F. solaris has an allodiploid genome structure, and activation of lipid accumulation and degradation metabolism pathways at the same time might underlie its simultaneous growth and oil accumulation.
Abstract
Oleaginous photosynthetic organisms such as microalgae are promising sources for biofuel production through the generation of carbon-neutral sustainable energy. However, the metabolic mechanisms driving high-rate lipid production in these oleaginous organisms remain unclear, thus impeding efforts to improve productivity through genetic modifications. We analyzed the genome and transcriptome of the oleaginous diatom Fistulifera solaris JPCC DA0580. Next-generation sequencing technology provided evidence of an allodiploid genome structure, suggesting unorthodox molecular evolutionary and genetic regulatory systems for reinforcing metabolic efficiencies. Although major metabolic pathways were shared with nonoleaginous diatoms, transcriptome analysis revealed unique expression patterns, such as concomitant upregulation of fatty acid/triacylglycerol biosynthesis and fatty acid degradation (β-oxidation) in concert with ATP production. This peculiar pattern of gene expression may account for the simultaneous growth and oil accumulation phenotype and may inspire novel biofuel production technology based on this oleaginous microalga.
INTRODUCTION
Sustainable energy production without massive CO2 release is a critical issue to be addressed in the 21st century, when we face exhaustion of fossil fuel reserves as well as their negative effects on climate. Bioenergy production from the photosynthetic systems of land plants has been recognized as a promising solution, but current systems are often in competition with food production and require vast areas of cultivated land to accommodate the low productivity of traditional annual crops. Identification of alternative energy producers is therefore an important objective. Microalgae provide one such alternative source of bioenergy, with their high rates of CO2 fixation, great biomass yields, and the fact that they do not compete with food crops for resources (Smith et al., 2010). Recent studies have identified candidate species for biofuel production within several microalgal groups, including Chlorophyta, Heterokontophyta (including Bacillariophyceae [diatoms] and Eustigmatophyceae), Haptophyta, Rhodophyta, and Dinophyceae (Fuentes-Grünewald et al., 2009; Oh et al., 2009; Rodolfi et al., 2009; Mahapatra et al., 2013).
We recently reported the characterization of the oleaginous diatom Fistulifera solaris JPCC DA0580 from our marine microalgal culture collection (Matsumoto et al., 2010, 2014). Its high neutral lipid content (40 to 60%, w/w) and growth rate are beneficial for biodiesel production. Another critical advantage is the temporal overlap of lipid accumulation and cell growth during the logarithmic phase (Satoh et al., 2013), a feature that is absent in more conventional oleaginous microalgae such as Nannochloropsis sp (Radakovits et al., 2012) or Neochloris oleoabundans (Rismani-Yazdi et al., 2012), which tend to accumulate oil during the stationary phase. F. solaris is therefore one of the most promising biodiesel feedstock candidates for use with repeated batch culture technology, in which cells are maintained in logarithmic growth by repetitive culture dilution to improve biomass production (Sato et al., 2014).
To fully exploit the potential of this strain, we must understand the molecular underpinnings of its ability to simultaneously grow and accumulate oil and use this mechanism to improve lipid productivity by metabolic engineering (Muto et al., 2013a). With this goal in mind, we used next-generation sequencing technology for whole-genome and transcriptome analyses. Our study revealed an allodiploid genome structure that, to our knowledge, has never been observed in microalgae. We also identified the major metabolic pathways and characterized their transcriptional regulators, allowing us to propose a mechanism by which F. solaris achieves simultaneous growth and lipid accumulation. Because omics data from oleaginous microalgae are sparse (Radakovits et al., 2012; Rismani-Yazdi et al., 2012), our integrative study represents a significant contribution to the field of biodiesel production.
RESULTS
Genome Sequencing and Assembly
The genome of F. solaris was sequenced on an FLX System, which generated ∼1.24 gigabases in reads (Supplemental Table 1). Assembly of these sequences generated 3913 contigs, which were assembled into 297 scaffolds ranging from 21 kb to 1.6 Mb. The scaffolds were assembled with LASTZ and LALIGN to yield a draft genome sequence corresponding to 24.8-fold redundant sequence coverage. At this stage, a chloroplast genome (135 kb) (Tanaka et al., 2011) and a mitochondrial genome (>38.6 kb) were identified; however, we failed to determine the chromosome structures from the alignment blocks in the draft nuclear genome.
During the preliminary assembly, we noticed a curious result: most BLAST searches of genes of interest from the draft genome yielded duplicate genes with highly conserved but not identical DNA sequences. This prompted a more careful analysis of the alignment blocks generated by LASTZ and LALIGN, bringing us to the important discovery that many blocks have counterparts with sequence similarity and well-conserved synteny. These conserved (but not identical) nucleotide and gene arrangements helped fill the gaps between blocks that could not be connected by the assembly algorithms. Through this assembly procedure, the sequenced blocks eventually converged into 84 hypothetical chromosomes, all of which form pairs based on sequence similarity (Figure 1; Supplemental Figure 1). Note that unassembled blocks remained (Supplemental Data Set 1), and the assembly of 84 chromosomes remains tentative. Telomeric repeats (CCCTAA) (Bowler et al., 2008) were found at the ends of 36 of the 84 chromosomes (Supplemental Data Set 1). Throughout the genome, the chromosome pairs showed near-perfect conservation of synteny (Supplemental Figure 2). Sequence variation between chromosome pairs ranged from 14 to 38% (25% on average). This amount of sequence variation in the genome of F. solaris is much higher than that other diatom genomes that are believed to be diploid (0.85% in Phaeodactylum tricornutum [Bowler et al., 2008] and 0.75% in Thalassiosira pseudonana [Armbrust et al., 2004]) and is comparable to allodiploid yeasts (∼15 to 30%) (Horinouchi et al., 2010; Louis et al., 2012; Morales and Dujon, 2012). Such variation could cause the formation of distinct chromosome assemblies. Thus, we hypothesized that F. solaris may also be an allodiploid, defined as a diploid organism containing two distinct subgenomes resulting from interspecies hybridization of haploids.
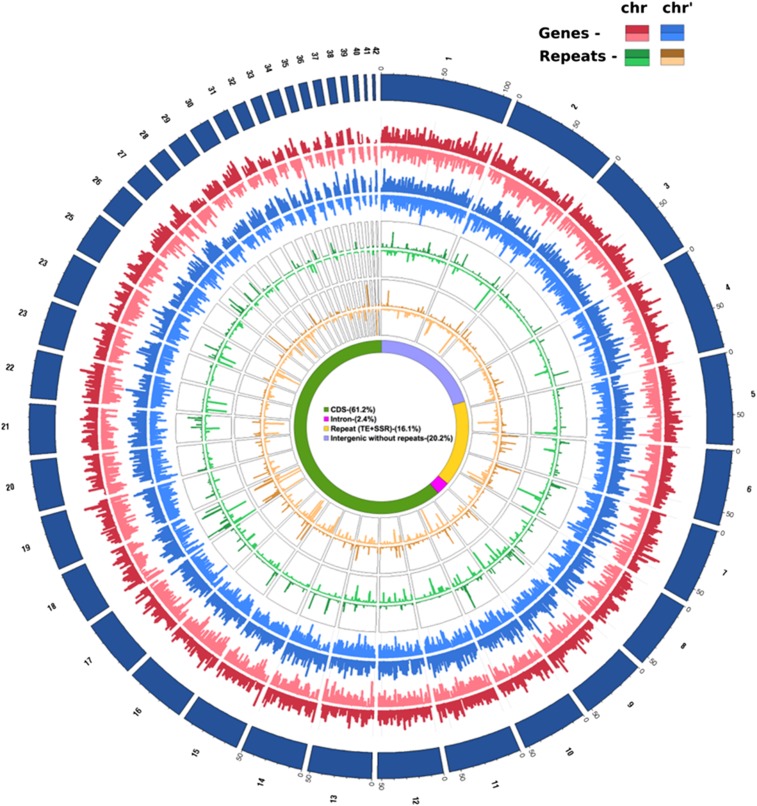
Figure 1.
Circular View of the Genomic Landscape of F. solaris JPCC DA0580.
Forty-two chromosome pairs are represented with genes (red for positive strand and light red for negative strand) and repeat elements (green for positive strand and light green for negative strand). The chromosome numbers (black) are marked on the outermost circle (dark blue). Within each chromosome pair, the chromosome with higher GC content is defined as chrX (X is the chromosome number) and the other as chrX′. The inner four tracks (blue and orange) of genes and repeats are for the second pair of 42 chromosomes. The innermost circle is a pie chart showing the percentage of each genomic feature in diploid genome coverage.
A summary of genome structure and gene content is shown in Table 1 and Supplemental Data Set 1. The total nuclear genome assembly with gaps was 49.7 Mb, which is the total length of scaffolds, including all 84 homoeologous chromosomes (defined as chromosome pairs duplicated by interspecies hybridization, as in the example of allodiploid yeast [Louis et al., 2012]). By contrast, the genome sizes of P. tricornutum and T. pseudonana were reported in their haploid state. Therefore, for better comparison, the genome of F. solaris was considered to be half the total length of the scaffolds: 24.9 Mb. Compared with the genomes of the other two diatoms, the F. solaris genome is closer to that of P. tricornutum (27.4 Mb) than to that of T. pseudonana (32.4 Mb). Indeed, both F. solaris and P. tricornutum are pennate diatoms, while T. pseudonana is a centric diatom. The F. solaris genome has a relatively low GC content (46%). A total of 20,455 predicted protein-coding genes were identified using AUGUSTUS version 2.5.5 customized for F. solaris (Nemoto et al., 2014), including the 9007 homoeologous gene pairs (defined as gene pairs duplicated by interspecies hybridization; see also Methods section) double-counted as distinct genes. For better comparison, this double-counting was eliminated; thus, the number of predicted genes in F. solaris was considered to be 11,448, comparable to other sequenced diatoms. More than 80% (9007/11,448) of the genes have homoeologous counterparts.
