Significance
The advance of biotechnology opens up greater possibilities of bioterror and bioerror. Here, we propose multiplexed safeguard switches rooted in the development of foundational genomic, regulatory, and metabolic technologies. Safeguard switches can be regulated by submicromolar small molecule(s) and combined in a modular fashion. The resulting safeguard strains show high fitness and low reversion rates. Moreover, two distinct classes of safeguard switches are orthogonal, providing a potential fail-safe mechanism. The safeguard technologies provide a practical and generic approach to containing engineered microbes within defined laboratory and/or industrial environments, and can in principle be used in the field as well.
Keywords: biotechnology, biosafety, Saccharomyces cerevisiae
Abstract
Biocontainment may be required in a wide variety of situations such as work with pathogens, field release applications of engineered organisms, and protection of intellectual properties. Here, we describe the control of growth of the brewer’s yeast, Saccharomyces cerevisiae, using both transcriptional and recombinational “safeguard” control of essential gene function. Practical biocontainment strategies dependent on the presence of small molecules require them to be active at very low concentrations, rendering them inexpensive and difficult to detect. Histone genes were controlled by an inducible promoter and controlled by 30 nM estradiol. The stability of the engineered genes was separately regulated by the expression of a site-specific recombinase. The combined frequency of generating viable derivatives when both systems were active was below detection (<10−10), consistent with their orthogonal nature and the individual escape frequencies of <10−6. Evaluation of escaper mutants suggests strategies for reducing their emergence. Transcript profiling and growth test suggest high fitness of safeguarded strains, an important characteristic for wide acceptance.
The rapid development of biotechnology depends heavily on engineered microbes. In particular, the booming field of synthetic biology (1) demonstrates the feasibility of de novo synthesis of viral genomes (2–5), bacterial genomes (6, 7), and eukaryotic chromosomes (8, 9). The technologies underpinning synthetic biology are advanced DNA synthesis and assembly (10), genome editing (11, 12), and computational assisted designs (13–15), which are all becoming commoditized and thus increasingly available to the public. The advance of synthetic biology promises to ultimately improve human living conditions through a better understanding of fundamental sciences as well as a multitude of practical applications. However, biosafety mechanisms should be carefully considered to minimize or prevent dual use (16). Professionals have chemically synthesized infectious virus in the absence of natural templates (2) and reconstitute infectious human retroviruses (3) and the 1918 “Spanish” influenza virus (5). At the same time, relative amateurs are trying to engineer microbes in a do-it-yourself fashion (www.diybio.org). Proactive measures are warranted to minimize both bioterror (e.g., the anthrax attack in the United States in 2000) and “bioerror” (accidental environment releases or self-infection by laboratory-adapted microbes as in the case of a laboratory infection of an individual with hemochromatosis, where the victim scientist’s high iron levels caused by hemochromatosis complemented the natural iron requirement of attenuated Yersinia pestis) (17). Intrinsic biocontainment can also be used to prevent industrial espionage by protecting the intellectual property of biotechnology companies. In light of the wide accessibility of whole-genome sequencing, we envision in the future embedding “decoy” circuits into the safeguard strains and the combinatorial complexity will likely to defeat anyone who wants to decode the safeguard strains.
Previous work on biocontainment has largely focused on auxotrophic mutations or inducible lethality based on toxin–antitoxin pairs in bacteria (18–21). There is also recent work to reduce unintended plasmid propagation in bacteria (22). Existing biocontainment technologies usually depend on a single-cellular mechanism. Such “uniplex” approaches lack redundancy, and the leakiness of toxin genes often compromises fitness and normal behavior of the engineered microbes, reducing the likelihood that they will be accepted by researchers or industrial biotechnologists. Also, ongoing selection for the toxin gene is required. Reduced fitness has three undesirable features: (i) it reduces their usefulness as models for the behavior of the natural organism, (ii) it may reduce their “performance” in industrial applications, and (iii) it may increase the frequency of escape mutants (revertants able to grow in the absence of the compound). Finally, auxotrophic strains rely on supplementation of corresponding nutrients at micromolar concentrations, rendering them too costly for industrial scale-up. We have developed multiplex safeguard switches regulated by a combination of small molecules, some of which are active at submicromolar concentrations. In the future, one can envision supplying a blend of such compounds as a kind of “special sauce” to keep a designer microbe alive. This too could be laced with decoy compounds to defy biohackers. Each safeguard switch technology targets a different cellular process and can be combined in a multiplex fashion. With safeguard switches, we are able to regulate engineered microbes with small molecules and contain them in defined laboratory conditions to prevent escape to the environment. The safeguard switch concept also provides a potentially effective way to protect intellectual property biotechnology, by containing the engineered strains with defined combinations of small molecules.
