeast strain TM5 (green). The inset shows the enlargement of the chromatogram between 25.4 and 28.4 min for strain TM50. The authentic standards of δ-amyrin, β-amyrin, and α-amyrin are shown in purple, blue, and red, respectively.
(B) Overlay of GC-MS chromatograms of N. benthamiana leaves transiently expressing OSC2 (black) and untransformed control leaves (green). The inset (gray) shows the overlay of GC-MS chromatograms for the same samples reanalyzed with the DB-1 ht capillary column to separate δ-amyrin from the coeluting peak. Panels on the right show the trimethylsilylated structure and EI-MS spectra of δ-amyrin, β-amyrin, and α-amyrin, eluting at retention times of 26.2, 26.5, and 27.9 min, respectively.
To verify the enzymatic activity of OSC2 observed in S. cerevisiae, we transiently expressed A. annua OSC2 from a binary pEAQ vector (Sainsbury et al., 2009) in N. benthamiana leaves using Agrobacterium tumefaciens-mediated infiltration. GC-MS chromatograms of organic extracts from leaves 3 d postinfiltration were compared with noninfiltrated leaves from the same plant. Three unique peaks corresponding to α-amyrin, β-amyrin, and δ-amyrin were observed in the tobacco leaves expressing OSC2, but not in the control leaves (Figure 3B), confirming the multifunctionality of this enzyme. Like in yeast, N. benthamiana OSC2 also exhibited the preferential cyclization of 2,3-oxidosqualene to α-amyrin. For relative quantification, the N. benthamiana samples were reanalyzed to better separate δ-amyrin from the coeluting peak, and the accumulation of α-amyrin:β-amyrin:δ-amyrin was found to be in the ratio 8.1:1.8:0.1 (Figure 3B, inset).
A. annua Plant Triterpene Profile
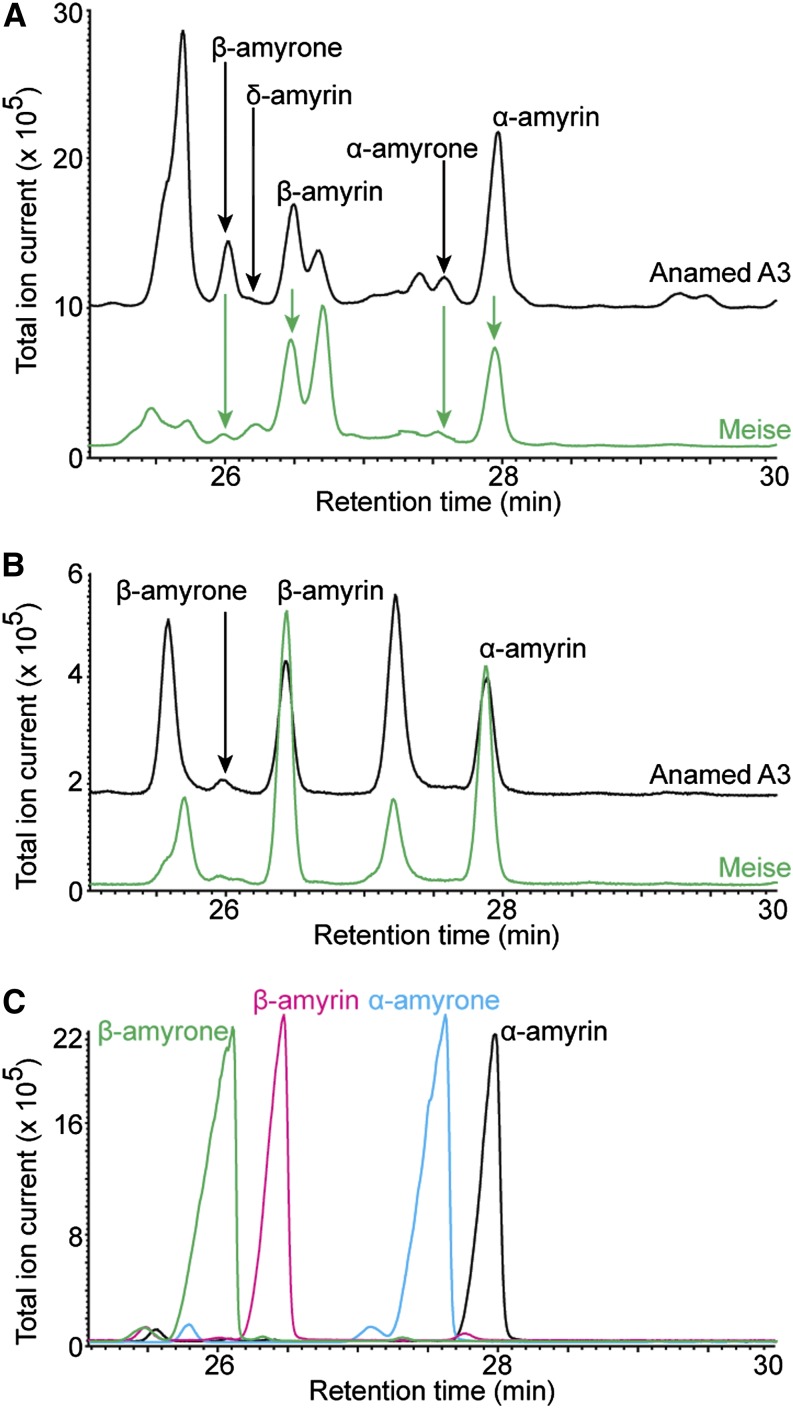
A. annua OSC2 is a multifunctional OSC when expressed in heterologous yeast and plant hosts. To validate the physiological relevance of this finding, we determined the triterpene content of greenhouse-grown A. annua plants. Pentacyclic triterpenoids, mainly derived from α-amyrin, β-amyrin, and lupeol, often occur in the leaf, flower, and fruit cuticular wax layers of many plants (Bringe et al., 2006; Murata et al., 2008; Wang et al., 2011; Szakiel et al., 2013). Therefore, we analyzed the triterpene content of the cuticular wax layer from flower buds and leaves of two A. annua cultivars: Anamed A3, the cultivar from which OSC2 was isolated, and Meise, a low artemisinin-producing cultivar that we previously used to study phytohormonal regulation of artemisinin biosynthesis (Maes et al., 2011).
The flower buds and leaves were harvested separately and dipped in hexane to extract the hydrophobic metabolites from the cuticular wax layer. The extracts were analyzed by GC-MS and the organ-specific specialized triterpene profiles of the two cultivars were compared. From the triterpene profiles, the following general observations could be drawn. First, the triterpene profiles of the wax layer of flower buds and leaves were different. The flower buds demonstrated more diversity in their accumulating triterpenes than the leaves (Figures 4A and 4B). Second, α-amyrin and β-amyrin accumulated in abundance in both the Anamed A3 and Meise cultivars. Two distinct peaks corresponding to these compounds were observed in the GC-MS chromatograms of both flower buds (Figure 4A; Supplemental Figures 1A and 1B) and leaves (Figure 4B; Supplemental Figures 1C and 1D). Third, δ-amyrin accumulated only in trace amounts. A trace peak was observed in the GC-MS chromatogram of flower buds from the Anamed A3 cultivar (Figure 4A; Supplemental Figure 1A), whereas in that of flower buds from the Meise cultivar and of leaves of both cultivars no distinct peak could be visualized. However, trace amounts of the most abundant m/z representative of δ-amyrin could be detected in the EI-MS data of the Meise flower buds and leaves from both cultivars (Supplemental Figures 1A to 1D). Fourth, the relative accumulation of α-amyrin, β-amyrin, and δ-amyrin varied between the cultivars and organ types. Both cultivars showed very low accumulation of δ-amyrin, irrespective of the organ type, whereas major differences were observed in the accumulation of α-amyrin and β-amyrin. The Meise cultivar showed accumulation of similar amounts of α-amyrin and β-amyrin in flower buds and leaves, while in the Anamed A3 cultivar the flower buds showed higher accumulation of α-amyrin, contrary to the leaves, in which β-amyrin accumulated the most (Figures 4A and 4B). Detection of all three triterpenes in the wax layers of different aerial organs of different A. annua cultivars confirms the physiological relevance of our finding and supports the multifunctional cyclization activity of OSC2.
Figure 4.
Triterpenoid Profile of A. annua Anamed A3 and Meise Cultivars.
(A) and (B) Overlay of GC-MS chromatograms of extractions from flower buds (A) and leaves (B) of Anamed A3 (black) and Meise (green) cultivars.
