Unconventional modes of microtubule nucleation exist in the moss cytoplasm, representing a broader spectrum of nucleation mechanisms in plants.
Abstract
The mechanism underlying microtubule (MT) generation in plants has been primarily studied using the cortical MT array, in which fixed-angled branching nucleation and katanin-dependent MT severing predominate. However, little is known about MT generation in the endoplasm. Here, we explored the mechanism of endoplasmic MT generation in protonemal cells of Physcomitrella patens. We developed an assay that utilizes flow cell and oblique illumination fluorescence microscopy, which allowed visualization and quantification of individual MT dynamics. MT severing was infrequently observed, and disruption of katanin did not severely affect MT generation. Branching nucleation was observed, but it showed markedly variable branch angles and was occasionally accompanied by the transport of nucleated MTs. Cytoplasmic nucleation at seemingly random locations was most frequently observed and predominated when depolymerized MTs were regrown. The MT nucleator γ-tubulin was detected at the majority of the nucleation sites, at which a single MT was generated in random directions. When γ-tubulin was knocked down, MT generation was significantly delayed in the regrowth assay. However, nucleation occurred at a normal frequency in steady state, suggesting the presence of a γ-tubulin-independent backup mechanism. Thus, endoplasmic MTs in this cell type are generated in a less ordered manner, showing a broader spectrum of nucleation mechanisms in plants.
INTRODUCTION
Although the centrosome is the dominant microtubule (MT) generation site in animal somatic cells, acentrosomal MTs also play critical roles in many aspects of cell biology, such as cell division and polarization, in a variety of cell types (Bartolini and Gundersen, 2006; Goshima and Kimura, 2010). Land plants, which have lost centrosomes during evolution, generate MTs independent of centrosomes. Therefore, they serve as an ideal model system for studying the mechanism of acentrosomal MT generation.
The best studied MT network is the cortical MT array in flowering plants such as Arabidopsis thaliana and tobacco (Nicotiana tabacum), specifically bright yellow-2 cells. The MTs, which are required for cellulose synthase deposition, are nucleated at the cortex, bundled in parallel, bound to the plasma membrane, and arranged transverse to the elongation axis of the cell (Dixit and Cyr, 2004; Lloyd and Chan, 2008). Live-cell microscopy, gene perturbation experiments, and mathematical modeling have elucidated a detailed mechanism for this process (Murata et al., 2005; Chan et al., 2009; Nakamura and Hashimoto, 2009; Wasteneys and Ambrose, 2009; Nakamura et al., 2010; Fishel and Dixit, 2013; Lindeboom et al., 2013a). MT-dependent MT nucleation (also called branching nucleation) is the dominant mode of MT nucleation, in which a new MT (“daughter” MT) is nucleated from the lattice of an existing MT (“mother” MT). This reaction requires γ-tubulin and associated subunits, which transiently localize on the lattice of the mother MT. In the cortical MT network, daughter MTs are nucleated preferentially at an angle of ∼40° or 0° from the mother, suggesting the presence of a mechanism that roughly maintains parallel orientation of the MTs (the 0° branching nucleation is sometimes distinctly called “parallel nucleation”). Katanin-mediated MT severing is also a critical process in cortical MT generation. Recent live imaging has shown that katanin preferentially severs crossing MTs, which have a 15 to 20% probability of regrowth. Thus, this simple reaction increases MT numbers (Lindeboom et al., 2013b; Zhang et al., 2013). Branching nucleation requires additional regulatory factors, such as the protein phosphatase TONNEAU2/FASS (TON2). In the ton2 mutant, mother-daughter angles of 0° predominate (Kirik et al., 2012).
The other well-studied acentrosomal MT networks in plant cells are the mitotic spindle and phragmoplast. Recent RNA interference (RNAi)- and mutant-based studies in the moss Physcomitrella patens and in Arabidopsis have shown that augmin-dependent branching MT nucleation plays a dominant role in MT generation (Hotta et al., 2012; Nakaoka et al., 2012). In this mechanism, a conserved eight-subunit complex, augmin, binds to spindle/phragmoplast MTs, recruits the γ-tubulin ring complex (γ-TuRC), and thereby nucleates new MTs. Studies in animal systems have shown that daughter MTs are nucleated predominantly at <20° angles from mother MTs during this process, indicating that this mechanism is suitable for generating the parallel MT arrays commonly seen in the spindle and phragmoplast (Kamasaki et al., 2013; Petry et al., 2013). A recent report has shown that augmin also plays a role in MT-dependent MT nucleation by recruiting the γ-TuRC in the cortical MT arrays (Liu et al., 2014).
Although cortical and spindle/phragmoplast MTs have been the main focus of studies investigating the mechanism of MT generation, certain cell types in land plants, such as endosperms, pollen tubes, or root hairs, have MT networks that differ from conventional cortical MTs (Brown et al., 1994; Wasteneys and Galway, 2003; Wasteneys and Ambrose, 2009). In P. patens, germinated spores develop into a tissue called “protonemata,” in which two types of cells, chloronemata and caulonemata, have been identified. Immunofluorescence microscopy and live imaging of MTs have shown that these cells do not form conventional cortical MT arrays but rather form “endoplasmic MTs” that run throughout the cytoplasm (Doonan et al., 1985; Hiwatashi et al., 2014). Endoplasmic MTs play a critical role in protonemal cell growth (Doonan et al., 1985, 1988), but how they are generated is unknown.
In this study, we used protonemal cells of P. patens, in particular the subapical chloronemal cells, as a model for studying noncortical MT generation. P. patens has an unusually high rate of homologous recombination, such that gene disruption and green fluorescent protein (GFP) tagging of endogenous protein can be performed in a reasonable period (∼3 months) (Cove, 2005). We recently established a conditional RNAi system, in which any gene of interest can be knocked down in protonemal cells, including genes that are essential for viability and for which gene disruptants cannot therefore be obtained (Nakaoka et al., 2012; Kosetsu et al., 2013; Miki et al., 2015; Naito and Goshima, 2015). Taking advantage of these tools and a newly developed MT generation visualization assay, we observed and quantified MT generation dynamics and quantitatively assessed the contribution of several factors to the process. We identified cytoplasmic nucleation and branching nucleation as the two dominant mechanisms of MT generation. Numerical modeling confirmed that these nucleation mechanisms were sufficient to reproduce the MT dynamics characterized in the experiments. Interestingly, neither mode of nucleation appeared to be a well-ordered process; new MTs were formed at unassigned locations and oriented in random directions. In addition, independent nucleation appeared to occur in the absence of γ-tubulin. Based on these results, we have proposed a model for endoplasmic MT formation through multiple mechanisms.
RESULTS
Identification of Three Modes of MT Generation in the Cytoplasm of P. patens
Initially, we used confocal microscopy to observe MT generation in protonemal cells, which is typically used to visualize nucleation and severing events in flowering plants (Murata et al., 2005; Nakamura and Hashimoto, 2009; Lindeboom et al., 2013b; Zhang et al., 2013). However, partly because of the high background signals derived from autofluorescence, we were not able to confidently identify MT generation events. To reduce background fluorescence, we selectively illuminated limited space in the focal plane near the cell cortex with a laser beam that was tilted away from the optical axis of the objective lens, near the critical angle of the glass-water interface (oblique illumination fluorescence microscopy; Konopka and Bednarek, 2008; Tokunaga et al., 2008). This method allowed for clear visualization of the dynamics of individual endoplasmic MTs in the protonemata (Figure 1A; Supplemental Movie 1).
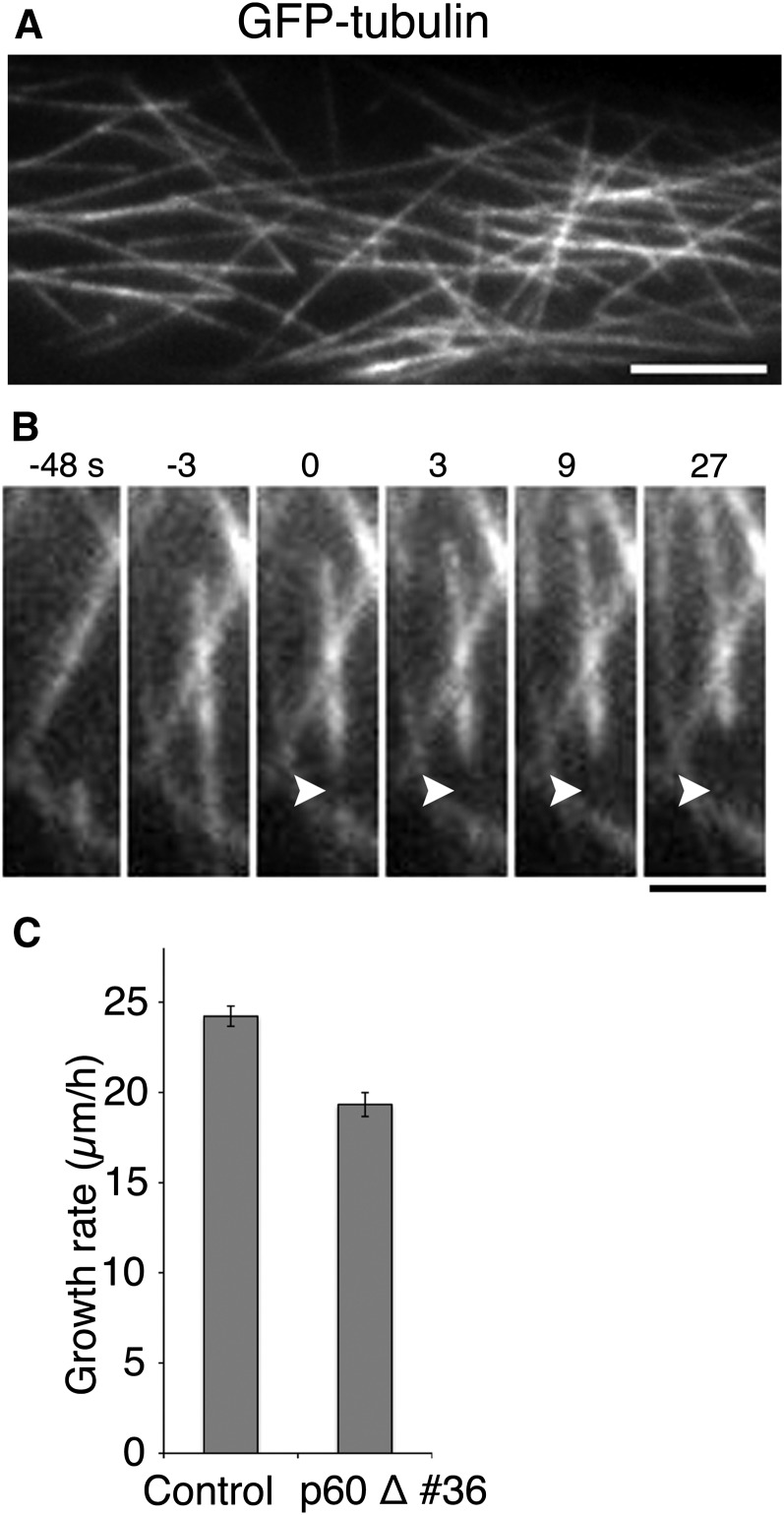
Figure 1.
