Significance
Human IgM+IgD+CD27+ B lymphocytes represent a large subpopulation of the human B-cell pool, but their generation is debated and their immunological functions are poorly understood. This work shows that these lymphocytes possess typical memory B-cell expression patterns, enabling them to differentiate rapidly into plasma cells upon restimulation. Moreover, we reveal unique features of these IgM memory B cells, their potential to reenter germinal center reactions, and their specific interaction with immunomodulatory neutrophils in early inflammatory responses. Thus, key characteristics and functions of a major human B-cell subset are elucidated.
Keywords: IgM memory B-cell functions, germinal center reentry, early inflammatory response
Abstract
The generation and functions of human peripheral blood (PB) IgM+IgD+CD27+ B lymphocytes with somatically mutated IgV genes are controversially discussed. We determined their differential gene expression to naive B cells and to IgM-only and IgG+ memory B cells. This analysis revealed a high similarity of IgM+(IgD+)CD27+ and IgG+ memory B cells but also pointed at distinct functional capacities of both subsets. In vitro analyses revealed a tendency of activated IgM+IgD+CD27+ B cells to migrate to B-cell follicles and undergo germinal center (GC) B-cell differentiation, whereas activated IgG+ memory B cells preferentially showed a plasma cell (PC) fate. This observation was supported by reverse regulation of B-cell lymphoma 6 and PR domain containing 1 and differential BTB and CNC homology 1, basic leucine zipper transcription factor 2 expression. Moreover, IgM+IgD+CD27+ B lymphocytes preferentially responded to neutrophil-derived cytokines. Costimulation with catecholamines, carcinoembryonic antigen cell adhesion molecule 8 (CEACAM8), and IFN-γ caused differentiation of IgM+IgD+CD27+ B cells into PCs, induced class switching to IgG2, and was reproducible in cocultures with neutrophils. In conclusion, this study substantiates memory B-cell characteristics of human IgM+IgD+CD27+ B cells in that they share typical memory B-cell transcription patterns with IgG+ post-GC B cells and show a faster and more vigorous restimulation potential, a hallmark of immune memory. Moreover, this work reveals a functional plasticity of human IgM memory B cells by showing their propensity to undergo secondary GC reactions upon reactivation, but also by their special role in early inflammation via interaction with immunomodulatory neutrophils.
Antigen-activated naive B cells migrate along chemokine (C-C motif) ligand 19 (CCL19) / chemokine (C-C motif) ligand 21 (CCL21) gradients to the periphery of B-cell follicles (1), where they interact with stimulating T cells and expand in primary focus reactions (2). The majority of their progeny differentiate into short-lived, mostly IgM+, antibody-secreting cells (ASCs) that contribute a first wave of selected, but not affinity-matured, antibodies to the immune reaction. A minor fraction of these B-cell clones migrate along a chemokine (C-X-C motif) ligand 13 (CXCL13) chemotactic axis into the follicle center (3) and initiate a germinal center (GC) reaction by interaction with follicular helper T cells and dendritic cells (4). The decision to become a (short-lived) plasma cell (PC) or a GC B cell is governed by the B-cell lymphoma 6 (BCL6) and PR domain containing 1 (PRDM1) transcription factors (5). B lymphocytes up-regulating BCL6 induce a GC B-cell differentiation program, whereas B cells up-regulating PRDM1 at the expense of BTB and CNC homology 1, basic leucine zipper transcription factor 2 (BACH2) expression (6) become ASCs. In the GC, B cells undergo T cell-dependent (TD) affinity maturation and often class switching, resulting in improved and selected B-cell receptors (BCRs) that are expressed on post-GC memory B cells or secreted by long-lived PCs (7). Egress of these cells to the lymph node medullary chords, red pulp, or bone marrow is mediated by chemokine (C-X-C motif) ligand 12 (CXCL12) / chemokine (C-X-C motif) receptor 4 (CXCR4) (8).
Specificity and memory represent the hallmarks of adaptive immunity. Memory B cells are generated in adaptive immune responses and confer enhanced and improved reactivity against recurring antigen. The capability of a memory B cell to contribute to secondary immune responses is granted by longevity, clonal expansion, increased ability to be activated, and a propensity to differentiate into PCs. In mice, the memory B-cell compartment is multilayered, including an early GC-independent IgV gene-unmutated and mostly IgM+ B-cell population (9–11), as well as post-GC memory B cells with affinity-matured and either class-switched or IgM BCRs (12, 13). IgG+ memory B cells efficiently differentiate into PCs (14, 15) and IgM+ memory B cells frequently reenter GC reactions upon secondary challenge (12). Improved immune responses against T cell-independent (TI) antigens also exist (16, 17), but these B cells are not long lived and their IGV mutation load is very low.
In humans, the memory B-cell compartment is less well understood. Traditionally, class-switched B cells with mutated IgV genes and surface CD27 expression were regarded as “prototypical” memory B cells (5), although more recent data have shown that CD27 is a sufficient, but not essential, memory B-cell marker, because a small subset of IgG+CD27− and potentially also IgM+CD27− memory B cells has been identified (18–20). However, the adult human peripheral blood (PB) B-cell pool also includes about 15% IgM+IgD+ and 5% IgM-only (expressing little or no IgD) B lymphocytes with mutated V gene rearrangements and CD27 expression (21). Although both of these IgM+ subsets fulfill functional requirements of conventional memory B cells (22) and frequently derive from GC reactions (23), the developmental and immunological characteristics of IgM+IgD+CD27+ B cells are debated and only poorly understood. The presence of few and lowly mutated IgM+IgD+CD27+ B cells in cord blood (24, 25), as well as in patients with X-linked hyper-IgM syndrome with a deficiency of the CD40 ligand, and hence supposedly of GC reactions (26), and their reduced frequency in asplenic patients, correlating with impaired immune response to encapsulated bacteria (27), were interpreted in favor of a GC-independent generation. This generation could occur either by primary antigen-independent Ig gene diversification mechanisms or by TI immune responses involving somatic hypermutation. A specific function of IgM+IgD+CD27+ B cells in TI type II immune reactions was further suggested by their high phenotypical and functional similarity to human marginal zone (MGZ) B cells (28, 29), which are important players in immune responses against blood-borne TI pathogens (30).
Human MGZ B cells have been reported specifically to interact with neutrophils that reside in close vicinity to the MGZ and to show B cell-helper function and induce Ig gene diversification via secretion of tumor necrosis factor superfamily members 13 and 13B and interleukin 21 (31), although a recent study contradicts these findings (32). Neutrophils are recruited to epithelial cells in injured or infected tissue by secretion of early cytokines. In this early inflammatory response, neutrophils become activated and participate in a multilayered immunomodulatory network, recruiting and directing local responses by secretion of catecholamines and multiple cytokines, including chemokine (C-C motif) ligand 2 (CCL2), soluble carcinoembryonic antigen cell adhesion molecule 8 (sCEACAM8), and IFN-γ (33–35).
This study aims at defining specific functions of human IgM+IgD+CD27+ and IgG+CD27+ B cells. We discovered that human PB IgM+IgD+CD27+ B cells, despite sharing a high phenotypical similarity with IgG+CD27+ memory B lymphocytes, show a high potential to be stimulated by activated neutrophils early in inflammation. Moreover, we show that human IgM+IgD+CD27+ B cells share with murine IgM memory B cells a propensity to B-cell follicle homing and GC B-cell differentiation upon BCR (re)stimulation.
Results
Human IgM+CD27+ and Class-Switched B-Cell Subsets Share Typical Memory B-Cell Characteristics.
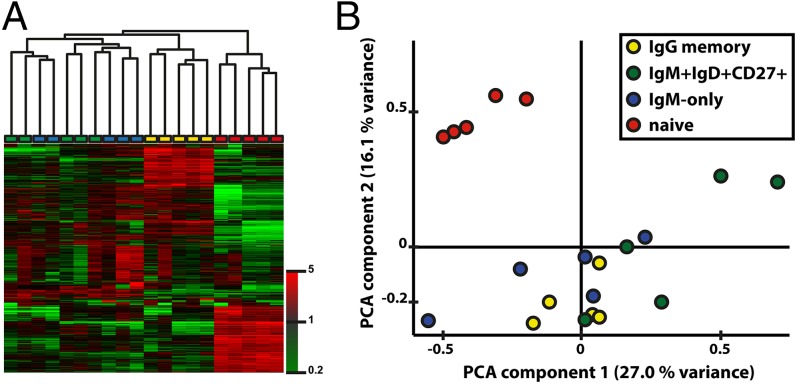
We explored the transcriptome patterns of human PB naive and CD27+ memory B-cell subsets to reveal shared and unique features of four major human PB B-lymphocyte subsets [i.e., naive B cells (IgM+IgDhighCD27−), IgM-only (IgM+IgDlow/−CD27+), IgM+IgD+CD27+, and class-switched (IgG+CD27+) memory B cells]. The comprehensive analysis of 21,000 genes with the highest SDs revealed a significant similarity of human IgM-only, IgM+IgD+CD27+, and class-switched memory B cells, and a clear separation of these three subsets from naive B cells, as determined by unsupervised hierarchical clustering and principal component analysis (PCA) (Fig. 1 A and B), thus arguing for a common generation and functional similarity of CD27+ B-cell subsets. Indeed, gene set enrichment analysis (GSEA) identified shared gene signatures functionally associated with a “memory B-cell” phenotype, including enhanced antigen response, increased metabolic turnover, a (homeostatic?) proliferation signature, and a propensity to plasmablast differentiation, although this latter feature was more pronounced in class-switched memory B cells than in IgM memory B cells (Fig. S1 A–G).
Fig. 1.