(C) Overlay of GC-MS chromatograms of authentic standards of β-amyrone (green), β-amyrin (pink), α-amyrone (blue), and α-amyrin (black). The abundant triterpenes and triterpenoids are indicated on the chromatograms, and their EI-MS spectra are given in Supplemental Figure 1. The GC retention times of β-amyrone, δ-amyrin, β-amyrin, α-amyrone, and α-amyrin are 26, 26.2, 26.5, 27.5, and 27.9 min, respectively.
Finally, we also observed peaks derived from α-amyrin and β-amyrin, with a GC retention time and EI-MS corresponding to the triterpenoids α-amyrone and β-amyrone (Figure 4; Supplemental Figure 1), in which the C-3 hydroxyl group of the ursane and oleanane backbones of α-amyrin and β-amyrin, respectively, are oxidized to a carbonyl group (Figure 1B). Flower buds and leaves of the two cultivars showed variable relative triterpenoid accumulation. Although the triterpenoids were present in low amounts and could be detected in both organ types, they were more readily detected in the flower buds than in the leaves of the same cultivar (Figures 4A and 4B).
Mining for A. annua P450s
In ursane and oleanane triterpenoid biosynthesis, oxidation reactions of the triterpene backbone are most often catalyzed by cytochrome P450-dependent monooxygenases (P450s) and include single- or multi-step reactions, such as the addition of a hydroxyl group that can be oxidized further to a carbonyl or carboxyl group. The accumulation of oxidized derivatives of α-amyrin and β-amyrin (α-amyrone and β-amyrone, respectively) suggests the existence of P450s in A. annua that are capable of catalyzing these oxidation reactions. However, to date, no P450 involved in ursane or oleanane triterpenoid biosynthesis has been isolated from A. annua; furthermore, no P450 with an oxidase activity for carbon 3 of ursane or oleanane triterpenes has been identified from any plant species. Therefore, we screened the transcriptome data of the A. annua Anamed A3 cultivar to identify candidate P450s that might be involved in specialized triterpenoid biosynthesis.
Since the contigs corresponding to triterpene biosynthesis genes, including OSC2, were expressed at higher levels in filamentous trichomes, we mined for P450 contigs that were significantly overrepresented in this trichome type. Five P450s represented by comp8932, comp9232, comp10696, comp22290, and comp27086 were identified. Only for comp8932 and comp9232 could full-length gene sequences be retrieved from our transcriptome data set, and they were annotated as CYP704A87 and CYP81B58, respectively, by standard P450 nomenclature. CYP704A87 is only expressed in filamentous trichomes, whereas CYP81B58 is 105-fold more abundant in filamentous trichomes than in glandular trichomes (Supplemental Table 1).
In addition to this expression pattern-based gene mining, we also screened for P450s that belong to the CYP716 family of proteins. The CYP716 family represents the most common clade of P450s that have been characterized to be involved in specialized triterpenoid or sapogenin biosynthesis. To date, nine CYP716 family proteins with oxidase activity on α-amyrin, β-amyrin, lupeol, or dammarenediol have been identified. CYP716A12 from Medicago truncatula (Carelli et al., 2011), CYP716A15 and CYP716A17 from Vitis vinifera (Fukushima et al., 2011), CYP716AL1 from C. roseus (Huang et al., 2012), CYP716A52v2 from P. ginseng (Han et al., 2013), and CYP716A75 from Maesa lanceolata (Moses et al., 2015) are C-28 oxidases of α-amyrin, β-amyrin, and lupeol. CYP716Y1 from Bupleurum falcatum is a C-16α oxidase of α-amyrin and β-amyrin (Moses et al., 2014), and CYP716A47 and CYP716A53v2 from P. ginseng are C-12 and C-6 oxidases of dammaranes, respectively (Han et al., 2011, 2012). The CYP716A12 protein was used to screen for candidate CYP716 family P450s in the A. annua transcriptome data set. Two hits with 54% and 63% sequence similarity to CYP716A12 were identified and annotated as CYP716D22 and CYP716A14v2, respectively. Compared with the glandular trichomes, the contigs corresponding to CYP716D22 and CYP716A14v2 were 7- and 19-fold more abundantly expressed, respectively, in the filamentous trichomes (Supplemental Table 1).
The full-length coding sequences of all candidate P450s were cloned from leaves of the A. annua Anamed A3 cultivar, and their encoding amino acid sequences were used for a phylogenetic analysis with all known and functionally characterized P450s involved in specialized triterpenoid (saponin) biosynthesis. CYP716D22 and CYP716A14v2 clearly grouped with the other CYP716 family proteins, whereas CYP81B58 grouped with other members of the CYP71 clan, and CYP704A87 (CYP86 clan) branched as a single clade separate from the CYP71 and CYP72 clans (Supplemental Figure 2 and Supplemental Data Set 2).
Expression and Regulation of Candidate P450s
Secondary metabolite biosynthesis in plants is tightly controlled. Various mechanisms have been recognized to be involved in its regulation, including the transcriptional activation of entire pathways by a common trigger, which allows the synchronized action of the pathway proteins (Maes et al., 2011; De Geyter et al., 2012; Moses et al., 2013). Therefore, coexpression and coregulation of candidate genes can be used as an indication of their possible involvement in a common biosynthetic process.
We first determined the expression of our candidate P450s, OSC2, and BAS in leaves and flower buds of both the A. annua cultivars, Anamed A3 and Meise. All four candidate P450s (CYP716D22, CYP716A14v2, CYP704A87, and CYP81B58) and both OSC genes were expressed in the flower buds and leaves of both cultivars (Supplemental Figure 3). Furthermore, no significant differences in expression levels were observed between the A. annua cultivars and organ types, although large variations between the biological samples were noted (Supplemental Figure 3).
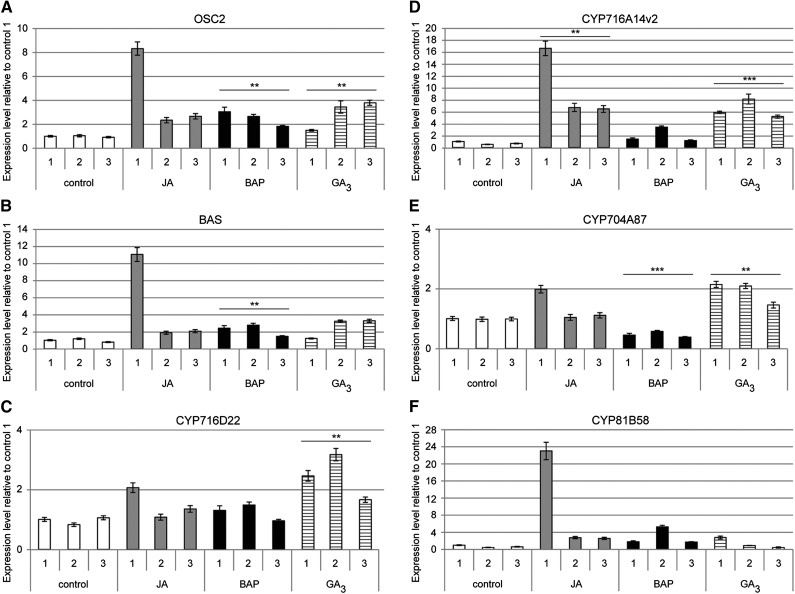
Next, we determined whether expression of the candidate P450s was similarly affected by phytohormonal cues as that of the OSCs. This was performed by analyzing their expression profile in a sample set generated in previous research (Maes et al., 2011), in which seedlings of the A. annua Meise cultivar were treated with different hormones, namely, jasmonate, the synthetic cytokinin 6-benzylaminopurine (BAP), and gibberellic acid (GA3). Two general observations could be drawn from this analysis (Figure 5). First, both OSCs showed similar responses to the hormone treatments. All three hormones induced OSC expression by 2- to 3-fold for both OSC2 and BAS (Figures 5A and 5B). Second, the P450 genes showed different responses to the different phytohormone treatments. CYP716D22 was significantly induced by GA3 only (∼2.5-fold), whereas CYP716A14v2 was significantly induced by both jasmonate and GA3 (over 5-fold) and also showed induction following BAP treatment (∼2-fold). CYP704A87 was significantly induced by GA3 (∼2-fold) and repressed by BAP (∼2-fold). The expression of CYP81B58 was unfortunately difficult to assess in this sample set (Figure 5).
Figure 5.
Regulation of Triterpene Biosynthesis Genes and Candidate P450s upon Phytohormone Treatment in Leaves of the A. annua Meise Cultivar.
The expression of OSC2 (A), BAS (B), CYP716D22 (C), CYP716A14v2 (D), CYP704A87 (E), and CYP81B58 (F) was verified for three biological samples by qPCR in leaves treated or not with jasmonate (JA), BAP, or GA3. Error bars are se of mean for n = 3. Statistical significance was determined by Student’s t test (**P ≤ 0.01; ***P ≤ 0.001).