Table 1. General Features of Sequenced Diatom Genomes.
| F. solaris JPCC DA0580 | P. tricornutum | T. pseudonana | |
|---|---|---|---|
| Nuclear genome | |||
| Estimated genome size (Mb) | 24.9 (49.7)a | 27.4 | 32.4 |
| Chromosome | 42 (84)b | 33 | 24 |
| Predicted protein-coding genes | 11,448 (20,455)c | 10,402 | 11,776 |
| GC content (%) | 46 | 49 | 47 |
| Proportion of TE (%) | 16 | 6.4 | 7.3 |
| Chloroplast genome | |||
| Estimated genome size (kb) | 135 | 117 | 129 |
| Predicted protein-coding genes | 132 | 130 | 127 |
| GC content (%) | 32 | 33 | 31 |
| Mitochondria genome | |||
| Estimated genome size (kb) | >38.6d | 77.4 | 43.8 |
| Predicted protein-coding genes | 31 | 32 | 35 |
| GC content (%) | 28 | 35 | 30 |
The total nuclear genome assembly with gaps was 49.7 Mb, which is equal to the total read length. An allodiploid genome structure could cause an increase in genome size in comparison to other microalgae. Since the sizes of P. tricornutum and T. pseudonana were described as haploid, the genome size of F. solaris was determined as half the total length of the scaffolds (24.9 Mb) for better comparison.
Scaffold assembly resulted in the formation of 42 homoeologous chromosome pairs; thus, 84 types of chromosomes were confirmed.
Estimated number of homoeologous gene pairs: 9007.
Mitochondrial genome of F. solaris has repeat sequences that are unreadable by the high-throughput DNA sequencing method employed here. The mitochondrial genome did not include the repeat sequences.
We also identified 135 chloroplast genes and 31 mitochondrial genes (the chloroplast genes were previously reported [Tanaka et al., 2011]). No homoeologous gene pairs were found in the mitochondrial or chloroplast genomes. The homoplasmic nature of organelle genomes was consistent with the mitochondrial genome of allopolyploid yeast (Sipiczki, 2008). It should be noted that the mitochondrial genome assembly is incomplete, possibly due to the large inverted repeats commonly found in the mitochondrial genomes of diatoms (Oudot-Le Secq and Green, 2011). The sequenced region of the mitochondrial genome with no long repeat sequences is 38.6 kb, similar in size to the nonrepeat regions of the mitochondrial genomes of T. pseudonana (38.8 kb with 5.0-kb repeat region) and P. tricornutum (42.0 kb with 35.4-kb repeat region).
Allodiploid Genome Structure
We continued to explore the possibility that F. solaris is an allodiploid by analyzing the 18S rRNA gene (rDNA) sequence. In general, individual organisms have unique 18S rDNA sequences, and interspecies hybrids (such as allodiploids) have multiple sequences derived from the parental species. Partial 18S rDNA was PCR amplified from the F. solaris genome and sequenced, revealing abundant polymorphisms (Supplemental Figure 3A). The data suggest the amplified fragments include two distinct types of 18S rDNA, and their abundance was ∼1:1, as indicated by the similar heights of the merged DNA sequence peaks. To separate the different 18S rDNA fragments and obtain pure sequence information for each type, the PCR products were cloned and resequenced. Sequencing of 10 clones revealed five each of two types of homoeologous 18S rDNA (Supplemental Figures 3B and 3C), suggesting that two types of 18S rDNA coexist in the genome.
Although the homoeologous 18S rDNA pair supports the allodiploid genome structure of F. solaris, a critical drawback of the PCR method is the inability to exclude the possibility of cellular contaminants, as the template DNA was extracted from a cell pellet. To address this issue, we attempted to detect distinct homoeologous gene pairs from a single cell using the microcavity array technique (Hosokawa et al., 2009). A unique feature of this technique is the rapid and efficient array of single cells at even intervals on a planar substrate with small holes (3 μm), which allows the recovery of single cells (Supplemental Figures 4A and 4B). After arraying the single cells, several were carefully recovered with a micromanipulator and used as a template for PCR detection of the homoeologous gene set. Because the amount of nucleic acid in a single cell is extremely low for PCR, we targeted the mRNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene that is stably and strongly expressed during all growth phases. Following RT-PCR with specific primer pairs for each of the homoeologous GAPDH genes (g19411 and g12876), PCR products for both genes were detected in several single cell samples (Supplemental Figure 4C). Amplification specificity was confirmed by DNA sequencing. The results indicated that two different types of the GAPDH gene coexist and are expressed in a single cell.
These experiments provide strong evidence that F. solaris is an allodiploid possessing homoeologous gene pairs. This led us to ask whether these homoeologous gene pairs display similar or different expression patterns. We performed a genome-wide transcriptome analysis and compared transcript abundance in each homoeologous gene pair. cDNA libraries were prepared from 48-h (linear phase), 96-h (late-linear phase), and 144-h (stationary phase) cultures and analyzed with the Genome Analyzer IIx system. Transcript abundance is expressed as reads per kilobase of exon model per million mapped reads (RPKM; see Methods). The expression patterns were statistically analyzed, classified as either synchronized or differential expression, and summarized in terms of each chromosome pair (Supplemental Figure 5; see Methods). This comparison revealed that ∼80% of the gene pairs were differentially expressed and that only ∼20% of the gene pairs were expressed synchronously. Interestingly, this ratio was conserved in all but the shortest two chromosome pairs (Chr41/41′ and Chr42/42′).
Repeats and Transposable Elements
Transposable elements (TEs) are an important component of genome structure in many organisms; they influence molecular evolution, gene expression, and epigenetic regulation. Varied types and proportions of TEs are found in diatoms. Class I (retrotransposon) and class II (DNA transposon) TEs are present in considerable numbers in both pennate and centric diatoms (Maumus et al., 2009); the percentage of each genome occupied by repetitive elements is 6.4% in P. tricornutum and 1.9% in T. pseudonana. Within the long-terminal repeat class-I TEs, diatom-specific Ty1/Copia-like elements (denoted CoDi) have been found in both diatoms and may contribute to the rapid diversification rates observed in these organisms (Maumus et al., 2009).
Sixteen percent of the F. solaris genome comprised simple sequence repeats, suggesting F. solaris has more TEs than do the other two diatoms. Furthermore, the most abundant repetitive elements were long-terminal repeat TEs (2.6%), predominantly Copia-type elements (1.1%). A hidden Markov model (HMM) of the reverse transcriptase domain was used to search against other Copia-type elements to classify the CoDi groups. The predicted 65 sequences of RT domains in F. solaris were used to construct a phylogenetic tree with the known CoDi sequences from P. tricornutum and T. pseudonana. Among the seven CoDi groups reported earlier, five groups (CoDi2, 3, 4, 5, and 6) were also found in F. solaris (Supplemental Figure 6). As reported earlier, CoDi4, 5, and 6 are shared among all diatoms (Maumus et al., 2009); in the case of F. solaris, the CoDi6 group is very diverse and includes 19 sequences. The CoDi2 and CoDi3 groups, originally thought to be specific to P. tricornutum, were also found in F. solaris. CoDi1 and CoDi7 were not found and therefore could be specific to P. tricornutum. A new CoDi group specific to F. solaris was also detected (denoted CoDi8, which could be further divided into subclasses; Supplemental Figure 6).
Comparative Genomics
To characterize the uniqueness of the F. solaris genome, its genes were compared with other typical eukaryotes, including other sequenced diatoms (P. tricornutum, T. pseudonana, Thalassiosira oceanica, and Fragillariopsis cylindrus) (Supplemental Figure 7). Of the genes encoded in the F. solaris genome, the numbers of diatom-, pennate-, and F. solaris-specific genes were 844, 231, and 1381, respectively (diatom-specific genes were present in all analyzed diatoms but not elsewhere; pennate-specific genes were present only in P. tricornutum, F. cylindrus, and F. solaris; F. solaris-specific genes were found only in the genome of F. solaris).
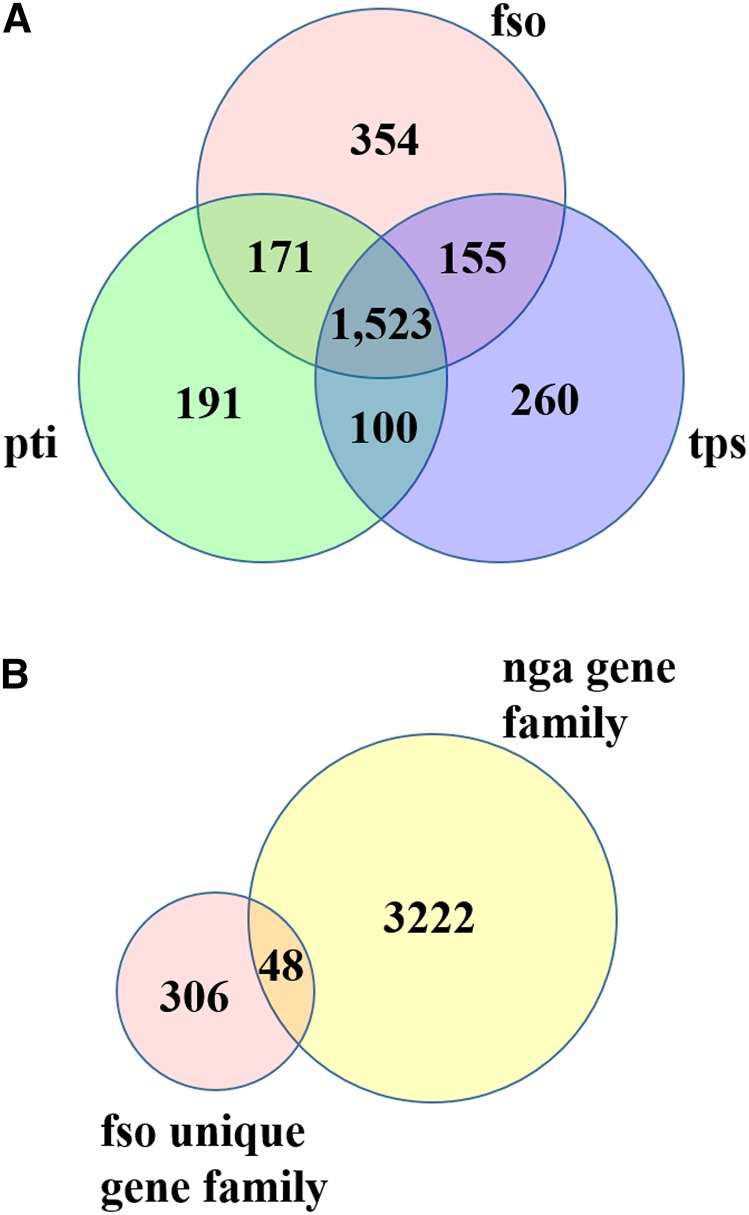
To characterize protein functions, the gene families (retrieved from the domain database Pfam-A) were compared with those found in the two best characterized, albeit nonoleaginous, diatom genomes, from P. tricornutum and T. pseudonana. Of the 2203 gene families of F. solaris, 1523 were found to belong to the “core gene families in diatoms” shared by all sequenced diatoms. Common pennate diatom gene families amounted to a total of 171. By contrast, 354 gene families belong to the group of F. solaris-specific families (Figure 2A). Then, the 354 F. solaris-specific gene families were compared with gene families in the other sequenced oleaginous alga Nannochloropsis gaditana (Radakovits et al., 2012) (Eustigmatophyceae). A total of 48 gene families were shared between these two oleaginous algae (Figure 2B; Supplemental Table 2). Among the shared families, Scs3p (PF10261) was found. This is a family of transmembrane proteins previously annotated as inositol phospholipid synthesis proteins (Hosaka et al., 1994), and a member of this family (afterward referred to as fat storage-inducing transmembrane protein [FIT]) was reported to be involved in lipid metabolism (Kadereit et al., 2008). In this study, we identified a FIT-like protein from the F. solaris genome and examined its subcellular localization using the yeast expression system, with which we previously succeeded in heterologously expressing a transmembrane protein derived from F. solaris (Muto et al., 2013b). By fusing the diatom FIT-like protein to green fluorescent protein, we found that it exhibited a strong accumulation within inner endoplasmic reticulum (ER) membrane structures (Drew et al., 2008) (Supplemental Figure 8).