Transcriptional similarity of human PB B-cell subsets. (A) Dendrogram is calculated by Manhattan clustering of 4,717 probe sets with the highest SDs. Subcluster stability was confirmed by a bootstrapping procedure (>70%). The color bar depicts normalized intensity values. The color boxes at the bottom of the dendrogram depict the B-cell subsets: green, IgM+IgD+CD27+ B cells; blue, IgM-only B cells; yellow, IgG+CD27+ B cells; red, naive B cells. (B) Unsupervised PCA shows a high similarity of PB IgM+IgD+CD27+, IgM-only, and class-switched B cells, but not naive B cells. The PCA is based on 29,969 probe sets with the highest SDs, explaining more than 43% of total variance. Axis scaling is according to mean centering and scaling. The color code is as in A.
Taken together, human PB IgM+IgD+CD27+ and IgM-only B cells closely resemble post-GC IgG+ memory B lymphocytes in their global gene expression pattern.
Human IgM+ Memory B-Cell Expression Patterns Reveal Specific Immunological Functions.
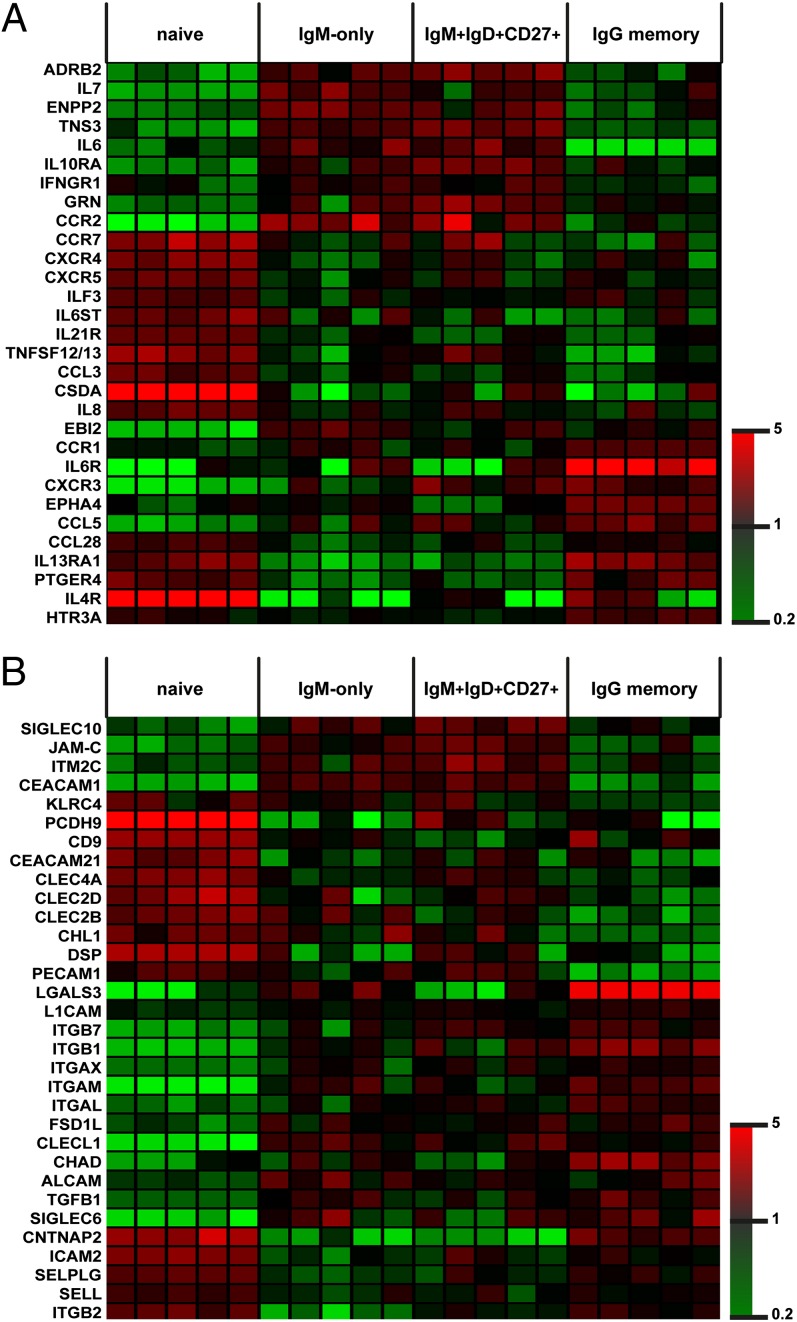
IgM-only and IgM+IgD+CD27+ B-cell subsets did not show statistically significant differentially expressed transcripts, except for IgD. Moreover, the two populations displayed highly similar surface receptor expression profiles, thus arguing for identical stimulation requirements or immune-sensing capacities (Fig. 2 A and B). From here on, IgM-only B cells are excluded from our analyses and we focus on IgM+IgD+CD27+ B cells (referred to as IgM memory B cells), IgG+CD27+ B cells (IgG memory B cells), and naive B cells only. Besides the significant overlap in transcription associated with IgG memory B cells, both hierarchical clustering and PCA indicated unique expression patterns in IgM memory B cells (Figs. 1 A and B and 2 A and B). Pairwise comparison revealed 422 genes that showed at least a twofold difference [P < 0.05 and false discovery rate (FDR) < 0.1] between IgM and IgG memory B cells, of which 214 annotated transcripts were also differentially expressed with at least twofold change between IgM memory and naive B cells (P < 0.05 and FDR < 0.1; Table S1). Fig. 2 A and B show selected genes from this comparison that were categorized as either “cytokines, hormones, growth factors, neurotransmitters, and their receptors” or as “cell adhesion molecules.” Fig. 2 A and B further includes manually selected transcripts of both categories with uniquely high or low expression in a single subset.
Fig. 2.
Transcriptional patterns in human PB B-cell subsets. (A) Expression heat map of normalized signal intensities from statistically significant (ANOVA, P < 0.05; Tukey post hoc, P < 0.05) differentially expressed cytokines, hormones, growth factors, and their receptors in naive, IgG+CD27+, IgM+IgD+CD27+, and IgM-only B cells. (B) Expression heat map of normalized signal intensities from statistically significant differentially expressed cell adhesion molecules in naive, IgG+CD27+, IgM+IgD+CD27+, and IgM-only B cells.
To interpret these patterns, we performed GSEA and identified enriched or decreased expression of gene sets involved in cytokine signaling [interleukin 6 (IL-6), IL-8, IFN], integrin expression, growth factor receptor signaling, nuclear factor kappa B or nuclear factor of activated T cells signaling, and PC differentiation (Fig. S2 A–M).
To identify key molecules with the potential to mediate specific IgM memory B-cell functions, we filtered for single, significantly over- or underrepresented genes and pathways with preferential expression in IgM memory B cells. In the following, we describe functional characteristics mediated by such genes.
Neurotransmitter Noradrenaline Is a Costimulator of IgM Memory B Cells.
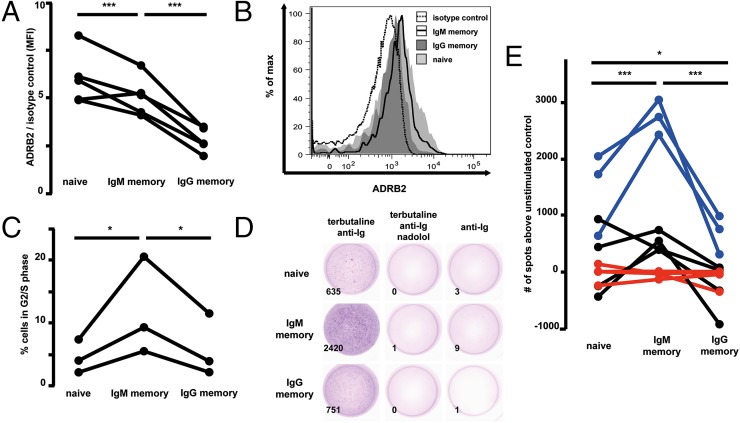
High expression of beta-2 adrenergic receptor (ADRB2) transcripts was detected on IgM memory cells but not on IgG+ or naive B cells (Fig. 2A). Flow cytometric analysis confirmed significantly higher ADRB2 expression on IgM memory cells than on IgG memory B cells; however, in contrast to transcriptional data, the highest protein levels were observed on naive B cells (Fig. 3 A and B). We assessed the stimulatory potential of ADRB2 on isolated human PB B-cell subsets by in vitro challenge with the selective ADRB2 agonist terbutaline and the nonselective beta-adrenergic receptor antagonist nadolol in combination with BCR triggering. In contrast to previous publications (36, 37), high levels of terbutaline (10−5 M) induced only marginal B-cell proliferation. However, the average number of cells in S/G2 phase, as determined by DNA content (Fig. 3C), was consistently higher in IgM memory B cells (12%, range: 5–21%) than in IgG memory cells (6%, range: 2–12%) or naive B cells (4%, range 2–7%). The differences observed were statistically significant (P < 0.05). In addition, we analyzed the Ig secretion capacity of human PB B-cell subsets upon stimulation by terbutaline by enzyme-linked immunospot (ELISpot) assays. We incubated a minimum of 25,000 sort-purified B cells per subset for 1 h in anti-IgM– or anti-IgG–coated ELISpot wells. The cells were stimulated in advance with 10−5 M terbutaline in combination with anti-Ig antibody or with anti-Ig alone. The differentiation into Ig-secreting cells upon treatment with terbutaline was higher in IgM memory B cells than in IgG memory or naive B cells, compared with anti-Ig stimulation alone. Notably, this effect could be markedly enhanced with a combination of BCR and ADRB2 stimulation, and it could be blocked by addition of the beta-adrenergic receptor antagonist nadolol, arguing for the specificity of the effects observed (Fig. 3 D and E).