Taken together, this expression analysis suggests that of the four candidate P450 genes, at least the expression of CYP716A14v2 is similarly modulated as that of OSC2 and BAS; therefore, it might correspond to the P450 involved in specialized triterpenoid biosynthesis.
Functional Characterization of Candidate P450s
To verify the outcome of the expression analysis and to determine which of the candidate P450s would be functional and oxidize β-amyrin and/or α-amyrin, we expressed the candidate genes together with OSC2 in yeast cells and tobacco leaves.
We generated five yeast strains TM51-TM55 (Supplemental Table 2) starting from the yeast strain TM50 expressing OSC2 and tHMG1. Strains TM51, TM52, TM53, and TM54 express the candidate P450s CYP716D22, CYP716A14v2, CYP704A87, and CYP81B58, respectively, together with a P450 reductase from Arabidopsis thaliana (At-ATR1), whereas strain TM55 is a control strain only expressing At-ATR1. Strains TM51-TM55 were cultured in the presence of MβCD, and extracts of the spent medium were compared by GC-MS analysis. Peaks corresponding to β-amyrone and α-amyrone were observed in the GC-MS chromatogram of strain TM52, but not of the control strain TM55 or any of the other yeast strains. The GC retention time and EI-MS of the oxidized products in strain TM52, expressing CYP716A14v2, matched authentic standards of β-amyrone and α-amyrone (Figure 6A).
Figure 6.
CYP716A14v2 Oxidizes Carbon 3 of Ursane and Oleanane Triterpenes.
(A) Overlay of GC-MS chromatograms showing the oxidation of β-amyrin and α-amyrin to β-amyrone and α-amyrone, respectively, by CYP716A14v2 in the yeast strain TM52 (black) but not in the yeast strains TM51, TM53, and TM54 (all in gray) and the control yeast strain TM55 (green). The authentic standards of β-amyrone and α-amyrone are shown in pink and blue, respectively. Their structures are depicted in the insets.
(B) Overlay of GC-MS chromatograms of N. benthamiana leaves transiently expressing OSC2 and CYP716A14v2 (black) or only OSC2 (green), showing the oxidation products of CYP716A14v2. Right panels show the EI-MS spectra of β-amyrone and α-amyrone eluting at 26.1 and 27.7 min, respectively.
To confirm the functional activity of CYP716A14v2 observed in yeast and to verify if any other candidate P450 might be functional in a plant host, we transiently expressed the candidate P450s together with OSC2 in tobacco leaves, analyzed extracts by GC-MS, and compared them to an extract from a leaf infiltrated only with OSC2. As observed in yeast, only leaves transiently expressing CYP716A14v2 together with OSC2 showed the presence of β-amyrone and α-amyrone (Figure 6B).
From these observations, we conclude that CYP716A14v2 is a functional P450 that oxidizes the hydroxyl group at carbon 3 of oleanane and ursane triterpenes to a carbonyl group (Figure 1B). Furthermore, the fact that only CYP716A14v2 was identified as a functional triterpene oxidase corroborates the expression analysis (Figure 5) and supports the concept of tight orchestration of triterpenoid biosynthesis in A. annua aerial organs.
Multifunctionality of CYP716A14v2
To explore the multifunctionality of CYP716A14v2, we expressed its gene in yeast strain TM6 (Moses et al., 2014), which produces the pentacyclic triterpenes lupeol and lupanediol. Yeast strain TM56 (Supplemental Table 2) was derived from strain TM6 by transforming the latter with CYP716A14v2 and At-ATR1. After culturing in the presence of MβCD, the extract from the spent medium of strain TM56 was compared with that of the control strain TM29 that was also derived from strain TM6 and expresses only At-ATR1. A peak coeluting with another peak and having an EI-MS corresponding to lupenone was observed at 28 min in the GC-MS chromatogram of strain TM56, but not of the control strain TM29 (Supplemental Figure 4A). The GC retention time and EI-MS of this peak were identical to those of an authentic lupenone standard (Supplemental Figure 4B), confirming the C-3 oxidation of the hydroxyl group to the carbonyl group by CYP716A14v2. In addition, two other peaks unique to strain TM56 were observed (Supplemental Figure 4A) that could not be identified because of their low abundance and consequently unclear EI-MS fragmentation pattern. Nonetheless, the detection of these new peaks displays the versatility of CYP716A14v2 to oxidize different triterpene backbones.
Silencing of OSC2 and CYP716A14v2 in A. annua
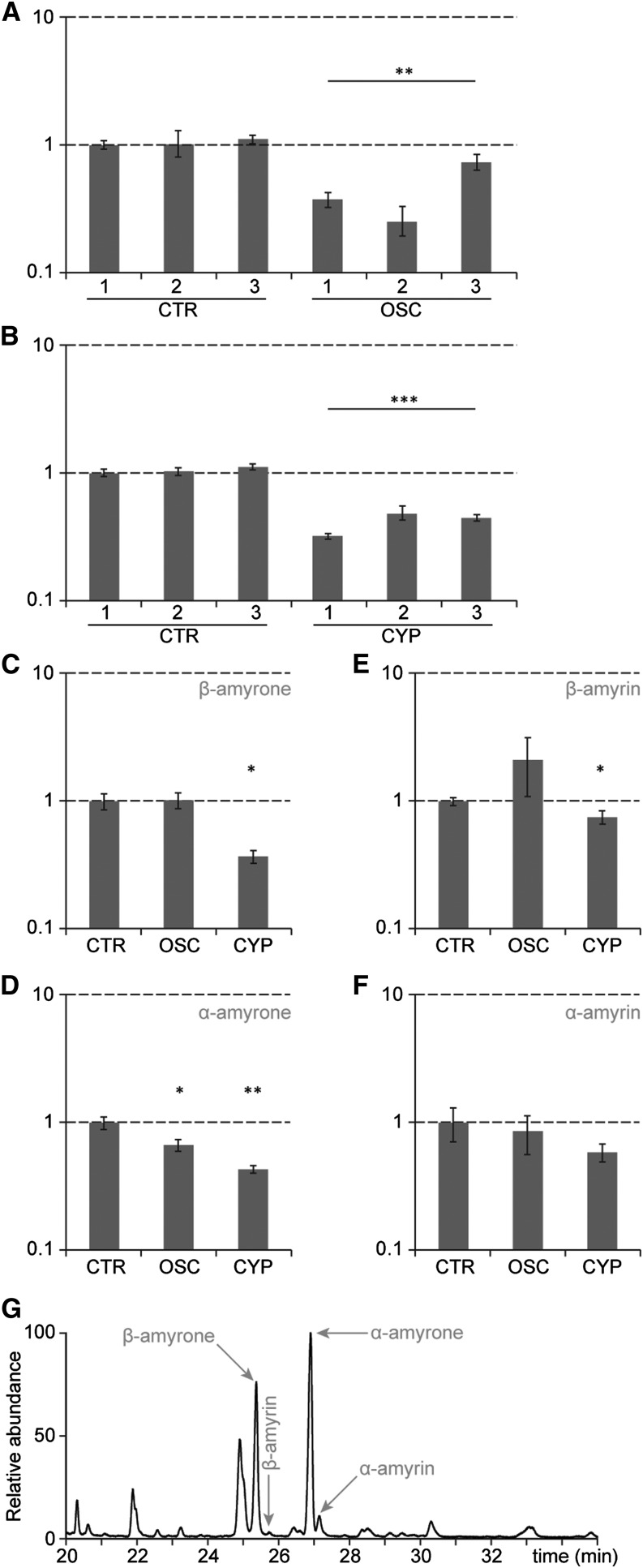
To assess the in planta role of OSC2 and CYP716A14v2, we generated transgenic A. annua plants in which the expression of OSC2 or CYP716A14v2 was silenced by means of hairpin RNA interference constructs. Gene silencing was confirmed by quantitative PCR (qPCR) (Figures 7A and 7B), and three independent transgenic plants were analyzed by GC-MS to quantify the accumulation of β-amyrin, α-amyrin, β-amyrone, and α-amyrone, the major products of the enzymatic reactions of OSC2 and CYP716A14v2. Compared with control plants, the β-amyrone levels were unaltered in OSC2-silenced plants but significantly decreased in CYP716A14v2-silenced plants (Figure 7C). Also, the levels of α-amyrone were significantly decreased in CYP716A14v2-silenced plants (Figure 7D), confirming the role of CYP716A14v2 in the biosynthesis of these A. annua triterpenoids.
Figure 7.
