Significance
Nostoc symbioses with plants represent one of the most versatile and ancient types of symbioses. The infection process is a tug-of-war between the plant host and the cyanobacterial symbiont, with a reciprocal influence of both partners on the differentiation of the infectious motile filaments, hormogonia. In the present study, we have uncovered a major hormogonium-repressing factor of Nostoc punctiforme, nostopeptolide. To our knowledge, the nonribosomal peptide is the first complex secondary metabolite of cyanobacteria for which a governing role during cellular differentiation could be demonstrated. Different plant partners were shown to strictly downregulate the factor in symbiosis, thereby unveiling a complex cross-talk network between plants and Nostoc.
Keywords: nonribosomal peptide, cell differentiation, symbiosis, MALDI imaging, signaling
Abstract
Nostoc punctiforme is a versatile cyanobacterium that can live either independently or in symbiosis with plants from distinct taxa. Chemical cues from plants and N. punctiforme were shown to stimulate or repress, respectively, the differentiation of infectious motile filaments known as hormogonia. We have used a polyketide synthase mutant that accumulates an elevated amount of hormogonia as a tool to understand the effect of secondary metabolites on cellular differentiation of N. punctiforme. Applying MALDI imaging to illustrate the reprogramming of the secondary metabolome, nostopeptolides were identified as the predominant difference in the pks2− mutant secretome. Subsequent differentiation assays and visualization of cell-type-specific expression of nostopeptolides via a transcriptional reporter strain provided evidence for a multifaceted role of nostopeptolides, either as an autogenic hormogonium-repressing factor or as a chemoattractant, depending on its extracellular concentration. Although nostopeptolide is constitutively expressed in the free-living state, secreted levels dynamically change before, during, and after the hormogonium differentiation phase. The metabolite was found to be strictly down-regulated in symbiosis with Gunnera manicata and Blasia pusilla, whereas other metabolites are up-regulated, as demonstrated via MALDI imaging, suggesting plants modulate the fine-balanced cross-talk network of secondary metabolites within N. punctiforme.
The filamentous cyanobacterium Nostoc punctiforme displays a very complex and unique life cycle (1). When experiencing changes in environmental conditions or in response to chemical cues, individual vegetative cells in the filaments or entire trichomes are capable of entering distinct differentiation routes. Specifically, N. punctiforme can develop vegetative cells and nitrogen-fixing heterocysts that are regularly spaced within filaments, resting stages designated as akinetes, and motile hormogonia (1) (SI Appendix, Fig. S1). Remarkably, hormogonia can be internalized by plants from three distant taxa; namely, the bryophytes Blasia and Anthoceros, the angiosperm Gunnera, and cycads such as Macrozamia spp. (2). A given strain of N. punctiforme is capable of infecting all distinct plant partners, yet the plant hosts undergo very different morphogenetic changes on symbiotic interactions (2). In hornworts and Gunnera species, the major changes occur in contact cells of specialized cavities (auricles) and glands, respectively, both of which are preexisting symbiotic organs. Cycads, in contrast, develop so-called coralloid roots on infection (3). These symbiont-induced changes on the plant side indicate a rather long coevolution process, yet no gene transfer, genome reduction, or dual-partner gene products have been documented for plant–Nostoc symbioses (3). The partnership is considered mutualistic or commensal, with plants providing fixed carbon in exchange for fixed nitrogen from the cyanobacteria (3).
Still, the interaction is only facultative for Nostoc, suggesting that under specific conditions, the benefit gained is outweighed by the costs. Hence, the infection process needs to be tightly balanced. The tug-of-war between plants and symbiotic cyanobacteria is exemplified by the reciprocal influence of both partners on hormogonia differentiation (4). Plant hosts secrete yet-unidentified hormogonium-inducing factors (HIF) (5) to stimulate development of the infectious filaments that remain motile for 48–72 h (6). Once the Nostoc culture has completed a cycle of HIF-induced hormogonium formation, there is a lengthy delay before hormogonium formation can occur again (6). Meeks has introduced the term “immunity period” for this phase, during which no secondary infection can be initiated, even in the presence of HIFs (5). Nostoc apparently contributes autogenic secretory factors to this hormogonia repression, as an exchange of growth medium stimulates hormogonia formation (7). Similarly, hormogonia are repressed inside plant hosts by an unidentified hormogonium-repressing factor (HRF), whereas vegetative filaments with a high abundance of heterocysts are maintained.