Fig. 3.
ADRB2 is a costimulator of human IgM memory B cells. (A) FACS analysis of CD19-enriched human PB B-lymphocyte subsets, defined by IgD, IgG, and CD27 expression. ADRB2 surface expression of B-cell subsets from five healthy subjects is depicted as the mean fluorescence intensity (MFI), normalized to an isotype control. Lines between the values link the samples derived from a given donor. (B) Representative FACS histogram of ADRB2 expression on naive (light gray), IgG memory (dark gray), and IgM memory (black line) B cells and an isotype control (dashed line). (C) Frequency of B-cell subsets in the G2/S phase of the cell cycle, as determined by propidium iodide (PI) staining, after BCR stimulation and treatment with 10−5 M terbutaline (normalized to BCR stimulation alone) for three blood donors. Cells were stimulated for 16 h. (D) One representative ELISpot analysis out of three experiments with sort-purified B-cell subsets, stimulated with combinations of 10−5 M terbutaline, 10 mg/L anti-Ig antibody, and 10−3 M nadolol or anti-Ig antibody alone. Numbers in the lower left corners give the total number of spots counted by the ELISpot reader. (E) Overview of ELISpot analyses of ADRB2-stimulated B-cell subsets as in D: blue, 10−5 M terbutaline and 10 mg/L anti-Ig antibody; black, 10−3 M terbutaline alone; red, 10−5 M terbutaline, 10 mg/L anti-Ig antibody, and 10−3 M nadolol. Shown are the number of spots per well after subtraction of anti-Ig stimulation alone. (A, C, and E) *P < 0.05; ***P < 0.001, significance values in E refer to blue data points.
We conclude that IgM memory B cells preferentially respond in vitro to BCR costimulation with the noradrenaline/adrenaline-mimicking factor terbutaline via ADRB2 by low-level proliferation and efficient differentiation into Ig-secreting cells.
Human IgM Memory B Cells Show Migration and Differentiation Capacity Toward Specific Early Inflammatory Cytokines.
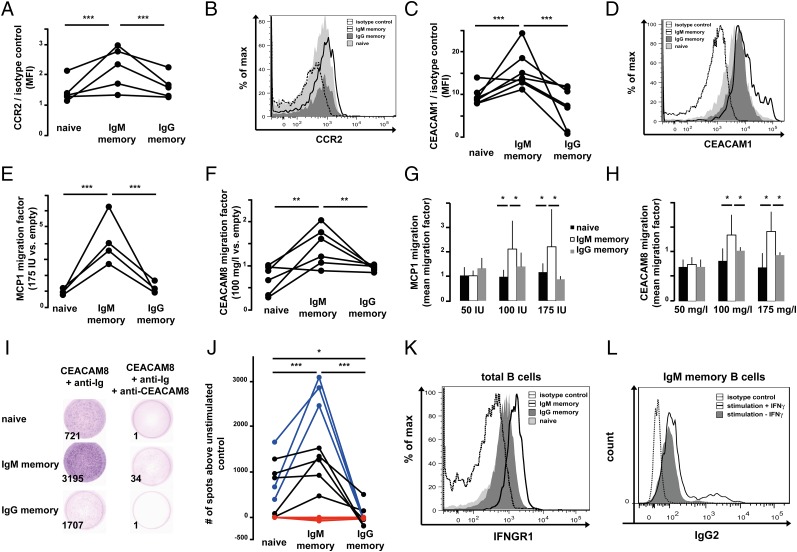
We screened cytokine receptors and immunomodulatory molecules with higher expression in IgM memory B cells than in IgG memory or naive B cells (Fig. 2A). Carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1) and chemokine (C-C motif) receptor 2 (CCR2), both molecules that were previously reported to be expressed on a fraction of bulk CD19+ B cells (38–41), turned out to be predominantly detectable on the surface of IgM memory B cells (Fig. 4 A–D). CCR2 is involved in the recruitment of macrophages to inflammatory sites along a macrophage chemoattractant protein 1 (MCP1) gradient. CEACAM1 belongs to a family of intermediate-affinity cell adhesion molecules involved in a variety of immune modulatory functions, including cell–cell contact, immune cell activation, and survival (35, 42–44). In vitro chemotactic assays with MCP1 and sCEACAM8, the natural ligands for CCR2 and CEACAM1, respectively, showed that IgM memory B cells possess chemotactic activity toward both ligands (P < 0.001 and P < 0.01, respectively; Fig. 4 E and F). In line with a specific biological responsiveness, this chemotactic activity was dose-dependent (Fig. 4 G and H) and significantly lower, if at all detectable, in IgG memory or naive B cells.
Fig. 4.
Analysis of CCR2, CEACAM1, and IFNGR1 expression and function on PB naive and memory B cells. (A) CCR2 surface expression of B cells from five healthy subjects is depicted as MFI, normalized to isotype control. (B) Representative FACS histogram of CCR2 expression on naive (light gray), IgG memory (dark gray), and IgM memory B (black line) cells and isotype control (dashed line). max, maximum. (C) Surface CEACAM1 expression as in A. (D) Representative CEACAM1 expression histogram; B-cell populations are depicted as in B. (E and F) Relative numbers of sort-purified B-cell subsets that migrated toward MCP1 (175 IU) and sCEACAM8 (100 mg/L), respectively. (G and H) Mean migration factors (migrated cells in stimulation divided by unstimulated migrated cells) of sort-purified B-cell subsets toward 50, 100, and 175 IU of MCP1 and 50, 100, and 175 mg/L sCEACAM8, respectively. Error bars depict SD of five donors analyzed. (I) Representative ELISpot analysis out of three experiments with sorted B-cell subsets, stimulated with 100 mg/L sCEACAM8 and 10 mg/L anti-Ig antibody or 10 mg/L anti-CEACAM8 and 10 mg/L anti-Ig antibody. Numbers in the lower left corners give the total number of spots counted. (J) Overview of ELISpot analyses of CEACAM8-stimulated B-cell subsets: blue, 100 mg/L CEACAM8 and 10 mg/L anti-Ig antibody; black, 100 mg/L CEACAM8 alone; red, 10 mg/L anti-CEACAM1 and 10 mg/L anti-Ig antibody. Shown are the number of spots per well after subtraction of anti-Ig stimulation alone. Significance values refer to blue data points. (K) Representative IFNGR1 expression histogram; populations are depicted as in B. (L) FACS histogram of IgG2 expression on IgM memory B cells activated for 6 days with 1 mg/L each of anti-Ig antibody, PWM, and BAFF alone (dark gray) or with 1 mg/L IFN-γ in addition (black line). The isotype control is shown as a dashed line. Data are representative of four independent donors (*P < 0.05; **P < 0.01; ***P < 0.001).
We analyzed the costimulatory effect of sCEACAM8 on the Ig secretion capacity of human PB B-cell subsets by ELISpot assays. The sort-purified B-cell subsets were stimulated with 10 ng/μL sCEACAM8 alone or in combination with anti-Ig antibody. Blocking of sCEACAM8 binding to transmembrane-anchored CEACAM1 with anti-CEACAM8 antibody served as a negative control. The differentiation into Ig-secreting cells was higher for IgM memory B cells than for IgG memory or naive B cells compared with anti-Ig stimulation alone. This effect was significantly enhanced when combined with BCR stimulation (Fig. 4 I and J).
Finally, we observed higher expression of IFN-gamma receptor 1 (IFNGR1) on transcriptional and protein levels on IgM memory B cells in comparison to IgG memory or naive B cells (Figs. 2A and 4K). In line with previously published data (45), the in vitro challenge of sort-purified B-cell subsets with IFN-γ led to the weak induction of Cγ2 switch transcripts in IgM memory B cells in two of four donors, an effect that was enhanced by BCR cross-linking (Fig. S3A). Cγ2 switch transcripts were also detectable in class-switched B cells (two of six donors) or, at a lower level, when IgM or IgG memory B cells were stimulated with anti-Ig antibody alone. However, we never observed the induction of switch transcripts of Ig subclasses other than Cγ2 in IgM memory B cells, and we could not detect any induction in naive B cells independent of time or type of stimulation. Because the induction of Cγ2 switch transcripts alone may not induce class switch recombination (CSR) to Cγ2 but seems to require additional costimulation (46), we measured the amount of Cγ2 CSR in sort-purified B-cell subsets of four donors stimulated with pokeweed mitogen (PWM), BAFF, and anti-Ig antibody with or without IFN-γ. Indeed, the addition of IFN-γ enhanced CSR significantly in IgM memory B cells (P < 0.05; Fig. 4L and Fig. S3 B–E). Although Cγ2 CSR could also be observed in naive B cells costimulated with IFN-γ in two of four donors (Fig. S3D), the difference from controls treated with PWM, BAFF, and anti-Ig alone did not reach statistical significance (Fig. S3B). Finally, we did not measure significant changes in the fraction of IgG2 class-switched B cells after stimulation conditions in three donors analyzed. However, we observed a high interindividual variance of the IgG2-positive B-cell fraction in healthy adults, ranging from 6% to 30% of PB IgG+ B cells (Fig. S3 F and G).
We conclude that IgM memory B cells express a pattern of surface cytokine receptors that enables them to respond preferentially in vitro to stimulation with specific cytokines. The functional response ranges from enhanced migratory capacity to differentiation into ASC and Ig subclass-switching capacity. The differentiation-inducing effects tested here are significantly enhanced by BCR ligation. Finally, we show here that IFN-γ contributes to Cγ2 CSR direction in human B cells, preferentially in the IFN-γ receptor high-expressing IgM memory B cells.