Silencing of OSC2 and CYP716A14v2 in Transgenic A. annua Plants.
(A) and (B) qPCR analysis of transgenic A. annua plants revealed silencing of OSC2 (OSC) (A) and CYP716A14v2 (CYP) (B). Control plants (CTR) were transgenic plants transformed with a nonfunctional β-glucuronidase construct. The y axis is the expression ratio relative to the normalized transcript levels of CTR line 1 in log scale.
(C) to (F) GC-MS analysis revealed altered metabolite levels in transgenic A. annua plants. The amounts of β-amyrone (C), α-amyrone (D), β-amyrin (E), and α-amyrin (F) in OSC2 (OSC) and CYP716A14v2 (CYP) silenced lines relative to the amounts in the control (CTR) lines are plotted. Error bars are se of mean for n = 3. Statistical significance was determined by Student’s t test (*P ≤ 0.1; **P ≤ 0.01; ***P ≤ 0.001).
(G) GC chromatogram of the A. annua cultivar used for silencing, showing it mainly accumulates β-amyrone and α-amyrone and only trace amounts of β-amyrin and α-amyrin.
In the OSC2-silenced plants, the α-amyrone levels were significantly reduced (Figure 7D), pointing toward a role of OSC2 in the synthesis of A. annua ursanes. The levels of β-amyrone were unchanged, possibly due to the redundant activity of the inherent β-amyrin synthase BAS (Kirby et al., 2008) and, hence, silencing of OSC2 alone, which preferentially produces α-amyrin in yeast and tobacco, is not sufficient to decrease the oleanane levels in planta.
The effect of OSC2-silencing on the levels of the amyrins was less clear (Figures 7E and 7F). In the A. annua cultivar ‘Chongqing’ used for silencing (Zhang et al., 2009), over 90% of the accumulating triterpenoids correspond to the amyrones, whereas the amyrins accumulate only in trace amounts (Figure 7G). Likely, CYP716A14v2 readily converts the amyrins to the amyrones, thereby maintaining very low levels of amyrins in the plant and a further reduction in their amounts is difficult to detect, and as such the effects of silencing of OSC2 are prominently observed in the levels of the amyrones. The levels of δ-amyrin and δ-amyrone were not assessed, as these metabolites only accumulated in trace amounts.
Additional A. annua OSCs
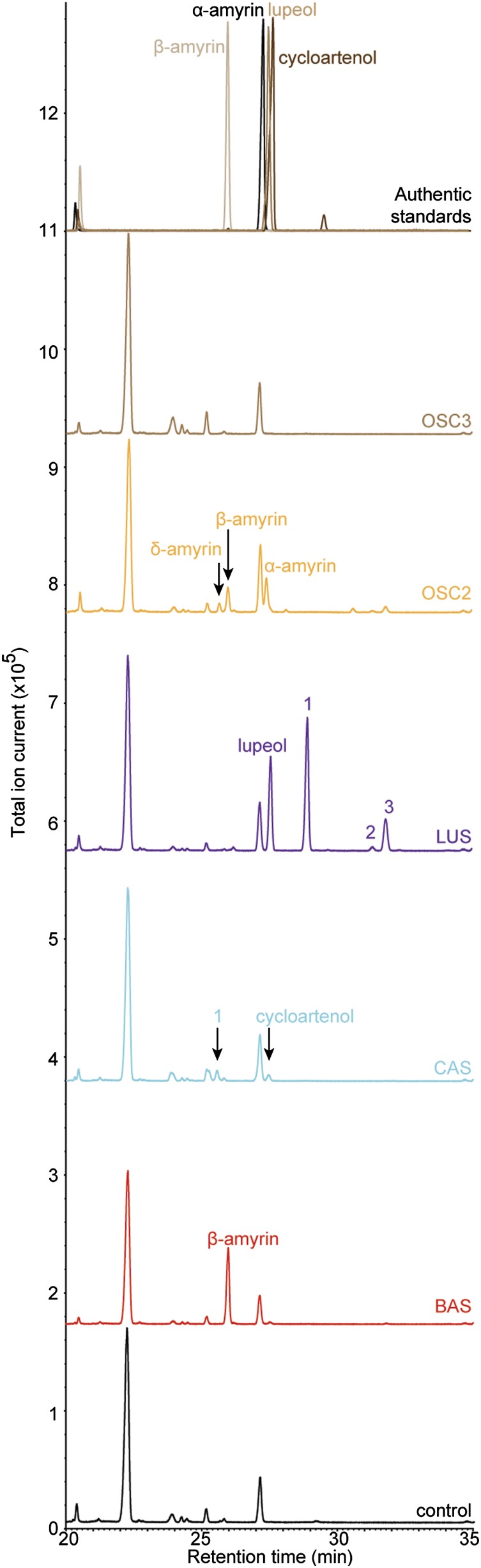
To investigate whether, in addition to OSC2 and the already characterized BAS, other OSCs might contribute to the triterpenoid compendium of A. annua, we screened publicly available Artemisia RNA-seq data and identified three additional full-length OSC sequences. Based on the clustering of their translated protein sequences in a maximum likelihood phylogenetic analysis (Figure 2), the first and second additional OSCs were named CAS and LUS, for cycloartenol synthase and lupeol synthase, respectively. The third OSC showed 80% nucleotide and protein sequence similarity to the previously characterized BAS and was named OSC3.
To assess the function of these additional OSCs, yeast strains TM106-TM110 were generated from strain TM5 by transforming the plasmid pAG423GAL harboring BAS, CAS, LUS, OSC2, or OSC3, respectively. In addition, strain TM111 containing an empty vector control was also generated (Supplemental Table 2). These yeast strains were cultured for 72 h in synthetic medium containing galactose and MβCD as described (Moses et al., 2014), and the resulting spent medium was used for metabolite extraction and GC-MS analysis.
Compared with the control strain TM111, the chromatogram of strain TM106 expressing BAS showed the presence of a single major peak corresponding to β-amyrin (Figure 8), thereby confirming the previously reported β-amyrin synthase activity of this gene (Kirby et al., 2008). Trace amounts of an additional peak were consistently observed at 27.5 min in strain TM106, which was absent in control strain TM111. However, due to its very low abundance, the EI-MS pattern of this peak could not be used for compound identification (Supplemental Figure 5). Therefore, we determined the ratio of accumulation of β-amyrin and this compound in strain TM106 using the area under the curve for each compound. The accumulation of β-amyrin:peak at 27.5 min was found to be 9.98:0.02. In a previous study on OSCs of Arabidopsis, the ratio between accumulating products was used to define enzyme accuracy and multifunctionality. This study suggested that the higher the ratio between the primary product and the second most abundant product or the total products, the greater the accuracy of the enzyme to cyclize 2,3-oxidosqualene to the primary product (Lodeiro et al., 2007). Considering the markedly higher accumulation of β-amyrin in strain TM106 compared with the peak at 27.5 min and the previous characterization of BAS as a single product β-amyrin synthase, it is highly probable that this enzyme cyclizes 2,3-oxidosqualene to only a single compound, β-amyrin.
Figure 8.
Functional Characterization of A. annua OSCs.
Overlay of GC-MS chromatograms showing the cyclization of 2,3-oxidosqualene to various triterpene backbones by the identified OSCs. Supplemental Figure 5 shows the comparison of EI-MS spectra of the compounds produced in yeast with those of authentic standards.
In the chromatogram of strain TM107 expressing CAS, a peak corresponding to cycloartenol was detected (Figure 8), pointing toward cycloartenol synthase activity of this gene. An additional peak that could not be identified and unique to this strain was also observed in strain TM107, suggesting that either CAS is a multifunctional OSC or that a native yeast enzyme further converts the accumulating cycloartenol. Likewise, in the chromatogram of strain TM108 expressing LUS, not only lupeol, but several additional peaks were uniquely present (Figure 8). Like strain TM50, strain TM109 expressing OSC2 accumulated δ-, β-, and α-amyrin. In the chromatogram of strain TM110 expressing OSC3, no unique peaks were identified (Figure 8). Hence, despite the high sequence similarity between OSC3 and BAS, the function of the former remains unknown.
As α-amyrone is the major triterpenoid present in the wax of A. annua, an OSC leading to the ursane α-amyrin is needed for the cyclization of 2,3-oxidosqualene. In spite of thorough searches in various A. annua RNA-seq data accessions, no additional OSCs that catalyze the formation of α-amyrin from 2,3-oxidosqualene were discovered. Hence, OSC2 remains the only enzyme that can catalyze the formation of α-amyrin in A. annua.