Figure 2.
Venn Diagrams of Shared/Unique Gene Families of F. solaris JPCC DA0580.
Numbers of gene families belonging to each area are indicated.
(A) Comparison of whole gene families between F. solaris JPCC DA0580 (fso), T. pseudonana (tps), and P. tricornutum (pti).
(B) Comparison of F. solaris-specific gene families with whole gene families of the oleaginous Eustigmatophyte, N. gaditana (nga).
In the F. solaris genome, we identified gene sets for many essential pathways and could predict the putative subcellular localizations of their gene products. The oleaginous diatom F. solaris and nonoleaginous diatoms T. pseudonana and P. tricornutum had approximately the same number of genes in these fundamental pathways (Supplemental Table 3). An interesting feature of diatom genomes is the presence of genes encoding a complete urea cycle, which may allow diatoms to prosper in aquatic environments (Armbrust et al., 2004; Allen et al., 2011). The completeness of the pathway was also confirmed in F. solaris. Overall, our results suggest that there are no F. solaris-specific pathways responsible for its outstanding oil accumulation capabilities.
Although the gene sets in the fundamental pathways generally tend to be well conserved among the three diatoms, some genes differ between T. pseudonana and P. tricornutum (Kroth et al., 2008), such as the predicted localizations of carbonic anhydrase (EC 4.2.1.1) and enolase (EC:4.2.1.11). Furthermore, T. pseudonana has only β-glucosidase (EC3.2.1.21) family 1, while P. tricornutum has families 1 and 3. To add F. solaris into these comparisons, we established a pipeline for predicting protein localization with some modifications (Sunaga et al., 2015) (see also Supplemental Methods). We found that the pennate diatoms (F. solaris and P. tricornutum) exhibited identical features in all comparisons (localization of carbonic anhydrase and enolase, and the β-glucosidase family) and thus differed from the centric diatom T. pseudonana, suggesting that pennate diatoms have conserved metabolic frameworks, as well as the features of individual gene components.
We compared the numbers of transcription factors in each of the three diatoms and found that F. solaris contains 160 transcription factors, of which the heat shock factors are most abundant (61, ∼38%). This is consistent with what has been found in P. tricornutum and T. pseudonana (Rayko et al., 2010).
We also identified a large number of genes encoding highly divergent cyclins in the F. solaris genome. Cyclins regulate the cell cycle in eukaryotes; they are expanded in P. tricornutum and T. pseudonana, and novel diatom-specific cyclins have been discovered (Huysman et al., 2010). These abundant cyclins are believed to contribute to the ability of diatoms to adapt and survive under highly fluctuating environments. The F. solaris genome contains 29 cyclins (of which 25 have homoeologous counterparts; the percentage of cyclin genes among all 11,448 genes is 0.25%). The abundance of cyclin genes is also considerably higher than in other typical organisms and is comparable to that found in P. tricornutum (0.27%) (Huysman et al., 2010). Further phylogenetic analysis demonstrated that F. solaris has 17 diatom-specific cyclins (of which 15 genes have homoeologous counterparts), as do P. tricornutum and T. pseudonana (Supplemental Figure 9 and Supplemental Data Set 2) (Bowler et al., 2008; Huysman et al., 2010). Thus, diatoms seem to utilize well-conserved metabolic pathways, transcription factors, and cell cycle regulators.
Transcriptome Analysis
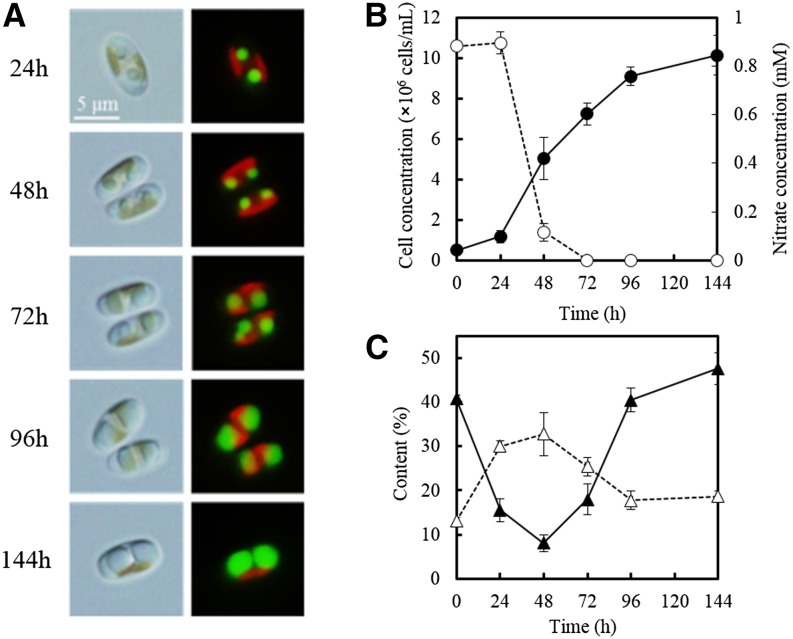
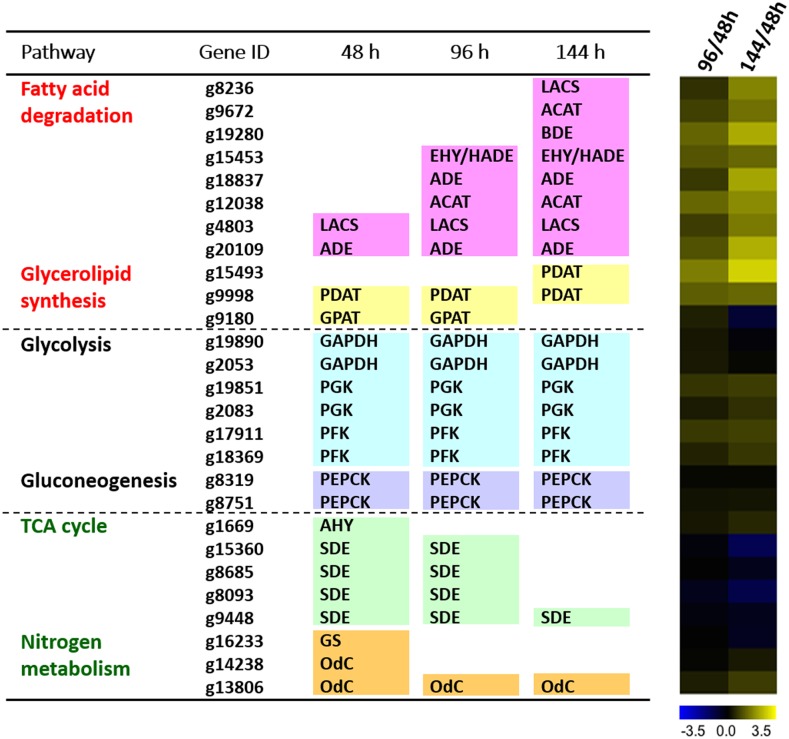
We monitored gene expression in 48-h (linear phase), 96-h (late-linear phase), and 144-h (stationary phase) cultures. Meanwhile, oil accumulation and growth were simultaneously observed (Figure 3). After 96 h, oil accumulation reached 40% (weight/dry cell weight). The 20 most strongly expressed genes (RPKM) and their expression dynamics are summarized in Figure 4. Genes involved in glycerolipid synthesis were upregulated as predicted; however, we were surprised to find that fatty acid degradation was also strongly upregulated. In comparison to lipid metabolism, the tricarboxylic acid (TCA) cycle and nitrogen metabolism tended to be downregulated, whereas glycolysis and gluconeogenesis were stable. It should be noted that genes encoding TCA cycle components remained at high RPKM levels, although their expression was reduced during the culture period. Glycerol-3-phosphate acyltransferase (GPAT) gene expression also followed this trend.
Figure 3.
Monitoring Neutral Lipid Accumulation in F. solaris JPCC DA0580.
(A) Differential interference microscopy (left) and fluorescence microscopy (right) of F. solaris cells stained with BODIPY 505/515. Red and green represent chloroplast and oil bodies.
(B) Growth (closed circles) and nitrate concentrations in the culture medium (open circles). Data are means of triplicate experiments ± sd.
(C) Neutral lipid content (closed triangles) and carbohydrate content (open triangles).
Figure 4.
Differential Gene Expression during Oil Accumulation.
The 20 genes with the highest RPKM values in the major pathways (i.e., glycolysis, gluconeogenesis, TCA cycle, nitrogen metabolism, fatty acid degradation, glycerolipid biosynthesis, and Calvin-Benson cycle) are shown at each time point (48-, 96-, and 144-h culture). Transcription profiles are depicted as heat maps, where yellow and blue indicate up- and downregulation, respectively, as shown in the scale bar (log2 ratio). GPAT (g9180) in glycerolipid synthesis ranked off from the list at 144 h because of a number of genes in fatty acid degradation ranked in it, although GPAT remained in high RPKM.