Coculture of IgM Memory B Cells and Neutrophils Induces PC Differentiation and Cγ2 Class Switch Transcription.
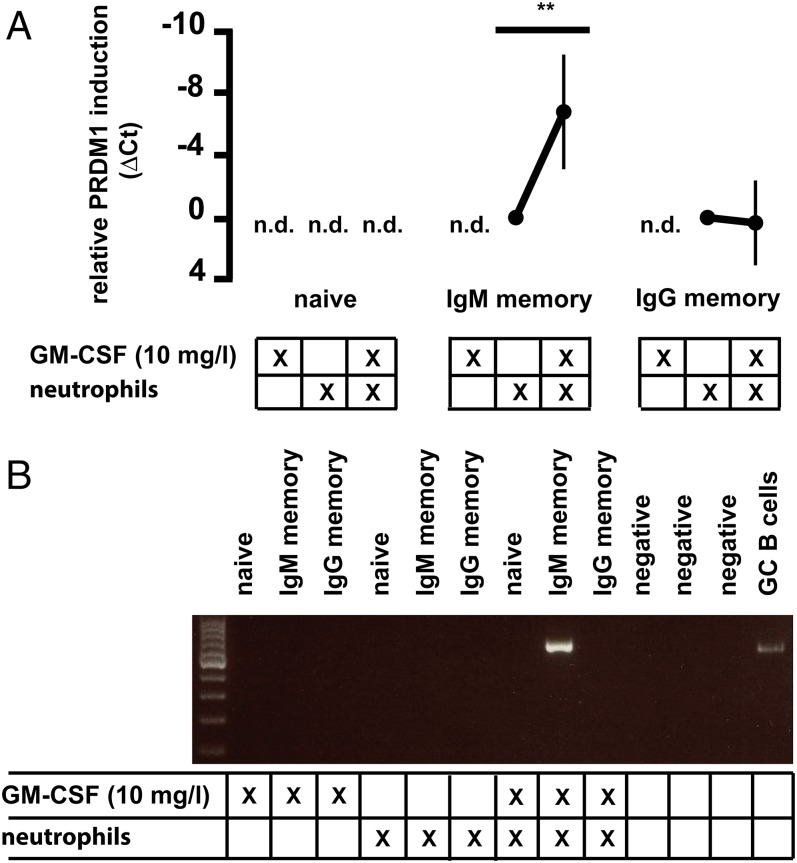
MCP1, sCEACAM8, and IFN-γ are secreted within minutes by immunomodulatory neutrophils in an early, priming phase of inflammation (33, 47). These molecules help in recruiting leukocytes and establishing a proinflammatory microenvironment. Importantly, the secretion of norepinephrine by activated neutrophils induces a paracrine or autocrine stimulatory effect leading to enhanced intracellular signaling and secretion of cytokines (48, 49). We tested whether the expression of ADRB2 and specific cytokine receptors enabled IgM memory B cells to interact preferentially with activated neutrophils. We determined the PC differentiation and Ig secretion potential of B-cell subsets in coculture with neutrophils stimulated with granulocyte-macrophage colony stimulating factor (GM-CSF) for 16 h. Fig. 5A shows the relative induction of PRDM1 transcripts in B-cell subsets after neutrophil coculture, normalized to the coculture condition without GM-CSF. GM-CSF alone did not induce PRDM1 transcription in B-cell subsets. However, cocultures of IgM memory B cells and GM-CSF–activated neutrophils consistently showed up-regulation of PRDM1 in five donors analyzed (Fig. 5A). Notably, no PRDM1 transcripts were detected in cultured IgG memory and naive B cells.
Fig. 5.
Human IgM memory B cells differentiate into PCs upon interaction with activated neutrophils. (A) Coculture of human PB B-cell subsets with GM-CSF–activated neutrophils induces PRDM1 transcription in IgM memory B cells, compared with coculture conditions excluding GM-CSF. The cycle threshold (ΔCt) of PRDM1 vs. GAPDH is shown (i.e., negative values indicate increased PRDM1 expression). GM-CSF treatment alone does not induce PRDM1 transcription in B lymphocytes. n.d., not detectable (**P < 0.01). (B) Cγ2 switch transcript induction is detectable in IgM memory B cells cocultured with GM-CSF–activated neutrophils. One representative analysis of three is shown.
Previous studies showed that human neutrophils produce and secrete IFN-γ upon stimulation (50). The coculture of GM-CSF–activated neutrophils and IgM memory B cells also induced IFN-γ production in neutrophils, but not B cells, as determined by ELISpot assays (Fig. S4). These coculture conditions also led to detectable levels of Cγ2 switch transcripts in two of three donors (Fig. 5B). No switch transcript induction was observed in controls or cocultures that included IgG memory or naive B cells. Switch transcripts of Ig subclasses other than Cγ2 were not detectable.
Taken together, the coculture of activated neutrophils with B cells reproduced the efficient differentiation potential of IgM memory B cells stimulated with terbutaline, sCEACAM8, and IFN-γ in combination with anti-Ig antibody. This finding indicates that activated neutrophils in early stages of inflammation may attract and modulate IgM memory B cells specifically.
Human PB IgM Memory B Cells Possess B-Cell Follicle Homing Capacity.
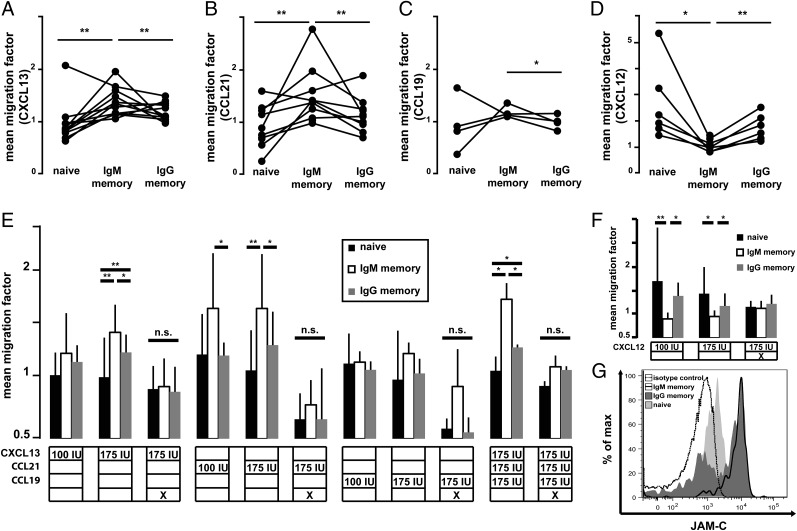
The transcription profiles of naive and memory B-cell subsets included several differentially expressed chemokine receptors and ligands (Fig. 2A), as well as G protein-signaling molecules (Table S1), that may have an influence on migratory properties of leukocytes. In line with murine data (51), the expression of the B-cell follicle homing molecule chemokine (C-X-C motif) receptor 5 (CXCR5) was higher in naive than post-GC B cells (Fig. S5 E and F). Surprisingly, when we tested the migration capacity of sort-purified PB B-cell subsets toward CXCL13, IgM memory B cells showed the highest migration factors among the B-cell subsets tested from eight of 10 donors (Fig. 6A). Similarly, the migration capacity mediated by chemokine (C-C motif) receptor 7 (CCR7) toward the B-cell follicle border homing chemokine CCL21 was found to be higher in IgM memory B cells than in naive or IgG+ B cells in six of nine samples (Fig. 6B), in line with a higher surface expression level of this receptor on these cells (Fig. S5 A and B). Migration efficiency toward CCL19, the second chemoattractant to the B-cell follicle border, showed a similar tendency of IgM memory, but not IgG memory or naive B cells, to respond preferentially to CCR7-dependent chemotaxis in three of four donors. However, the effect was milder compared with CCL21 challenge, and statistically significant differences could only be observed between IgM and IgG memory B cells at a CCL19 concentration of 175 international units (IU) (Fig. 6C). Lower concentrations (100 IU) of CXCL13, CCL21, and CCL19 induced migration in a similar pattern (i.e., IgM memory B cells showed the highest migration capacity). However, statistical significance was only reached for CCL21 (Fig. 6E). The combination of the three B-cell follicle (border) homing chemoattractants did not show an enhanced effect on the migration pattern of IgM memory B cells, suggesting that the observed pattern is mostly determined by CCL21 (Fig. 6E). Notably, when the B cells were costimulated for 1 h with the Toll-like receptor 7/8 agonist R848 and PWM to induce PC differentiation, all subsets showed significantly reduced migratory potential and IgM memory B cells lost their predominant migration capacity toward CXCL13 and CCL21 in six of six and four of four donors analyzed, respectively (Fig. 6E). Importantly, upon challenge with CXCL12, a chemokine that guides lymphocytes toward bone marrow and other PC homing sites, IgM memory B cells showed a lower migration capacity than naive or IgG memory B cells. This difference was lost upon PC differentiation, because IgM memory B cells gained (P < 0.05) and naive and IgG memory B cells reduced their migratory potential toward CXCL12 (Fig. 6 D and F). Similar to CXCR5, the different CXCR4 surface expression between IgM and IgG memory B cells did not reflect their migratory capacity (Fig. S5 C and D).
Fig. 6.