DISCUSSION
Triterpenoids are ubiquitous in eukaryotes as primary sterol metabolites. However, higher land plants also synthesize and accumulate specialized triterpenes, triterpenoids, and saponins as defense molecules against biotic and abiotic stresses. Here, we profiled the secondary triterpenoid spectrum from the medicinal plant A. annua and identified the two genes involved in their biosynthesis, the multifunctional amyrin synthase OSC2 and the P450 oxidase CYP716A14v2.
Occurrence of Triterpenoids in the Cuticle of A. annua Organs
The outermost surfaces of aerial parts of terrestrial higher plants are covered with a hydrophobic, waxy layer called the cuticle. The cuticle performs multiple functions in protecting the plant from biotic and abiotic stress. In addition to being the first line of defense against plant pathogens, the cuticle acts as a physical barrier to prevent mechanical damage, infiltration of xenobiotics and harmful irradiance, dehydration of the plant from nonstomatal water loss, and loss of organic and inorganic compounds by leaching (Szakiel et al., 2012). The cuticle is composed of the fatty acid-derived structural matrix cutin and wax. The wax is both intracuticular, embedded in the cutin, and epicuticular, forming a continuous layer on the top (Jetter et al., 2000). Triterpenes and triterpenoids form components of the wax layer in many plants and are located preferentially or entirely in the intracuticular layer (Buschhaus and Jetter, 2011).
The extraction of the triterpenes, α- and β-amyrin, and the triterpenoids, α- and β-amyrone, from the leaves and flower buds of A. annua cultivars by dipping in an organic solvent suggests the accumulation of these compounds in the cuticular wax layer of the plant. The differences in the relative triterpene and triterpenoid levels between the cuticular wax layer of leaves and flower buds points to their differential accumulation in these organs and suggests a possible variable role of these pentacyclic compounds in defense or protection in different plant organs. Conversely, the similarity between the composition of the cuticular wax layer in the Anamed A3 and Meise cultivars indicates the general nature of this triterpenoid composition in A. annua plants and underscores their potential utility.
Terpenoid Biosynthesis Is Differentially Regulated in Trichomes of A. annua
The flower, leaf, and stem surfaces of A. annua plants are covered with two main kinds of trichomes, glandular and filamentous. The glandular trichomes in A. annua are composed of five pairs of cells: three pairs of secretory cells, two pairs of which are rich in chloroplasts, and a pair each of stalk cells and basal cells. The cuticle of the six secretory cells forms a subcuticular sac on the top of the trichome and is thought to function as a storage compartment for secondary metabolites (Olsson et al., 2009). The filamentous trichomes, on the other hand, are made up of five cells that are arranged in a T-shape, of which the top is formed by one elongated cell and the remaining four cells form the stalk (Figure 1A) (Soetaert et al., 2013). The filamentous trichomes are assumed to function as physical barriers to herbivores and have until now not been explored for their metabolic capacities.
In A. annua, the expression of monoterpenoid, sesquiterpenoid, and flavonoid biosynthesis genes have been localized to the glandular trichomes, making them a rich source of secondary metabolites (Wang et al., 2009). In addition, the glandular trichomes have been proposed to be the primary site for artemisinin biosynthesis and storage in the plant (Duke et al., 1994; Van Nieuwerburgh et al., 2006; Olsson et al., 2009; Wang et al., 2013). In our comparative transcriptome analysis, transcripts corresponding to putative or known monoterpenoid, diterpenoid, and (most) sesquiterpenoid biosynthesis genes were overrepresented in the glandular trichomes, as opposed to the triterpenoid transcripts, which were abundant in the filamentous trichomes (Soetaert et al., 2013). This segregation of the different terpenoid synthesis pathways in two different trichome types suggests the existence of a spatiotemporal regulation for terpenoid biosynthesis in A. annua.
The overrepresentation of monoterpenoid, diterpenoid, and (most) sesquiterpenoid transcripts in the glandular trichomes highlights the significance of synthesizing these metabolites in the secretory cells, so they can be released into the subcuticular sac of the glandular trichomes. Conversely, the higher expression of triterpenoid transcripts in the filamentous trichomes could suggest a possible spatial regulation in A. annua to prevent competition between the secondary sesqui- and triterpenoid biosynthesis pathways for the isopentenyl pyrophosphate precursor source derived from the mevalonate pathway. Alternatively, however, the specific nature and physiological function of the two trichome types might demand different cuticles or wax compositions, empowering rigidity for physical defense in the filamentous trichomes or enabling accumulation of bioactive compounds in the glandular sac, respectively.
In this regard, the activity of CYP716A14v2 is of particular relevance. The general fate of many specialized plant triterpenes like β-amyrin and α-amyrin is to be oxidized multiple times before being glycosylated into the amphipathic molecules called saponins (Moses et al., 2013). Across the plant kingdom, typical saponin glycosylations occur at the C-3 and/or C-28 positions through an O-linkage, implying the significance of the hydroxyl and carboxyl groups at the C-3 and C-28 positions, respectively, in rendering a triterpenoid accessible to UDP-dependent glycosyltransferase reactions (Augustin et al., 2011). CYP716A14v2 oxidizes the C-3 hydroxyl group of triterpenes like β-amyrin and α-amyrin to a carbonyl group and consequently blocks further enzymatic modifications like glycosylation on this position. This may have pronounced effects on the fate and role of the compounds, as it may ensure that they can now accumulate in a hydrophobic environment, such as the cuticle of plant aerial organs like leaves and filamentous trichomes, where they can exert particular protective or physiological functions, instead of in the vacuoles where the amphipathic glycosylated saponins presumably accumulate (Kesselmeier and Urban, 1983; Mylona et al., 2008).
The differential response of triterpenoid and sesquiterpenoid synthesis genes to different hormone treatments further supports the existence of different spatiotemporal regulatory cues. Triterpenoid genes were found to be induced by jasmonate, cytokinin, and gibberellin (this study), whereas artemisinin genes were induced only by jasmonate and repressed by both cytokinin and gibberellin (Maes et al., 2011). Notably, all three phytohormones can also induce the formation of filamentous trichomes, whereas only jasmonate can stimulate the formation and maturation of the glandular trichomes (Maes et al., 2011), which correlates well with the respective enrichment of the triterpenoid and artemisinin transcripts in these organs.
A Two-Enzyme Process to Generate C-3 Carbonyl
In nature, several tetra- and pentacyclic triterpenes with a carbonyl group at the C-3 position have been identified. For instance, shionone is the major triterpenoid isolated from roots of the medicinal plant Aster tataricus (Ng et al., 2003; Sawai et al., 2011). Shionone is a tetracyclic triterpene containing a carbonyl group at the C-3 position and is cyclized from 2,3-oxidosqualene in a single enzymatic reaction catalyzed by the triterpene cyclase shionone synthase (Sawai et al., 2011). Likewise, friedelin, a pentacyclic triterpene with a carbonyl group at the C-3 position, is cyclized from 2,3-oxidosqualene by the triterpene cyclase friedelin synthase (Wang et al., 2010).
In contrast to this, the carbonyl group at the C-3 position of the triterpenoids accumulating in the leaf surface wax of A. annua is generated via two enzymatic steps. First, 2,3-oxidosqualene is cyclized to the triterpenes α-, β-, and δ-amyrin by the triterpene cyclase OSC2; subsequently, the amyrins are oxidized to the corresponding amyrones by the P450 enzyme CYP716A14v2. As such, this two-step enzymatic conversion provides an alternative biosynthesis route to triterpenoids with a C-3 carbonyl group.
The major difference between both biosynthesis routes is the molecular formula of the resulting products. Shionone and friedelin are C30H50O isomers, whereas the A. annua amyrones are C30H48O isomers. Thus, it could be speculated that the biosynthesis of triterpenoids with a molecular formula of C30H50O, such as lupan-3-one, would occur through a single enzymatic step via the cyclization of 2,3-oxidosqualene, whereas two enzymatic steps would be needed for the synthesis of C30H48O isomers, such as lupenone. The latter compound is an oxidation product of lupeol and was produced by the expression of the A. annua CYP716A14v2 in a lupeol-producing yeast strain, underscoring the two-step production of such compounds.
Reconstitution of the A. annua Triterpenoid Spectrum in Heterologous Hosts
The expression of OSC2 and CYP716A14v2 in S. cerevisiae and N. benthamiana resulted in the production and accumulation of the A. annua triterpenoids in these heterologous hosts, underscoring the utility of these genes for synthetic biology programs.