Here, we report two orthogonal safeguard switches, with one targeting transcriptional control and another one based on recombination-induced lethality; in both cases, one or more essential genes are targeted, instead of autotrophic markers and/or toxin genes to minimize physiological impacts on cell fitness. Transcriptome profiling and growth tests confirm that fitness of several safeguard strains is close to the wild type, and performance on multiple media types suggests robustness to varying conditions. The escape rates for individual safeguards are 10−8 to 10−6. Analysis of escape variants (revertants) sheds useful light to guide the design of next-generation safeguard development.
Results
Goals and Overall Design Principles.
To establish design principles for generic genomic safeguards, we evaluated several designs based on the premise that the desired combination of high fitness, normal physiology, and tight control by a small molecule could be obtained by putting essential genes under multiple forms of chemical control. Here, we show that, by putting both the presence/absence of the gene and its transcription under chemical control, it is possible to produce yeast strains that live or die according to the presence/absence of the ligand, and show high-fitness and low-escape frequencies. Analysis of escapers (revertants) provides insight into the mechanisms of escape that can be used for a subsequent round of engineering to reduce those escape frequencies. Here, we evaluated two essential histone genes for performance as genome safeguards for Saccharomyces cerevisiae in various contexts. As each of the two histones studied were themselves encoded by two gene copies, the studies were performed in strains in which both native copies of histone H3 and both native copies of histone H4 genes had been deleted from the genome (Fig. S1). We also screened other essential genes for performance as safeguards.
Plasmid-Based Chemically Regulated Transcriptional Switches.
As a proof of principle, we first constructed a chemically regulated transcriptional switch based on two essential synthetic histone genes called HHTS and HHFS (encoding histone H3 and H4, respectively; the “S” refers to the synthetic nature of the genes) described in detail in ref. 23. In the safeguard strains described here, the HHTS and HHFS genes are regulated by GAL1 and GAL7 promoters, respectively, and each histone transcriptional unit is flanked by a pair of loxP sites (see following sections for detail). Thus, the transcription of the same genes can be regulated by the presence of galactose or glucose in the growth medium, or related methods, and the presence/absence of the genes can be regulated by the expression of Cre recombinase activity. This histone-based triplex safeguard was cloned into a pRS415 centromeric (episomal) vector (24) to construct plasmid pPC012 (Fig. 1).
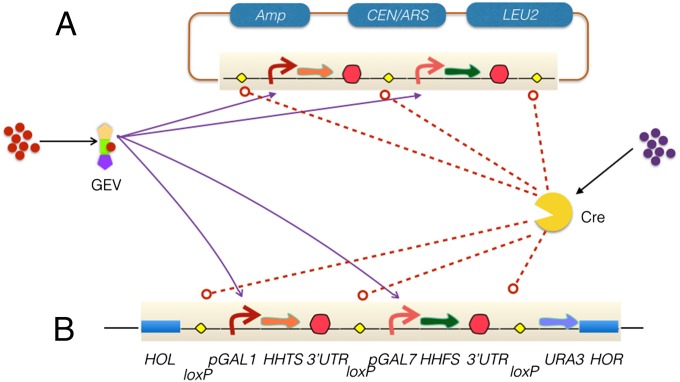
Fig. 1.
The structure of triplex histone switch. Duplex integrated safeguard based on a pair of histone genes. Histone H3 gene HHTS is regulated by a galactose promoter pGAL1, and the histone H4 gene HHFS is controlled by another galactose promoter pGAL7. Each histone switch is flanked by a pair of loxP sites. The duplex histone safeguard switches can either be on a CEN/ARS plasmid (A) or integrated into a genomic locus (B). Either the native Gal4 protein or an engineered tribrid protein called GEV turns on the transcription of the histone genes in the presence of their ligands, such that the engineered yeast survives. The third level of redundancy is use of an orthogonal control mechanism, site-specific recombination. Any pair of loxP sites (yellow diamonds) on the construct will recombine and delete one or both histone genes upon Cre, activation, leading to inviability. Both proteins can be independently controlled by small molecule(s). Hooked arrows, promoters; filled arrows, histone genes; blue lozenges, standardized vector components; HOL, HOR, sequences to Left and Right of HO gene, where the safeguard was integrated.
To evaluate ability to function as a genome safeguard, we generated yeast strain bearing the construct, PCy230 [his3∆200 leu2∆1 lys2∆202 trp1∆63 ura3-52 hht1-hhf1∆::natMX4 hht2-hhf2∆::hygMX4 (pRS415-loxPWT-HHTS-loxPWT-HHFS-loxpWT); Materials and Methods]. Strain PCy230 was subsequently plated on both permissive (galactose) and restrictive (glucose) conditions, and the escape (reversion) rate was calculated according to Luria and Delbruck fluctuation analysis using the method of the median (25) (Table S1). Although the escape rate was quite favorable at 10−7, we observed a significant fitness defect by comparing the colony size of PCy230 with that of a control strain (Fig. S2). Subsequent transcriptome profiling indicated that a large number of genes were dysregulated, possibly as a result of the combined effects of variations in plasmid copy number and the accumulation of a fraction of cells that had lost plasmid under permissive conditions (Fig. S3).