Katanin-Dependent MT Severing in Moss.
(A) Endoplasmic MTs labeled with GFP-tubulin. Individual MTs in the cytoplasm were clearly visible with oblique illumination fluorescence microscopy. See also Supplemental Movie 1. Bar = 5 μm.
(B) MT severing. A single MT was severed at time 0 (arrowhead). See also Supplemental Movie 2. Bar = 2 μm.
(C) A mild decrease in the growth rate of the tip in caulonemal apical cells in the absence of the katanin p60 subunit (±se). Control, n = 94; p60 disruptant, n = 83 (P < 0.0001).
We also prepared a transgenic moss line to examine γ-tubulin localization during MT nucleation. We utilized a γ-tubulin-b (TubG2)-Citrine replacement line in a previous study, where one of the two γ-tubulin paralogs was tagged with Citrine (Nakaoka et al., 2012) (Citrine is a GFP variant). In this study, we also deleted the γ-tubulin-a (TubG1) gene through homologous recombination, ensuring that all γ-tubulin proteins in the cells are tagged with Citrine (Supplemental Figure 1). MTs were visualized by mCherry-tubulin expression. The protonemal cells of the established mCherry-tubulin/γ-tubulin-b-Citrine/γ-tubulin-aΔ line displayed growth similar to that of wild-type cells, indicating the functionality of the tagged γ-tubulin in protonemata (Supplemental Figure 1D).
Katanin-Dependent Severing
From time-lapse imaging of >28 cells (3-s intervals, 10 to 12 min), we identified the instance at which new MTs emerged. MT generation was classified into three modes. First, we observed events in which the lattice of a MT was severed and two MTs emerged (Figure 1B; Supplemental Movie 2). The minus end generated by severing was quickly stabilized or slowly depolymerized (<1.5 μm/min, n = 47) in 76% of the cases, while the remaining 24% of minus-ends expressed faster depolymerization rates (>1.5 μm/min, n = 15). Treadmilling behavior, in which the plus-ends grew with a corresponding shrinkage of the minus-ends, was observed in some cases. On the other hand, the majority of the plus-ends of severed MTs were initially depolymerized. We detected rescue and regrowth of 22% of those MTs. The other MTs (78%) disappeared from the imaging field, largely due to movement out of the imaging field or focal plane and partly due to complete depolymerization. Therefore, severing may result in a modest increase in MT number.
However, the occurrence of severing was not frequent (63 severing events were identified in 28 cells) compared with that of other MT generation modes described below. Nevertheless, we speculated that katanin was involved in this process because it is a MT severing protein active on cortical MTs (Lindeboom et al., 2013b; Zhang et al., 2013). We disrupted two paralogous genes encoding the p60 subunit of katanin and obtained viable lines (Supplemental Figures 2A to 2C and Supplemental Data Set 1). We detected no severing events in the protonemal cells of the p60 disruptant (n = 10 cells), indicating the occurrence of katanin-dependent severing in the normal cells. We examined the effect of p60 disruption on caulonemal apical cell division and tip growth, two processes that require an intact MT cytoskeleton (Doonan et al., 1985), using time-lapse microscopy. We observed a slight increase in the duration of prometaphase/metaphase (10.7 min from nuclear envelope breakdown to the onset of anaphase [n = 10] compared with the 8.0 min required by the control [n = 8]) and a decrease in the rate of tip growth (Figure 1C). However, the immunofluorescence images indicated that MT organization was not detectably skewed in the absence of p60 (Supplemental Figure 3). These results are consistent with the hypothesis that katanin-dependent severing makes only a modest contribution to the increase of MT in protonemal cells.
In contrast, we observed severe defects in the gametophore (shoot with sexual organs) structure in the p60 disruptant, such as smaller leaves and abnormally swollen cells with disorganized MTs (Supplemental Figures 2D to 2F). MT severing was scarcely observed in p60Δ gametophore cells, indicating active katanin-dependent severing in the gametophore (we observed 34 severing events in six control cells, but only one in eight disruptant cells).
Based on these results, we conclude that katanin-dependent severing plays a role in MT generation in moss. However, the extent to which katanin contributes differs in protonemal (relatively minor) and gametophore (active) cells.
Branching Nucleation
In the second MT generation mode, we observed branching nucleation, in which MTs emerged from the wall of existing MTs (Figure 2A; Supplemental Movie 3; 182 events were identified in 28 cells). We observed the colocalization of γ-tubulin-Citrine with the nucleation site in most cases (Figure 2E). Branching nucleation has been predominantly observed in cortical MTs with 0° or ∼40° angles (Murata et al., 2005; Chan et al., 2009; Nakamura and Hashimoto, 2009; Wasteneys and Ambrose, 2009; Nakamura et al., 2010; Fishel and Dixit, 2013; Lindeboom et al., 2013a). We measured the angle between mother and daughter MTs in 2D. Similar to cortical MTs, we identified branching nucleation at 0° (parallel nucleation; ∼11%) and 20° to 60° (∼36%) angles. However, we also observed various angles of branching nucleation, including those above 90° (Figures 2C and 2D; in our measurements, ∼90° branching might have been underestimated because of the difficulty in unambiguously distinguishing branching nucleation from the crossing of two MTs at this particular angle). We concluded that the angle between mother and daughter MTs was more variable than in cortical MTs.
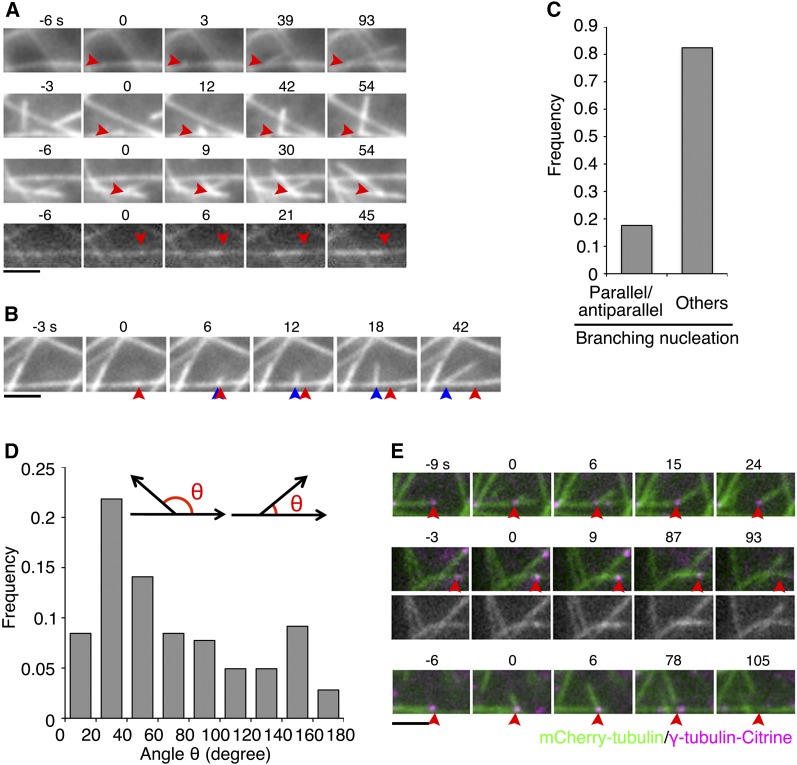
Figure 2.
Atypical Branching Nucleation in the Endoplasm.
(A) Four examples of branching MT nucleation. Arrowheads indicate the branching sites. Branching occurred at time 0. The plus ends of mother MTs appear on the right in each image sequence. Antiparallel nucleation is displayed in the bottom (the plus-end of the mother MT was located in the left). See also Supplemental Movie 3.
(B) An example of a moving branching point along the mother MT. The red arrowhead indicates the nucleation site, and the blue arrowheads show the migrating branching point. The plus ends of mother MTs appear on the right in the sequence.
(C) Frequency of different branching types.
(D) Angles between the orientation of mother and daughter MTs. Parallel and antiparallel nucleation was excluded from this graph. The angle θ was measured at 9 to 15 s after the appearance of daughter MTs. A wide range of angles was observed, including those above 90°.
(E) Branching nucleation and fate of daughter MTs (three types), visualized using mCherry-tubulin (green) and γ-tubulin-b-Citrine (magenta). Time 0 corresponds to the time at which daughter MT nucleation occurs. A punctate signal of γ-tubulin-b-Citrine was localized at the branching point (arrowheads). Top: Daughter MT was released as a result of mother MT depolymerization. Middle: Daughter MT was completely depolymerized while the mother MT remained intact (an example of antiparallel nucleation). Bottom: Daughter MT was released (at 78 s) from the mother MT concomitantly with γ-tubulin dissociation. Bars = 2 μm.