Triterpenes of the oleanane, lupane, or ursane type are all derived from the tetracyclic dammarenyl cation (Supplemental Figure 6) and triterpene cyclases, specifically producing β-amyrin or lupeol as the sole product in heterologous hosts, have been identified. So far, no α-amyrin-specific triterpene cyclase has been isolated from any plant species, but multifunctional triterpene cyclases, producing α-amyrin as a major or minor product in heterologous hosts, have been isolated from apple (Brendolise et al., 2011), Kandelia candel (Basyuni et al., 2006), Olea europaea (Saimaru et al., 2007), pea (Pisum sativum; Morita et al., 2000), tomato (Wang et al., 2011), Arabidopsis (Ebizuka et al., 2003), C. asiatica (Moses et al., 2014), and C. roseus (Huang et al., 2012). The OSC2 isolated in this study is also such a multifunctional OSC that synthesizes α-amyrin as the major product.
The CYP716 family of plant P450 enzymes (Nelson et al., 2008) has been identified only in dicots and constitutes of over 60 proteins, of which nine are functionally characterized to be involved in triterpenoid biosynthesis. These characterized P450s catalyze single- or multi-step oxidations on the dammarane, oleanane, ursane, and/or lupane backbones and show significant functional diversity with respect to their substrate of choice, the carbon position on the substrate where they catalyze the oxidation reaction, and the nature of the oxidation reaction they catalyze (Moses et al., 2013). Hydroxylations are the most commonly observed modifications catalyzed by the CYP716 family, but enzymes that can further oxidize the hydroxyl group to a carboxyl group through an aldehyde intermediate also exist. Here, the reported novel P450 enzymatic activity of CYP716A14v2 represents an important expansion of the existing synthetic biology toolkit for the engineering of pharmaceutically valuable triterpenoids in heterologous hosts. The oxidation of the hydroxyl group at C-3 to a carbonyl group is a highly relevant modification for the synthesis of synthetic triterpenoids, like 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid (CDDO) and its derivatives, which are potent anti-inflammatory agents (Sporn et al., 2011). For the chemical synthesis of CDDO, oleanolic acid is subjected to multiple chemical modifications, including the oxidation of the C-3 hydroxyl to a C-3 carbonyl group. The presence of a C-3 carbonyl group in the starter molecule could lead to a shorter chemical synthesis route for CDDO. Therefore, the C-3 oxidative activity of CYP716A14v2 consents improved perspectives for designing novel triterpenoids in synthetic biology or plant metabolic engineering programs.
METHODS
Chemicals
β-Amyrin, α-amyrin, and lupenone were purchased from Extrasynthese; β-amyrone (olean-12-en-3-one) from Sigma-Aldrich; α-amyrone (urs-12-en-3-one) from Chiron AS; and MβCD from CAVASOL. δ-Amyrin was extracted from tomato wax according to Wang et al. (2011).
Plant Material
Two different Artemisia annua cultivars, ‘Anamed A3’ (http://www.anamed.net) and ‘Meise’ (National Botanic Garden of Belgium) were used for all experiments but transformation. Plants were grown and maintained in soil at 21°C, under an 8-h-day and 16-h-night regime. When maintained in these conditions, Anamed A3 cultivar starts flowering after ∼6 months (Soetaert et al., 2013), whereas in the Meise cultivar, flowering was induced earlier (Ferreira et al., 1995).
Cloning of OSCs and Candidate P450s
As the full-length coding regions of OSC2 and the candidate P450s CYP704A87 and CYP81B58 were not present in the assembled RNA-seq data set of Soetaert et al. (2013), additional A. annua RNA-seq data were downloaded from the NCBI Short Read Archive (accession number SRA009227; Graham et al., 2010), and the FASTQ files containing the read sequences and quality scores were extracted using the NCBI SRA Toolkit version 2.1.7. The reads were quality filtered and assembled using default settings of the CLC Genomics Workbench 4.8. BLAST searches with the available sequences revealed the full-length coding regions of OSC2, CYP704A87, and CYP81B58. BLAST searches with the Medicago truncatula CYP716A12 (GenBank accession number FN995113) in the newly assembled database revealed full-length CYP716D22 and CYP716A14v2. BLAST searches with A. annua OSC2 in this database revealed the other full-length OSC genes A. annua CAS, LUS, and OSC3.
OSC2 was amplified with primers P1+P2 (Supplemental Table 3) and Gateway recombined into pDONR207. The candidate P450s CYP716D22, CYP716A14v2, CYP704A87, and CYP81B58 were amplified with primers P5+P6, P7+P8, P9+P10, and P11+P12, respectively, and Gateway recombined into pDONR221 and pDONR207. BAS, CAS, LUS, and OSC3 were amplified with primers P13+P14, P15+P16, P17+P18, and P19+P20, respectively, and Gateway recombined into pDONR207. For silencing by hairpin RNA interference, fragments of OSC2 (348 bp) and CYP716A14v2 (349 bp) were amplified with primers P21+P22 and P23+P24, respectively, and Gateway recombined into pDONR207.
Phylogenetic Analysis
The protein sequences of the OSCs and P450s were retrieved from GenBank and aligned with ClustalW. The phylogenetic tree was generated with MEGA 5.10 software (Tamura et al., 2011), according to the maximum likelihood method and bootstrapping with 10,000 replicates. The evolutionary distances were computed with the Poisson correction method, and all positions containing gaps and missing data were eliminated from the data set (complete deletion option).
Generation of Plasmid Vectors
For yeast expression, OSC2 was reamplified with primers P3+P4, cloned into the Gateway vector pDONR221 and sequence verified prior to insertion into MCS2 of pESC-URA[GAL10/tHMG1] to generate pESC-URA[GAL10/tHMG1; GAL1/OSC2]. For transient expression in Nicotiana benthamiana, the entry vector of OSC2 in pDONR207 was Gateway recombined into the binary vector pEAQ-HT-DEST1 (Sainsbury et al., 2009).
For the candidate P450s, the entry clones in pDONR221 and pDONR207 were further recombined into the yeast expression vector pAG423GAL-ccdB (Addgene plasmid 14149; Alberti et al., 2007) and the binary pEAQ-HT-DEST1 vector for expression in N. benthamiana, respectively.
At-ATR1 (AT4G24520) was cloned into the yeast expression vector pAG415GAL-ccdB (Addgene plasmid 14145) as described by Moses et al. (2014).
The BAS, CAS, LUS, and OSC3 entry clones were recombined into the yeast expression vector pAG423GAL-ccdB (Addgene plasmid 14149; Alberti et al., 2007).
For gene silencing using hairpin RNA interference, the entry clones containing short fragments corresponding to the gene were recombined into the binary vector pK7GWIWG2D(II) (Karimi et al., 2002).
Generation and Cultivation of Yeast Strains
All yeast strains (Supplemental Table 2) were generated from strain TM1 and cultivated as described (Moses et al., 2014). Briefly, precultures were grown at 30°C with agitation for 18 to 20 h in synthetic defined medium containing glucose with appropriate dropout supplements (Clontech). For cultivation, gene expression was induced by inoculating washed precultures into synthetic defined Gal/Raf medium containing galactose and raffinose with appropriate dropout supplements (Clontech) to an OD600 of 0.25 on day 1. The induced cultures were incubated for 24 h, and on day 2, methionine and MβCD were added to 1 and 5 mM, respectively. After further 24 h incubation, MβCD was added once again to 5 mM on day 3, and on day 4, all cultures were extracted for metabolite analysis.
N. benthamiana Leaf Infiltrations
The constructs for expression in N. benthamiana were individually transformed into the Agrobacterium tumefaciens strain C58C1, carrying the pMP90 helper plasmid. Agrobacterium was grown for 2 d in a shaking incubator (150 rpm) at 28°C in 5 mL yeast extract broth medium, supplemented with 25 μg/mL kanamycin and 20 μg/mL gentamycin. After incubation, 500 μL of bacterial culture was used to inoculate 9.5 mL of yeast extract broth medium supplemented with antibiotics and containing 10 mM MES (pH 5.7) and 20 μM acetosyringone. After overnight incubation in a shaking incubator (150 rpm) at 28°C, bacteria for transient coexpression were mixed, collected via centrifugation, and resuspended in 5 mL of infiltration buffer (100 μM acetosyringone, 10 mM MgCl2, and 10 mM MES, pH 5.7). The amount of bacteria harvested for each construct was adjusted to a final OD600 of 1.0 after resuspension in the infiltration buffer. After 2 to 3 h incubation at room temperature, the bacteria mixtures were infiltrated to the abaxial side of fully expanded leaves of 3- to 4-week-old N. benthamiana plants grown at 25°C in a 14-h/10-h light/dark regime. The infiltrated plants were incubated under normal growth conditions for 5 d prior to metabolite analysis.