Chromosomal Integration of Histone Switches Restores Fitness.
We hypothesized that the fitness decrease of PCy230 may reflect variations in histone abundance in individual cells as the copy number of centromeric plasmids can fluctuate between one and three copies. Alternatively, it was possible that the expression of the two histone genes would give rise to aberrant histone ratios that differed from the native situation, or deviations in expression dynamics between galactose inducible and native histone promoters. Integration of the histone-based safeguard into the genome would ensure stable single copy. Thus, we integrated the histone safeguard (tagged with URA3) into the HO locus of PCy230, and after spontaneous mitotic loss of the original pPC012 plasmid (tagged with LEU2) on YP-galactose medium, we constructed integrated safeguard strains PCy596–599. These integrated safeguard strains have similar low escape rates as PCy230 on glucose (Table S1), yet the fitness and gene expression profiles were significantly improved as determined by measuring colony sizes (Fig. S4A), doubling time (Fig. S4B), and transcriptome profiling (Fig. S4C).
Regulating Transcriptional Switches with 30 nM Estradiol.
For many applications, a low ligand concentration, and a nonnative ligand unlikely to alter cell physiology are desirable. To demonstrate ability to regulate the cell viability using nonnative small molecules at nanomolar concentration, we transformed a second plasmid pHCA/GAL4(1-93).ER.VP16 (henceforth called GEV) that produces a GAL4 DBNA binding domain–estrogen receptor–VP16 tribrid protein (GAL4DB.ER.VP16; GEV) (26), into safeguard strain PCy599 to construct strain NAy236. This safeguard strain can be regulated not only by galactose, used at millimolar concentrations, but also by estradiol, a small molecule nonnative to yeast able to maintain viability at submicromolar (30 nM) concentrations. The GEV localizes into nucleus and binds to Gal4p consensus sequences to activate transcription of the targeted gene, upon induction by β-estradiol. The GEV protein is retained in the cytoplasm by Hsp90 in the absence of estradiol, resulting in fairly tight control of gene expression. We were able to regulate the histone safeguard strains (PCy230 and PCy599) with 30 nM estradiol on normal glucose-containing yeast growth medium, with an escape reversion frequency of 10−7 to 10−9 (Fig. 2 and Table S1).
Fig. 2.
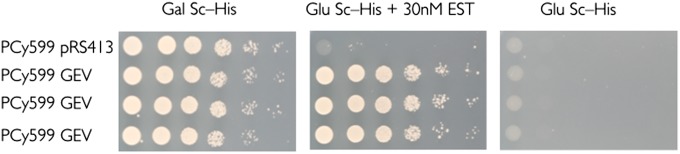
Regulating histone-based safeguard switches with 30 nM estradiol. The GEV-regulated histone safeguard strains grow well on permissive medium and die under restrictive conditions. The histone strain PCy599 with GEV plasmid grows well on both galactose medium and estradiol-supplemented glucose medium, and cannot survive on glucose medium without estradiol.
Escape Mutant Analysis: GEV-Regulated GAL-Histone Transcriptional Safeguards.
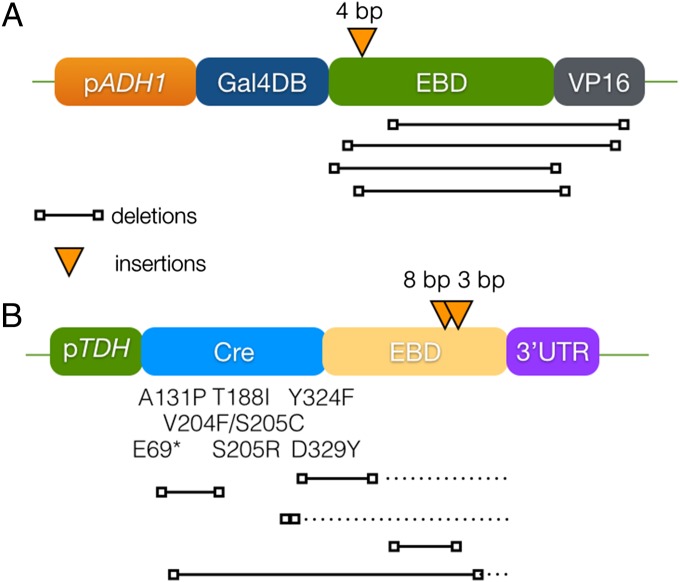
We isolated five independent escape mutants of the GEV-regulated histone safeguard strains, which grow on glucose media in absence of the estradiol. The GEV plasmids from those five escapers were recovered from yeast into Escherichia coli, transformed into the parental strain PCy599, and tested for their ability to recapitulate the “growth in absence of estradiol” phenotype (Fig. S5A), and sequenced with the same primers (Table S2). Interestingly, many of the escape mutants seemingly grow less well than the parental strain in the presence of estradiol, suggesting there will be selective pressure against their accumulation (Fig. S5A). All five plasmids that recapitulated the phenotype had mutations, mostly large internal deletions in the human estrogen receptor (hER) domain of the gene that delete the estradiol-responsive domain but remain in-frame with the downstream VP16 (Fig. 3A and Table S3). Similarly, we tested that reintroducing a fresh GEV plasmid restored estradiol dependence (Fig. S5B).