Occasionally, the branching point was not fixed during the elongation or shrinkage of daughter MTs and moved alongside the mother MTs (9.3%, n = 182 branching events; Figure 2B; Supplemental Movie 3, right). The branching points migrated at a rate of 2.6 ± 1.7 μm/min (n = 17). In 13 out of the 14 cases where the polarity of mother MTs could be assigned, the daughter MTs were observed to migrate toward the minus-end alongside the mother MTs, suggesting the influence of a minus-end-directed motor in the transport of daughter MTs (Figure 2B). This behavior is somewhat reminiscent of the poleward movement of minus-end-bound γ-tubulin in human spindles and plant phragmoplasts (Murata et al., 2013; Lecland and Lüders, 2014). Interestingly, the angle of daughter-mother orientation became shallower by migration, in 11 out of 13 cases (Supplemental Figure 4). In contrast, only 13% of the nonmigrating daughter MTs changed the branch angle. Therefore, daughter MT transport might be a mechanism that assists in its orientation parallel to the mother MT. To our knowledge, this behavior has not been previously reported in cortical MT branching.
During the branching nucleation process in flowering plants, the daughter MTs are frequently released from the mother by katanin-mediated severing near the minus-end (Nakamura et al., 2010). In protonemal cells, we examined the release mechanism of daughter MTs from the mother MTs and investigated the involvement of katanin in this process. Using the γ-tubulin-b-Citrine/γ-tubulin-aΔ/mCherry-tubulin line, we traced 34 daughter MTs for ∼12 min after they appeared at the mother lattice. We categorized the behavior of the daughter MTs into three types (Figure 2E). In 21 cases, daughter MTs were released by depolymerization of the mother MTs (Figure 2E, top). In 12 cases, we observed the complete depolymerization of the daughter MTs, while the mother MTs were still present (Figure 2E, middle). Another three daughter MTs were released from the mother MTs. However, a clear MT severing event was not detected during release; the entire daughter MT appeared to be released from the mother together with γ-tubulin (Figure 2E, bottom). However, the possibility cannot be excluded that the detachment was mediated by severing that occurred very close to the minus-end of the daughter MT. The daughter MT behavior was also analyzed in the katanin disruptant cells. Of the 30 daughter MTs traced, 20 were released by depolymerization of the mother and nine were completely self-depolymerized. We observed only one case in which a daughter MT was released from the mother MT. Although γ-tubulin was not marked in this transgenic line, the entire release of the daughter MTs suggests that this release was also independent of severing. These results suggest that the daughter MTs in P. patens protonemata are released predominantly by mother MT depolymerization and indicate the occurrence of atypical branching nucleation in P. patens protonemal cells.
Cytoplasmic Nucleation
Finally, we frequently observed the emergence of new MTs at seemingly random locations in a MT-free space in the cytoplasm (Figure 3A; Supplemental Movie 4; 310 events were identified in 28 cells). We called this event “cytoplasmic nucleation.” In this case, a punctate GFP-tubulin signal first appeared, followed by extension of a single MT from the spot. One caveat in interpreting the origin of the GFP-tubulin spot was that the observed spot might have represented a growing MT end (e.g., a MT nucleated out of the focal plane grew and entered the focal plane) rather than a nucleation point. In this study, we identified true nucleation events using the criteria described in Supplemental Figure 5.
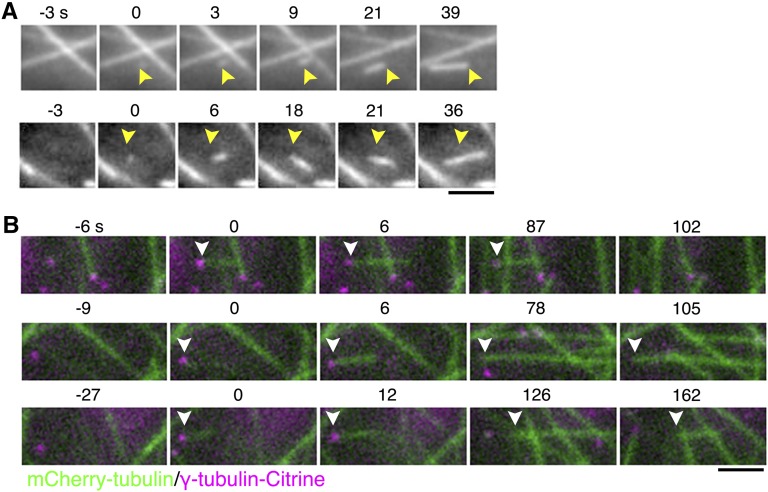
Figure 3.
Cytoplasmic Nucleation in the Endoplasm.
(A) Two examples of cytoplasmic MT nucleation. A new MT emerged at time 0 at a place that lacked other MTs (arrowheads). See also Supplemental Movie 4.
(B) Cytoplasmic nucleation and three fates of minus-ends (arrowheads), visualized using mCherry-tubulin (green) and γ-tubulin-b-Citrine (magenta). Time 0 corresponds to the time at which nucleation occurs. The MT was nucleated from the site where a punctate signal of γ-tubulin-b-Citrine was observed. Top: The nucleated MT grew and then shrank, while the γ-tubulin remained associated with the minus-end. Middle: γ-Tubulin was dissociated from the nucleated MT, but the minus-end remained stable. Bottom: γ-Tubulin was dissociated from the nucleated MT, and the minus-end was depolymerized. Bars = 2 μm.
We observed that cytoplasmic nucleation often occurred at the site where the γ-tubulin-b-Citrine spot was detected in the γ-tubulin-b-Citrine/γ-tubulin-aΔ/mCherry-tubulin line (Figure 3B). About 51% of the γ-tubulin-Citrine signals were observed persistently until the complete depolymerization of the MTs (n = 33) (Figure 3B, top). In the remaining 49%, the γ-tubulin signal was dissociated from the minus-end of the MT 2 min (on average) after nucleation (n = 32). Following dissociation of the γ-tubulin, 47% of the minus-ends were quickly depolymerized (Figure 3B, bottom), while the remaining 53% were stabilized (Figure 3B, middle) or slowly depolymerized.
Development of an Assay to Visualize and Quantify MT Generation Events
When observing normally growing cells, the MTs were so crowded that not all MT generation events could be unambiguously identified. To thoroughly detect MT generation, we assessed MT generation using the MT depolymerization-regrowth assay, in which drug treatment to induce MT depolymerization was followed by drug washout (Figure 4).
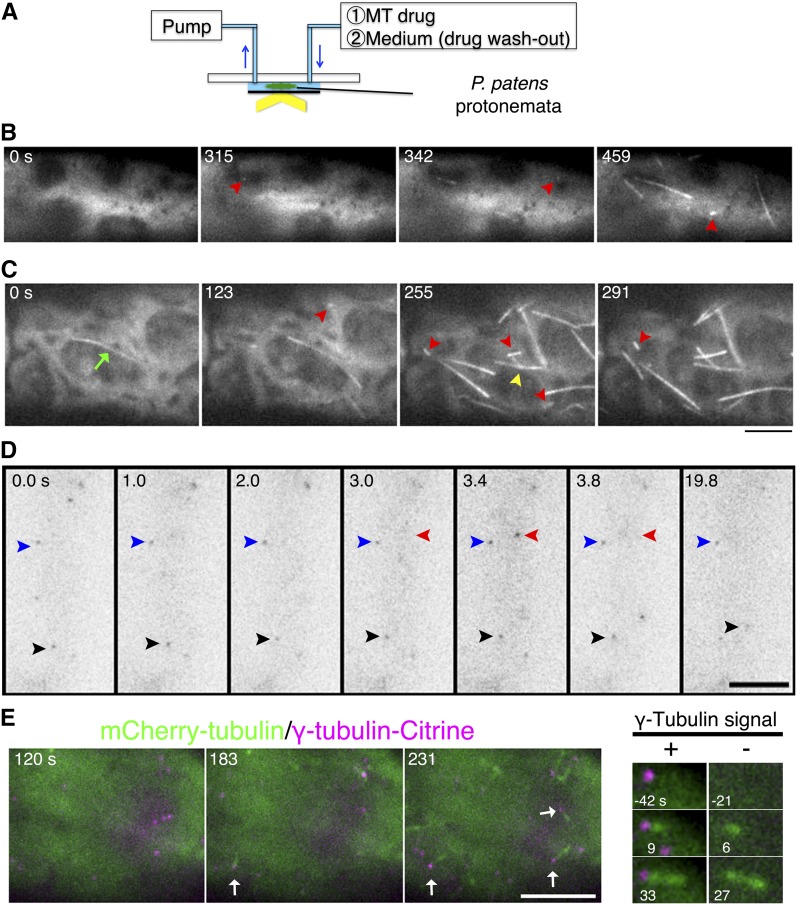
Figure 4.
MT Depolymerization-Regrowth Assay to Visualize MT Nucleation.
(A) Scheme of the MT regrowth assay. See Supplemental Figure 7 for a detailed explanation of the setup.
(B) and (C) MT depolymerization-regrowth assay using oryzalin. New MTs emerged after ∼5 min of oryzalin washout. MTs in most of the cells were completely depolymerized (B), whereas residual MTs (green arrow) were occasionally present after oryzalin treatment (C). In each case, cytoplasmic nucleation (red arrowheads) dominated over branching nucleation (yellow arrowhead). See also Supplemental Movies 5 and 6.
(D) γ-Tubulin imaging. γ-Tubulin-b-Citrine was imaged at 0.2-s intervals for 20 s. Some signals remained at the same location (blue arrowheads), whereas others displayed diffusive motion (red arrowheads, transiently appeared at the focal plane; black arrowheads, moved in two dimensions). See also Supplemental Movie 8.
(E) Left: γ-Tubulin localization during MT regrowth. Some γ-tubulin signals became the sites of cytoplasmic MT nucleation (arrows). However, in 18% of the cases, cytoplasmic MT nucleation was observed at sites where γ-tubulin was not detected (magnified images are shown on the right). Oryzalin was washed out at time 0. See also Supplemental Movie 9. Bars = 5 μm.