Follicle homing capacity of human IgM memory B cells. Relative number of B cells per sort-purified subset that migrated toward 175 IU of B-cell follicle (border) homing chemokines CXCL13 (A), CCL21 (B), and CCL19 (C), to which IgM memory B cells show the highest migration capacity, and PC site homing cytokine CXCL12 (D), where naive or class-switched memory B cells respond more intensively. (E) Dose dependency of follicle homing cytokines CXCL13, CCL21, and CCL19 on purified B-cell subsets. Significantly (P < 0.05) enhanced migration of IgM memory B cells is marked by black bars. “PC induction” marks assays, where the B cells were treated with mitogens (R848 and PWM) before the migration assay to induce PC development, blocking follicle homing potential. Due to the milder CCL19-dependent chemotaxis, only two donors were analyzed; an effect of plasmablast differentiation was detectable in one donor. n.s., not significant. (F) Dose dependency of PC site homing cytokine CXCL12. PC differentiation induces a PC site homing potential in IgM memory B cells. (G) Representative FACS histogram of JAM-C surface expression on naive (light gray), IgG memory (dark gray), and IgM memory (black line) B cells and an isotype control (dashed line). Similar results were obtained for a total of five donors tested. *P < 0.05; **P < 0.01.
The junctional cell adhesion molecule C (JAM-C) has been shown to direct transendothelial migration from the blood stream into B-cell follicles (52, 53). Interestingly, IgG+ B cells showed a biphasic expression of JAM-C, with about 50% of cells showing high levels, whereas the remaining cells had no detectable JAM-C surface expression (Fig. 6G), thus lacking an important molecule for lymph node entry. In contrast, IgM memory B cells consistently expressed JAM-C, in line with their higher lymph node homing capacity.
Taken together, IgM memory exceeded IgG memory and naive B cells in B-cell follicle homing capacity due to a higher sensitivity toward CXCR5- and CCR7-dependent chemotaxis and presumably JAM-C surface expression. In contrast, naive and IgG memory B cells showed a homing propensity along CXCL12 gradients (i.e., toward PC sites) and, on average, lower JAM-C surface expression.
Memory B-Cell Subsets Differ in Their Differentiation Propensity upon TD Restimulation.
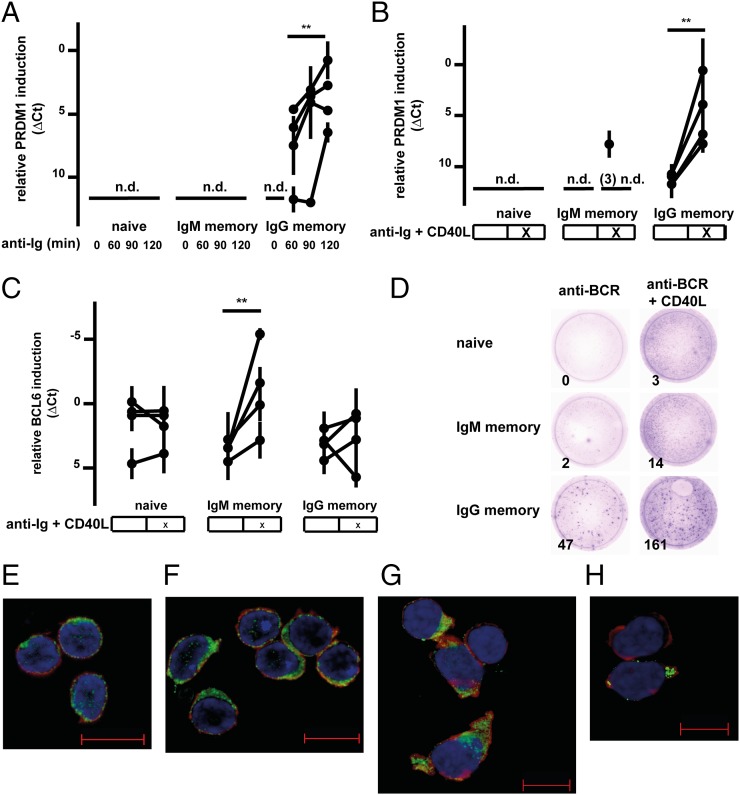
The higher migration capacity of IgM memory B cells toward B-cell follicles suggested a propensity to reenter GC reactions. Therefore, sort-purified PB B-cell subsets were subjected to in vitro activation assays mimicking a TD or TI activation/reactivation of naive and IgM or IgG memory B cells. Upon short-time challenge (2–3 h), we measured the induction of typical GC B-cell or PC differentiation markers by quantitative RT (QRT)-PCR. PRDM1 transcription was below the detection limit in untreated B-cell samples. In line with previously published results on the PC differentiation-enhancing role of the IgG BCR (54, 55), IgG memory B cells rapidly up-regulated PRDM1 upon TI or TD stimulation in contrast to naive and IgM memory B cells (Fig. 7 A and B). The latter, however, showed induction of PRDM1 transcription in one of four donors upon TD stimulation (Fig. 7B).
Fig. 7.
GC B-cell differentiation capacity of human IgM memory B cells. (A–C) Relative induction of transcripts, measured by QRT-PCR and normalized to GAPDH, associated with PC or GC B-cell differentiation in sorted PB B-cell subsets upon in vitro anti-Ig antibody treatment (TI stimulation) or in combination with CD40L (TD-like stimulation) costimulation. **P < 0.01; n.d.: not detected. TI (A) and TD (B) stimulation induced PRDM1 transcription in IgG memory B cells, but only in one-quarter of IgM memory B cells and to no detectable amount in naive B cells. (C) However, TD stimulation significantly up-regulated BCL6 in IgM memory B cells, but not in IgG memory B cells and not detectably in naive B cells. (D) Representative ELISpot analysis out of three experiments with sort-purified B-cell subsets, stimulated with 1 mg/L anti-Ig antibody, 2 mg/L recombinant human CD40-HA, and 1 mg/L anti-HA (TD) and anti-Ig antibody alone (TI). Numbers in the lower left corners give the total number of spots counted by the ELISpot reader. BACH2 fluorescence microscopy before and after 16 h of anti-Ig treatment of naive (E), IgM memory (F), and IgG memory (G) B cells is shown. Although BACH2 nuclear export was detectable in ca. 50% of IgM memory B cells, it occurred more efficiently in IgG memory B cells upon stimulation (G), showing cytosolic BACH2 degradation at the PC differentiation stage (H). Data are representative of three donors tested. Green, BACH2; blue, DAPI; red, phalloidin. (Scale bars: 10 μm.) (Also Fig. S6 B–I and Table S2.)
The GC B-cell transcription factor BCL6 was transcribed at low levels in untreated naive and IgM or IgG memory B cells. However, in each of four donors, IgM memory B cells rapidly up-regulated BCL6 transcription upon TD stimulation, whereas activated naive B cells showed, on average, no change and IgG memory B cells showed slightly increased BCL6 levels in three of four donors (Fig. 7C). To address the differentiation potentials of IgM and IgG memory B cells on the protein level, sort-purified B-cell subsets from three healthy donors were TI- or TD-stimulated and measured by ELISpot assays. In line with the efficient transcriptional up-regulation of PRDM1, IgG memory B cells showed an increased Ig secretion capacity in comparison to IgM memory B cells in both TI and TD activation conditions (Fig. 7D). However, this effect was stronger in the TD stimulation assays (P < 0.05; Fig. S6A).
Because BACH2 down-regulation precedes PC differentiation (56, 57), we performed a fluorescence microscopic analysis to clarify whether differential changes in protein level or subcellular distribution of BACH2 might occur. We observed nuclear and cytoplasmic BACH2 expression in all three B-cell subsets analyzed (Fig. 7 E–H and Fig. S6 B–I). On average, nuclear BACH2 expression was high in unstimulated naive and IgM memory B cells but clearly lower in IgG memory B cells (Fig. S6 B, D, and F and Table S2). Upon BCR stimulation with anti-Ig antibody for 16 h, naive B cells did not show significant differences in nuclear BACH2 expression (Fig. 7E and Table S2). In contrast, IgM memory B cells showed significantly reduced nuclear BACH2 expression levels (Fig. 7F). Notably, about 60% of activated IgG memory B cells lost nuclear BACH2 expression (Fig. 7H), often paralleled by extensive cell growth and PC formation. Moreover, in 15% of BCR-stimulated IgG memory B cells, nuclear and also cytoplasmic BACH2 levels were below the detection limit, arguing for efficient nuclear export and subsequent cytosolic degradation of BACH2 during PC formation. These features may cause the efficient PC differentiation potential of the IgG memory B cells (Fig. 7 G and H). In line with the lack of PRDM1 induction after short-time BCR stimulation (Fig. 7 A and B), IgM memory B cells showed reduced and delayed BACH2 nuclear export after 16 h of BCR challenge in comparison to IgG memory B cells.
Finally, we tested PB B-cell subsets for differential expression of four interleukin receptors (IL-4R, IL-6R, IL-10R, and IL-13R) because the transcription pattern of these receptors suggested a specific sensoring capability of single subsets (Fig. 2A). However, IL-4R, IL-10R, and IL-13R were not statistically significant differentially expressed between IgM memory and IgG memory lymphocytes (Fig. S7 A–F), and thus not further followed up on. IL-6R, in contrast, showed significantly higher expression levels on IgG memory B cells (Fig. S7 G and H). However, IL-6 treatment of sort-purified B-cell subsets did not lead to preferential differentiation into plasmablasts of either population analyzed (Fig. S7I).
Our data indicate that human IgG memory B cells primarily differentiate into PCs upon reencounter of antigen, independent of a TI or TD type of stimulation, guided by rapid PRDM1 induction and nuclear export and cytosolic degradation of BACH2. In contrast, upon TD stimulation, IgM memory B cells show a preferential tendency to adopt a pre-GC B-cell phenotype, as indicated by up-regulation of BCL6 transcripts. However, upon TI stimulation, IgM memory B cells also showed PC differentiation capacity, as indicated by BACH2 down-regulation and adoption of PC morphology, albeit at a lower rate and efficiency than IgG memory B cells.