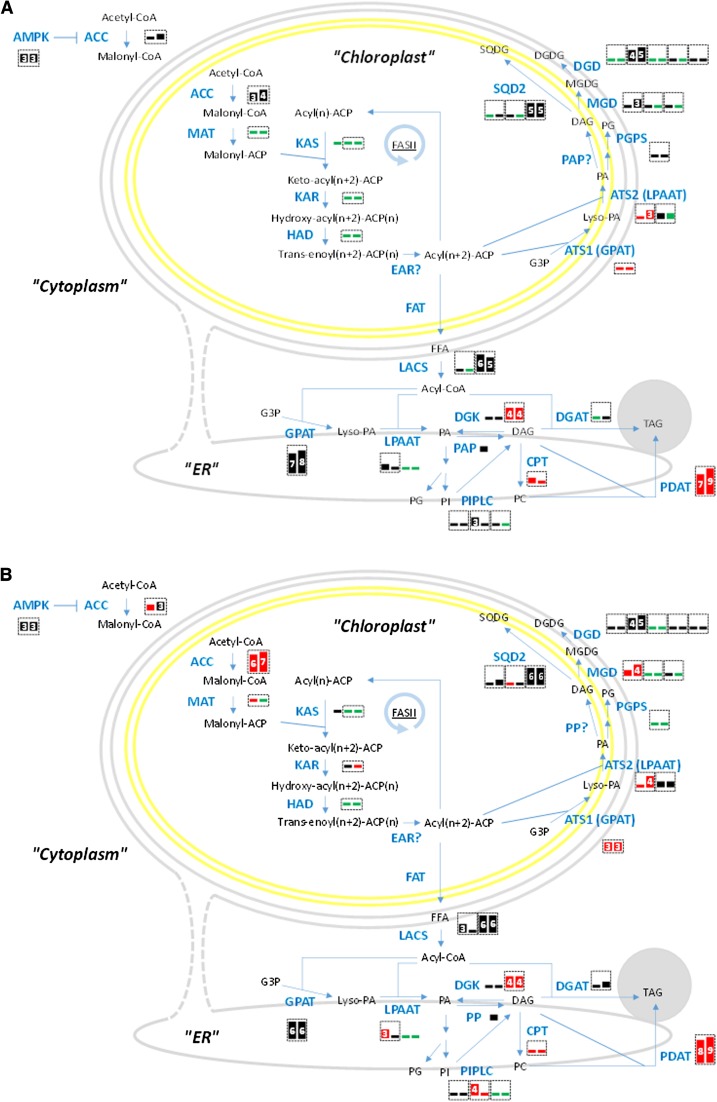
The dynamic behavior of glycerolipid biosynthesis gene expression at 96 and 144 h is depicted in Figures 5A and 5B, respectively, as are the predicted protein localizations of their gene products. We detected genes coding for the cytoplasmic and the chloroplast acetyl-CoA carboxylases (ACC), the chloroplast malonyl-CoA:ACP transacylase, and the components of the fatty acid synthase of type II (FASII), i.e., the 3-ketoacyl-ACP synthase, the 3-ketoacyl-ACP reductase, and the hydroxyacyl-ACP dehydratase. We could not detect any homologs for genes encoding the thioesterase that releases acyl-ACPs from FASII. Fatty acids can be used for glycerolipid biosynthesis at two locations, either in the chloroplast itself or in the ER. In both locations, glycerol-3-phosphate is acetylated in two steps, generating lysophosphatidic acid and phosphatidic acid (PA), either by ATS1 and ATS2 in the chloroplast membranes, using acyl-ACP as substrates or by GPAT and lysophosphatidic acid acyltransferases (LPAAT) in the ER, using acyl-CoA as substrates. Acyl-ACPs produced in the chloroplast need therefore to be converted into free fatty acids, which should then be transferred to the cytoplasmic by an unknown mechanism and esterified to coenzyme A by long-chain acyl-CoA synthetase (LACS) enzymes. PA, and its dephosphorylated form, diacylglycerol (DAG), are then at the origin of all membrane glycerolipids. In the chloroplast, PA is a precursor for phosphatidylglycerol, via a chloroplast phosphatidylglycerol phosphate synthase, and DAG is a precursor for the production of the main chloroplast lipids, i.e., monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol, synthesized by MGD, DGD, and SQD2 enzymes, respectively. In the ER, PA is also precursor for phosphatidylglycerol via the action of a phosphatidylglycerol phosphate synthase, but also for phosphoinositides, whereas DAG is substrate for other membrane lipids including phosphatidylcholine (PC), via cholinephosphotranseferase (CPT) and for the biosynthesis of triacylglycerol (TAG) following the canonical Kennedy pathway, by the action of DAG acyltransferase (DGAT). We could also detect a DAG kinase (DGK) catalyzing the reverse phosphorylation of DAG back into PA and a phospholipid:DAG acyltransferase (PDAT) allowing the synthesis of TAG from PC. We detected putative phosphoinositide-specific phospholipases C (PILPC) that might hydrolyze phospholipids into DAG. Concerning the possible utilization of chloroplast lipids for the production of TAG, as described in Chlamydomonas reinhardtii (Li et al., 2012), we did not detect any PGD1 homolog, but do not exclude that such lipid conversion might occur. Involvement of a route from chloroplast glycerolipids to TAG would then be suggested by a specific increase of the expression of MGD, DGD, or SQD2 and by the correlated detection of chloroplast glycerolipid fatty acids in TAG.
Figure 5.
Expression of Genes Encoding Fatty Acid and TAG Biosynthesis Pathway Components in F. solaris JPCC DA0580.
The 96-h (A) and 144-h (B) culture. Each box represents an isoform. Box sizes and numbers represent magnitudes of RPKM values [3: (8)∼16, 4: ∼32, 5: ∼64, 6: ∼128, 7: ∼256, 8: ∼512, 9: ∼1024, 10: 1024∼]. Red, black, and green boxes represent log2 fold changes in RPKM values (96 h/48 h or 144 h/48 h) >1, ≤−1 to 1, and <−1, respectively. Enzymes not predicted to localize in subcellular compartments are indicated by question marks. Homoeologous gene pairs are surrounded by dashed boxes.
During lipid accumulation, ATS1 (homoeologous pair; g15974 and g3622) and ATS2 (g4237), involved in glycerolipid biosynthesis in the chloroplast, were slightly upregulated at 96 h and even more so at 144 h. Downstream genes for two enzymes, MGD (homoeologous pair; g14744 and g2622) and to a lesser extent SQD2 (g3830) were also slightly upregulated but at a later stage, at 144 h. Over the same period, in the ER, the strongly expressed GPAT was unchanged, whereas LPAAT (g1029) was slightly upregulated at 144 h. PILPC (homoeologous genes; g11063 and g3585) were upregulated at 144 h, suggesting a possible production of DAG by a hydrolysis of phospholipids. Concerning DAG utilization, on the one hand, DGK (homoeologous pair; g802 and g8966) and CPT (homoeologous pair; g10699 and g19399) were strongly upregulated at 96 and 144 h, suggesting an intense production of phospholipids, including PC and, on the other hand, DGAT increased slightly at 144 h, supporting a TAG production from DAG by the Kennedy pathway. Consistent with an upregulation of PC biosynthesis as an alternative route toward TAG, genes encoding the ER-localized PDAT (homoeologous pair; g15493 and g9998) were strongly activated, with higher RPKM values at 96 and 144 h. Fueling the whole system in carbon, the genes that encode the chloroplast ACC (homoeologous pair; g10373 and g467) and mediate chloroplast fatty acid biosynthesis were upregulated ∼10-fold at 144 h, while upregulation of cytoplasmic ACC (homoeologous pair; g1072 and g13145) was slight. The genes that encode AMP-activated kinase (AMPK) were not significantly upregulated, and the localization was predicted to be only in the cytoplasm.
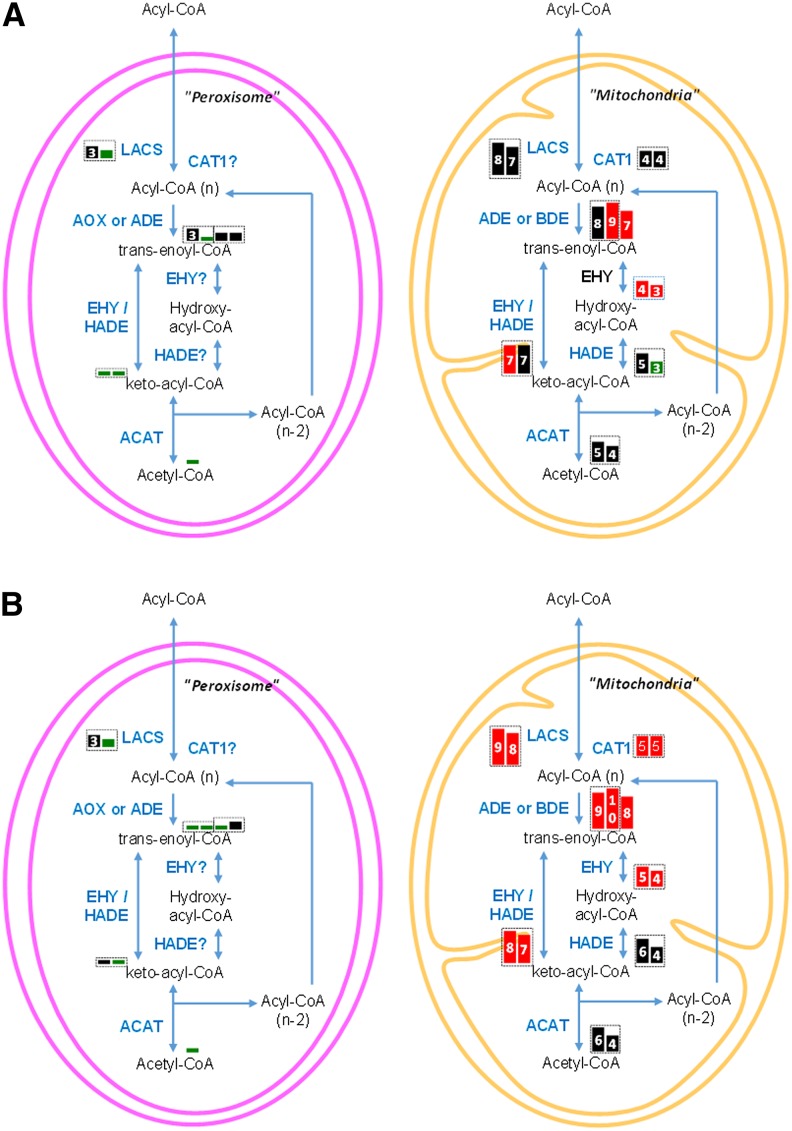
Figure 6 depicts the fatty acid degradation pathway (or β-oxidation) and its expression variation (Figures 6A and 6B correspond to 96 and 144 h, respectively). In F. solaris, fatty acid degradation pathways are predicted to exist in the peroxisome and mitochondria, as in other diatoms (Armbrust et al., 2004). Transcriptome data demonstrated upregulation of only those genes encoding mitochondrial components during neutral lipid accumulation; by contrast, most peroxisome components were slightly downregulated. Genes encoding mitochondrial carnitine acyltransferase I (CAT1, homoeologous pair; g7339 and g19974), acyl-CoA dehydrogenase (ADE, g20109), butyryl-CoA dehydrogenase (BDE, g19280), enoyl-CoA hydratase (EHY, homoeologous pair; g19794 and g2387), and enoyl-CoA hydratase/β-hydroxyacyl-CoA dehydrogenase (EHY/HADE) complex (homoeologous pair; g15453 and g18758) were upregulated at 96 h and further increased at 144 h. Mitochondrial LACS (homoeologous pair; g4803 and g8236) and another ADE gene (g18837) were specifically upregulated at 144 h. Changes in fatty acid degradation were rapid in comparison to the more gradual changes in fatty acid and glycerolipid biosynthesis pathways, for which upregulation was moderate at 96 h and only clearly apparent after 144 h.