The key to this assay is robust MT depolymerization. A previous study in tobacco bright yellow-2 and Arabidopsis cells suggested that some “seed” MTs were present, even when MTs were challenged with a high concentration of a MT destabilization drug and GFP-tubulin signals consequently disappeared (Lindeboom et al., 2013a). We treated cells with three MT destabilization drugs, propyzamide, cremart, and oryzalin, and confirmed that MT signals disappeared after each treatment (Supplemental Figure 6A). However, when we visualized Citrine-tagged XMAP215 (MOR1)-a (for Xenopus microtubule-associated protein 215 kD [MICROTUBULE ORGANIZATION1]), which binds to MT ends and the lattice, punctate XMAP215-a-Citrine signals were clearly observed when cells were treated with propyzamide and to a lesser extent when cells were treated with cremart (Supplemental Figure 6A). On the other hand, such signals completely disappeared after oryzalin treatment. A plausible explanation for this result is that residual MTs were present after propyzamide or cremart treatment, but nearly absent after oryzalin treatment. Consistent with this interpretation, regrowth of MTs started more quickly when propyzamide was used (1 to 2 min for propyzamide treatment versus 4 to 6 min for oryzalin treatment; Supplemental Figure 6B).
We treated GFP-tubulin expressing cells with a high dose of oryzalin, confirmed the disappearance of MTs, washed out oryzalin by medium exchange, and observed MT recovery with oblique fluorescence illumination (Figure 4A; see Supplemental Figure 7 for the experimental setup). We observed MT reappearance 4 to 6 min after drug washout (Figure 4B; Supplemental Movie 5). Similar to the cytoplasmic nucleation in the normal cell, a bright GFP spot first emerged, followed by the generation of a single MT. The majority of the generation events were cytoplasmic nucleation in the first 7 min after drug washout (87%, n = 28 cells, 121 events). Branching nucleation was infrequently observed (13%). Therefore, we rarely observed fan-like MT amplification, as observed during augmin-dependent MT amplification in vitro (Petry et al., 2013). Furthermore, we occasionally encountered a case in which a stabilized MT remained after oryzalin treatment. However, even in these cells, cytoplasmic nucleation predominated over branching nucleation from residual stable MTs (Figure 4C; Supplemental Movie 6). MT severing was never observed at these early time points. We concluded that cytoplasmic nucleation was the dominant mode of initial MT generation when assessed with this assay.
To test if cytoplasmic nucleation occurred at the surface of a specific organelle, as has been shown or suggested in other systems (the Golgi apparatus [Chabin-Brion et al., 2001; Efimov et al., 2007], mitochondrion [Noguchi et al., 2011], peroxisome [Zekert et al., 2010], nucleus [Shimamura et al., 2004; Murata et al., 2013], and chloroplast [Shimamura et al., 2004]), we prepared cell lines expressing mCherry-tagged (or RFP-tagged) organelle markers and GFP-tubulin. Although some GFP-tubulin signals were close to organelles, we also observed many nucleation events that were not organelle associated (Supplemental Figure 8 and Supplemental Movie 7). These data indicate that nucleation does not always occur at a specific organelle.
γ-Tubulin Is Required for Efficient Nucleation in the MT Regrowth Assay
We used the regrowth assay to assess the effects of depleting possible regulators of MT generation (Figure 5). γ-Tubulin forms a large complex with several other subunits and acts as the major MT nucleator in virtually every cell type (Moritz and Agard, 2001). Augmin recruits γ-tubulin onto the spindle, phragmoplast, or cortical MTs for new MT generation. In protonemal cells of P. patens, the augmin-γ-tubulin machinery predominates in spindle and phragmoplast MT nucleation, as shown with the Aug3 RNAi line (Nakaoka et al., 2012). XMAP215 is the major MT polymerase, but it is also required for the MT nucleating-activity of centrosomes in vitro (Popov et al., 2002). Katanin function was assessed using the aforementioned p60 disruptant.
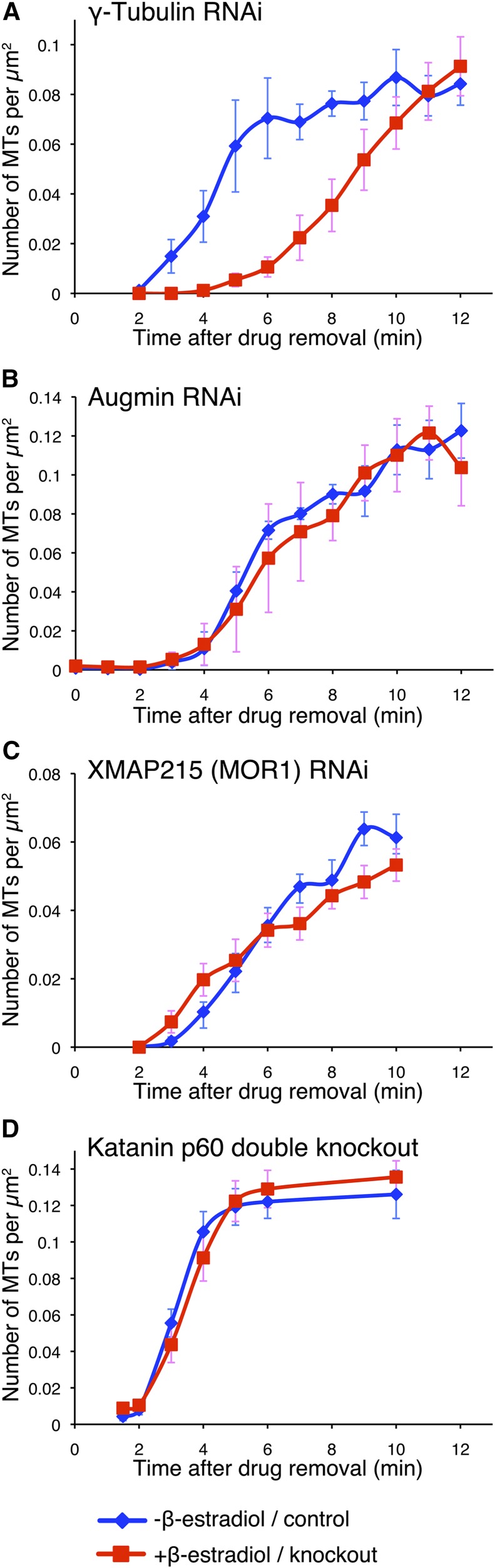
Figure 5.
γ-Tubulin Is Required for Rapid MT Nucleation in the Regrowth Assay.
The MT regrowth assay was used with inducible RNAi lines targeting γ-tubulin (A), the augmin subunit Aug3 (B), and XMAP215/MOR1 (C) and with the knockout line targeting the katanin p60 subunit (D). The number of MTs per area (μm2) was quantified every 1 to 4 min, and the MT density was plotted with the se (each ∼300 μm2 was analyzed). RNAi was induced by adding β-estradiol to the culture medium (red). Untreated cells served as the control (blue). In the case of katanin p60, the GFP-tubulin #14 line (GTU14) (Hiwatashi et al., 2008) was used as the control. Oryzalin alone was used for (A) to (C). In (D), oryzalin was washed out with propyzamide (200 μM)-containing medium, and propyzamide was subsequently washed out. (This experiment was performed when we were unsure of the robustness of oryzalin treatment. We believe that this double treatment is unnecessary given the results described in Supplemental Figure 6.) n = 8 and 7 ([A], control and RNAi, respectively), 4 and 4 ([B], control and RNAi, respectively), 8 and 10 ([C], control and RNAi, respectively), and 22 and 21 ([D], control and knockout).
We induced RNAi for 12 to 17 d, verified the prevalence of cell growth (XMAP215) or elongated spindle (γ-tubulin and Aug3) phenotypes, which are indicators of robust RNAi knockdown (Nakaoka et al., 2012), and performed the MT regrowth assay using oryzalin. Quantification of MT numbers at 1-min intervals indicated that MT generation was significantly delayed by γ-tubulin RNAi, but not by other treatments (Figure 5). In each case, the majority of the initial MT generation events involved cytoplasmic nucleation, as in control cells. We concluded that γ-tubulin was responsible for cytoplasmic MT nucleation in this assay, whereas katanin-mediated severing contributed little to MT production. Furthermore, although the study did not rule out that residual proteins suffice for function, the RNAi results strongly suggest that augmin and XMAP215 make, at most, minor contributions.
γ-Tubulin Often, but Not Always, Colocalizes with MT Nucleation Points
We imaged γ-tubulin-b-Citrine in the presence of oryzalin. Punctate γ-tubulin signals were observed throughout the imaging area, and the majority of the signals displayed a similar intensity (Figure 4D). Time-lapse acquisition at short intervals (∼200 ms) indicated that some signals were mobile, suggesting diffusion in the cytoplasm, whereas other signals were static for >20 s, suggesting that they were anchored at certain structures (Figure 4D; Supplemental Movie 8).
We next acquired images every 3 s after oryzalin washout (Figure 4E; Supplemental Movie 9). As expected, MTs labeled with mCherry-tubulin showed regrowth in the cytoplasm, and cytoplasmic nucleation was detected as mCherry-tubulin spots. A majority of the spots colocalized with the γ-tubulin-Citrine signals (82%, n = 148). The converse was not true; not all γ-tubulin-Citrine signals colocalized with mCherry-tubulin spots during the 12-min imaging period. On the other hand, we failed to detect γ-tubulin signals in 18% of the total nucleation events (Figure 4E, right). As for branching nucleation, γ-tubulin-Citrine colocalized with the nucleation site in 17 of 18 cases, suggesting that most of the events were also γ-tubulin-dependent.
The Three Modes of MT Generation Numerically Account for the Quantified MT Dynamics in a Simple Theoretical Model
We asked whether the three modes of MT generation observed in the cells were sufficient to account for the MT generation dynamics quantified in the MT regrowth assay. For this purpose, we constructed a numerical model of MT generation dynamics, which incorporated all three modes of MT generation with 11 unfixed parameters (Figure 6A; see Methods; Supplemental Table 1). We set up a computational algorithm to search for a set of parameters that reproduced the results of the MT regrowth assay in control cells without any prior knowledge of these parameters (see Methods).