Fig. 3.
Escape mutant analysis. Both GEV and Cre-EBD plasmids of escape mutants were isolated and analyzed. (A) Most of the plasmids bore in-frame deletions in the hER region that retained the C-terminal VP16 activator sequence. One plasmid had a 4-bp insertion, and the protein is predicted to have an out-of-frame C-terminal tail of 26 aa, of which 5 are acidic residues, a sequence that might serve as a transcriptional activator. See also Table S3. (B) The mutations found in the Cre-EBD constructs are mostly missense mutations in the Cre coding region or deletions (deletions that are also out of frame are indicated by a stippled line for the out-of-frame segment) in the Cre and/or EBD domains. See also Table S3.
Chemically Regulated Site-Specific Recombination Switches.
In our previous work on synthetic yeast chromosomes (8, 9), we have adapted a chemically regulated Cre (27) and shown that it very effectively kills yeast with a semisynthetic genome containing numerous loxP sites flanking a number of essential genes. The specialized Cre protein from ref. 27 is tightly regulated both transcriptionally and posttranslationally. A daughter-cell–specific promoter, pSCW11, ensures expression only in the daughter cell state. Furthermore, the Cre is fused to the estrogen binding domain (EBD), which is unfolded in the absence of estradiol, causing it to be retained in the cytoplasm by binding to the Hsp90 chaperone. In the following proof-of-principle experiments, we induced Cre with estradiol, leading to excision of the histone gene(s) and loss of viability. Ultimately, it will be desirable to “reverse” this logic, so that the small molecule maintains the viability of the cell.
We transformed the pSCW11-Cre-EBD plasmid into safeguard strain PCy230 to construct PCy251, and induced killing on Gal plates (galactose is required to maintain histone gene expression) with 1 µM estradiol, intended to delete the histone genes, despite their expression. However, in this context, the killing effect was not as complete as expected. We hypothesized that this was due to SCW11 promoter strength/expression properties, and thus we constructed a set of Cre-EBD plasmids by replacing the SCW11 promoter with various constitutive yeast promoters (Materials and Methods). After transforming the set of constructed Cre-EBD plasmids into PCy230 and screening on Gal-estradiol plates, pTDH3-Cre-EBD turned out to be the best candidate for our application, as it effectively killed the safeguard strain upon induction but remained inactive in the absence of estradiol. Upon estradiol induction, the Cre protein localizes into the nucleus and excises the histone transcription unit(s) residing between the three loxP sites. The reversion rate of this site-specific recombination-based safeguard was systematically measured and is less than 10−6.
Escape Mutant Analysis: CRE-EBD Sensitivity of GAL-Histone Recombinational Safeguards.
We evaluated the site-specific recombination regulation of the original GAL-histone safeguard in the presence of a CRE-EBD plasmid by studying escape mutants. We isolated 21 independent escape mutants of the safeguard strains that grew in the restrictive condition of Cre expression, which in this case is galactose containing 1 µM estradiol. The Cre-EBD plasmids were recovered and retransformed into the parent strain PCy599, and the resulting strains were found to be insensitive to estradiol induction, confirming that the responsible mutation resided on the plasmid. The Cre-EBD mutant plasmids were sequenced to identify the mutations leading to the phenotype (Fig. 3B and Table S3), and we observed a wide variety of mutations predicted to lead to a loss of function. Of the 21 plasmids, 2 had insertions (of 3 and 8 bp), 7 had missense or nonsense mutations in the Cre coding region, 5 had various deletions in the Cre and/or EBD regions, and finally 7 of them complete lost the Cre-EBD insert, instead containing either a single loxP site or loxP sites flanking the histone expression cassette (unlike the parental plasmid, which had none). The latter category of mutant can be explained by assuming there were two copies of the histone plasmid. We propose that homologous recombination between one copy of the safeguard plasmid (based on pRS415-LEU2) and the pTdh-Cre-EBD plasmid (based on the largely identical pRS413-HIS3) occurred, producing a Cre-EBD-LEU2 plasmid and a histone HIS3 plasmid. Upon Cre-EBD induction, an “empty” HIS3 plasmid containing just the loxP site would be produced at high frequency. Subsequent mitotic loss of the Cre-EBD plasmid, a frequent event, would produce the observed configurations.
Combined Escape of Transcriptional and Recombinational Safeguard Switches Not Observed.