Metabolite Extraction for GC-MS Analysis
Organic extracts of yeast cultures were prepared for identification of triterpenes and triterpenoids using GC-MS. One milliliter of yeast culture was extracted thrice with hexane and the organic extracts were pooled, vaporized to dryness, and trimethylsilylated for GC-MS analysis.
For metabolite analysis of infiltrated N. benthamiana leaves, infiltrated leaves were harvested and ground to a fine powder in liquid nitrogen. Then, 0.4 g of leaf material was extracted with 1 mL of methanol for 10 min and centrifuged for 10 min at 20,800 rpm. The resulting organic extract was evaporated to dryness under vacuum and subsequently resuspended in 1 mL of water. The resulting mixture was extracted thrice with hexane and the organic extracts were pooled, vaporized to dryness, and trimethylsilylated for GC-MS analysis.
To analyze the triterpene profile of A. annua plants, flower buds and leaves of each cultivar were harvested separately from the same plant and dipped in hexane for a few minutes. The hexane extract was concentrated by vaporizing the solvent to dryness and trimethylsilylated prior to GC-MS analysis. For quantitative analysis, plant material was harvested by quenching in liquid nitrogen, ground to a fine powder in liquid nitrogen and subsequently lyophilized. Ten milligrams of each sample was extracted thrice with 0.5 mL methanol. The methanol extracts were pooled and evaporated to dryness under a vacuum, and the dry pellet was resuspended in 1 mL of water. The mixture was extracted thrice with hexane and the organic extracts pooled, vaporized to dryness, and trimethylsilylated for GC-MS analysis.
GC-MS Analysis
GC-MS was performed on a GC model 6890 and MS model 5973 (Agilent). Analyses were performed on a VF-5ms capillary column (Varian CP9013; Agilent) or a DB-1 ht capillary column (J&W; Agilent) at a constant helium flow of 1 mL/min, using a 1-μL aliquot, which was injected in splitless mode. After injection, the oven was held at 80°C for 1 min, ramped to 280°C at a rate of 20°C/min, held at 280°C for 45 min, ramped to 320°C at a rate of 20°C/min, held at 320°C for 1 min, and finally cooled to 80°C at a rate of 50°C/min at the end of the run. The injector was set to 280°C, the MS transfer line to 250°C, the MS ion source to 230°C, and the quadrupole to 150°C, throughout. A full EI-MS spectrum was generated for each sample by scanning the m/z range of 60 to 800 with a solvent delay of 7.8 min. For relative quantification, the peak areas were calculated using the default settings of the AMDIS software (version 2.6; NIST).
Sample Preparation and qPCR Analysis
Flower buds and leaves from the same plant of each cultivar were harvested and frozen in liquid nitrogen. RNA was extracted with the RNeasy Plant Mini Kit (Qiagen). Samples from hormone-treated Meise seedlings were developed by Maes et al. (2011) and RNA prepared as described therein. cDNA was prepared from 1 μg total RNA using an iScript cDNA Synthesis Kit (Bio-Rad).
qPCR was performed with a Lightcycler 480 (Roche) and SYBR Green QPCR Master Mix (Stratagene). Gene-specific primers (Supplemental Table 4) were adopted from Soetaert et al. (2013). For comparative analysis between leaves and flower buds, the transcript levels were normalized to two reference genes, Protein phosphatase 2A subunit A3 (PP2AA3; homolog of AT1G13320) and Pentatricopeptide repeat superfamily protein (PPRl homolog of AT5G55840). For comparative analysis between hormone-treated Meise seedlings, Actin (homolog of AT3G18780) was used as a reference gene. All reactions were performed in triplicate and relative quantifications calculated using qBase (Hellemans et al., 2007).
Generation of A. annua RNA Interference Lines
The silencing constructs were transformed into the A. tumefaciens strain EHA105, and the resulting strains were used for the transformation of the A. annua cultivar ‘Chongqing’ as described (Zhang et al., 2009). Transformed plants were generated, cultured, and analyzed for gene silencing as described (Zhang et al., 2009).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: KF309252 (OSC2), KF309248 (CYP704A87), KF309249 (CYP81B58), KF309250 (CYP716D22), KF309251 (CYP716A14v2), KM670093 (CAS), KM670094 (LUS), and KM670095 (OSC3).
Supplemental Data
Supplemental Figure 1. Identity of A. annua Triterpenoids.
Supplemental Figure 2. Phylogenetic Analysis of A. annua Candidate P450s and Other P450s Involved in Specialized Triterpenoid Biosynthesis.
Supplemental Figure 3. Organ-Specific Expression of Triterpene Biosynthesis Genes and Candidate P450s in A. annua Cultivars.
Supplemental Figure 4. CYP716A14v2 Is a Multifunctional P450.
Supplemental Figure 5. Identification of the Products of A. annua OSCs.
Supplemental Figure 6. Schematic of the 2,3-Oxidosqualene Cyclization Cascade Mediated by Oxidosqualene Cyclases to Generate Oleanane, Ursane, and Lupane Triterpene Backbones.
Supplemental Table 1. RNA-Seq Triterpene Contig Counts for Glandular and Filamentous Trichomes.
Supplemental Table 2. Yeast Strains Generated in This Study.
Supplemental Table 3. Primers Used for Cloning.
Supplemental Table 4. Primers Used for qPCR Analysis.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny in Figure 2.
Supplemental Data Set 2. Alignments Used to Generate the Phylogeny in Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank David Nelson for naming the P450 enzymes and Annick Bleys for help with preparing the article. This work was supported by the Research Foundation-Flanders through the project GA13111N, the European Union Seventh Framework Program FP7/2007-2013 under grant agreement numbers 222716-SMARTCELL and 613692-TriForC, the Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant ‘Understanding and Exploiting Plant and Microbial Secondary Metabolism’ (BB/J004561/1), the John Innes Foundation, and the Engineering and Physical Sciences Research Council (EP/H019154/1). T.M. is indebted to the VIB International PhD Fellowship Program for a predoctoral fellowship. J.P. and S.S. are post- and predoctoral fellows of the Research Foundation Flanders, respectively. J.R. is funded by a Norwich Research Park PhD studentship.
AUTHOR CONTRIBUTIONS
T.M., J.P., and A.G. designed the research. T.M., J.P., Q.S., S.S., J.R., M.-L.E., and R.V.B. performed the research. T.M., J.P., S.S., F.C.W.V.N., K.T., and A.G. analyzed the data. T.M., J.P., A.O., J.M.T., D.D., and A.G. wrote the article.