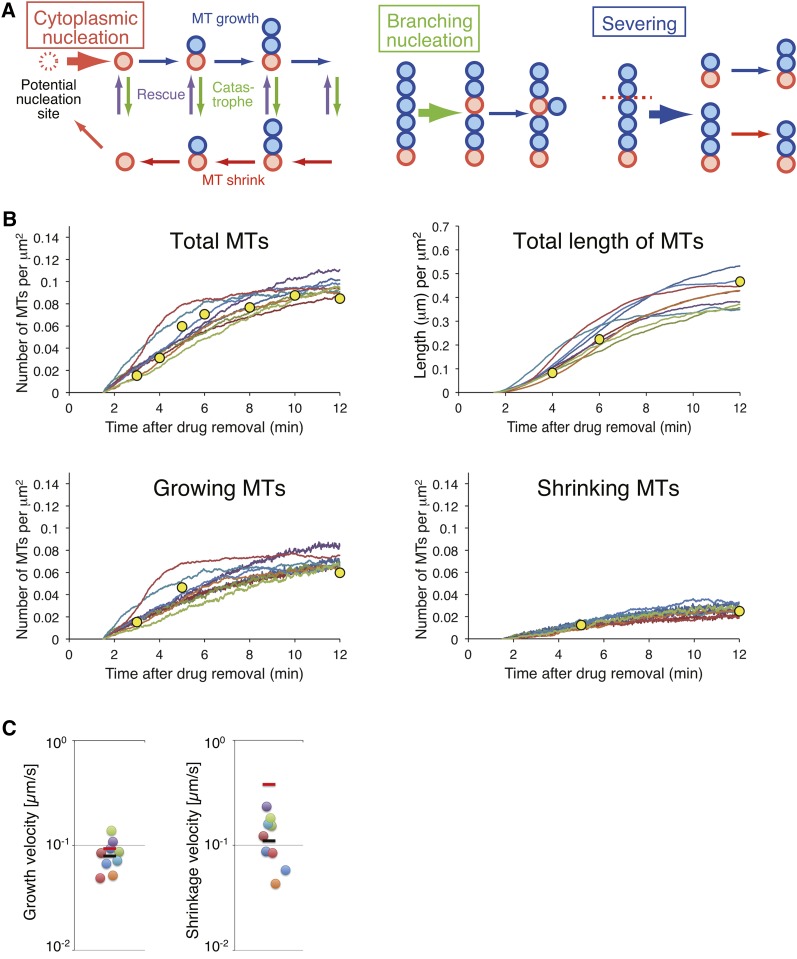
Figure 6.
Numerical Modeling and Fitting to the Experimental Data.
(A) Scheme for the numerical modeling of the nucleation and growth/shrinkage of MTs (see Methods for details). Cytoplasmic nucleation occurs from a potential nucleation site (red dotted circle) in a stochastic manner. Once a nucleation event occurs (red circle), a MT grows and shrinks from the nucleation site according to dynamic instability. Branching nucleation and severing occur stochastically with a rate proportional to the length of a MT. For simplicity, we assumed that severed minus-ends are immediately stabilized.
(B) The number of MTs, the total length of MTs, and the number of growing/shrinking MTs, measured in the MT regrowth assay was reproduced in the numerical model with the parameters estimated through the computational search. The parameter sets that reproduce dominance in cytoplasmic nucleation are displayed (nine independent trials; Supplemental Figure 9). Yellow circles indicate the experimental results (Supplemental Table 2).
(C) The estimated values of growth and shrinkage velocity. Each spot shows the estimated value for each fitting trial (n = 9). The black bar indicates the average value. The red bar shows the experimental value. The vertical axes are shown in logarithmic scales.
Thirty-one sets of parameters that quantitatively reproduced the results of the MT regrowth assay were obtained (Figure 6B; Supplemental Figure 9). Among these, we observed that cytoplasmic nucleation dominated over other generation modes in nine sets of parameters (Supplemental Figure 9). As cytoplasmic nucleation was dominant in cells, we selected these nine sets for further analysis.
The parameter values from the nine sets were used to predict the mean growth and shrinkage velocity of individual MTs (Table 1). The experimental velocity of growth (0.093 μm/s [n = 30]) and shrinkage (0.39 μm/s [n = 30]) was observed to be in the order of magnitude of the estimated parameters (Table 1, Figure 6C, red bars; experimental data, circles; estimated parameter values, black bars; the average of the estimated values). The good agreement between the in vivo results and numerical model indicated that the three modes of MT generation were sufficient to account for the quantified MT dynamics.
Table 1. The Rate of Nucleation and Dynamic Instability of MTs Estimated in Our Numerical Modeling.
| Parameter | Mean (1σ-range)a |
|---|---|
| MT growth velocity (μm/s)b | 0.079 (0.058–0.11) |
| MT shrinkage velocity (μm/s) | 0.11 (0.065–0.19) |
| Catastrophe frequency (/s) | 0.041 (0.0097–0.17) |
| Rescue frequency (/s) | 0.071 (0.011–0.47) |
| Cytoplasmic nucleation (×10−4/s μm2)b | 3.2 (2.1–5.0) |
| Branching nucleation (×10−4/s μm2)c | 0.49 (0.074–3.3) |
| Severing (×10−4/s μm2)c | 1.4 (0.57–3.4) |
n = 9.
See Methods for the calculation of the mean and 1σ(sd)-range.
The maximum value at the onset of simulation when the monomer concentration is the highest.
The maximum value at the end of simulation (12 min after the washout) when the total MT length is the largest.
Nucleation in the Steady State May Not Require γ-Tubulin or Augmin
The MT regrowth assay is an artificial experimental system that does not represent the steady state conditions of protonemal cells. We further analyzed MT nucleation and severing in the steady state, in the absence of γ-tubulin or augmin. Unexpectedly, we observed that the depletion of γ-tubulin or augmin did not significantly affect the rate and mode of MT generation (Tables 2 and 3). The branch angle distribution was also not significantly altered (Supplemental Figure 10; P > 0.2 using the Mann-Whitney U-test). This may not be due to failure in RNAi induction, as the presence of defective spindles in the same samples was confirmed each time prior to image acquisition. The results of MT immunostaining were also consistent with these data; no reduction or organizational defects were observed in endoplasmic MTs, although abnormal mitotic spindles were identified in the same sample (Supplemental Figure 3).
Table 2. Frequency of Three Modes of MT Generation in the Steady State (per μm2).
| Moss Line | Frequency (×10−4/s μm2) (P Value) |
n | Total Area (µm2) | ||
|---|---|---|---|---|---|
| Cytoplasmic Nucleation | Branching Nucleation | Severing | |||
| Control | 0.33 ± 0.19 | 0.21 ± 0.18 | 0.12 ± 0.19 | 21 Cells | 11,223 |
| 0.34 ± 0.16 | 0.20 ± 0.12 | 0.11 ± 0.10 | 6 Experiments | ||
| γ-Tubulin RNAi | 0.24 ± 0.19 (0.12) | 0.15 ± 0.16 (0.24) | 0.10 ± 0.12 (0.76) | 21 Cells | 9,421 |
| 0.26 ± 0.19 (0.47) | 0.19 ± 0.19 (0.92) | 0.08 ± 0.04 (0.58) | 4 Experiments | ||
| Augmin RNAi | 0.29 ± 0.19 (0.59) | 0.22 ± 0.16 (0.95) | 0.05 ± 0.09 (0.26) | 12 Cells | 4,583 |
| 0.30 ± 0.19 (0.76) | 0.24 ± 0.06 (0.64) | 0.04 ± 0.01 (0.29) | 3 Experiments | ||
Values are means ± sd. sd represents either cell-to-cell (top rows) or experiment-to-experiment (bottom rows) variability.
Table 3. Frequency of Branching Nucleation and Severing in the Steady State (per 1-μm MT).
| Frequency (×10−4/s μm) (P Value)a |
n | Total MT Length (μm) | ||
|---|---|---|---|---|
| Branching Nucleation | Severing | |||
| Control | 0.22 ± 0.18 | 0.14 ± 0.22 | 21 Cells | 9705 |
| 0.21 ± 0.12 | 0.12 ± 0.10 | 6 Experiments | ||
| γ-Tubulin RNAi | 0.15 ± 0.14 (0.17) | 0.10 ± 0.11 (0.53) | 21 Cells | 8956 |
| 0.19 ± 0.17 (0.85) | 0.08 ± 0.05 (0.54) | 4 Experiments | ||
| Augmin RNAi | 0.28 ± 0.21 (0.42) | 0.06 ± 0.08 (0.25) | 12 Cells | 3269 |
| 0.31 ± 0.07 (0.21) | 0.05 ± 0.01 (0.33) | 3 Experiments | ||
Values are means ± sd. sd represents either cell-to-cell (top rows) or experiment-to-experiment (bottom rows) variability.
The total MT length of the first and last frames of the image sequence was averaged.
DISCUSSION
This study aimed to elucidate the mechanism of endoplasmic MT generation using protonemal cells of P. patens. Unlike cortical MT arrays, in which fixed-angled branching nucleation and severing efficiently generate parallel bundled MTs, cytoplasmic nucleation and variably oriented branching nucleation were frequently observed at seemingly random locations. Imaging and RNAi analyses suggested that γ-tubulin is the central player in the nucleation process, but also suggested the presence of alternative machinery. Overall, this study presents novel mechanisms of MT generation in plants beyond those observed in cortical MT arrays and further suggests robustness in endoplasmic MT production (Figure 7).
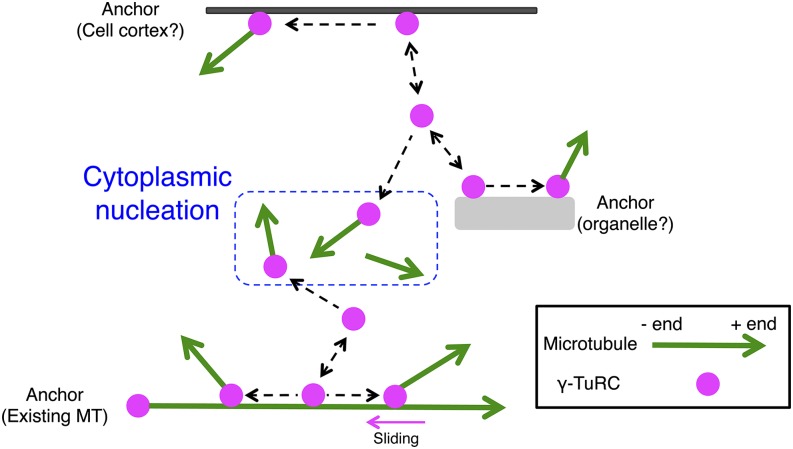
Figure 7.