Figure 6.
Expression of Genes Encoding Fatty Acid Degradation Pathway Components in F. solaris JPCC DA0580.
The 96-h (A) and 144-h (B) culture. Each box represents an isoform. Box sizes and numbers represent magnitudes of RPKM values [3: (8)∼16, 4: ∼32, 5: ∼64, 6: ∼128, 7: ∼256, 8: ∼512, 9: ∼1024, 10: 1024∼]. Red, black, and green boxes represent log2 fold changes in RPKM values (96 h/48 h or 144 h/48 h) >1, ≤−1 to 1, and <−1, respectively. The enzymes that are not predicted to localize in the subcellular compartments are marked by question marks. The homoeologous gene pairs are surrounded by dashed boxes.
As we assessed the expression patterns of homoeologous gene pairs in pathways for fatty acid/TAG biosynthesis and fatty acid degradation, it became strikingly clear that many homoeologous pairs demonstrated synchronized expression patterns. Genome-wide transcriptome analysis revealed synchronous expression patterns in only ∼20% of homoeologous gene pairs; by contrast, three of the seven homoeologous pairs in the mitochondrial fatty acid degradation pathway and 12 of the 33 pairs in the pathway for fatty acid/TAG biosynthesis were synchronized (Figures 5 and 6).
DISCUSSION
Allodiploid Genome of F. solaris JPCC DA0580
To further the aim of understanding and improving the oleaginous phenotype of F. solaris JPCC DA0580, we obtained its draft genome sequence with next-generation sequencing. While next-generation sequencing has advantages for ultra-high throughput of vast numbers of short reads, it is beset with the inevitable disadvantage of difficult assembly, despite the development of sophisticated algorithms. This issue is typically overcome with complementary methods such as Sanger sequencing (Singh et al., 2013), although these alternative sequencing strategies tend to be time-consuming and costly, thus negating some of the advantages of next-generation sequencing. Instead of using these cumbersome methods during assembly, we performed the assembly based on the unexpectedly discovered hypothesis that F. solaris might be an allodiploid organism. This hypothesis is supported by several lines of evidence: Genome sequence assembly resulted in the formation of chromosome pairs with conserved synteny but high levels of sequence variation; a homoeologous 18S rDNA pair was confirmed, and homoeologous GAPDH gene pairs were identified in single cells. The assembling process generated tentative 84 chromosomes, in which a number of telomeric repeats were found. This result supports the accuracy of our sequence assembly. Telomeric repeats are not normally identified in next-generation sequencing studies (Radakovits et al., 2012; Nakamura et al., 2013), with one exception (Smith et al., 2011), suggesting that the unorthodox genome structure in this alga facilitated their identification.
Interestingly, TEs are much more abundant in F. solaris than in the other two sequenced diatoms. Polyploidization has generally been assumed to induce a burst of transposition that results in genome reorganization and altered gene expression (Riddle and Birchler, 2003; Parisod et al., 2010). These mechanisms are believed to play an important role in the stabilization of hybrid genomes. Thus, the presence of abundant TEs, including the species-specific CoDi8 group in F. solaris, also supports an allodiploid genome structure in this strain. Direct observation of chromosome structure such as by karyotyping with genome in situ hybridization is necessary to validate this hypothesis (Hizume, 1994). It would also be interesting to identify the progenitors of the allodiploid F. solaris, as has been done with allodiploid yeasts (Kodama et al., 2005; Nakao et al., 2009; Louis et al., 2012). However, such studies are currently hampered by the lack of genome information for related diatoms.
Gene expression systems in allopolyploid (including allodiploid) organisms have been described in yeasts (Kodama et al., 2005; Horinouchi et al., 2010) and angiosperms (e.g., Coffea arabica allotetraploid [Bardil et al., 2011], Arabidopsis allotetraploids [Wang et al., 2006], and Gossypium allotetraploids [Chaudhary et al., 2009]). The allopolyploid transcriptome is not likely to be a simple mixture of the parent species but a result of interactions between the regulatory systems derived from each parental species (Riddle and Birchler, 2003; Pignatta and Comai, 2009), which may facilitate adaptation to fluctuating environments. The transcriptome analysis of F. solaris revealed that most (∼80%) homoeologous gene pairs differentially contribute to the transcriptome. This expression variation is consistent with other allopolyploid (or allodiploid) organisms (Adams et al., 2003; Kodama et al., 2005; Chaudhary et al., 2009; Horinouchi et al., 2010). Such complexity in transcriptional regulation is also believed to lead to functional plasticity, which could contribute to phenotypic novelty and better adaptation to variable environments. Indeed, there are a lot of successful crops with allopolyploidy in conventional agriculture (e.g., wheat [Triticum aestivum], canola [Brassica napus], and cotton [Gossypium hirsutum]) (Leitch and Leitch, 2008). The finding of the allodiploidy in F. solaris genome could be important to promote the development of farming diatom cultivars in the future.
Genomic Features Revealed by the Comparative Genomics
Comparative genomics of sequenced diatoms and the oleaginous N. gaditana (Eustigmatophyceae) identified the 48 gene families, which are expected to play an important role in lipid production and accumulation. Among them, FIT was found. FITs are involved in TAG droplet formation. A heterologous expression study of mouse FIT proteins demonstrated their specific localization to the ER in HEK293 cells (Kadereit et al., 2008). The yeast expression experiment confirmed that the FIT-like protein of F. solaris also exhibited ER-specific localization. Although the precise functions of the FIT-like protein remained obscure, it could be an interesting target for future functional analysis, which might provide insight to the metabolic engineering.
More specifically, comparison of the genes involved in the fundamental pathways (including fatty acid and glycerolipid synthesis) between three diatoms (Supplemental Table 3) suggests that the oleaginous phenotype of F. solaris is likely to be attributable to neither a greater number of those genes nor species-specific pathways. Likewise, F. solaris has well-conserved cell cycle regulators (cyclins) within the diatoms. Given these comparisons, it would be reasonable to hypothesize that any specific expression patterns of the genes encoding the common metabolic pathways could be responsible for its unique feature of concomitant oil accumulation and cell growth. We performed transcriptome analyses to test this hypothesis.
Transcription Profiles of Genes Encoding Components of Primary Metabolism
As we combined the genome and transcriptome data, we identified the major metabolic pathways (Figure 7). During lipid accumulation, significant changes in ACC expression were observed. ACC catalyzes the first-step conversion from acetyl-CoA to malonyl-CoA, which is the rate-limiting step in fatty acid biosynthesis. Chloroplast ACCs thus play a key role in fatty acid biosynthesis. Because only moderate upregulation of the ACC genes has been reported during prolonged lipid accumulation in Chlamydomonas (Miller et al., 2010) and P. tricornutum (Valenzuela et al., 2012), steadily expressed ACC may be sufficient to supply fatty acid moieties for the moderate lipid accumulation observed in these species. By contrast, oleaginous microalgae such as F. solaris produce much higher concentrations of lipids, thus requiring the biosynthesis of more fatty acids. Intensive upregulation of ACC gene expression has been observed in other oleaginous microalgae (Roessler et al., 1994; Guarnieri et al., 2011), which could be responsible for the abundant supply of fatty acids, leading to high glycerolipid accumulation.
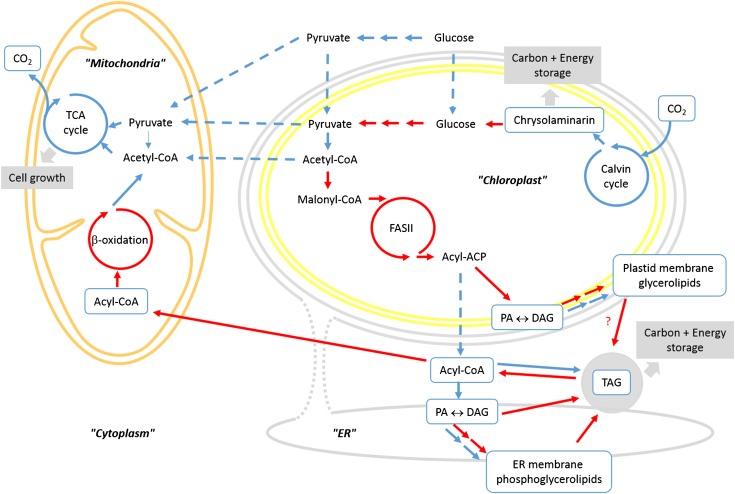
Figure 7.
Proposed Metabolic Scheme for TAG Accumulation in F. solaris JPCC DA0580.
Highly upregulated pathways are depicted by red arrows. They include pathways for FA and glycerolipid biosyntheses in the chloroplast. TAG accumulation is supported by the upregulation of parallel pathways, i.e., neosynthesis by the Kennedy pathway and conversion of ER phosphoglycerolipids combined with conversion of chloroplast glycerolipids by an unknown mechanism (?). Catabolism of acyl-CoAs is also upregulated in the mitochondrion.