Sequencing of N. punctiforme ATCC (American Type Culture Collection) 29133 has paved the way for a substantial molecular analysis of the complex lifestyle of this symbiotic strain (8). Array data showing the response to different environmental stimuli have provided insight into the complex regulatory network of N. punctiforme (9, 10). Several gene loci were shown to modulate the differentiation response of N. punctiforme to plant signals and the infection process itself (11–13). Notably, a large part of the 9-Mbp genome of N. punctiforme ATCC 29133 is devoted to the synthesis of a multitude of secondary metabolites of the nonribosomal peptide (NRPS) and polyketide (PKS) classes (14). Two of the nonribosomal peptide synthetase gene clusters of the strain could be assigned to known peptides from cyanobacteria; namely, nostopeptolide (15) (NpF2181–NpF2188) and anabaenopeptin (16) (NpF2459–NpF2465). Whereas anabaenopeptins are widespread in different genera of planktonic freshwater cyanobacteria (17), nostopeptolide was originally described for the terrestrial strain Nostoc sp. GSV 224 (18). Related gene clusters were predominantly detected in different Nostoc isolates, including strains isolated from lichen (19). This could be an indication that nostopeptolides (and related peptides) are specifically connected with the Nostoc lifestyle. The cyclic nonapeptide containing a butyryl side chain is synthesized by a hybrid NRPS/PKS complex comprising the NRPS NosA, NosC, and NosD and the PKS NosB (18). The two tailoring enzymes NosE and NosF are implicated in the biosynthesis of the noncanonical (2S, 4S) 4-methylproline moiety (18). The gene cluster further encodes the ABC transporter NosG presumably involved in nostopeptolide export (18). Although the identity of other PKS and NRPS-derived metabolites from N. punctiforme is unknown, there are some hints that point to an involvement of such factors in cellular differentiation. Knock-out mutagenesis of a cryptic pks gene cluster (pks2) led to an increased abundance of hormogonia and short filaments with end-standing heterocysts (14) (SI Appendix, Fig. S2). The phenotype could be complemented after the addition of wild-type exudate, indicating the involvement of a secreted factor (SI Appendix, Fig. S2). An effect of secondary metabolites on cellular differentiation has also been revealed for other microbial taxa that feature complex life cycles, including myxobacteria (20), the social amoeba Dictyostelium discoideum (21), and actinobacteria (22).
Here, we have systematically addressed the role of secondary metabolites in the differentiation process of Nostoc. Using a combination of MALDI imaging, complementation experiments, and reporter assays, we provide evidence for a governing role of nostopeptolides in the complex life cycle of N. punctiforme in the free-living state, but not in symbiosis with Gunnera manicata and Blasia pusilla.
Results
Nostopeptolide Constitutes a Major Difference in the Secretome of N. punctiforme ATCC29133 and the pks2− Mutant Under Diazotrophic Conditions.
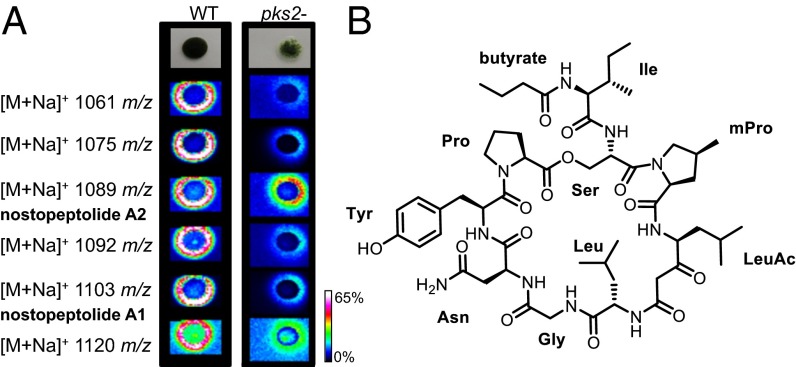
We used the recently constructed pks2− mutant that predominantly accumulates hormogonia and short filaments with end heterocysts (primordia) under diazotrophic conditions (SI Appendix, Fig. S2) (14) as a tool to evaluate the effect of secondary metabolites on the differentiation of N. punctiforme. N. punctiforme wild-type and the pks2− mutant were grown on solid agar and subjected to MALDI imaging analysis. The analysis revealed a large number of metabolic differences between the two strains (SI Appendix, Fig. S3). Although most of these differences were quantitative, a number of compounds were exclusively present either in the wild-type strain or the pks2− mutant (SI Appendix, Fig. S3). Data-dependent high-resolution tandem mass spectrometry analysis of crude extracts from both N. punctiforme strains was conducted, and structurally related metabolites were visualized using molecular network analysis, as described in ref. 23 (SI Appendix, Fig. S4). The network enables a fast dissection of metabolite families present in both wild-type and mutant from those that are exclusively present in either of the strains (SI Appendix, Fig. S4). Metabolite families solely occurring in the wild-type could potentially include the cryptic product of the pks2 pathway. However, none of the wild-type-specific fragment spectra could be correlated with the pks2 pathway (14). We were further interested in differences of the secretome that could potentially complement the pks2− phenotype. We identified a group of compounds with m/z 1061–1120 that were predominantly secreted in the wild-type strain and strongly reduced in the pks2− mutant (Fig. 1 and SI Appendix, Fig. S5). The mass range is characteristic for nostopeptolides (15). To test whether the entire group of metabolites belongs to the nostopeptolide family, nostopeptolide A was purified from this strain (SI Appendix, Fig. S6) and used as a reference for MS network analysis (SI Appendix, Fig. S5). A family of nine metabolites clustered with the nostopeptolide reference, and thus could be assigned to the nostopeptolide family. The lower amount of nostopeptolide produced (Fig. 1A) by the pks2− mutant is in agreement with the lower transcript accumulation of the nostopeptolide biosynthesis gene nosA in the pks2− mutant under diazotrophic conditions, as assessed by RT-PCR (SI Appendix, Fig. S7). Notably, the transcriptional differences were more pronounced in liquid cultures compared with cultures grown on solid agar. This finding may reflect the different diffusion properties on solid surface that may lead to differences in the dynamics of the cross-talk between the pks2 product and nostopeptolide.