Model of Endoplasmic MT Generation.
We suggest that cytoplasmic nucleation is the basic mechanism of MT nucleation in protonemal cells. In this model, a γ-tubulin ring complex (γ-TuRC) at the cell cortex, organelle surface, existing MTs, or in the cytoplasm spontaneously nucleates a single MT. We have no direct evidence supporting the model that γ-tubulin is anchored at the cell cortex or organelle surface. However, a subpopulation of γ-tubulin was static for >20 s, suggesting that it anchored to certain structures.
Mechanism of Cytoplasmic Nucleation
Cytoplasmic nucleation, which is rarely detected in cortical or mitotic MTs (1 to 2% of total nucleation events in cortical MT arrays; Nakamura et al., 2010), was observed at the highest frequency in our plant model. On the basis of live-cell imaging, four conclusions were made concerning cytoplasmic nucleation.
First, cytoplasmic nucleation was mostly initiated at the γ-tubulin spot. Each γ-tubulin-Citrine signal was similar in intensity and radial nucleation was rarely observed, suggesting that the spots represent a single γ-TuRC containing 13 to 15 γ-tubulin subunits (Choi et al., 2010; Kollman et al., 2010). We propose a simple view of cytoplasmic nucleation, in which each individual γ-TuRC at unassigned sites nucleates a single new MT. This simple nucleation scheme has been observed in the in vitro reaction of γ-TuRC and tubulin (Choi et al., 2010). However, it differs from the mechanism occurring at centrosomes and other acentrosomal MT organizing centers, such as those found in mice oocytes (Schuh and Ellenberg, 2007). It is possible that MT nucleation by a single γ-TuRC was overlooked in the past because of the limited visualization techniques utilized previously. As demonstrated in this study, oblique illumination fluorescence microscopy combined with the well-controlled drug treatment system may enable visualization of individual nucleation events in a wide variety of plant and animal cell types.
Second, cytoplasmic nucleation is a dynamic event that takes place at seemingly random locations and continues in random directions in this cell type. Some γ-tubulin-Citrine signals were immobile for >20 s, indicating that they are associated with a static structure, such as an organelle surface, the existing MT, or the cell cortex. However, other mobile γ-tubulin can also nucleate MTs. Thus, nucleation may be randomly distributed throughout the cytoplasm. This view differs from that recently observed for the cortical MT array assembled in tobacco cells (Lindeboom et al., 2013a). In this prior study, prepatterned MT remnants also appear to serve as new MT polymerization sites after cell division, efficiently establishing MT polarity in the cortical MT array.
Third, cytoplasmic nucleation appeared to be, to some extent, regulated by the cell cycle. Although cytoplasmic nucleation was frequently observed independent of augmin during interphase, this nucleation mechanism does not compensate for the reduction in MT count following augmin RNAi during mitosis (Nakaoka et al., 2012). Therefore, we hypothesize the occurrence of interphase-specific activation and/or mitosis-specific deactivation mechanisms. Additional research must be conducted to elucidate the regulation of these mechanisms.
Finally, some MTs were nucleated at sites with undetectable levels of γ-tubulin-Citrine. We do not believe this is due to limited microscope sensitivity as we have detected weaker γ-tubulin-b-Citrine signals when the paralogous protein γ-tubulin-a (untagged) was also expressed. Two more likely possibilities are that a subpopulation of MTs is nucleated independent of γ-tubulin or that γ-tubulin was only transiently localized at the very initial phase of nucleation/elongation at which time the MTs were undetectable. We favor the former hypothesis, as cytoplasmic nucleation in the steady state was not affected by RNAi knockdown of γ-tubulin (further discussed below). In either case, the γ-tubulin-Citrine-independent MTs were stable, indicating that the minus-ends must be protected from depolymerization by other proteins. Recently, the CAMSAP protein family has been identified as the minus-end stabilizers in animals (Meng et al., 2008; Goodwin and Vale, 2010). Although CAMSAP homologs have not been found in plants, functionally equivalent proteins might be present.
Mechanism of Branching Nucleation
Branching nucleation was another common mechanism observed in this study, supporting the idea that this mode of nucleation is a general mechanism used to amplify MTs in the acentrosomal system (Murata et al., 2005; Goshima and Kimura, 2010). However, unlike other cases, the branch angle utilized in this cell type was highly variable. Branching nucleation with a fixed angle has been thought to be an excellent cellular approach to not only amplify MT numbers, but also polarize the MT network (Janson et al., 2005; Murata et al., 2005; Kamasaki et al., 2013; Petry et al., 2013). Our observation suggests that protonemal cells utilize this mode of MT nucleation simply to increase MT numbers. In this regard, the branching nucleation in this system might be a variation of cytoplasmic nucleation; cytoplasmic-like nucleation occasionally occurs on MTs, rather than in the cytoplasm. Longitudinal alignment of endoplasmic MTs might occur after MT nucleation, for example, through MT transport (Figure 2B) or cross-linking (Doonan et al., 1985; Hiwatashi et al., 2014).
γ-Tubulin was detected at the majority of the branching sites. In cortical MT arrays, γ-TuRC appears to be much more effective during new MT nucleation when it is bound to the MT compared with being directly bound to the cell cortex, implicating the presence of an adaptor/activator protein on the MT in this system (Nakamura et al., 2010). Recently, augmin was identified as a candidate for the adaptor; in its absence, the recruitment of γ-TuRC and branching nucleation were severely compromised (Liu et al., 2014). However, the observation of nucleation occurring at various angles, even after augmin RNAi in the regrowth assay, suggests that protonemal cells utilize a distinct type of adaptor/activator protein in this process.
Role of MT Severing by Katanin
We assessed the loss-of-function phenotype of katanin for the first time in bryophytes. We demonstrated that katanin-dependent severing is critical for cortical MT organization in the gametophore. However, we also observed that it only modestly contributes to MT severing and amplification in the protonemal cells. This reinforces the idea that the strategy for the production of endoplasmic MTs is different from that for the cortical MTs.
Endoplasmic MT Generation in the Absence of γ-Tubulin
The lack of clear defects in the steady state following γ-tubulin RNAi was unexpected, considering the dominant role of γ-tubulin in MT nucleation in all eukaryotic cells and the fact that RNAi-treated cells showed a clear phenotype at early time points in the MT regrowth assay (Figure 5) or during mitosis (Nakaoka et al., 2012). We cannot exclude the possibility that the residual γ-tubulin after RNAi (∼15%; Nakaoka et al., 2012) could be sufficient for MT nucleation at a normal rate. However, an alternative interpretation of these results would be that, although γ-tubulin-dependent nucleation is a kinetically dominant mechanism when it is present, unknown backup machinery promotes MT nucleation in the absence of γ-tubulin. Indeed, we detected the nucleation of 18% of MTs independent of γ-tubulin association, even when γ-tubulin was present in the cells (Figure 4E). The γ-tubulin-independent mechanism could possibly compete with γ-tubulin for free tubulin in the normal cells. Notably, an identical result has been obtained in Drosophila melanogaster S2 cells, where exclusively acentrosomal MTs were generated in the cytoplasm during interphase (Rogers et al., 2008). In this study, γ-tubulin proteins were almost completely removed from the cell by RNAi (undetectable by immunoblotting). Although MT regrowth after depolymerization was delayed and mitosis was impaired, the steady state interphase MT levels remained unchanged (Rogers et al., 2008). Similarly, in Caenorhabditis elegans, astral MT nucleation occurs partly through a γ-tubulin-independent mechanism, in which aurora-A kinase is involved in a manner that is still unknown (Motegi et al., 2006; Toya et al., 2011). Our study provides yet another possible system in which the mysterious γ-tubulin-independent nucleation mechanism could be pursued.
METHODS
Moss Lines
Physcomitrella patens lines and PCR primers used in this study are listed in Supplemental Tables 3 and 4, respectively. Conditional RNAi lines were selected following the procedure described by Nakaoka et al. (2012). In brief, RNAi constructs cloned into the inducible RNAi vector pGG626 were transformed into the transgenic line expressing GFP-tubulin and histone H2B-mRFP. Transformation was performed with the polyethylene glycol-mediated method using the protoplast generated by Driselase (a gift of Kyowa Hakko Kogyo) treatment. Citrine was fused to the endogenous TubG2 and XMAP215-a genes using the plasmids with ∼1-kb C-terminal and 3′-untranslated region sequences of the genes. TubG1 was disrupted via homologous recombination in the γ-tubulin-b-Citrine-expressing line. Two katanin p60 genes (p60-a and p60-b) were deleted in the GFP-tubulin line. To observe RNAi phenotypes, we cultured protonemata for 12 to 17 d in the presence of 1 μM β-estradiol (Nakaoka et al., 2012). To compare protonemal growth and gametophore development, a section of protonemata was inoculated onto the BCDAT agar medium and cultured at 25°C under continuous light conditions for >3 weeks. Images were acquired using the Nikon SMZ1500 microscope attached to the Digital Sight DS-Fi2 camera.