Glossary
- OSC
oxidosqualene cyclase
- MβCD
methyl-β-cyclodextrin
- GC-MS
gas chromatography-mass spectrometry
- EI-MS
electron ionization-mass spectrum
- P450
P450-dependent monooxygenase
- BAP
6-benzylaminopurine
- GA3
gibberellic acid
- qPCR
quantitative PCR
- CDDO
2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid
References
- Alberti S., Gitler A.D., Lindquist S. (2007). A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J.M., Kuzina V., Andersen S.B., Bak S. (2011). Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72: 435–457. [DOI] [PubMed] [Google Scholar]
- Basyuni M., Oku H., Inafuku M., Baba S., Iwasaki H., Oshiro K., Okabe T., Shibuya M., Ebizuka Y. (2006). Molecular cloning and functional expression of a multifunctional triterpene synthase cDNA from a mangrove species Kandelia candel (L.) Druce. Phytochemistry 67: 2517–2524. [DOI] [PubMed] [Google Scholar]
- Bhakuni R.S., Jain D.C., Sharma R.P., Kumar S. (2001). Secondary metabolites of Artemisia annua and their biological activity. Curr. Sci. 80: 35–48. [Google Scholar]
- Bora K.S., Sharma A. (2011). The genus Artemisia: a comprehensive review. Pharm. Biol. 49: 101–109. [DOI] [PubMed] [Google Scholar]
- Brendolise C., Yauk Y.K., Eberhard E.D., Wang M., Chagne D., Andre C., Greenwood D.R., Beuning L.L. (2011). An unusual plant triterpene synthase with predominant α-amyrin-producing activity identified by characterizing oxidosqualene cyclases from Malus × domestica. FEBS J. 278: 2485–2499. [DOI] [PubMed] [Google Scholar]
- Bringe K., Schumacher C.F., Schmitz-Eiberger M., Steiner U., Oerke E.C. (2006). Ontogenetic variation in chemical and physical characteristics of adaxial apple leaf surfaces. Phytochemistry 67: 161–170. [DOI] [PubMed] [Google Scholar]
- Buschhaus C., Jetter R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62: 841–853. [DOI] [PubMed] [Google Scholar]
- Carelli M., et al. (2011). Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23: 3070–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17: 349–359. [DOI] [PubMed] [Google Scholar]
- Duke M.V., Paul R.N., Elsohly H.N., Sturtz G., Duke S.O. (1994). Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int. J. Plant Sci. 155: 365–372. [Google Scholar]
- Ebizuka Y., Katsube Y., Tsutsumi T., Kushiro T., Shibuya M. (2003). Functional genomics approach to the study of triterpene biosynthesis. Pure Appl. Chem. 75: 369–374. [Google Scholar]
- Ferreira J.F., Simon J.E., Janick J. (1995). Developmental studies of Artemisia annua: flowering and artemisinin production under greenhouse and field conditions. Planta Med. 61: 167–170. [DOI] [PubMed] [Google Scholar]
- Fukushima E.O., Seki H., Ohyama K., Ono E., Umemoto N., Mizutani M., Saito K., Muranaka T. (2011). CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 52: 2050–2061. [DOI] [PubMed] [Google Scholar]
- Graham I.A., et al. (2010). The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327: 328–331. [DOI] [PubMed] [Google Scholar]
- Han J.Y., Hwang H.S., Choi S.W., Kim H.J., Choi Y.E. (2012). Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53: 1535–1545. [DOI] [PubMed] [Google Scholar]
- Han J.Y., Kim H.J., Kwon Y.S., Choi Y.E. (2011). The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52: 2062–2073. [DOI] [PubMed] [Google Scholar]
- Han J.Y., Kim M.J., Ban Y.W., Hwang H.S., Choi Y.E. (2013). The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 54: 2034–2046. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Li J., Ye H., Li C., Wang H., Liu B., Zhang Y. (2012). Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta 236: 1571–1581. [DOI] [PubMed] [Google Scholar]
- Jetter R., Schäffer S., Riederer M. (2000). Leaf cuticular waxes are arranged in chemically and mechanically distinct layers: evidence from Prunus laurocerasus L. Plant Cell Environ. 23: 619–628. [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kesselmeier J., Urban B. (1983). Subcellular-localization of saponins in green and etiolated leaves and green protoplasts of oat (Avena sativa L). Protoplasma 114: 133–140. [Google Scholar]
- Kim O.T., Lee J.W., Bang K.H., Kim Y.C., Hyun D.Y., Cha S.W., Choi Y.E., Jin M.L., Hwang B. (2009). Characterization of a dammarenediol synthase in Centella asiatica (L.) Urban. Plant Physiol. Biochem. 47: 998–1002. [DOI] [PubMed] [Google Scholar]
- Kirby J., Romanini D.W., Paradise E.M., Keasling J.D. (2008). Engineering triterpene production in Saccharomyces cerevisiae-β-amyrin synthase from Artemisia annua. FEBS J. 275: 1852–1859. [DOI] [PubMed] [Google Scholar]
- Lodeiro S., Xiong Q., Wilson W.K., Kolesnikova M.D., Onak C.S., Matsuda S.P. (2007). An oxidosqualene cyclase makes numerous products by diverse mechanisms: a challenge to prevailing concepts of triterpene biosynthesis. J. Am. Chem. Soc. 129: 11213–11222. [DOI] [PubMed] [Google Scholar]
- Maes L., Van Nieuwerburgh F.C., Zhang Y., Reed D.W., Pollier J., Vande Casteele S.R., Inzé D., Covello P.S., Deforce D.L., Goossens A. (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189: 176–189. [DOI] [PubMed] [Google Scholar]
- Morita M., Shibuya M., Kushiro T., Masuda K., Ebizuka Y. (2000). Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum) new alpha-amyrin-producing enzyme is a multifunctional triterpene synthase. Eur. J. Biochem. 267: 3453–3460. [DOI] [PubMed] [Google Scholar]
- Moses T., Pollier J., Almagro L., Buyst D., Van Montagu M., Pedreño M.A., Martins J.C., Thevelein J.M., Goossens A. (2014). Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. USA 111: 1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses T., Pollier J., Faizal A., Apers S., Pieters L., Thevelein J.M., Geelen D., Goossens A. (2015). Unraveling the triterpenoid saponin biosynthesis of the African shrub Maesa lanceolata. Mol. Plant 8: 122–135. [DOI] [PubMed] [Google Scholar]
- Moses T., Pollier J., Thevelein J.M., Goossens A. (2013). Bioengineering of plant (tri)terpenoids: from metabolic engineering of plants to synthetic biology in vivo and in vitro. New Phytol. 200: 27–43. [DOI] [PubMed] [Google Scholar]
- Murata J., Roepke J., Gordon H., De Luca V. (2008). The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P., Owatworakit A., Papadopoulou K., Jenner H., Qin B., Findlay K., Hill L., Qi X., Bakht S., Melton R., Osbourn A. (2008). Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Ming R., Alam M., Schuler M.A. (2008). Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 1: 216–235. [Google Scholar]
- Ng T.B., Liu F., Lu Y., Cheng C.H., Wang Z. (2003). Antioxidant activity of compounds from the medicinal herb Aster tataricus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 136: 109–115. [DOI] [PubMed] [Google Scholar]
- Olsson M.E., Olofsson L.M., Lindahl A.L., Lundgren A., Brodelius M., Brodelius P.E. (2009). Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70: 1123–1128. [DOI] [PubMed] [Google Scholar]
- Saimaru H., Orihara Y., Tansakul P., Kang Y.H., Shibuya M., Ebizuka Y. (2007). Production of triterpene acids by cell suspension cultures of Olea europaea. Chem. Pharm. Bull. (Tokyo) 55: 784–788. [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Sawai S., Uchiyama H., Mizuno S., Aoki T., Akashi T., Ayabe S., Takahashi T. (2011). Molecular characterization of an oxidosqualene cyclase that yields shionone, a unique tetracyclic triterpene ketone of Aster tataricus. FEBS Lett. 585: 1031–1036. [DOI] [PubMed] [Google Scholar]
- Soetaert S.S.A., Van Neste C.M.F., Vandewoestyne M.L., Head S.R., Goossens A., Van Nieuwerburgh F.C.W., Deforce D.L. (2013). Differential transcriptome analysis of glandular and filamentous trichomes in Artemisia annua. BMC Plant Biol. 13: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn M.B., Liby K.T., Yore M.M., Fu L., Lopchuk J.M., Gribble G.W. (2011). New synthetic triterpenoids: potent agents for prevention and treatment of tissue injury caused by inflammatory and oxidative stress. J. Nat. Prod. 74: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakiel A., Niżyński B., Pączkowski C. (2013). Triterpenoid profile of flower and leaf cuticular waxes of heather Calluna vulgaris. Nat. Prod. Res. 27: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Szakiel A., Pączkowski C., Pensec F., Bertsch C. (2012). Fruit cuticular waxes as a source of biologically active triterpenoids. Phytochem. Rev. 11: 263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansakul P., Shibuya M., Kushiro T., Ebizuka Y. (2006). Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 580: 5143–5149. [DOI] [PubMed] [Google Scholar]
- Thimmappa R., Geisler K., Louveau T., O’Maille P., Osbourn A. (2014). Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65: 225–257. [DOI] [PubMed] [Google Scholar]
- Van Nieuwerburgh F.C., Vande Casteele S.R., Maes L., Goossens A., Inzé D., Van Bocxlaer J., Deforce D.L. (2006). Quantitation of artemisinin and its biosynthetic precursors in Artemisia annua L. by high performance liquid chromatography-electrospray quadrupole time-of-flight tandem mass spectrometry. J. Chromatogr. A 1118: 180–187. [DOI] [PubMed] [Google Scholar]
- Wang H., Han J., Kanagarajan S., Lundgren A., Brodelius P.E. (2013). Trichome-specific expression of the amorpha-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion. Plant Mol. Biol. 81: 119–138. [DOI] [PubMed] [Google Scholar]
- Wang W., Wang Y., Zhang Q., Qi Y., Guo D. (2009). Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics 10: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Guhling O., Yao R., Li F., Yeats T.H., Rose J.K., Jetter R. (2011). Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol. 155: 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yeats T., Han H., Jetter R. (2010). Cloning and characterization of oxidosqualene cyclases from Kalanchoe daigremontiana: enzymes catalyzing up to 10 rearrangement steps yielding friedelin and other triterpenoids. J. Biol. Chem. 285: 29703–29712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Thamm A.M., Reed D., Villa-Ruano N., Quesada A.L., Gloria E.L., Covello P., De Luca V. (2013). Functional characterization of amyrin synthase involved in ursolic acid biosynthesis in Catharanthus roseus leaf epidermis. Phytochemistry 91: 122–127. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jing F., Li F., Li M., Wang Y., Wang G., Sun X., Tang K. (2009). Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Biochem. 52: 199–207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.