We performed a combined escape experiment by plating yeast strain PCy599 carrying TDH3-Cre-EBD by growing cells permissively, and then plating nearly 1010 cells on restrictive medium (glucose plus estradiol). Even after replica plating (i.e., two rounds of growth on restrictive medium), no escaper colonies were observed. We estimate that the cells went through at least one doubling after hitting the selective plates, possibly more. Thus, we conclude that the reversion frequency was <10−10, consistent with the predicted frequency of about 10−12.
Construction of Additional Safeguard Switches.
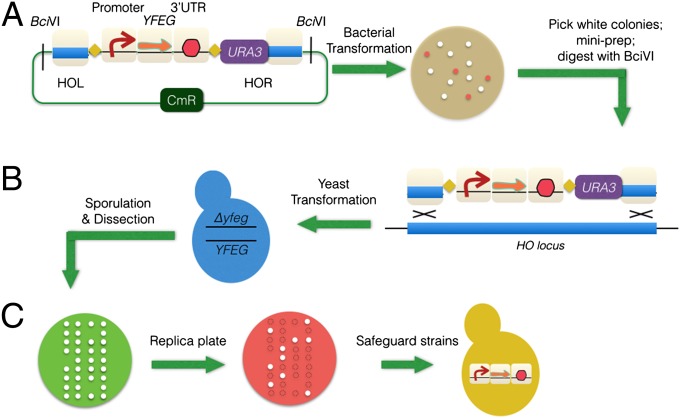
To screen for additional essential genes suitable as candidate safeguards, we used a variant of the Golden Gate DNA assembly method (28) to efficiently design and construct a systematic set of safeguard switches (Fig. 4). A library of parts (promoters, genes and 3′-UTRs), along with a set of acceptor vectors, was constructed and sequence verified. Using a Golden Gate reaction, we assembled 17 essential genes under three different galactose promoters (pGAL1, pGAL7, and pGAL10), respectively.
Fig. 4.
Construction of multiplex integrated safeguard strains. A Golden Gate assembly-based method was developed to construct various combinations of safeguards. A selected essential gene [your favorite essential gene (YFEG)] can be quickly assembled with a given promoter and a 3′-UTR into an acceptor vector [A; where red colonies are due to vector reassembly (they express RFP) and white colonies are the right assemblies] and integrate into the corresponding yeast mutant (B) to test the behavior of the safeguard strain under both permissive and restrictive conditions (C).
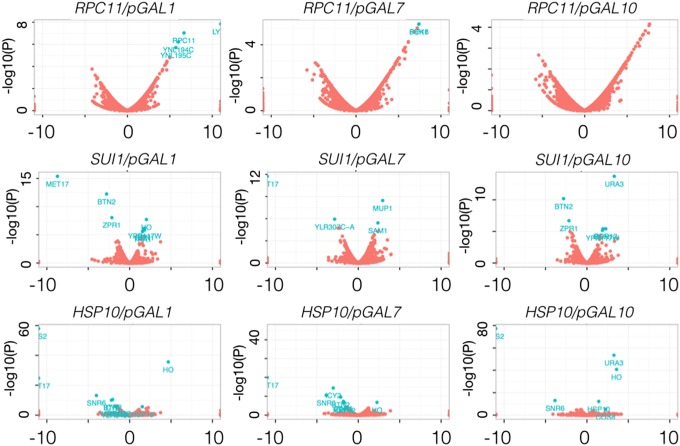
These were tested individually in the corresponding heterozygous diploid yeast deletion mutant. Diploids transformed with the appropriate test plasmid were sporulated, and tetrads were dissected on permissive agar plates, and resulting haploid spore clones were replica plated onto restrictive plates to identify potential safeguard strains. We identified three essential genes, namely SUI1, HSP10, and RPC11, as preferred gene candidates by this method. The selection criteria were as follows: the safeguard strain (i) should grow well on the permissive medium and (ii) should have a low escape rate under restrictive conditions. Transcriptome profiling was carried out on the constructed safeguard strains to identify potential fitness defects (Fig. 5). The GEV plasmid and CreEBD plasmid were separately transformed into such an RPC11 safeguard strain, to construct estradiol-regulated transcriptional switch safeguard strain and site-specific recombination-regulated safeguard strains, respectively.
Fig. 5.
Transcriptome profiling of safeguard strains. Transcriptome profiling of various safeguard strains. The graph is organized by gene/promoter pairs. The blue dots in the volcano plots represent statistically significant dysregulated genes (see Table S4 for lists of genes affected). The transcriptome profiling shows limited transcriptome changes to the safeguard strains compared with the wild type, with GAL10-RPC11 showing the best performance.
Reducing Leakiness of Safeguard Switches.