Microscopy
Protonemal cells grown on BCD medium for 5 to 7 d were used. Cells were homogenized, sandwiched between two sheets of cellophane (a gift from Futamura Chemical Industries), and plated on the BCD agar medium for 5 to 7 d. Sandwiching ensured flat growth of protonemal cells, which was suitable for imaging and drug treatment in the following method. Typically, the protonemal cells and ∼20 μL BCDATG medium were placed on a glass slide and directly covered with a cover slip. Double-sided tape was occasionally used to ensure sufficient space between the cover slip and slide glass. The syringe pump setup for medium exchange is described in Supplemental Figure 7. Briefly, the protonemal cells and ∼1 μL BCDATG medium were placed on a slide glass in which two holes had been made and covered by a cover slip. Medium with or without drug was supplied to the cells using a Harvard Apparatus Pump 11 Elite Syringe Pump (50 μL/min flow). A fluorescence microscope developed for total internal reflection fluorescence imaging (Ti, 100× 1.49 NA lens; Nikon) attached with an EMCCD camera Evolve (Roper) or iXON (DU888E; Andor) was used with oblique illumination fluorescence. Imaging was performed at 24 to 25°C. The endoplasm of the subapical cells (or in rare cases apical cells) that was most closely located to the cover slip was in focus. The microscope was controlled by the Micromanager software or NIS-Elements (Nikon). To observe protonemal cell growth, the cells cultured on BCD agar medium in the glass-bottom plate were imaged. A wide-field microscope (Ti; Nikon) attached to an EMCCD camera iXON (DU888E; Andor) or Evolve (Roper) was used for this purpose. Images were acquired at 3-min intervals from multiple sites. The TE2000 microscope (Nikon), equipped with a spinning-disc confocal unit CSU-X (Yokogawa) and an EMCCD camera ImageEM (Hamamatsu), was used for observing gametophore MTs and immunofluorescence samples. Immunostaining of MTs was performed as previously described (Nakaoka et al., 2012), with minor modifications. Briefly, the cells were cultured for 6 d on a thin layer of BCD agar medium placed in a glass-bottom plate and MTs were stained with a DM1A antibody (Sigma-Aldrich; 1:500).
Image Analysis
Obtained images were manually analyzed with ImageJ and the 3DMOD program included in the IMOD software package (Kremer et al., 1996). In the program, we marked individual MTs, such that the number and length information was obtained (note that actual MT length should be larger than what was measured, since some MTs went beyond the image field). Area, branch angle, and growth/shrinkage rates were measured with ImageJ. Branching nucleation was identified only when the daughter MT end was detected precisely on the mother MT; otherwise, we considered them as crossing MTs. Since the majority of the dynamic MT ends are expected to be plus-ends, the polarity of mother MTs was determined when their dynamic ends were detected. Unless the ends were detected, we did not measure the branch angle.
Biochemistry
Three moss lines were used for immunoprecipitation followed by immunoblotting using anti-γ-tubulin antibody: mCherry-tubulin, mCherry-tubulin/γ-tubulin-b-Citrine, and mCherry-tubulin/γ-tubulin-b-Citrine/γ-tubulin-aΔ. The protonemata cultured on two BCDAT plates for 5 to 6 d were flash frozen with liquid nitrogen, crushed using a mortar and pestle, and suspended in 1.7 mL of HB100 (50 mM HEPES-KOH, pH 7.6, 100 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 1% Nonidet P-40, and protease inhibitors) (Nakaoka et al., 2012). After centrifugation at 22,000g for 30 min at 4°C, the supernatant was mixed with the anti-GFP-conjugated beads (10 μL; MBL) and rotated for 4 h at 4°C. The immunoprecipitants were washed with 4 mL of HB100 and boiled with 100 μL of SDS-containing buffer. Immunoblotting was performed using the G9 antibody, which recognizes P. patens γ-tubulin (1:2000; a gift from Tomohiro Akashi [Nagoya University] and Tetsuya Horio [Nippon Sport Science University] (Horio et al., 1999).
Computer Simulation
Numerical modeling was performed for the nucleation, severing, and dynamic instability of MTs. Our model comprised three potential modes of MT generation (cytoplasmic nucleation, branching nucleation, and severing), elongation and shrinkage of MTs, and 11 parameters (Supplemental Table 1). We used the model to output the number and length of MTs in the system (Figure 6A). At every step of the simulation (Δt = 0.5 s/step), cytoplasmic nucleation occurred stochastically with a probability of NucTot × NucRate × (FreeTub/TubTot)NucAlpha [/s area]. The NucTot parameter denoted the number of potential nucleation sites per unit area and was included to consider the effect of area size. In our model, the unit area corresponded to 1120 μm2 (=100,000 pixels2) in the actual microscopic observation. TubTot denoted the number of total tubulin monomers per unit area, and 1 unit of TubTot corresponded to 0.25 μm of MT. FreeTub was calculated through subtracting the total length of MTs from TubTot. NucRate and NucAlpha defined the rate of cytoplasmic nucleation. Since the nucleation of MTs is not always proportional to the concentration of tubulin monomers (Howard, 2001), we introduced the exponent NucAlpha as an unknown parameter.
Branching nucleation occurred on each MT with a probability of MDMNRate × (FreeTub/TubTot)MDMNAlpha × 4 [/s μm of MT]. Again, we introduced an exponent parameter (MDMNAlpha) to account for the dependency on tubulin monomer concentration. Severing-mediated MT generation occurred on each MT with a probability of SevRate × 4 [/s μm of MT], assuming that the severing probability was simply proportional to the length of the MT.
Post nucleation, the size of MTs changed according to dynamic instability (Mitchison and Kirschner, 1984; Nédélec, 2002; Kimura and Onami, 2010). After cytoplasmic and branching nucleation, newly generated MTs entered the growing phase wherein the MT elongated by 1 unit (= 0.25 μm) with a probability of Kgrowth × (FreeTub/TubTot) [/s] at its plus-end at every simulation step. For severing, the newly generated plus-end of the MT started shrinking. In the shrinking phase, the MT shrank by 1 unit (= 0.25 μm) with a probability of Kshrink [/s] at its plus-end. In the model, the minus-ends were assumed to be in a stable state. Whereas a MT in the growing phase transformed into shrinking phase with a probability of FreqCat [/s], a MT in the shrinking phase entered the growing phase with a probability of FreqRes [/s]. If a MT with a length of 1 unit shrank, the MT disappeared from the model.
For one simulation cycle, the process of nucleation, growth/shrinkage, and phase transition was repeated over 1260 steps, which corresponded to 10.5 min. On the basis of cellular observation, we assumed that the first nucleation occurred 1.5 min after washout of the MT destabilizing drug in the MT regrowth assay.
Estimation of parameter values by automated fitting of modeling results to experimental results was performed as follows. For a single estimation trial, 96 sets of the 11 parameters were randomly selected, where the parameter values fell within the search range indicated in Supplemental Table 1. For each set of parameters, a simulation was run. We evaluated how well the simulation results agreed with the experimental data by calculating the evaluation score, S, defined as follows: S = exp(-4/3×Σi=1N(1-Silico(i)/Vivo(i))2). Vivo(i) (i=1,2,…N) denoted the experimental value of the number or length of MTs (as shown in Supplemental Table 2) and Silico(i) denoted the corresponding value in the simulation.
Next, each parameter value was changed by multiplying the original value by 2α, where α was a standard normal random variable. The simulation was run again with the new parameter sets and the score, S, was calculated. If the score of the new parameter set (Spost) was greater than that of the old parameter set (Spri), the new set was adopted in place of the old set. If Spost < Spri, the new parameter set was adopted as the revised parameter set with a probability of Spost/Spri. Otherwise, the old parameter set and its values remained for the next round of parameter change. This procedure is similar to the Markov chain Monte Carlo (MCMC) method (Madras, 2002), which is expected to converge to a parameter set with the highest score.
The MCMC procedure was repeated for 200 cycles and then resampling was performed. The importance weight (W(j), j = 1,2,…96) of the 96 parameter sets was calculated by normalizing their latest scores (S(j)) as W(j) = S(j)/Σj = 196 S(j), such that Σj = 196 W(j) = 1. The 96 parameter sets were revised by assigning the original j-th parameter set to the integer nearest to 96 × W(j) among 96. This procedure is similar to the resampling process in particle filter, which can concentrate parameter sets with higher scores to the next round (Nakamura et al., 2009).
After the initial round of resampling, the MCMC procedure was repeated for another 20 cycles before resampling again. The process of 20 rounds of MCMC followed by resampling was repeated over 120 cycles for each fitting trial. The final 96 sets of parameters were averaged proportional to the weight (W(j)) of each set to give the final output.
A manual simulation was performed using the parameter values from the final output and the nucleation events were counted for the time window corresponding to 0 to 7 min of the regrowth assay. Among 31 independently estimated sets of parameters, nine sets reproduced the dominance of cytoplasmic nucleation during this period, as observed in the experiment (Supplemental Figure 9). These nine sets of parameters were used for further analyses.
The mean and sd (σ) of the estimated values of the nine sets were calculated (Table 1). Because the parameters were searched by changing the values by multiplying the original value with 2α as described above, the logarithm of the parameter values was used to calculate the mean and σ. More specifically, for the estimated parameters x1, x2, …, xn (n = 9), the mean, and sd of log(x1), log(x2), …, log(xn) were first calculated. Using this mean value (‘m’) and sd (‘s’), the final mean value was calculated as exp(m) and 1σ-range as from exp(m - s) to exp(m + s) in Table 1. The vertical axes of Figure 6C are therefore shown in logarithmic scale.
Accession Numbers
P. patens genes used in this study are as follows:γ-tubulin-a (Pp1s234_17), γ-tubulin-b (Pp1s266_15), XMAP215/MOR1-a (Pp1s66_170), katanin p60-a (Pp1s213_118), katanin p60-b (Pp1s99_113), and Man1 (Pp1s387_34). These sequences are found in Phytozome (http://www.phytozome.net/physcomitrella.php). Gene IDs used for the phylogenetic analysis are as follows: At-KTN1 (AT1G80350), Selaginella moellendorffii (Sm)-katanin p60 (78692), and Homo sapiens (Hs)-katanin p60 subunit A1 (O75449).
Supplemental Data
Supplemental Figure 1. Construction of the γ-Tubulin-Citrine Replacement Line.
Supplemental Figure 2. Construction of the Katanin p60 Disruptant.
Supplemental Figure 3. Orientation of Endoplasmic MTs Is Not Significantly Changed in γ-Tubulin RNAi- or Augmin (Aug3) RNAi-Treated Cells.
Supplemental Figure 4. Summary of Daughter MT Migration after Branching Nucleation.