Discussion
It is debated whether human PB IgM+IgD+CD27+ B cells are post-GC memory B cells or derive from an antigen-independent or TI developmental pathway (26–28, 58). The existence of consistently mutated IgV genes of human PB IgM+IgD+CD27+ B cells, as well as their increased responsiveness to mitogenic stimulation in comparison to naive B cells (59, 60), argues that these lymphocytes are antigen-experienced, post-GC memory B cells. We recently provided further genetic evidence that at least a large fraction of these PB B cells in adult humans are derived from GC reactions, because they frequently show BCL6 mutations (occurring only in GC B cells) and often show a common derivation with IgG memory B cells from diversified GC B-cell clones (23). This latter point was recently validated by an independent study on memory B-cell subtypes generated in defined immune responses (61). The current work further supports this idea. First, we demonstrate that IgM+IgD+CD27+ B cells show a much higher similarity in their global gene expression with classical IgG+ memory B cells than with naive B cells, including key features of typical memory B cells, encompassing enhanced responsiveness, metabolism, proliferation, and a propensity to plasmablast differentiation, together arguing for these cells being antigen-experienced. Second, in various functional activation assays, the IgM+IgD+CD27+ B cells showed a higher and faster reactivity than naive B cells, which is a key feature of memory B cells. Third, IgM+IgD+CD27+ B cells reacted upon stimulations mimicking a TD immune response (as discussed below), an additional feature that is in line with their derivation from and involvement in TD immune reactions and has also been described for IgM+IgD+CD27+ B cells from human spleen, which are presumably a mixture of circulating IgM+IgD+CD27+ and splenic MGZ B cells (62, 63). Earlier expression profiling analyses (28, 58, 64, 65) did not include subsets like naive, IgM-only, or class-switched B cells, or studied IgM+CD27+ B cells from the spleen only (62), thus preventing the determination of similarities in circulating memory B cells in a comparative analysis.
Considering the IgM memory B-cell subsets analyzed, we did not detect any significantly differentially expressed genes (except IgD) between IgM+IgD+CD27+ and IgM-only B cells, and we thus conclude that a comparison on the transcriptional level cannot help to explain previously published results on a differential class-switching capacity (66) or different generation pathways (26, 67) of these two subsets.
However, a set of about 400 genes was specifically expressed or underrepresented in IgM memory B cells in comparison to IgG memory and also partially naive B cells. To understand these IgM memory B cell-specific gene signatures and translate them into immunological functions further, we focused on genes that may provide a specific sensoring capability to these resting B lymphocytes, preparing them to react specifically and quickly under particular reactivation conditions. Indeed, a large number of surface molecules, such as cytokine receptors, cell adhesion molecules, and BCR signaling components, as well as downstream signaling molecules, such as kinases and regulators of G protein signaling, that were expressed specifically and homogeneously by the IgM memory B-cell population were revealed. To our surprise, we observed expression of the noradrenaline/adrenaline binding receptor ADRB2 on naive and IgM memory B cells. Although an influence of neurotransmitters on B lymphocytes is known (36), we describe here the preferential responsiveness of an IgM+CD27+ memory B-cell subset and show the preferential proliferation and differentiation potential of these cells upon ADRB2 stimulation in vitro. Similarly, higher expression of CCR2 and CEACAM1, two molecules that provide chemotactic activity toward MCP1 and sCEACAM8, respectively, was observed on IgM memory B cells. Moreover, CEACAM1 represents a potent costimulatory molecule for a subset of human B cells that is preferentially composed of human IgM memory B cells. Finally, we show that IFN-γ (most efficiently in combination with further costimulation) can also serve as an IgG2 class switch-enhancing factor in humans, an issue that has been debated (46). Although we also observed IgG2 CSR upon IFN-γ stimulation in naive B cells (detectable but not significant in total), we assume that the high levels of IFNGR1 expression on post-GC IgM memory B cells provide these cells with an increased potential to differentiate into IgG2-expressing PCs upon costimulation with IFN-γ.
Interestingly, the cytokines, adhesion molecules, and neurotransmitters that convey the observed features of IgM memory B cells have in common that they are secreted by neutrophils in early phases of inflammation (33). To test whether activated neutrophils indeed have the potential to stimulate IgM memory B cells to differentiate and secrete IgM or a class switch to IgG2, we performed coculture assays of neutrophils and B-cell subsets, and could confirm such an interaction in vitro. Because we did not assess the impact of single molecules on stimulation or differentiation of IgM memory B cells in coculture with GM-CSF–stimulated neutrophils, we cannot conclude whether our observations indeed result from secreted cytokines or may rather derive from cell–cell contact. Indeed, there is evidence that CEACAM1 interaction with membrane-bound ligands also promotes CSR in murine B cells (41).
Recent studies in the mouse have indicated that the memory B-cell pool is more heterogeneous than previously thought, and that distinct memory B-cell subsets have largely distinct functions (12, 13, 68). Besides the indication that a proportion of memory B cells in TD immune responses are generated before the onset of the GC reaction (9), several studies have indicated that IgM and IgG memory B cells have specific functions, with IgM memory B cells preferentially differentiating into GC B cells upon reactivation, whereas IgG memory B cells showed a propensity to differentiate directly into PCs in recall responses (12, 13). However, this view was recently challenged because it was reported that murine memory B-cell subsets defined by expression patterns of CD80 and programmed cell death 1 ligand 2 (PD-L2) better distinguish memory B-cell subsets with preferential PC differentiation or GC reentry (69).
Whether the human IgM+ and IgG+ memory B cells show distinct functions in recall responses was not previously known. Here, we provide data that human memory B-cell subsets may indeed show a similar dichotomy (i.e., IgM memory B cells might have the capacity to home to B-cell follicles upon restimulation and more easily undergo GC B-cell differentiation, as indicated by the facilitated transcript induction of the GC B-cell transcription factor BCL6 in vitro). This behavior is in contrast to that of IgG memory B cells, which show a migration potential toward CXCL12, transcriptional down-regulation and nuclear exclusion of BACH2, up-regulation of PRDM1, and PC differentiation, in line with the signaling and differentiation-enhancing role of the class-switched BCR (54, 55). Moreover, the adhesion molecule JAM-C plays an important role in lymph node homing and is expressed on virtually all IgM memory cells but only on a fraction of IgG memory B cells, allowing GC reentry preferentially for the former population.
Several key questions clearly remain to be clarified. These questions include the relationship between PB IgM+IgD+CD27+ B cells and splenic MGZ B cells, which, at least in young individuals, may derive from another developmental pathway (58). Moreover, it remains to be clarified whether further subsets among IgM and IgG memory B cells exist, in line with the recent study on murine memory B cells (69), although there is no indication for differential expression of CD80 and PD-L2 by human memory B cells.
We conclude that human IgM+IgD+CD27+ B cells represent memory B cells generated in TD immune responses. Besides their potential ability to reenter GC reactions upon reencounter of antigen, which apparently distinguishes them from IgG memory B cells, these unique B cells show distinct differentiation potentials, depending on their specific activation and interaction with immune cells. These findings indicate an unexpected functional plasticity of human IgM memory B cells.
Materials and Methods
Cell Separation.
Buffy coats of healthy adult blood donors were obtained from the Institut für Transfusionsmedizin of the University Hospital Essen. Donors for full-blood donations were recruited from the University Medical School. The study protocol was approved by the Internal Review Board of the University Hospital Essen. Samples were collected with informed consent of the donors. PB mononuclear cells were isolated by Ficoll–Paque density centrifugation (Amersham). CD19+ B cells were enriched to >98% by magnetic cell separation using the MACS system (Miltenyi Biotech) or by negative selection using the EasySep Human B-Cell Enrichment Kit (Stemcell). Neutrophils were isolated by Ficoll–Paque density centrifugation of blood samples, sediment resuspension with PBS/BSA, and twofold erythrocyte lysis for 1 min with 10 mL of distilled water and neutralization with 10× PBS (Gibco), centrifugation, and subsequent CD16+ MACS (Miltenyi Biotech) enrichment of neutrophils (ca. 95% purity).
Flow Cytometry and Cell Sorting.
For Human Genome U133 2.0 Plus Array (Affymetrix UK Ltd.) analysis, B-cell suspensions were stained with anti-CD27–allophycocyanin (APC) (eBiosciences), anti-IgD–phycoerythrin (PE)–Cy7, anti-IgM–fluorescein isothiocyanate (FITC), and anti-IgG–PE or anti-CD21–PE antibodies [all from Becton Dickinson (BD) Biosciences] and sorted with a FACSDiva cell sorter (BD Biosciences) as IgG memory (IgG+CD27+), IgM memory (IgM+IgD+CD27+), IgM-only (IgM+IgD−/lowCD27+), or naive (IgM+IgD+CD27−) B cells. Purity was >99% for each population as calculated by reanalysis on a FACSCanto flow cytometer (BD Biosciences) in combination with FACSDiva software. For functional analysis, B-cell subsets were stained with anti-IgD–APC, anti-CD27–FITC, and anti-IgG–PE or anti-CD16–FITC antibodies (all from BD Biosciences) and sorted as IgG memory (IgG+CD27+), IgM memory (IgD+CD27+), and naive (IgD+CD27−) B cells and neutrophils [forward scatter (FSC)high/side scatter (SSC)highCD16+] at >98% purity. For FACS analysis, enriched B cells were stained with anti-CD27–APC, anti-IgD–FITC, anti-CCR2–PE, anti-IFNGR1–PE, anti-IgD–PE, anti-CXCR4–PE, anti-CXCR5–FITC, anti–IL-4R–PE, anti–IL-10R–PE, anti–IL-13R–PE, anti–IL-6R–PE (all from BD Biosciences), anti-ADRB2–PE (Santa Cruz Biotechnology), anti–JAM-C–PE, anti-CCR7–PE (both from R&D Systems), or anti-IgG2–AF488 (Biozol), and anti-CEACAM1–FITC antibodies (mAb B3-17) (35).