Fig. 1.
Differential metabolome analysis of N. punctiforme wild-type and pks2− mutant. (A) False color intensity representations of nostopeptolide production and secretion for wild-type and pks2− mutant. Nostopeptolide A1 and A2 are indicated. Please see SI Appendix, Fig. S5 for MS/MS fragmentation spectra of further nostopeptolides. Intensities are reflected by a heat map (Right). (B) Chemical structure of nostopeptolide A.
Nostopeptolide Addition Complements the Hormogonium Phenotype of the pks2− Mutant and Represses Hormogonia Formation in the Wild-Type.
To test whether nostopeptolide addition can complement the pks2− phenotype, precultures of N. punctiforme wild-type and pks2− mutant were either grown under diazotrophic conditions or with ammonium supplementation before the medium was exchanged to fresh nitrogen-depleted (BG110) medium either supplemented with 500 ng/mL nostopeptolide A or kept as an untreated control (Fig. 2A and SI Appendix, Table S2 and Fig. S8A). The pks2− mutant showed an ongoing accumulation of hormogonia and primordia, as described previously (14) (SI Appendix, Fig. S8A). Nostopeptolide addition to the pks2− mutant led to a decreasing amount of hormogonia and primordia starting at day 2, paralleled by an increasing amount of long vegetative filaments with intercalary heterocysts (Fig. 2A and SI Appendix, Fig. S8). At day 14, more than 90% of the filaments were long vegetative filaments, as in the wild-type (Fig. 2A and SI Appendix, Table S2 and Fig. S8). This finding suggests that the lack of the hormogonium immunity period observed in the pks2− mutant is primarily a result of the strong decrease in nostopeptolide production. Remarkably, nostopeptolide addition also had an effect on cellular differentiation of the wild-type. The amount of hormogonia was clearly reduced compared to the untreated controls (SI Appendix, Table S2 and Fig. S8). To get more quantitative insights into this HRF activity, N. punctiforme wild-type cultures precultured under diazotrophic conditions or with ammonium supplementation were analyzed after exchange of the medium to fresh BG110 or BG110+NH4, respectively, with or without the addition of 100–1,500 ng/mL nostopeptolide A. Medium exchange generally triggered hormogonium formation with a maximum after 2 d, where up to 75% of the filaments were hormogonia in the untreated cultures (Fig. 2B and SI Appendix, Fig. S8). The addition of nostopeptolide led to a gradually increasing hormogonium repression effect, and the addition of 1,500 ng/mL nostopeptolide A completely abolished hormogonium formation in all cultures. In diazotrophic medium, the HRF activity was clearly visible with 500 ng/mL nostopeptolide A, whereas in cultures grown with ammonium, 1,000 ng/mL nostopeptolide A was required to get a noticeable repression effect. These data provide very clear evidence that nostopeptolide represents an autogenic HRF.
Fig. 2.
Comparison of microscopic features of N. punctiforme wild-type and pks2− mutant with and without nostopeptolide addition under diazotrophic conditions (liquid culture). (A) Microscopic pictures of pks2− mutant (Left), wild-type (Right), and pks2− mutant treated with 500 ng/mL nostopeptolide (+Nos, mid) after 14 d of cultivation. H, hormogonia; V, vegetative cells; P, primordia; arrows, heterocysts. (B) Graphic representation of the median percentage of hormogonia after exchange of medium with and without addition of 100–1,500 ng/mL nostopeptolide A in three replicates of N. punctiforme wild-type. Shown are quantitative data for cultures grown under diazotrophic conditions (N2) or with ammonium as nitrogen source (NH4). Please see SI Appendix, Table S2 and Fig. S8 for statistics on further filament types and representative micrographs obtained with 1,000 ng/mL nostopeptolide, respectively.
To test whether nostopeptolide addition can complement the pks2− phenotype on diazotrophic agar plates, a pks2− colony was placed in 1 cm distance to another pks2− colony, a wild-type colony, or a paper disk soaked with different concentrations of nostopeptolide A. When a pks2− colony or a paper disk containing only solvent was placed at 1 cm distance, pks2− colonies showed a very even distribution of hormogonia surrounding the colonies (Fig. 3 A and C). Placing a wild-type colony in the neighborhood of a pks2− colony, in contrast, led to a reduction of the amount of hormogonia in the pks2− mutant on the side facing the wild-type colony (Fig. 3B). Similarly, when 50 µg nostopeptolide A was spotted on the paper disk, the hormogonia differentiation and spreading on the side facing the nostopeptolide paper disk was reduced. In addition, the colony showed a darker appearance compared with the untreated control (Fig. 3E). Strikingly, spotting of only 10 µg nostopeptolide A on the paper disk led to a chemoattractive response, with hormogonia moving toward the paper disk (Fig. 3D). When N. punctiforme wild-type or pks2− mutant were directly grown on top of paper discs soaked with nostopeptolide A, 10 µg was sufficient to repress hormogonium formation over a period of up to 6 wk (SI Appendix, Fig. S9). These results substantiate the hypothesis that nostopeptolide is a major hormogonium-repressing factor. We also aimed to construct a knockout strain completely impaired in nostopeptolide production. However, numerous attempts with two independent constructs targeting the nosA and the nosC genes did not yield a single mutant clone.