The glycerolipid synthesis pathways in F. solaris are thought to be localized to the chloroplast and the ER like in all photosynthetic eukaryotes studied to date (Petroutsos et al., 2014), and we could indeed predict genes encoding enzymes involved in these two pathways, targeting accordingly to each organelle. The precursors for all membrane glycerolipids, PA, can be either produced in the chloroplast by ATS1 and ATS2 or in the ER by GPAT and LPAAT. Transcriptome analysis in F. solaris revealed that GPAT expression was high and that LPAAT was slightly upregulated, whereas the chloroplast ATS1 and ATS2 were upregulated, indicating a strong contribution of the chloroplast for the assembly of glycerolipids. Downstream of ATS2, sulfolipid and galactolipid biosynthesis genes are also upregulated, suggesting that these lipids might be subsequently converted into TAG via an unknown mechanism. This result is consistent with our lipidome analysis predicting the glycerolipid biosynthesis pathway based on the positional distribution of fatty acid moieties in the glycerolipid backbone (Liang et al., 2015). The complete set of genes for de novo TAG biosynthesis does not exist in the chloroplast; thus, the pathway that converts glycerolipid backbones into TAGs (Figures 5 and 7) remains unknown. PDATs catalyze TAG synthesis by transferring a fatty acid moiety from polar lipids to DAG. This acyl-CoA-independent pathway could be an alternative contributor to high-level lipid accumulation.
Fatty acid degradation is the process by which energy is extracted from lipids. Our transcriptome analysis during lipid accumulation demonstrated that the genes involved in β-oxidation were highly upregulated in the mitochondria (Figure 6). We believe this expression pattern could be a proliferation strategy by which the advantages of high-level lipid accumulation are exploited for cell replication. Specifically, F. solaris may activate β-oxidation pathways to utilize the massive amounts of lipid available for cell growth through the TCA cycle, the genes of which are downregulated but remain at high RPMK values (Figure 7).
Concomitant activation of fatty acid biosynthesis and degradation seems paradoxical because it is well known that the ACC-mediated regulation system coordinates the two processes. ACC tends to be upregulated for the activation of fatty acid biosynthesis in eukaryotic cells (Brady et al., 1993; Kahn et al., 2005); consequently, generation of malonyl-CoA is enhanced. The resulting malonyl-CoA is used for fatty acid biosynthesis, as well as serving as an inhibitor of carnitine acyltransferase I (CAT1; also known as carnitine parmitoyltransferase I), which is needed for transportation of acyl-CoA into mitochondria (Figure 6) followed by the suppression of β-oxidation (or vice versa). ACC activity is inhibited by AMPK to coordinate the balance of fatty acid biosynthesis and degradation (Kahn et al., 2005). These regulation systems were believed to play a role in microalgae as well (Guarnieri et al., 2011; Wan et al., 2012). However, genome information with protein localization data and transcriptome information of F. solaris indicate that only chloroplast ACCs were significantly upregulated during the oil accumulation. It means that excess malonyl-CoA would be produced only in the chloroplast and that in cytoplasm might be slight. Since diatom chloroplasts are surrounded by four membranes, it is unlikely that malonyl-CoA in the chloroplast migrates to the mitochondria and inhibits CAT1 and subsequent β-oxidation. Additionally, AMPKs were found only in the cytoplasm; thus, ACCs in the chloroplast would not be suppressed by AMPKs. These data led us to conclude that the ACC-mediated regulation of fatty acid biosynthesis and degradation might not be strict in microalga due to the specific organelle compartmentalization; thus, simultaneous activation of fatty acid biosynthesis (and subsequent lipid assembly) and degradation would be possible.
Indeed, cell growth and lipid accumulation occurred simultaneously in this strain (Figure 3), a feature that has not been found in other microalgae (Tonon et al., 2002; Popovich et al., 2012; Praveenkumar et al., 2012; Radakovits et al., 2012; Valenzuela et al., 2012). Moreover, the upregulation of the β-oxidation pathway has not been reported in other microalgae during oil accumulation (Radakovits et al., 2012; Rismani-Yazdi et al., 2012), suggesting the close connection between β-oxidation pathway activation and cell growth.
While most homoeologous gene pairs showed differential expression patterns with respect to each other (Supplemental Figure 5), a number of gene pairs were synchronously expressed in the pathways for fatty acid/TAG biosynthesis and fatty acid degradation. In other words, the differential expression patterns of most homoeologous gene pairs likely contributes to genome plasticity (Riddle and Birchler, 2003) and the expression patterns of gene pairs involved in lipid metabolism may be synchronized to maximize fatty acid/TAG biosynthesis and fatty acid degradation.
In conclusion, we analyzed the whole genome of the oleaginous diatom, F. solaris, and found that it has allodiploid genome structure. We also propose that activation of both lipid accumulation and degradation metabolism pathways could result in simultaneous growth and oil accumulation, which is a great advantage for efficient oil production. The molecular insights generated in this study will open doors to the development of metabolically engineered strains for biofuel production.
METHODS
Culture Conditions
The marine diatom Fistulifera solaris JPCC DA0580 was isolated from the junction of Sumiyo River and Yakugachi River, Kagoshima, Japan (28°15′N, 129°24′E) (Matsumoto et al., 2010, 2014). F. solaris was cultured in f/2 medium (Guillard and Ryther, 1962) (75 mg NaNO3, 6 mg Na2HPO4·2H2O, 0.5 μg vitamin B12, 0.5 μg biotin, 100 μg thiamine HCl, 10 mg Na2SiO3·9H2O, 4.4 mg Na2-EDTA, 3.16 mg FeCl3·6H2O, 12 μg CoSO4·5H2O, 21 μg ZnSO4·7H2O, 0.18 mg MnCl2·4H2O, 70 μg CuSO4·5H2O, and 7 μg Na2MoO4·2H2O per liter of artificial seawater). Cultures were bubbled with sterile air at 20°C under 140 μmol/m2/s continuous illumination.
DNA Sequencing
Cells in late logarithmic growth were collected by centrifugation at 10,000g for 10 min at 4°C. Cell pellets were frozen in liquid nitrogen until required, then resuspended in 15 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, pH 8.0, 1% SDS, 10 mM DTT, and 10 mg/mL proteinase K) and incubated at 50°C for 30 min (Bowler et al., 2008). Genomic DNA was stained with Hoechst33258 dye (0.1 mg/mL; Dojindo) and purified by caesium chloride centrifugation (Armbrust et al., 2004).
The whole genome of F. solaris was sequenced using a GS FLX Titanium DNA pyrosequencer (Roche 454 Life Sciences). The library was constructed using the GS FLX Titanium General Library Preparation Kit and amplified onto DNA capture beads by emulsion PCR according to the manufacturer’s instructions.
Assembly of Nuclear Genome and Prediction of Gene Regions
The beads were quantified and the genome was sequenced using a GS Titanium Sequencing Kit XLR70 and Genome Sequencer FLX System. Nucleotide sequences were assembled using GS De Novo Assembler version 2.3 (Newbler, Roche) using default parameters, and 297 scaffolds were generated.
To determine chromosome structure, scaffold sequences were first passed through LASTZ (Harris, 2007) to identify scaffolds that could be represented as the best global alignment according to sequence similarity. The scaffolds were then passed to LALIGN (Huang and Miller, 1991) to generate local alignments with the aid of information generated by LASTZ. Other scaffolds were removed from the assembly as false positives. Finally, the scaffolds were connected according to an alignment pattern where two or more scaffolds were aligned to one scaffold, and MUMmer (Kurtz et al., 2004) was used to evaluate the results.
Gene regions were predicted using AUGUSTUS version 2.5.5. The manual suggests using sequence information for ∼100 genes to train the software with species-specific parameters. We constructed and sequenced a cDNA library from F. solaris and found 99 distinct full-length genes. These sequences were used to train the AUGUSTUS program to predict F. solaris gene regions.
In this study, homoeologous chromosomes are defined as chromosome pairs duplicated by interspecies hybridization. More specifically, we identified homoeologous chromosomes as those with high sequence similarity and synteny (Supplemental Figures 1 and 2). Within each homoeologous chromosome pair, the chromosome with higher GC% was defined as chrX and the other as chrX′. Homoeologous genes are defined as gene pairs duplicated by interspecies hybridization. We identified each homoeologous pair as genes located on equivalent homoeologous chromosomes and which share the best homology within the F. solaris genome (a “best to best” relationship).
Methods for repeat identification, transcription factor detection, and prediction of pathway localization are described in Supplemental Methods.
Comparative Studies of Gene Families
We performed a comparative analysis of protein families in F. solaris, Thalassiosira pseudonana, Phaeodactylum tricornutum, and Nannochloropsis gaditana. Sets of HMM profiles assigned as “family” were retrieved from the domain database Pfam-A. Protein-encoding genetic sequences were passed to the HMM search program to identify the sets of domains in each genome. Then, every hit with an e-value <10−3 was stored as a family with respect to each genome. Protein family-based comparative genomics was used to identify family sets specific to each diatom or shared between several diatoms. Families specific to F. solaris were compared with the whole protein families of another oleaginous Eustigmatophyte, N. gaditana. Prediction of subcellular localization was performed by combining Wolf (www.genscript.com/psort/wolf_psort.html), SignalP, and ChloroP (www.cbs.dtu.dk) tools. Chloroplast localization was supported by the presence of both a cleavable signal peptide (Sp) and a chloroplast-like transit peptide (Ctp) and in some cases by a Ctp only. The method for comparative studies of genes is described in the supplemental information.
Single-Cell Analysis
Single-cell entrapment and manipulation of F. solaris were conducted on a microcavity array. The device was microfabricated as described (Hosokawa et al., 2009, 2012). In brief, a poly(ethylene terephthalate) plate (20 × 20 mm, thickness = 38 µm) was used as the substrate. The distance between each cavity was 30 µm, with 44,100 cavities arranged in a 210 × 210 array on a typical microscope glass slide. A poly(dimethylsiloxane) structure equipped with a vacuum microchannel (i.d. = 500 μm) was fitted directly beneath the microcavity array to apply negative pressure for cell entrapment. The vacuum microchannel was connected to a peristaltic pump, and the cell entrapment setup was placed onto the computer-operated motorized stage of an upright microscope. Experimental procedures for cell trapping, recovery, and RT-PCR are described in Supplemental Methods.