Fluorescence Microscopy.
For immunofluorescence microscopy, B cells were sorted as described above and stained with the BD Cytofix/Cytoperm Kit (BD Biosciences) according to the manufacturer’s instructions. Expression of BACH2 was determined by intracellular staining with an anti-BACH2 antibody (rabbit Cγ2 antibody; Biozol), an anti-rabbit–Cy2 antibody (Jackson ImmunoResearch), Phalloidin-TRITC (Sigma–Aldrich), and Hoechst 33258 (Roche). Fluorescence microscopy was performed on a Zeiss Axio Observer.Z1 fluorescence microscope equipped with the respective filter sets and an apotome (Carl Zeiss MicroImaging). Image acquisition was performed via a Plan-Apochromat 63×/1.40 oil objective lens (1.46 N.A.) and a Zeiss AxioCam MRm camera from cell suspensions in fluorescent mounting medium (S3023; Biozol) at 23 °C. Images were processed with Zeiss AxioVision Rel. 4.8 software.
Sample Preparation for Gene Expression Profiling.
RNA (250 ng) was extracted from sort-purified B cells by Gentra Purescript (Gentra). RNA integrity was assessed using an Agilent 2100 Bioanalyzer. Samples with an RNA integrity number >9.0 were processed by means of a MessageAmp II Biotin Enhanced Kit (Ambion). For the Human Genome U133 2.0 Plus gene expression profiling analysis, data were generated in two batches, equally composed of naive, IgM-only, IgM+IgD+CD27+, and IgG memory B cells. After variance stabilizing normalization, data were corrected for batch effect by ComBat software (www.bu.edu/jlab/wp-assets/ComBat) (70). Arrays were scanned with a GeneChip Scanner 3000 7G (Affymetrix). GeneChip data have been submitted to the Gene Expression Omnibus database under accession number GSE64028.
Statistical Analysis.
Data were analyzed with GeneSpring GX software (Affymetrix) and R software for statistical computing (ISBN 3-900051-07-0, R Development Core Team; www.R-project.org.) Probe sets with a minimum raw signal of 50 and at least four present calls in at least one condition, according to Mas5 software, were used for further analysis. Multivariate data analysis was performed with ANOVA and Tukey post hoc testing procedures, and pairwise comparisons were tested for statistical significance by t test (P < 0.05). The Benjamini–Hochberg method was used for multiple testing corrections. GSEA was performed with GSEA2 software. Each GSEA plot (Figs. S1 and S2) shows the name of the respective gene set in the header, the enrichment score profile (green graph), and the hits of single genes of the respective gene set (black bars) along the ranking metric scores (gray bars) that are used for the running sum statistics as described (71). Gene sets are derived from the Molecular Signature Database (C2, curated gene sets; C7, immunological signatures) (71). The significance of surface molecule expression on IgM memory B cells was determined by ANOVA and Tukey post hoc testing, and the significance of differential Ig secretion, proliferation, or migration potential of single B-cell subsets was calculated by paired t testing procedures. Each statistical test was performed on all available data points, including outliers.
In Vitro Activation of B-Cell Subsets.
Naive and memory B cells were sort-purified and cultured in RPMI 1640/10% (vol/vol) FCS/1% P/S (Pan Biotech) with 10 mg/L goat anti-human Ig antibody (Dianova) alone or with 10−5 to 10−7 M terbutaline and/or 10−5 to 10−7 M nadolol (Sigma–Aldrich) or 50 mg/L soluble CEACAM8-Fc with or without anti-CEACAM8 blocking antibody mAb 6/40c (35) for 3 or 6 h at 37 °C and 5% (vol/vol) CO2. ELISpot assays were performed with the human IgM and IgG ELISspot PLUS system (Mabtech) as recommended by the manufacturer. In short, 25,000 sort-purified and preactivated B cells were incubated in 100 μL of PBS per well for 1 h, if not stated otherwise. Readout of ELISpot plates was performed automatically by the ELISpot Reader System ELRFL06 (AID GmBH). Cells in G2/S phase were determined by FACS DNA content measurement with propidium iodide (Sigma–Aldrich). Migration assays were performed in 5-μm ChemoTx System migration assays (Neuro Probe) as recommended by the manufacturer. In short, the fraction of 50,000 sort-purified B cells that passed through a 5-μm filter membrane toward 50, 100, or 175 IU of MCP1, CXCL13, CXCL12, CCL19, or CCL21 (all from Immunotools) or 50 or 100 mg/L sCEACAM8-Fc (35) was determined by FACS counting after 16 h. Plasmablast/PC differentiation was induced by preincubation of B cells with 100 mg/L R848 and PWM (both from Mabtech) for 1 h. Switch transcript induction was determined by incubation of B cells alone or in neutrophil coculture (discussed below) with 1 mg/L IFN-γ (Immunotools) overnight. IgG2 CSR was performed by stimulation with 1 mg/L PWM and BAFF (Immunotools) and anti-Ig antibody (Dianova) with or without 1 mg/L IFN-γ for 6 d. RNA was extracted using an RNeasy Micro Kit (Qiagen) and after DNase I digestion reversely transcribed with Sensiscript RT (Qiagen). For QRT-PCR analysis, the following TaqMan assays (Life Technologies) were used according to the manufacturer’s instructions: BACH2 (HS00222364_m1), CD80 (HS00175478_m1), IFNG (HS00175238_m1), BCL6 (HS00153368_m1), and PRDM1 (HS00153357_m1). Expression was normalized to GAPDH (HS02758991_g1). Switch transcripts were amplified as described previously (72), with the exception that an IGHV family 3 FR1 primer instead of an FR3 primer was used (23). For each PCR assay, equal amounts of cDNA generated from equal cell numbers were used. For TI activation, 250,000 sort-purified B cells were incubated with 10 mg/L goat anti-human Ig for 0, 60, 90, and 120 min in medium. For TD activation, sort-purified B cells were incubated with 2 mg/L recombinant human CD40L with HA-tag plus 1 mg/L anti-HA antibody (both from R&D Systems) and 10 mg/L goat anti-human Ig for 1 h, followed by a washing step and anti-Ig antibody treatment alone for 2 h.
In Vitro Coculture of B-Cell Subsets and Neutrophils.
A total of 5 × 105 B cells per subset were sort-purified and cultured with 106 sort-purified neutrophils from the same donors in RPMI 1640/10% (vol/vol) FCS/1% P/S with 10 mg/L GM-CSF (Immunotools) for 16 h at 37 °C and 5% (vol/vol) CO2. After coculture, B cells were resorted according to FSC/SSC criteria into Qiazol (Qiagen) and RNA was extracted. cDNA was prepared using the Sensiscript protocol (Qiagen), and switch transcripts were amplified or QRT-PCR analysis of PRDM1 transcripts was performed as described above.
Supplementary Material
Acknowledgments
We thank Julia Jesdinsky-Elsenbruch for excellent technical assistance and Klaus Lennartz for his valuable engineering support. We thank the Institut für Transfusionsmedizin of the University Hospital Essen and the Imaging Center Essen for their support. This work was supported by the Deutsche Forschungsgemeinschaft through Grants Ku1315/8-1, SE1885/2-1, and GKR1431.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The transcriptome profiling data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE64028).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416276112/-/DCSupplemental.