Fig. 3.
Comparison of macroscopic features of N. punctiforme wild-type and the pks2− mutant with or without nostopeptolide addition under diazotrophic conditions on plate. pks2− mutant colonies were spotted at a 1 cm distance to (A) a second pks2− colony and (B) a wild-type colony. (C–E) Paper discs soaked with 0–50 µg nostopeptolide A, as indicated.
Nostopeptolide Is Not Differentially Regulated on the Transcriptional Level, but on the Level of Secretion During the Differentiation Cycle.
Cell-type-specific expression of nostopeptolide biosynthesis genes was evaluated using a transcriptional reporter construct in which the 500-bp-long 5′ UTR of the nosA gene was fused to a cyan fluorescence protein (CFP) gene (P-nosA-CFP) and expressed from an autonomously replicating plasmid. The N. punctiforme wild-type strain revealed a low background in the CFP channel (Fig. 4). The P-nosA-CFP mutant yielded specific fluorescence signals in vegetative cells, hormogonia, and akinetes; however, it did not do so in heterocysts (Fig. 4 A–I). No major differences could be observed in the intensity of P-nosA-CFP signals in different stages of the differentiation cycle. This is in agreement with microarray data that have evaluated transcriptional variations of the nosA-G genes during the hormogonium differentiation phase (10). To test the hypothesis that nostopeptolide is regulated on the level of secretion, nostopeptolide A was quantified from cell extracts and supernatants. In agreement with the transcriptional data, the amount of cell-bound nostopeptolides (constituting up to 99% of the nostopeptolide pool) was constantly increasing parallel to growth (Fig. 4J). Subcellular fractionation clearly demonstrated that this cell-bound nostopeptolide portion was localized in the extracellular sheath (SI Appendix, Fig. S10). In contrast, the amount of released nostopeptolide A showed clear variations with a minimum between day 2 and 5 after a transfer to fresh diazotrophic medium (hormogonium differentiation phase). The concentration of secreted nostopeptolide A increased to around 500 ng/mL at day 7 and thereafter (Fig. 4K), equaling the amount that had resulted in the hormogonium repression effect (Fig. 2B). This could indicate that the typical immunity period after an initial round of hormogonium formation is safeguarded by secreted nostopeptolides. Further support for a regulation of nostopeptolides on the level of secretion comes from MALDI imaging analyses. When wild-type colonies were placed in the direct neighborhood of pks2− colonies under either nitrogen-deplete (−N) or nitrogen-replete (+N) conditions (Fig. 4L), nostopeptolides showed asymmetric secretion patterns around the wild-type colony. Under −N conditions, secretion of nostopeptolides was increased at the colony side facing the pks2− colony, and under +N conditions, secretion of nostopeptolides was more pronounced on the side opposite the pks2− colony. When two wild-type colonies were placed in direct neighborhoods, nostopeptolides showed symmetric secretion patterns around the colonies (SI Appendix, Fig. S11). This can be taken as an indication that, depending on the availability of solute nitrogen, metabolites released by the pks2− mutant are either stimulating or repressing nostopeptolide secretion.
Fig. 4.
Fluorescence micrographs of a P-nosA-CFP transcriptional reporter strain under diazotrophic conditions and dynamics of nostopeptolide production and secretion. (A and D) P-nosA-CFP strain visualized using phase contrast microscopy. (B and E) P-nosA-CFP strain visualized in the CFP channel. (C and F) Red autofluorescence of P-nosA-CFP strain. (G–I) Control micrographs of ATCC29133 wild-type. (H) Hormogonia; P, primordia; V, vegetative filaments; A, akinetes; arrows, heterocysts. (J) Quantitative amount of cell-bound nostopeptolide A (NPL A) detected in N. punctiforme at different stages of growth under diazotrophic conditions. Shown is the median of two biologically independent cultures. (K) Quantitative amount of secreted nostopeptolide A (NPL A) in the same cultures. (L, Top) Macroscopic picture of N. punctiforme wild type and pks2− mutant in coculturing experiments. (Middle) False color intensity representations of selected nostopeptolide analogs from MALDI imaging experiments showing asymmetric secretion patterns. −N diazotrophic conditions; +N, nitrogen-replete conditions.
Nostopeptolide Is Down-Regulated in Planta and in Response to Blasia Signals.