We discovered that the non–histone-based GEV-regulated safeguard strains grew under restrictive conditions and hypothesized that this resulted from leakiness of the galactose promoters (pGAL1/pGAL7/pGAL10) in this context, where lower amounts of the protein products might be required for viability than for histone genes. In previous work (31), various numbers (2–10) of Gal4 binding sites were fused to the SPO13 promoter, which contains a repressive upstream regulatory sequence (URS) sequence, to generate a set of tightly regulated galactose-dependent promoters called SPAL promoters (SPO/GAL). We retrieved this set of promoters from strains containing such promoters (Materials and Methods) and use them to construct a second generation of safeguard strains based on RPC11. The resulting safeguard strains had the appropriate growth phenotypes compared with those with native galactose promoters (Fig. S6).
Discussion
Engineering containment is anticipated to be more critical than ever as the world transitions from a petroleum economy to a bioeconomy for producing a wide variety of chemicals and fuels, and as the ease of constructing more and more complex biological systems needed to accomplish this grows. Containment will be critical for any field application of engineered organisms but is also extremely desirable for any work done on pathogenic organisms, from the perspectives of investigator safety and prevention of accidental or intentional release of harmful organisms from the laboratory environment. Finally, biotechnology companies make large financial investments in building specific strains of bacteria and yeast for large-scale fermentation. Preventing the theft of such organisms is a challenging problem, but genomic safeguards such as those described here have the potential to minimize the risk of such intellectual property.
In order for safeguards to be practically useful, they require several engineered features: redundancy, robustness, low escape frequencies, and low cost. The likelihood of their widespread adoption by experimentalists and industrial biotechnologists alike also requires strong evidence that the safeguard itself does not affect cellular metabolism or phenotype.
Redundancy.
We show here two different forms of redundancy. The first is to use two essential genes rather than just one. The second is to use two modes of dependency, one being transcriptional and the other, the presence/absence of the gene, i.e., recombinational. In the latter case, two distinct small molecules, galactose and estradiol, were used to control the switch.
Robustness.
Cells in the environment might face a wide variety of conditions. It would be useful to know that the safeguard has little or no effect on growth in the presence of the ligand but also that growth in the absence of the ligand was similarly absent under multiple conditions. With our histone safeguard, we showed that performance was excellent in both minimal and rich medium conditions, two environments routinely used in the laboratory. We also observed that our histone safeguard with the GEV system can function over a wide range starting at 5 nM estradiol concentration, and the system is positive up to at least 100 nM. This is advantageous in a complex biological environment such as a bioreactor or a fermenter, where levels of exogenous molecules may not be as homogenously distributed as in a well-controlled laboratory environment. It would be interesting to expand these studies to wider ranges of conditions.
Escape Frequencies.
Because each safeguard described here produces escapers at a frequency of about one in a million or in some cases significantly less, it is anticipated by duplexing them very favorable escape rates of 10−12 or lower might be obtained. In our experiment to evaluate the combined transcriptional and recombinational switches, we failed to identify escapers, consistent with this prediction. Interestingly, many of the observed revertants grow less well than the parental in the presence of the ligand, suggesting their persistence in population will be minimized. Similarly, triplex or higher-plex safeguards should produce increasingly stringent levels of control.
Cost.
The key to low cost is to identify ligands that are both relatively inexpensive per gram and that operate at low concentrations, preferably micromolar or even nanomolar. Our work demonstrates that the estradiol-based GEV-based safeguard operates at a concentration of 30 nM with escape rate characteristics. Because estradiol is a commodity that can be purchased cheaply, the cost of treating a 1,000-L fermentor is estimated at US$0.13.
Minimal Phenotype Effects.
We evaluated the impact of the safeguards on fitness by examining colony size. We hypothesize that the use of essential genes, rather than toxins, to control cell growth is inherently more favorable to overall growth properties and lack of perturbation of cellular systems. Indeed, the histone switch, once integrated in single copy, was surprisingly fit. As a first evaluation of the impact of the safeguards we built, we examined the transcriptome effects of both the essential genes used in safeguards and ligands commonly used to keep them in the “on” state. Although the systems in use were not perfect in that effects were observed on gene expression, the extent of these perturbations varied significantly across different essential genes and small molecules. This helps with prioritization of the most promising designs.
Escaper (revertant) analyses suggest means to reduce escape frequencies. For the Cre-EBD and GEV constructs, escapers affecting the function of these proteins were isolated. For the Cre-EBD, the mutations isolated were loss-of-function mutations. This frequency could be greatly lowered by simultaneously expressing a second recoded copy of the same gene, because mutations of this type should be recessive. The GEV mutants could similarly be reduced in frequency by incorporating two ligand-binding domains into the same protein.