Lipid, Carbohydrate, and Nitrate Quantification during F. solaris Growth
To visualize intercellular neutral lipids, cultured cells were stained with BODIPY 505/515 (1:99 v/v; Molecular Probes; 25 μg/mL, 2% DMSO [v/v] in water) and incubated for 10 min at room temperature. The stained cells were observed by fluorescence microscopy (BX51; Olympus).
To quantify the neutral lipid content, cells were collected by centrifugation (8500g, 10 min, 25°C), washed twice with deionized water, and lyophilized for 24 h. To extract neutral lipids, the lyophilized cells (∼100 mg) were fractured using mortars and muddlers, and then resuspended in n-hexane (10 mL). The cell debris was deposited by centrifugation (1000g, 3 min, 25°C), and the supernatant was collected. The extraction was repeated until the supernatant became colorless. After the collected supernatants were combined and lyophilized, total extraction yields were determined gravimetrically.
Carbohydrate content was quantified by the phenol-sulphuric acid method (Granum and Myklestad, 2002). Cells were collected by centrifugation (8500g, 3 min, 25°C), and 0.05 M H2SO4 (5 mL) was added. After incubation (10 min, 60°C), the cell debris was deposited by centrifugation (15,000g, 3 min, 25°C) and the collected supernatant represented a glucan extract (2 mL), to which 3% aqueous phenol (0.5 mL) and concentrated H2SO4 (5 mL) were added. After vortexing and incubation (30 min, 25°C), absorbance was measured at 485 nm. The amount of reducing sugar was calculated using glucose as a standard.
Nitrate concentrations in the algal cultures were determined by high-performance liquid chromatography (Li et al., 2008). Supernatants were collected after centrifugation (8500g, 10 min, 25°C) and filtered (DISMIC 13HP020CN; Toyo Roshi Kaisha). High-performance liquid chromatography was performed on a Waters instrument (e2695 and e2489) loaded with a TSKgel IC-Anion-PW (#06837; Tosoh). The nitrate concentration in each sample was calculated by measuring the peak area and comparing it to samples with known nitrate concentrations. The mobile phase was maintained at 1.2 mL/min, and the UV detector was set at 220 nm. The mobile phase contained borate buffer/gluconate concentrate, methanol, acetonitrile, and deionized water at a ratio of 2:12:12:74 (v/v/v/v). The borate buffer/gluconate concentrate comprised 0.07 M sodium gluconate, 0.3 M H3BO3, 0.1 M Na2B4O7, and 3.8 M glycerol in deionized water (100 mL). The injection volume was 10 μL.
RNA Sequencing
Total RNA was extracted as described (Muto et al., 2013a). Briefly, cells were collected by centrifugation (8500g, 10 min, 4°C) after 48-, 96-, and 144-h culture. After the cell pellets were frozen in liquid nitrogen and fractured using mortars and muddlers, samples were treated with Plant RNA isolation reagent (Invitrogen) and then incubated with DNase I (Takara Bio) for 30 min at 37°C. Total RNA was purified with an RNeasy Mini kit (Qiagen). After purification, RNA quality was evaluated on an Agilent 2100 bioanalyzer (Agilent Technologies).
For RNA sequencing, cDNA sequencing libraries were constructed with the mRNA-Seq Library Prep Kit (Illumina) and Small RNA Sample Prep Kit (Illumina) according to directional mRNA-Seq library preparation protocol revision A (Illumina). Briefly, poly(A)+ RNA was isolated from 10 μg total RNA using oligo(dT) magnetic beads. The isolated poly(A)+ RNA was fragmented into small pieces under elevated temperature. After the fragmented RNA was treated with alkaline phosphatase (Takara Bio), the 5′ end of the fragmented RNA was phosphorylated using T4 polynucleotide kinase (Takara Bio). Subsequently, 5′ and 3′ adapters were ligated to the phosphorylated RNA using T4 RNA ligase 2, truncated (New England Biolabs). The adapter-ligated RNA was reverse-transcribed using the primers designed for the 3′ end adapter. The products of the reverse transcription reaction were amplified by 12 cycles of PCR. The prepared PCR products were size-fractionated by polyacrylamide gel electrophoresis to create a final cDNA sequencing library.
Transcriptome Analysis
The cDNA sequencing libraries were sequenced on a Genome Analyzer IIx (Illumina) according to the manufacturer’s instructions. Briefly, clusters were generated from the library on the surface of a flow-cell by bridge amplification using Cluster Station (Illumina) and Single-read Cluster Generation Kit v4 (Illumina). Sequencing was performed using Genome Analyzer IIx and TruSeq SBS kit v5 (Illumina). Illumina Sequence Control Software v2.8 with Real Time Analysis v1.8 was used for operation and data processing including image analysis and base calling.
Transcript abundances were expressed as RPKM (Mortazavi et al., 2008). Bowtie v0.12.7 (Langmead et al., 2009) was used to align the RNA-sequencing reads to the draft whole genome of F. solaris. ERANGE software v3.2 (Mortazavi et al., 2008) was used to calculate RPKM values. Log2 fold changes in RPKM values [log2 (RPKM96h,144h/RPKM48h)] were calculated to evaluate changes in gene expression. RPKM48h indicates values obtained from mRNA samples before TAG accumulation, while RPKM96h,144h indicate values obtained from mRNA samples during TAG accumulation.
To cluster the homoeologous gene pairs into synchronized or differentially expressed groups, we applied the pattern matching method (Pavlidis and Noble, 2001) to the transcriptome data. First, six vectors composed of a binary pattern (0 and 1) at three time points except for (0,0,0) and (1,1,1) were generated as reference expression patterns. The RPKM fluctuations of all genes were categorized in one of these reference expression patterns based on a comparison of correlation parameters. Finally, we compared the reference expression patterns in which the homoeologous gene pairs were categorized.
Accession Numbers
Genome and transcriptome data from this article can be found in the DDBJ Sequence Read Archive. Accession numbers for genome and transcriptome are DRA002403 and DRA002404, respectively.
Supplemental Data
Supplemental Figure 1. Schematic Representation of 84 Chromosomes in F. solaris JPCC DA0580.
Supplemental Figure 2. Conservation of Synteny between Homoeologous Chromosome Pairs in F. solaris JPCC DA0580.
Supplemental Figure 3. The Multiple 18S rDNA Sequences in the Genome of F. solaris JPCC DA0580.
Supplemental Figure 4. Single-Cell RT-PCR.
Supplemental Figure 5. Expression Patterns of homoeologous Gene Pairs throughout the 42 Chromosome Pairs.
Supplemental Figure 6. CoDi Classification in F. solaris JPCC DA0580 Based on the RT Domains.
Supplemental Figure 7. Venn Diagram of Shared/Unique Genes of F. solaris JPCC DA0580.
Supplemental Figure 8. Fluorescence Microscopy of Saccharomyces cerevisiae Cells Expressing FIT-Like Protein.
Supplemental Figure 9. Phylogenetic Analysis of the Cyclins of F. solaris JPCC DA0580.
Supplemental Table 1. Sequencing Analysis of F. solaris JPCC DA0580 by GS FLX Titanium DNA Pyrosequencing.
Supplemental Table 2. The List of Genes Shared between F. solaris JPCC DA0580-Specific Gene Families and the Entire N. gaditana Gene Family
Supplemental Table 3. The Number of Diatom Genes in the Major Metabolism Pathways.
Supplemental Data Set 1. Homoeologous Chromosome Pairs in the F. solaris JPCC DA0580 Allodiploid Genome.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 9.
Supplementary Material
Acknowledgments
This work was supported by JST, CREST. C.B. and A.V. acknowledge funding from the European Union (MicroB3 and the ERC Diatomite projects) and from the Agence Nationale de la Recherche (ANR DiaDomOil and Oceanomics projects). We thank Kyoko Osada (Tokyo University of Agriculture and Technology [TUAT]), Daisuke Nojima (TUAT), Yue Liang (TUAT), and Tadashi Matsunaga (TUAT) for fruitful discussions.
AUTHOR CONTRIBUTIONS
T. Tanaka and W.F. performed the overall experimental design and analysis. M. Muto performed the next-generation sequencing experiments. Y.S. and Y.F. performed the transcriptomics experiments. A.V., H.A., E.M., Michihiro Tanaka, T. Taniguchi, and S.A. performed bioinformatics analyses. M.N. analyzed the 18S rDNA sequences. Masayoshi Tanaka performed the single-cell analyses. M. Matsumoto monitored cell growth and lipid accumulation. All authors, including Y.M., C.B., T.Y., and P.S.W., analyzed data, participated in discussions, and prepared the article.