To gain insight into the expression of nostopeptolides and other secondary metabolites in symbiotic interactions, seedlings of G. manicata were infected with the wild-type and the pks2− strain and cocultivated for 9 mo. Thin slices of the shoot basis of G. manicata plants hosting either wild-type or mutant were analyzed by MALDI imaging (Fig. 5). In parallel, N. punctiforme grown in the free-living state was spotted on the same MALDI slide. The majority of metabolites detected in planta could be assigned to the cyanobacterial biomass in the glands. Whereas a considerable number of metabolites were expressed both in the free-living state and in planta, several metabolites were either up- or down-regulated (SI Appendix, Fig. S12). None of the up-regulated metabolites could be identified or assigned to a known metabolite family (Fig. 5 E and F). The specific masses and fragment ions of nostopeptolides could not be detected in G. manicata infected with N. punctiforme wild-type and pks2− mutant (Fig. 5D), indicating that the peptide is down-regulated in symbiosis or altered by plant cells in the direct proximity.
Fig. 5.
Effect of symbiotic interaction with G. manicata on the secondary metabolome of N. punctiforme and the pks2− mutant. (A) Macroscopic picture of the shoot basis of G. manicata infected with Nostoc 9 mo postinfection. (B) Microscopic picture of cyanobacteria inside host cells in the G. manicata glands. (C, Top) Macroscopic picture of symbiotic tissue of G. manicata harboring N. punctiforme wild-type and pks2− mutant. 1, N. punctiforme wild-type in BG110 liquid medium; 2, N. punctiforme wild-type in Gunnera gland; 3, pks2− mutant in BG110 liquid medium; 4, pks2− mutant in G. manicata gland. (D–F) MALDI imaging false color intensity representation of selected metabolites. Intensities are reflected by a heat map (Right). For a detailed overview, see SI Appendix, Fig. S12. (D) Selected nostopeptolides that are down-regulated in symbiosis. (E) Selected peptide only produced in symbiosis by the pks2− mutant. (F) Selected metabolites that are only produced in symbiosis. (G) Selected metabolites that are produced both in the free-living state and in symbiosis by both wild-type and pks2− mutant.
We selected B. pusilla as a second symbiosis host in our analysis. The fragile liverwort tissue, however, was not suitable for MALDI imaging experiments. As an alternative approach, the host was infected with the P-nosA-CFP reporter strain. No specific CFP signal could be detected inside the liverwort, even though the cyanobacterial autofluorescence was perfectly visible (Fig. 6 J–L). This is a clear hint that nostopeptolide is strictly down-regulated in symbiosis. A P-nosA-CFP culture grown in parallel in the free-living state showed the regular pattern of nostopeptolide gene expression (Fig. 6 G–I). To see whether B. pusilla exudate is sufficient to repress nostopeptolide expression, the P-nosA-CFP strain was cultivated in parallel either with or without B. pusilla medium supplementation. Remarkably, the P-nosA-CFP signal was strongly reduced in the presence of the B. pusilla medium (Fig. 6 D–F) compared with the control without supplementation (Fig. 6 A–C). Thus, there are different lines of evidence in two different symbiosis hosts, indicating that nostopeptolide is down-regulated in planta and in response to plant exudates.
Fig. 6.
Fluorescence micrographs of a P-nosA-CFP transcriptional reporter strain treated with supernatant of B. pusilla or in symbiosis. (A–F) Comparative analysis of P-nosA-CFP strain-treated Blasia exudate and untreated control. (A–C) Control micrographs showing P-nosA-CFP strain without Blasia exudate addition. (D–F) Micrographs showing P-nosA-CFP strain treated with Blasia exudate. (G–L) Comparative analysis of P-nosA-CFP strain inside and outside of symbiotic host. (G–I) Control micrographs of P-nosA-CFP strain grown in the free-living state. (J–L) Micrographs showing P-nosA-CFP strain in symbiosis with B. pusilla.
Discussion
It has long been suggested that the complex lifestyle of N. punctiforme is influenced by autogenic secretion factors that act at least partly antagonistic to plant factors that stimulate the infection process (1). The present study uncovers the identity of one of those factors and demonstrates that nostopeptolide plays a crucial role in the orchestration of cellular differentiation under diazotrophic conditions.
Remarkably, different concentrations of released nostopeptolide had completely different effects on cellular differentiation and the direction of motility of N. punctiforme. A possible explanation for this observation is that other factors produced by N. punctiforme interfere with the signaling process triggered by nostopeptolides, and that the ratio between the different factors finally determines the route of differentiation and the direction of motility. Evidence for an interconnection of secondary metabolites in N. punctiforme comes from the metabolomic comparison of the wild-type and the pks2− mutant strains, using MALDI imaging. Loss of the cryptic pks2 product has led to a pronounced reprogramming of the secondary metabolome. A knockout mutant impaired in nostopeptolide production could have strengthened this view. However, despite numerous attempts, we could not generate a mutant. Although we cannot rule out the possibility that this failure is a result of technical problems, this could be a hint that nostopeptolide is essential for the strain in the free-living state. Further support for a metabolite cross talk in N. punctiforme is provided by the MALDI imaging analysis of neighboring wild-type and pks2− colonies. Nostopeptolide secretion was apparently influenced by other factors released by the pks2− mutant.