One key limitation to safeguard design and construction is the paucity of well-regulated transcriptional regulatory systems controlled by low-cost nontoxic small molecules that can work across prokaryotic and eukaryotic systems generically. Besides estradiol, there are several low-cost natural and synthetic hormone and hormone binding domains that have been proven to regulate protein activity in various systems including budding yeast, such as androgen (29), glucocorticoid (30), and progesterone/RU486 (31). Once carefully characterized, these small molecular/binding domain pairs can potentially be used to build orthogonal safeguard switches in the future. Also, prokaryotic strains have been engineered in parallel, using many of the same design philosophies (32, 33). We anticipate that many improvements can and will be made to safeguards in the coming years to anticipate the explosion of recombinant organisms under study in basic and industrial research laboratories, resulting in an increased focus on safety mechanisms integral to the systems under study.
Materials and Methods
Strains, Plasmids, and Oligonucleotides.
Yeast strains and the plasmids contained are listed in Table S5. Oligonucleotides used are listed in Table S2.
Plasmid Shuffling.
Plasmid pPC012 was transformed into a yeast strain, JDY6, from which all four genomic copies of histones H3 and H4 genes had been previously deleted (23) with viability maintained by plasmid pDM9, a plasmid carrying the URA3 marker and wild-type histone H3 and H4 genes HHT1 and HHF1. After introducing pPC012, a centromeric (CEN) LEU2 plasmid, into the strain by LioAc/SS/PEG transformation (34), plasmid shuffling on 5-fluoro-orotic acid [5-Foa (35)], 2% (wt/vol) galactose medium was successful, indicating that the GAL promoters could successfully express the two histone genes. More importantly, such 5-FoaR strains were unable to grow on glucose, consistent with effective shutoff of histone gene expression.
Golden Gate DNA Assembly.
The Golden Gate assembly protocol is described in ref. 28. Specifically, a 15-µL Golden Gate reaction containing 1.5 µL of 10× T4 DNA ligase reaction buffer (New England Biolabs), 0.15 µL of 100× BSA (New England Biolabs), 1 µL of 600 units/µL T4 DNA ligase (Enzymatics), 10 µL of H2O, 1 µL of acceptor vector DNA (∼100 ng/µL), and 0.5 µL of insert DNA (∼100 ng/µL). The following temperature cycles were used: 1 h at 37 °C, 5 min at 50 °C, 5 min at 80 °C followed by incubation at 4 °C. One microliter of the finished Golden Gate reaction was transformed into 100 µL of Top10/DH5α chemical-competent cells.
Plasmid Recovery from Yeast.
Plasmid recovery from yeast was carried out using a Zymoprep yeast plasmid miniprep kit (Zymo Research) following the manufacturer’s instructions.
Cloning and Sequencing of SPALX Promoters.
Twelve yeast MAV strains (36) containing various numbers of Gal4 binding sites were used as template to clone out SPAL promoters with varying numbers of Gal4 binding sites. Genomic DNAs of these yeast strains were extracted using phenol/chloroform isolation method. Two rounds of PCR were used to amplify the SPALX promoters from genomic DNA: first, F primer PC_oligo379 and Ura3 R primer PC_oligo380 were used; then the PCR product was diluted 1,000-fold. Primers PC_oligo401 and PC_oligo324 were used to amplify the SPAL promoter fragments with appropriate Golden Gate overhangs. The PCR products were cloned using a Zero Blunt Topo kit (Life Technologies). The resulting plasmids were sequenced to identify the exact number of Gal4 binding sites. These promoters were used in subsequent assembly of safeguard switches to identify tighter constructs.
β-Estradiol Induction of Cre Expression.
Safeguard strains were grown up on appropriate selective growth medium, then either plated or spotted on appropriate selective solid medium containing 1 µM β-estradiol, and incubated at 30 °C for 2–3 d.
β-Estradiol Induction of GEV System.
Safeguard strains were grown up in appropriate glucose selective growth medium containing 30 nM β-estradiol, then either plated or spotted on appropriate glucose selective solid medium containing 30 nM β-estradiol (or restrictive plates lacking estradiol), and incubated at 30 °C for 2–3 d.
Reversion Rate Measurement and Colony Picking for Sequencing.
Escape rates were calculated using the method of the median (25). For measurement rates in the 10−5 to 10−9 range, 5–12 independent cultures (each grown from a single parent colony) were inoculated into 20 mL of SC–His supplemented with 2% galactose in liquid cultures. In total, 108 and 107 cells were plated on restrictive medium. Viable titer was determined by plating 100 µL of a five to six serial 10-fold dilution on permissive medium. The reversion frequency was obtained by dividing colony-forming units on restrictive plates by colony-forming units on permissive plates. The median reversion frequency was then used to calculate the rate using the method of the median.
For each safeguard strain to be measured, five “escaper” revertant colonies were picked from independently grown cultures and grown up in 10 mL of permissive liquid culture (with plasmid selection if applicable), for 48 h at 30 °C. Tenfold serial dilutions were plated on restrictive and permissive agar plates, and incubated at 30 °C for 2–3 d until single colonies appeared. One colony was chosen per culture to assure independence.
RNASeq.