References
- 1.Okada T, Cyster JG. B cell migration and interactions in the early phase of antibody responses. Curr Opin Immunol. 2006;18(3):278–285. doi: 10.1016/j.coi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Lee SK, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208(7):1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatto D, Brink R. B cell localization: Regulation by EBI2 and its oxysterol ligand. Trends Immunol. 2013;34(7):336–341. doi: 10.1016/j.it.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 4.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 5.Tarlinton D, Good-Jacobson K. Diversity among memory B cells: Origin, consequences, and utility. Science. 2013;341(6151):1205–1211. doi: 10.1126/science.1241146. [DOI] [PubMed] [Google Scholar]
- 6.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 7.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 8.Hargreaves DC, et al. A coordinated change in chemokine responsiveness guides plasma cell movements. J Exp Med. 2001;194(1):45–56. doi: 10.1084/jem.194.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaji T, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209(11):2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. 2012;209(3):597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyama H, et al. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17(3):329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 12.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10(12):1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 13.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331(6021):1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan TG, et al. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203(11):2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey AM, Pierce SK. Intrinsic differences in the initiation of B cell receptor signaling favor responses of human IgG(+) memory B cells over IgM(+) naive B cells. J Immunol. 2012;188(7):3332–3341. doi: 10.4049/jimmunol.1102322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obukhanych TV, Nussenzweig MC. T-independent type II immune responses generate memory B cells. J Exp Med. 2006;203(2):305–310. doi: 10.1084/jem.20052036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defrance T, Taillardet M, Genestier L. T cell-independent B cell memory. Curr Opin Immunol. 2011;23(3):330–336. doi: 10.1016/j.coi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Wirths S, Lanzavecchia A. ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. Eur J Immunol. 2005;35(12):3433–3441. doi: 10.1002/eji.200535364. [DOI] [PubMed] [Google Scholar]
- 19.Weston-Bell N, Townsend M, Di Genova G, Forconi F, Sahota SS. Defining origins of malignant B cells: A new circulating normal human IgM(+)D(+) B-cell subset lacking CD27 expression and displaying somatically mutated IGHV genes as a relevant memory population. Leukemia. 2009;23(11):2075–2080. doi: 10.1038/leu.2009.178. [DOI] [PubMed] [Google Scholar]
- 20.Fecteau JF, Côté G, Néron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol. 2006;177(6):3728–3736. doi: 10.4049/jimmunol.177.6.3728. [DOI] [PubMed] [Google Scholar]
- 21.Klein U, Rajewsky K, Küppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188(9):1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tangye SG, Good KL. Human IgM+CD27+ B cells: Memory B cells or “memory” B cells? J Immunol. 2007;179(1):13–19. doi: 10.4049/jimmunol.179.1.13. [DOI] [PubMed] [Google Scholar]
- 23.Seifert M, Küppers R. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J Exp Med. 2009;206(12):2659–2669. doi: 10.1084/jem.20091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheeren FA, et al. T cell-independent development and induction of somatic hypermutation in human IgM+ IgD+ CD27+ B cells. J Exp Med. 2008;205(9):2033–2042. doi: 10.1084/jem.20070447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McWilliams L, et al. The human fetal lymphocyte lineage: identification by CD27 and LIN28B expression in B cell progenitors. J Leukoc Biol. 2013;94(5):991–1001. doi: 10.1189/jlb.0113048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weller S, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci USA. 2001;98(3):1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruetzmann S, et al. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med. 2003;197(7):939–945. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weller S, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104(12):3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weill JC, Weller S, Reynaud CA. Human marginal zone B cells. Annu Rev Immunol. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. [DOI] [PubMed] [Google Scholar]
- 30.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 31.Puga I, et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol. 2012;13(2):170–180. doi: 10.1038/ni.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagelkerke SQ, aan de Kerk DJ, Jansen MH, van den Berg TK, Kuijpers TW. Failure to detect functional neutrophil B helper cells in the human spleen. PLoS ONE. 2014;9(2):e88377. doi: 10.1371/journal.pone.0088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathan C. Neutrophils and immunity: Challenges and opportunities. Nat Rev Immunol. 2006;6(3):173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 34.Scapini P, et al. G-CSF-stimulated neutrophils are a prominent source of functional BLyS. J Exp Med. 2003;197(3):297–302. doi: 10.1084/jem.20021343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer BB, et al. Soluble CEACAM8 interacts with CEACAM1 inhibiting TLR2-triggered immune responses. PLoS ONE. 2014;9(4):e94106. doi: 10.1371/journal.pone.0094106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kin NW, Sanders VM. It takes nerve to tell T and B cells what to do. J Leukoc Biol. 2006;79(6):1093–1104. doi: 10.1189/jlb.1105625. [DOI] [PubMed] [Google Scholar]
- 37.Kohm AP, Sanders VM. Norepinephrine and beta 2-adrenergic receptor stimulation regulate CD4+ T and B lymphocyte function in vitro and in vivo. Pharmacol Rev. 2001;53(4):487–525. [PubMed] [Google Scholar]
- 38.Singer BB, et al. Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: Evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes. J Immunol. 2002;168(10):5139–5146. doi: 10.4049/jimmunol.168.10.5139. [DOI] [PubMed] [Google Scholar]
- 39.Frade JM, et al. Characterization of the CCR2 chemokine receptor: Functional CCR2 receptor expression in B cells. J Immunol. 1997;159(11):5576–5584. [PubMed] [Google Scholar]
- 40.Corcione A, et al. Chemotaxis of human tonsil B lymphocytes to CC chemokine receptor (CCR) 1, CCR2 and CCR4 ligands is restricted to non-germinal center cells. Int Immunol. 2002;14(8):883–892. doi: 10.1093/intimm/dxf054. [DOI] [PubMed] [Google Scholar]
- 41.Greicius G, Severinson E, Beauchemin N, Obrink B, Singer BB. CEACAM1 is a potent regulator of B cell receptor complex-induced activation. J Leukoc Biol. 2003;74(1):126–134. doi: 10.1189/jlb.1202594. [DOI] [PubMed] [Google Scholar]
- 42.Skubitz KM, Skubitz AP. Interdependency of CEACAM-1, -3, -6, and -8 induced human neutrophil adhesion to endothelial cells. J Transl Med. 2008;6:78. doi: 10.1186/1479-5876-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulton IC, Gray-Owen SD. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat Immunol. 2002;3(3):229–236. doi: 10.1038/ni769. [DOI] [PubMed] [Google Scholar]
- 44.Singer BB, et al. CEACAM1 (CD66a) mediates delay of spontaneous and Fas ligand-induced apoptosis in granulocytes. Eur J Immunol. 2005;35(6):1949–1959. doi: 10.1002/eji.200425691. [DOI] [PubMed] [Google Scholar]
- 45.Kitani A, Strober W. Regulation of C gamma subclass germ-line transcripts in human peripheral blood B cells. J Immunol. 1993;151(7):3478–3488. [PubMed] [Google Scholar]
- 46.Kawano Y, Noma T, Yata J. Regulation of human IgG subclass production by cytokines. IFN-gamma and IL-6 act antagonistically in the induction of human IgG1 but additively in the induction of IgG2. J Immunol. 1994;153(11):4948–4958. [PubMed] [Google Scholar]
- 47.Kuroki M, et al. Immunochemical analysis of carcinoembryonic antigen (CEA)-related antigens differentially localized in intracellular granules of human neutrophils. Immunol Invest. 1995;24(5):829–843. doi: 10.3109/08820139509060710. [DOI] [PubMed] [Google Scholar]
- 48.Flierl MA, et al. Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature. 2007;449(7163):721–725. doi: 10.1038/nature06185. [DOI] [PubMed] [Google Scholar]
- 49.Flierl MA, et al. Upregulation of phagocyte-derived catecholamines augments the acute inflammatory response. PLoS ONE. 2009;4(2):e4414. doi: 10.1371/journal.pone.0004414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ethuin F, et al. Human neutrophils produce interferon gamma upon stimulation by interleukin-12. Lab Invest. 2004;84(10):1363–1371. doi: 10.1038/labinvest.3700148. [DOI] [PubMed] [Google Scholar]
- 51.Gatto D, Wood K, Brink R. EBI2 operates independently of but in cooperation with CXCR5 and CCR7 to direct B cell migration and organization in follicles and the germinal center. J Immunol. 2011;187(9):4621–4628. doi: 10.4049/jimmunol.1101542. [DOI] [PubMed] [Google Scholar]
- 52.Doñate C, et al. Homing of human B cells to lymphoid organs and B-cell lymphoma engraftment are controlled by cell adhesion molecule JAM-C. Cancer Res. 2013;73(2):640–651. doi: 10.1158/0008-5472.CAN-12-1756. [DOI] [PubMed] [Google Scholar]
- 53.Zimmerli C, et al. Adaptive immune response in JAM-C-deficient mice: Normal initiation but reduced IgG memory. J Immunol. 2009;182(8):4728–4736. doi: 10.4049/jimmunol.0803892. [DOI] [PubMed] [Google Scholar]
- 54.Engels N, et al. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nat Immunol. 2009;10(9):1018–1025. doi: 10.1038/ni.1764. [DOI] [PubMed] [Google Scholar]
- 55.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3(2):182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 56.Xu Y, et al. No receptor stands alone: IgG B-cell receptor intrinsic and extrinsic mechanisms contribute to antibody memory. Cell Res. 2014;24(6):651–664. doi: 10.1038/cr.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol. 2008;20(3):453–460. doi: 10.1093/intimm/dxn005. [DOI] [PubMed] [Google Scholar]
- 58.Descatoire M, et al. Identification of a human splenic marginal zone B cell precursor with NOTCH2-dependent differentiation properties. J Exp Med. 2014;211(5):987–1000. doi: 10.1084/jem.20132203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agematsu K, et al. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur J Immunol. 1997;27(8):2073–2079. doi: 10.1002/eji.1830270835. [DOI] [PubMed] [Google Scholar]
- 60.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: Up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101(11):4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 61.Della Valle L, et al. The majority of human memory B cells recognizing RhD and tetanus resides in IgM+ B cells. J Immunol. 2014;193(3):1071–1079. doi: 10.4049/jimmunol.1400706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Good KL, Avery DT, Tangye SG. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. J Immunol. 2009;182(2):890–901. doi: 10.4049/jimmunol.182.2.890. [DOI] [PubMed] [Google Scholar]
- 63.Good KL, Bryant VL, Tangye SG. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J Immunol. 2006;177(8):5236–5247. doi: 10.4049/jimmunol.177.8.5236. [DOI] [PubMed] [Google Scholar]
- 64.Ehrhardt GR, et al. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205(8):1807–1817. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y, et al. Distinct gene expression profiles in different B-cell compartments in human peripheral lymphoid organs. BMC Immunol. 2004;5:20. doi: 10.1186/1471-2172-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werner-Favre C, et al. IgG subclass switch capacity is low in switched and in IgM-only, but high in IgD+IgM+, post-germinal center (CD27+) human B cells. Eur J Immunol. 2001;31(1):243–249. doi: 10.1002/1521-4141(200101)31:1<243::AID-IMMU243>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 67.Berkowska MA, et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118(8):2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takemori T, Kaji T, Takahashi Y, Shimoda M, Rajewsky K. Generation of memory B cells inside and outside germinal centers. Eur J Immunol. 2014;44(5):1258–1264. doi: 10.1002/eji.201343716. [DOI] [PubMed] [Google Scholar]
- 69.Zuccarino-Catania GV, et al. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15(7):631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 71.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerutti A, et al. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J Immunol. 2000;165(2):786–794. doi: 10.4049/jimmunol.165.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.