Strikingly, MALDI images suggested that the entire nostopeptolide fraction is located outside cells, contradicting the fact that nostopeptolides were primarily detected in cellular extracts. Subcellular fractionation studies, however, could solve this discrepancy and demonstrate that the cell-bound fraction is localized in the extracellular polysaccharide sheath. Together, these findings suggest that nostopeptolide is regulated neither on the level of transcription nor at the level of membrane transport in the free-living state. The dynamics observed are apparently the result of a dynamic release and reuptake of nostopeptolide from and into the sheath. The metabolite cross-talk indicated on the MALDI neighboring images may thus reflect a reciprocal influence of different secretion factors on nostopeptolide release from the extracellular polysaccharide matrix. The identity of the factors that bind nostopeptolides in this matrix remains elusive. Lectin-binding studies have revealed that the sugar composition of the N. punctiforme extracellular polysaccharide is changing during the hormogonium differentiation phase and the following immunity phase (24). Different types of extracellular polysaccharide may have different capacities to “hold” nostopeptolides, and may thus interfere with the secondary metabolite cross-talk.
Nostopeptolide was down-regulated in two completely different symbiotic hosts, as shown using MALDI imaging and a transcriptional reporter strain, respectively. This is particularly surprising, as nostopeptolide is expressed under all conditions in the free-living state. In sharp contrast, the exudate of the B. pusilla host led to complete down-regulation of the nos cluster. This could be an indication that nostopeptolide is one of the primary targets of the B. pusilla signal or signals. Apparently, nostopeptolide is not essential for the lifestyle of N. punctiforme in symbiosis. In contrast, several low-molecular-weight metabolites detected inside G. manicata were never observed in the free-living state. The “plant-induced” metabolites could potentially result from a biotransformation of N. punctiforme metabolites by plant cells in close proximity or may result from tissue effects that may lead to an altered ionization. The lack of nostopeptolides, however, corresponds well to the down-regulation of the biosynthetic gene cluster on the transcriptional level in the second symbiosis host. One may therefore speculate that other metabolite families are being up-regulated as response to plant factors. This observation can inspire the development of novel genome mining strategies for plant-induced secondary metabolites from Nostoc species. Similar observations were made for the interaction of Streptomycetes and fungi. Only the intimate physical interaction of the partners could stimulate the production of certain types of metabolites, including the archetypical metabolite lecanoric acid that was previously known from lichen (25).
Considering the complexity of the lifestyle of N. punctiforme and the multitude of distinct secondary metabolite families that can be estimated for the strain, one can speculate that nostopeptolide represents only the tip of an iceberg. Future studies have to show the nature of other factors balancing the effects of nostopeptolide, to solve the structure of the cryptic pks2 metabolite, and to identify the nostopeptolide-signaling cascade and the mechanism regulating nostopeptolide secretion. Understanding the lifestyle of symbiotic cyanobacteria and the role of secondary metabolites in the process can provide fascinating insight into one of the most versatile and ancient partnerships on earth.
Materials and Methods
Organisms and Cultivation Conditions.
Cyanobacteria were maintained under permanent white light at the illumination intensity of 30 µmol photons/M2S1 at 23 °C in 50 mL BG110 for diazotrophic growth or in BG11 for nitrogen-supplemented cultures (26). Ammonium chloride was added at a concentration of 2.5 mM when used as alternative nitrogen source. In all assays on agar plates, the media contained 0.8% agar. The medium for the pks2− mutant strain was supplemented with 12.5 µg/mL neomycin, and the P-nosA-CFP mutant was maintained on media with 2 µg/mL streptomycin. The antibiotics were omitted when strains were cocultivated with other strains or organisms. Cell density was measured by estimation of Chla content, according to ref. 27.
The hepatic B. pusilla was grown under the same light and temperature conditions in 1/5 BG11 or 1/5 BG110. To obtain active supernatant, the culture of B. pusilla was transferred to nitrogen-free medium and starved for 5 wk, as described earlier (28). G. manicata Linden was propagated from fresh seeds collected in the greenhouses of the Department of Botany, University of Stockholm, Sweden. The propagation procedure and media used were according to Johansson and Bergman (26). Seedlings with one true leaf were transferred to sterile plastic boxes filled with vermiculite soaked with the medium and were grown hydroponically under sterile conditions.
Symbiosis and Differentiation Assays.
A loopful of cyanobacterial culture was transferred to the nitrogen-starved B. pusilla liquid culture and grown for 3 wk. The effect of plant exudates was tested in medium composed of one part filtered plant-free supernatant from starved B. pusilla and one part BG110. Media for control cultures was made by mixing 1/5 BG110 and BG110. G. manicata seedlings were infected by the addition of 10 µL cyanobacterial suspension to the apex. Reinoculations were repeated every 4 wk. For differentiation assays, cyanobacterial cultures were grown to a cell density of ∼5 µg/mL Chla. The cultures were washed three times with fresh medium and then inoculated into the respective medium to a cell density of 0.2–0.3 µg/mL Chla. Nostopeptolide was added to a final concentration of 10–1,500 ng/mL. The respective volume of solvent [50% (vol/vol) methanol] was added to the control cultures. Samples for microscopic observations were taken after 2, 5, and 14 d. For each sample, 1,000 filaments were counted in focus plane in randomly taken snapshots. Microscopic analyses were performed on a Leitz DMRBE microscope equipped with Leica DFC420 camera and filters N2.1 for autofluorescence and CGFP for detection of CFP. Image acquisition was done in related LAS Version 3.4.1 software (Leica Microsystems).