A single colony was picked and grown up in 10 mL of permissive liquid medium, and incubated at 30 °C until the A600 was between 0.8 and 1.2, and RNA was isolated as described using a Qiagen RNAEasy kit using the manufacturer’s protocol. We performed RNASeq of strains integrating the histone, RPC11, SUI1, and HSP10 safeguards. mRNA was sequenced using an Illumina HiSeq and standard TruSeq preparation kits. For each strain, we obtained ∼12 million 50-bp single-end reads. Reads were mapped using TOPHAT (37) to the reference S. cerevisiae genome (S288c). Approximately 95% of the reads were mapped. For each gene, read counts were computed using HTSEQ (38) and analyzed for differential expression using DESEQ (39), with standard parameters and following the no-replicates scenario. For each gene, we obtained a raw P value and an adjusted P value using the standard Benjamini–Hochberg procedure (40). We used the 1% false-discovery rate (adjusted P value ≤ 0.01) to identify genes that are significantly differentially expressed. Differentially expressed genes are reported in Table S4.
Supplementary Material
Acknowledgments
We thank Qiuhui Lin and Bin Jia for help on Cre-EBD escaper analysis experiments and helpful discussions. Y.C., N.A., W.J.C., G.S., K.C., H.H., J.S.B., and J.D.B. were supported by the Defense Advanced Research Project Agency [N66001-12-C4020 (to J.D.B.)]. Y.C. and A.U. were supported by a Chancellor’s Fellowship from the University of Edinburgh, a start-up fund from Scottish Universities Life Sciences Alliance, and Biotechnology and Biological Sciences Research Council Grant BB/M005690/1 (to Y.C.). The open access charge is supported by Research Councils UK Open Access Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424704112/-/DCSupplemental.
References
- 1.Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: Where synthetic biology came from and where it needs to go. Cell. 2014;157(1):151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 2.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: Generation of infectious virus in the absence of natural template. Science. 2002;297(5583):1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 3.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3(1):e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: PhiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA. 2003;100(26):15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310(5745):77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 6.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319(5867):1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 7.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 8.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344(6179):55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477(7365):471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: From parts to pathways and beyond. Integr Biol (Camb) 2011;3(2):109–118. doi: 10.1039/c0ib00070a. [DOI] [PubMed] [Google Scholar]
- 11.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333(6040):348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23(20):2760–2767. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- 14.Czar MJ, Cai Y, Peccoud J. Writing DNA with GenoCAD. Nucleic Acids Res. 2009;37(Web Server issue):W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillson NJ, Rosengarten RD, Keasling JD. j5 DNA assembly design automation software. ACS Synth Biol. 2012;1(1):14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 16.Moe-Behrens GH, Davis R, Haynes KA. Preparing synthetic biology for the world. Front Microbiol. 2013;4:5. doi: 10.3389/fmicb.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank KM, Schneewind O, Shieh WJ. Investigation of a researcher’s death due to septicemic plague. N Engl J Med. 2011;364(26):2563–2564. doi: 10.1056/NEJMc1010939. [DOI] [PubMed] [Google Scholar]
- 18.Bej AK, Perlin MH, Atlas RM. Model suicide vector for containment of genetically engineered microorganisms. Appl Environ Microbiol. 1988;54(10):2472–2477. doi: 10.1128/aem.54.10.2472-2477.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes K, et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen SM, Karlström OH. Development of efficient suicide mechanisms for biological containment of bacteria. Appl Environ Microbiol. 1991;57(1):85–92. doi: 10.1128/aem.57.1.85-92.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulsen LK, Larsen NW, Molin S, Andersson P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol Microbiol. 1989;3(11):1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 22.Wright O, Delmans M, Stan G-B, Ellis T. GeneGuard: A modular plasmid system designed for biosafety. ACS Synth Biol 2014 doi: 10.1021/sb500234s. , 10.1021/sb500234s. [DOI] [PubMed] [Google Scholar]
- 23.Dai J, et al. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134(6):1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49(3):264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 26.McIsaac RS, et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22(22):4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindstrom DL, Gottschling DE. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183(2):413–422, 1SI–13SI. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden Gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4(5):e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moilanen A, Rouleau N, Ikonen T, Palvimo JJ, Jänne OA. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1997;412(2):355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- 30.Popovic N, et al. Site-specific and dose-dependent effects of glucocorticoid receptor phosphorylation in yeast Saccharomyces cerevisiae. Steroids. 2010;75(6):457–465. doi: 10.1016/j.steroids.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, O’Malley BW, Jr, Tsai SY, O’Malley BW. A regulatory system for use in gene transfer. Proc Natl Acad Sci USA. 1994;91(17):8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher RR, Patel JR, Interiano AL, Rovner AJ, Isaacs FJ. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res 2015 doi: 10.1093/nar/gku1378. , 10.1093/nar/gku1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 2015 doi: 10.1038/nature14095. , 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 35.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 36.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93(19):10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.