Supplemental Figure 5. Criteria for Identification of Cytoplasmic Nucleation.
Supplemental Figure 6. Effectiveness of Three MT Destabilization Drugs.
Supplemental Figure 7. MT Depolymerization-Regrowth Assay Using a Syringe Pump.
Supplemental Figure 8. Cytoplasmic Nucleation Does Not Occur at a Specific Organelle.
Supplemental Figure 9. Supplementary Data for Numerical Modeling.
Supplemental Figure 10. Branch Angle Is Unchanged after γ-Tubulin or Augmin RNAi Treatment.
Supplemental Table 1. Eleven Unfixed Parameters Used for Modeling.
Supplemental Table 2. The Number and Length of MTs Observed in the MT Regrowth Assay Used for Fitting with the Numerical Model.
Supplemental Table 3. List of Transgenic Lines Used in This Study.
Supplemental Table 4. List of PCR Primers Used in This Study.
Supplemental Data Set 1. Protein Sequences Used to Generate the Phylogeny Presented in Supplemental Figure 2A.
Supplemental Movie 1. Endoplasmic MT Dynamics.
Supplemental Movie 2. MT Severing.
Supplemental Movie 3. Branching MT Nucleation.
Supplemental Movie 4. Cytoplasmic MT Nucleation.
Supplemental Movie 5. MT Depolymerization-Regrowth Assay.
Supplemental Movie 6. MT Regrowth with a Stable MT.
Supplemental Movie 7. Organelle-Independent MT Nucleation.
Supplemental Movie 8. γ-Tubulin Dynamics.
Supplemental Movie 9. γ-Tubulin-Dependent MT Nucleation.
Supplementary Material
Acknowledgments
We thank Yuji Hiwatashi, Mitsuyasu Hasebe, Mamoru Sugita, Jeroen de Keijzer, Shu-Zon Wu, Magdalena Bezanilla, and Ken Kosetsu for plasmids and moss lines; Tomoko Nishiyama for use of her equipment; and Momoko Nishina and Rie Inaba for technical assistance. This work was supported by the Human Frontier Science Program, Grants-in-Aid for Scientific Research (MEXT), the Toray Science Foundation, the James A. and Faith Miller Memorial Fund, the Laura and Arthur Colwin Endowed Summer Research Fellowship Fund (to G.G.), and the Data Centric Science Research Commons of the Research Organization of Information and Systems, Japan (to A.K.). Y.N. is a recipient of the JSPS predoctoral scholarship.
AUTHOR CONTRIBUTIONS
Y.N., A.K., T.T., and G.G. designed the research. Y.N. performed experiments. A.K. conducted computer simulations. Y.N., A.K., and G.G. analyzed data. Y.N., A.K., and G.G. wrote the article.
Glossary
- MT
microtubule
- RNAi
RNA interference
- γ-TuRC
γ-tubulin ring complex
- MCMC
Markov chain Monte Carlo
Footnotes
Articles can be viewed online without a subscription.
References
- Bartolini F., Gundersen G.G. (2006). Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119: 4155–4163. [DOI] [PubMed] [Google Scholar]
- Brown R.C., Lemmon B.E., Olsen O.A. (1994). Endosperm development in barley: Microtubule involvement in the morphogenetic pathway. Plant Cell 6: 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. (2001). The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 12: 2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Sambade A., Calder G., Lloyd C. (2009). Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.K., Liu P., Sze S.K., Dai C., Qi R.Z. (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. (2005). The moss Physcomitrella patens. Annu. Rev. Genet. 39: 339–358. [DOI] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004). The cortical microtubule array: from dynamics to organization. Plant Cell 16: 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J.H., Cove D.J., Lloyd C.W. (1985). Immunofluorescence microscopy of microtubules in intact cell lineages of the moss, Physcomitrella patens. I. Normal and CIPC-treated tip cells. J. Cell Sci. 75: 131–147. [DOI] [PubMed] [Google Scholar]
- Doonan J.H., Cove D.J., Lloyd C.W. (1988). Microtubules and microfilaments in tip growth: evidence that microtubules impose polarity on protonemal growth in Physcomitrella patens. J. Cell Sci. 89: 533–540. [Google Scholar]
- Efimov A., et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12: 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel E.A., Dixit R. (2013). Role of nucleation in cortical microtubule array organization: variations on a theme. Plant J. 75: 270–277. [DOI] [PubMed] [Google Scholar]
- Goodwin S.S., Vale R.D. (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kimura A. (2010). New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 22: 44–49. [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y., Sato Y., Doonan J.H. (2014). Kinesins have a dual function in organizing microtubules during both tip growth and cytokinesis in Physcomitrella patens. Plant Cell 26: 1256–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi Y., Obara M., Sato Y., Fujita T., Murata T., Hasebe M. (2008). Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 20: 3094–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T., Basaki A., Takeoka A., Yamato M. (1999). Lethal level overexpression of gamma-tubulin in fission yeast causes mitotic arrest. Cell Motil. Cytoskeleton 44: 284–295. [DOI] [PubMed] [Google Scholar]
- Hotta T., Kong Z., Ho C.M., Zeng C.J., Horio T., Fong S., Vuong T., Lee Y.R., Liu B. (2012). Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell 24: 1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. (2001). Mechanics of Motor Proteins and the Cytoskeleton. (Sunderland, MA: Sinauer Associates). [Google Scholar]
- Janson M.E., Setty T.G., Paoletti A., Tran P.T. (2005). Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J. Cell Biol. 169: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., O’Toole E., Kita S., Osumi M., Usukura J., McIntosh J.R., Goshima G. (2013). Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 202: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Onami S. (2010). Modeling microtubule-mediated forces and centrosome positioning in Caenorhabditis elegans embryos. Methods Cell Biol. 97: 437–453. [DOI] [PubMed] [Google Scholar]
- Kirik A., Ehrhardt D.W., Kirik V. (2012). TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman J.M., Polka J.K., Zelter A., Davis T.N., Agard D.A. (2010). Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196. [DOI] [PubMed] [Google Scholar]
- Kosetsu K., de Keijzer J., Janson M.E., Goshima G. (2013). MICROTUBULE-ASSOCIATED PROTEIN65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in Physcomitrella patens. Plant Cell 25: 4479–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116: 71–76. [DOI] [PubMed] [Google Scholar]
- Lecland N., Lüders J. (2014). The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 16: 770–778. [DOI] [PubMed] [Google Scholar]
- Lindeboom J.J., Lioutas A., Deinum E.E., Tindemans S.H., Ehrhardt D.W., Emons A.M., Vos J.W., Mulder B.M. (2013a). Cortical microtubule arrays are initiated from a nonrandom prepattern driven by atypical microtubule initiation. Plant Physiol. 161: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., Emons A.M., Mulder B.M., Kirik V., Ehrhardt D.W. (2013b). A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342: 1245533. [DOI] [PubMed] [Google Scholar]
- Liu T., Tian J., Wang G., Yu Y., Wang C., Ma Y., Zhang X., Xia G., Liu B., Kong Z. (2014). Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 24: 2708–2713. [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646. [DOI] [PubMed] [Google Scholar]
- Madras N.N. (2002). Lectures on Monte Carlo Methods. (Providence, RI: American Mathematical Society. [Google Scholar]
- Meng W., Mushika Y., Ichii T., Takeichi M. (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135: 948–959. [DOI] [PubMed] [Google Scholar]
- Miki T., Nishina M., Goshima G. (2015). RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol. pii:doi/10.1093/pcp/pcv002. [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. (1984). Dynamic instability of microtubule growth. Nature 312: 237–242. [DOI] [PubMed] [Google Scholar]
- Moritz M., Agard D.A. (2001). Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11: 174–181. [DOI] [PubMed] [Google Scholar]
- Motegi F., Velarde N.V., Piano F., Sugimoto A. (2006). Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev. Cell 10: 509–520. [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7: 961–968. [DOI] [PubMed] [Google Scholar]
- Murata T., Sano T., Sasabe M., Nonaka S., Higashiyama T., Hasezawa S., Machida Y., Hasebe M. (2013). Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 4: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito H., Goshima G. (2015). NACK kinesin is required for metaphase chromosome alignment and cytokinesis in the moss Physcomitrella patens. Cell Struct. Funct. 40: doi/10.1247/csf.14016. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Yoshida R., Nagasaki M., Miyano S., Higuchi T. (2009). Parameter estimation of in silico biological pathways with particle filtering towards a petascale computing. Pac. Symp. Biocomput. 14: 227–238. [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070. [DOI] [PubMed] [Google Scholar]
- Nakaoka Y., Miki T., Fujioka R., Uehara R., Tomioka A., Obuse C., Kubo M., Hiwatashi Y., Goshima G. (2012). An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 24: 1478–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec F. (2002). Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Koizumi M., Hayashi S. (2011). Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 21: 805–814. [DOI] [PubMed] [Google Scholar]
- Petry S., Groen A.C., Ishihara K., Mitchison T.J., Vale R.D. (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov A.V., Severin F., Karsenti E. (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Curr. Biol. 12: 1326–1330. [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Peifer M., Rogers S.L. (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell 19: 3163–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130: 484–498. [DOI] [PubMed] [Google Scholar]
- Shimamura M., Brown R.C., Lemmon B.E., Akashi T., Mizuno K., Nishihara N., Tomizawa K., Yoshimoto K., Deguchi H., Hosoya H., Horio T., Mineyuki Y. (2004). Gamma-tubulin in basal land plants: characterization, localization, and implication in the evolution of acentriolar microtubule organizing centers. Plant Cell 16: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. (2008). Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5: 159–161. [DOI] [PubMed] [Google Scholar]
- Toya M., Terasawa M., Nagata K., Iida Y., Sugimoto A. (2011). A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat. Cell Biol. 13: 708–714. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Galway M.E. (2003). Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu. Rev. Plant Biol. 54: 691–722. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Ambrose J.C. (2009). Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19: 62–71. [DOI] [PubMed] [Google Scholar]
- Zekert N., Veith D., Fischer R. (2010). Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot. Cell 9: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fishel E., Bertroche T., Dixit R. (2013). Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23: 2191–2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.