Assays on Agar Plates.
For testing of mutual influence on motility and growth, 20 µL cell suspension from late exponential cultures was pipetted on the surface of agar at a 1-cm distance. The 6-mm filter paper disks with or without nostopeptolide were applied in the same way. Incubation time was 3 wk.
MALDI Imaging.
Thin-layer BG11 and BG110 agar plates of N. punctiforme wild-type and pks2− mutant monocultures or bacterial interactions were inoculated from fresh liquid cultures and incubated for 10 d (29). Sample preparation, matrix application, and dehydration were conducted as previously described (30). For MALDI imaging experiments of plant cyanobacteria symbiosis, G. manicata plants harboring the symbiotic glands were cut into thin slices. The stem slices were immobilized on conductive glass slides. Universal MALDI matrix in methanol was sprayed onto the plant samples. Data were acquired on a Bruker UltrafleXtreme MALDI TOF mass spectrometer equipped with smart beam technology in positive mode. Method optimization was conducted using FlexControl3.0 (Bruker Daltonic GmbH). Data were analyzed with FlexImaging3.0 (Bruker Daltonic GmbH). False colors were assigned to masses of interest and superimposed onto the optical reference picture taken before matrix application. Single spectra analysis on different spatial localizations for spectral comparison was performed using FlexAnalysis3.0 (Bruker Daltonic GmbH).
Generation of a Reporter Mutant for the Nostopeptolide Biosynthesis 5′ UTR.
The 5′ UTR of Npun_F2181 (P-nosA) was amplified by PCR, using the primer pair NosA_Prom_SacI_FW and NosA_Prom_NdeI_RV, thereby introducing a SacI and an NdeI site. The cfp gene was obtained from the pECFP-C1 vector (Clontech Laboratories) by PCR, using the primer pair E-C/YPF-Fw and ECFP-Rv, thereby introducing NdeI and BamHI sites for subsequent ligation into the vector pRL1049 and introducing a stop codon for the cfp gene. The resulting construct pRL1049-Npun_F2181CFP was transferred into N. punctiforme by electroporation at 1.5 kV in a Bio-Rad-MicroPulser. The reaction mix was immediately diluted with BG11 medium supplemented with 2.5 mM NH4Cl + 5 mM Mops + 20 mM MgCl2 and plated onto HATF filters (Millipore) on plates containing BG11 medium. After initial growth, the filters were transferred to selective plates containing 2 µg/mL streptomycin. The obtained mutant was tested by PCR analysis, using a primer pair targeting the construct-flanking region, and the resulting amplicon was sequenced.
Nostopeptolide Quantification.
Strains were precultured for 1 wk in BG110 with 2.5 mM NH4Cl and divided in cultures of 50 mL BG110. Cells were harvested by centrifugation after 0–14 d. Supernatants were purified on C18-Sep-Pak cartridges (Waters). Cell pellets were lysed by sonication and finally dissolved in methanol. HPLC was conducted on a Shimadzu HPLC unit comprising the system controller CBM-20A, the pump LC-20AD, the autosampler SIL-20AC HT, and the photo diode array detector SPD-M-20A. Separation was carried out on a SymmetryShield RP18 column (Waters). HPLC profiles were monitored at the wavelength 199 nm, and peaks were integrated and calculated using a quantitative nostopeptolide reference.
Supplementary Material
Acknowledgments
We are grateful to Professor Birgitta Bergman and Peter Litfors, Stockholm University, for seeds of G. manicata. We thank Florian Kloss for the design of a device for MALDI imaging Matrix application. This project was supported by a grant from the German Research Foundation (DFG Di910/5-1) and the Cluster of Excellence UniCAT (to E.D.). Travel grants to A.L. from Funksjonell Genomforskning i Nord-Norge (FUGE-Nord) are also acknowledged. This work was further supported by a scholarship from the German National Academic Foundation (to E.J.N.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419543112/-/DCSupplemental.