Glossary
- RPKM
reads per kilobase of exon model per million mapped reads
- TE
transposable element
- HMM
hidden Markov model
- ER
endoplasmic reticulum
- TCA
tricarboxylic acid
- PA
phosphatidic acid
- DAG
diacylglycerol
- TAG
triacylglycerol
- PC
phosphatidylcholine
- PDAT
phospholipid:DAG acyltransferase
- AMPK
AMP-activated kinase
References
- Adams K.L., Cronn R., Percifield R., Wendel J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A.E., Dupont C.L., Oborník M., Horák A., Nunes-Nesi A., McCrow J.P., Zheng H., Johnson D.A., Hu H., Fernie A.R., Bowler C. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207. [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- Bardil A., de Almeida J.D., Combes M.C., Lashermes P., Bertrand B. (2011). Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytol. 192: 760–774. [DOI] [PubMed] [Google Scholar]
- Bowler C., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. [DOI] [PubMed] [Google Scholar]
- Brady P.S., Ramsay R.R., Brady L.J. (1993). Regulation of the long-chain carnitine acyltransferases. FASEB J. 7: 1039–1044. [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R.M., Udall J.A., Verma N., Springer N.M., Wendel J.F. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D., Newstead S., Sonoda Y., Kim H., von Heijne G., Iwata S. (2008). GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 3: 784–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Grünewald C., Garcés E., Rossi S., Camp J. (2009). Use of the dinoflagellate Karlodinium veneficum as a sustainable source of biodiesel production. J. Ind. Microbiol. Biotechnol. 36: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Granum E., Myklestad S.M. (2002). A simple combined method for determination of beta-1,3-glucan and cell wall polysaccharides in diatoms. Hydrobiologia 477: 155–161. [Google Scholar]
- Guarnieri M.T., Nag A., Smolinski S.L., Darzins A., Seibert M., Pienkos P.T. (2011). Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS ONE 6: e25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard R.R., Ryther J.H. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 8: 229–239. [DOI] [PubMed] [Google Scholar]
- Harris R.S. (2007). Improved Pairwise Alignment of Genomic DNA. PhD dissertation (University Park, PA: The Pennsylvania State University; ). [Google Scholar]
- Hizume M. (1994). Allodiploid nature of Allium wakegi Araki revealed by genomic in situ hybridization and localization of 5S and 18S rDNAs. Jpn. J. Genet. 69: 407–415. [DOI] [PubMed] [Google Scholar]
- Horinouchi T., Yoshikawa K., Kawaide R., Furusawa C., Nakao Y., Hirasawa T., Shimizu H. (2010). Genome-wide expression analysis of Saccharomyces pastorianus orthologous genes using oligonucleotide microarrays. J. Biosci. Bioeng. 110: 602–607. [DOI] [PubMed] [Google Scholar]
- Hosaka K., Nikawa J., Kodaki T., Ishizu H., Yamashita S. (1994). Cloning and sequence of the SCS3 gene which is required for inositol prototrophy in Saccharomyces cerevisiae. J. Biochem. 116: 1317–1321. [DOI] [PubMed] [Google Scholar]
- Hosokawa M., Arakaki A., Takahashi M., Mori T., Takeyama H., Matsunaga T. (2009). High-density microcavity array for cell detection: single-cell analysis of hematopoietic stem cells in peripheral blood mononuclear cells. Anal. Chem. 81: 5308–5313. [DOI] [PubMed] [Google Scholar]
- Hosokawa M., Asami M., Nakamura S., Yoshino T., Tsujimura N., Takahashi M., Nakasono S., Tanaka T., Matsunaga T. (2012). Leukocyte counting from a small amount of whole blood using a size-controlled microcavity array. Biotechnol. Bioeng. 109: 2017–2024. [DOI] [PubMed] [Google Scholar]
- Huang X., Miller W. (1991). A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12: 337–357. [Google Scholar]
- Huysman M.J., Martens C., Vandepoele K., Gillard J., Rayko E., Heijde M., Bowler C., Inzé D., Van de Peer Y., De Veylder L., Vyverman W. (2010). Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 11: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., Silver D.L. (2008). Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA 105: 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn B.B., Alquier T., Carling D., Hardie D.G. (2005). AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1: 15–25. [DOI] [PubMed] [Google Scholar]
- Kodama Y., Kielland-Brandt M.C., Hansen J. (2006). Lager brewing yeast. In Comparative Genomics, Topics in Current Genetics, Vol. 15, Sunnerhagen P., Piškur J., eds (Berlin: Springer, pp. 145–164. [Google Scholar]
- Kroth P.G., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A.R., Leitch I.J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Li X., Moellering E.R., Liu B., Johnny C., Fedewa M., Sears B.B., Kuo M.H., Benning C. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Horsman M., Wang B., Wu N., Lan C.Q. (2008). Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl. Microbiol. Biotechnol. 81: 629–636. [DOI] [PubMed] [Google Scholar]
- Liang Y., Yoshino T., Maeda Y., Tanaka M., Matsumoto M., Tanaka T. (2015). Profiling of fatty acid methyl esters from the oleaginous diatom Fistulifera sp. strain JPCC DA0580 under nutrition-sufficient and -deficient conditions. J. Appl. Phycol., http://dx.doi.org/10.1007/s10811-014-0265-y. [Google Scholar]
- Louis, V.L., et al. (2012). Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda) 2: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra D.M., Chanakya H., Ramachandra T. (2013). Euglena sp. as a suitable source of lipids for potential use as biofuel and sustainable wastewater treatment. J. Appl. Phycol. 25: 855–865. [Google Scholar]
- Matsumoto M., Sugiyama H., Maeda Y., Sato R., Tanaka T., Matsunaga T. (2010). Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl. Biochem. Biotechnol. 161: 483–490. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Mayama S., Nemoto M., Fukuda Y., Muto M., Yoshino T., Matsunaga T., Tanaka T. (2014). Morphological and molecular phylogenetic analysis of the high triglyceride-producing marine diatom, Fistulifera solaris sp. nov. (Bacillariophyceae). Phycol. Res. 62: 257–268. [Google Scholar]
- Maumus F., Allen A.E., Mhiri C., Hu H., Jabbari K., Vardi A., Grandbastien M.A., Bowler C. (2009). Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics 10: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L., Dujon B. (2012). Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol. Mol. Biol. Rev. 76: 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Muto M., Fukuda Y., Nemoto M., Yoshino T., Matsunaga T., Tanaka T. (2013a). Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—a high triglyceride producer. Mar. Biotechnol. (NY) 15: 48–55. [DOI] [PubMed] [Google Scholar]
- Muto M., Kubota C., Tanaka M., Satoh A., Matsumoto M., Yoshino T., Tanaka T. (2013b). Identification and functional analysis of delta-9 desaturase, a key enzyme in PUFA Synthesis, isolated from the oleaginous diatom Fistulifera. PLoS ONE 8: e73507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., et al. (2013). The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS ONE 8: e57122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y., Kanamori T., Itoh T., Kodama Y., Rainieri S., Nakamura N., Shimonaga T., Hattori M., Ashikari T. (2009). Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 16: 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto M., Maeda Y., Muto M., Tanaka M., Yoshino T., Mayama S., Tanaka T. (2014). Identification of a frustule-associated protein of the marine pennate diatom Fistulifera sp. strain JPCC DA0580. Mar. Genomics 16: 39–44. [DOI] [PubMed] [Google Scholar]
- Oh S.H., et al. (2009). Lipid production in Porphyridium cruentum grown under different culture conditions. J. Biosci. Bioeng. 108: 429–434. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M.P., Green B.R. (2011). Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476: 20–26. [DOI] [PubMed] [Google Scholar]
- Parisod C., Alix K., Just J., Petit M., Sarilar V., Mhiri C., Ainouche M., Chalhoub B., Grandbastien M.A. (2010). Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 186: 37–45. [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Noble W.S. (2001). Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2: RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D., et al. (2014). Evolution of galactoglycerolipid biosynthetic pathways—from cyanobacteria to primary plastids and from primary to secondary plastids. Prog. Lipid Res. 54: 68–85. [DOI] [PubMed] [Google Scholar]
- Pignatta D., Comai L. (2009). Parental squabbles and genome expression: lessons from the polyploids. J. Biol. 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovich C.A., Damiani C., Constenla D., Martínez A.M., Freije H., Giovanardi M., Pancaldi S., Leonardi P.I. (2012). Neochloris oleoabundans grown in enriched natural seawater for biodiesel feedstock: evaluation of its growth and biochemical composition. Bioresour. Technol. 114: 287–293. [DOI] [PubMed] [Google Scholar]
- Praveenkumar R., Shameera K., Mahalakshmi G., Akbarsha M.A., Thajuddin N. (2012). Influence of nutrient deprivations on lipid accumulation in a dominant indigenous microalga Chlorella sp., BUM11008: Evaluation for biodiesel production. Biomass Bioenergy 37: 60–66. [Google Scholar]
- Radakovits R., Jinkerson R.E., Fuerstenberg S.I., Tae H., Settlage R.E., Boore J.L., Posewitz M.C. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 3: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayko E., Maumus F., Maheswari U., Jabbari K., Bowler C. (2010). Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188: 52–66. [DOI] [PubMed] [Google Scholar]
- Riddle N.C., Birchler J.A. (2003). Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19: 597–600. [DOI] [PubMed] [Google Scholar]
- Rismani-Yazdi H., Haznedaroglu B.Z., Hsin C., Peccia J. (2012). Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol. Biofuels 5: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolfi L., Chini Zittelli G., Bassi N., Padovani G., Biondi N., Bonini G., Tredici M.R. (2009). Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102: 100–112. [DOI] [PubMed] [Google Scholar]
- Roessler P.G., Bleibaum J.L., Thompson G.A., Ohlrogge J.B. (1994). Characteristics of the gene that encodes acetyl-CoA carboxylase in the diatom Cyclotella cryptica. Ann. N. Y. Acad. Sci. 721: 250–256. [DOI] [PubMed] [Google Scholar]
- Sato R., Maeda Y., Yoshino T., Tanaka T., Matsumoto M. (2014). Seasonal variation of biomass and oil production of the oleaginous diatom Fistulifera sp. in outdoor vertical bubble column and raceway-type bioreactors. J. Biosci. Bioeng. 117: 720–724. [DOI] [PubMed] [Google Scholar]
- Satoh A., Ichii K., Matsumoto M., Kubota C., Nemoto M., Tanaka M., Yoshino T., Matsunaga T., Tanaka T. (2013). A process design and productivity evaluation for oil production by indoor mass cultivation of a marine diatom, Fistulifera sp. JPCC DA0580. Bioresour. Technol. 137: 132–138. [DOI] [PubMed] [Google Scholar]
- Singh R., et al. (2013). Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. (2008). Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 8: 996–1007. [DOI] [PubMed] [Google Scholar]
- Smith C.R., et al. (2011). Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. USA 108: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V.H., Sturm B.S., Denoyelles F.J., Billings S.A. (2010). The ecology of algal biodiesel production. Trends Ecol. Evol. (Amst.) 25: 301–309. [DOI] [PubMed] [Google Scholar]
- Sunaga Y., Maeda Y., Yabuuchi T., Muto M., Yoshino T., Tanaka T. (2015). Chloroplast-targeting protein expression in the oleaginous diatom Fistulifera solaris JPCC DA0580 toward metabolic engineering. J. Biosci. Bioeng. 119: 28–34. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fukuda Y., Yoshino T., Maeda Y., Muto M., Matsumoto M., Mayama S., Matsunaga T. (2011). High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth. Res. 109: 223–229. [DOI] [PubMed] [Google Scholar]
- Tonon T., Harvey D., Larson T.R., Graham I.A. (2002). Long chain polyunsaturated fatty acid production and partitioning to triacylglycerols in four microalgae. Phytochemistry 61: 15–24. [DOI] [PubMed] [Google Scholar]
- Valenzuela J., Mazurie A., Carlson R.P., Gerlach R., Cooksey K.E., Peyton B.M., Fields M.W. (2012). Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol. Biofuels 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Han J., Sang M., Li A., Wu H., Yin S., Zhang C. (2012). De novo transcriptomic analysis of an oleaginous microalga: pathway description and gene discovery for production of next-generation biofuels. PLoS ONE 7: e35142 Retracted. PLoS ONE 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang J., Tian L., Lee H.S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.