References
- 1.Meeks JC, Elhai J. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol Rev. 2002;66(1):94–121. doi: 10.1128/MMBR.66.1.94-121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rai AN, Soderback E, Bergman B. Cyanobacterium-plant symbioses. New Phytol. 2000;147(3):449–481. doi: 10.1046/j.1469-8137.2000.00720.x. [DOI] [PubMed] [Google Scholar]
- 3.Chapman MJ, Margulis L. Morphogenesis by symbiogenesis. Int Microbiol. 1998;1(4):319–326. [PubMed] [Google Scholar]
- 4.Meeks JC, Campbell EL, Summers ML, Wong FC. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch Microbiol. 2002;178(6):395–403. doi: 10.1007/s00203-002-0476-5. [DOI] [PubMed] [Google Scholar]
- 5.Meeks JC. Symbiosis between nitrogen-fixing cyanobacteria and plants - The establishment of symbiosis causes dramatic morphological and physiological changes in the cyanobacterium. Bioscience. 1998;48(4):266–276. [Google Scholar]
- 6.Meeks JC. Molecular mechanisms in the nitrogen-fixing Nostoc-Bryophyte symbiosis. In: Overmann J, editor. Molecular Basis of Symbiosis. Springer; Heidelberg, Germany: 2006. pp. 165–190. [DOI] [PubMed] [Google Scholar]
- 7.Herdman M, Rippka R. Cellular differentiation: Hormogonia and baeocytes. Methods Enzymol. 1988;167:232–242. [Google Scholar]
- 8.Meeks JC, et al. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth Res. 2001;70(1):85–106. doi: 10.1023/A:1013840025518. [DOI] [PubMed] [Google Scholar]
- 9.Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol. 2007;189(14):5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell EL, Christman H, Meeks JC. DNA microarray comparisons of plant factor- and nitrogen deprivation-induced Hormogonia reveal decision-making transcriptional regulation patterns in Nostoc punctiforme. J Bacteriol. 2008;190(22):7382–7391. doi: 10.1128/JB.00990-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell EL, Brahamsha B, Meeks JC. Mutation of an alternative sigma factor in the cyanobacterium Nostoc punctiforme results in increased infection of its symbiotic plant partner, Anthoceros punctatus. J Bacteriol. 1998;180(18):4938–4941. doi: 10.1128/jb.180.18.4938-4941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell EL, Wong FC, Meeks JC. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in the differentiation of hormogonia. Mol Microbiol. 2003;47(2):573–582. doi: 10.1046/j.1365-2958.2003.03320.x. [DOI] [PubMed] [Google Scholar]
- 13.Chapman KE, Duggan PS, Billington NA, Adams DG. Mutation at different sites in the Nostoc punctiforme cyaC gene, encoding the multiple-domain enzyme adenylate cyclase, results in different levels of infection of the host plant Blasia pusilla. J Bacteriol. 2008;190(5):1843–1847. doi: 10.1128/JB.01321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liaimer A, et al. A polyketide interferes with cellular differentiation in the symbiotic cyanobacterium Nostoc punctiforme. Environ Microbiol Rep. 2011;3(5):550–558. doi: 10.1111/j.1758-2229.2011.00258.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunsucker SW, Klage K, Slaughter SM, Potts M, Helm RF. A preliminary investigation of the Nostoc punctiforme proteome. Biochem Biophys Res Commun. 2004;317(4):1121–1127. doi: 10.1016/j.bbrc.2004.03.173. [DOI] [PubMed] [Google Scholar]
- 16.Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria) Chem Biol. 2010;17(3):265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Welker M, von Döhren H. Cyanobacterial peptides - nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann D, Hevel JM, Moore RE, Moore BS. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene. 2003;311:171–180. doi: 10.1016/s0378-1119(03)00587-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, et al. 4-methylproline guided natural product discovery: Co-occurrence of 4-hydroxy- and 4-methylprolines in nostoweipeptins and nostopeptolides. ACS Chem Biol. 2014;9(11):2646–2655. doi: 10.1021/cb500436p. [DOI] [PubMed] [Google Scholar]
- 20.Meiser P, Bode HB, Müller R. The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation. Proc Natl Acad Sci USA. 2006;103(50):19128–19133. doi: 10.1073/pnas.0606039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Austin MB, et al. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol. 2006;2(9):494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traxler MF, Watrous JD, Alexandrov T, Dörrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4(4) doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schüssler A, Meyer T, Gehrig H, Kluge M. Variations of lectin binding sites in extracellular glycoconjugates during the life cycle of Nostoc punctiforme, a potentially endosymbiotic cyanobacterium. Eur J Phycol. 1997;32(3):233–239. [Google Scholar]
- 25.Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106(34):14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson C, Bergman B. Early events during the establishment of the Gunnera/Nostoc symbiosis. Planta. 1992;188(3):403–413. doi: 10.1007/BF00192808. [DOI] [PubMed] [Google Scholar]
- 27.Tandeau de Marsac N, Houmard J. Complementary chromatic adaptation:physiological conditions and action spectra. Methods Enzymol. 1988;167:318–328. [Google Scholar]
- 28.Watts SD, Knight CD, Adams DG. Characterisation of plant exudates inducing chemotaxis in nitrogen-fixing cyanobacteria. In: Peschek GA, Löffelhardt W, Schmetterer S, editors. The Photosynthetic Prokaryotes. Kluwer Academic/Plenum Publishers; New York: 1999. pp. 679–684. [Google Scholar]
- 29.Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35(2):171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang JY, et al. Primer on agar-based microbial imaging mass spectrometry. J Bacteriol. 2012;194(22):6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.