Abstract
OBJECTIVE
Neuregulin 1 (NRG1) is a multifunctional neurotrophin and a critical mediator of neurodevelopment and risk for schizophrenia. NRG1 undergoes extensive alternative splicing, and association of brain NRG1-IV isoform expression with the schizophrenia-risk polymorphism, rs6994992, is a potential molecular mechanism of risk. Novel splice variants of NRG1-IV (NRG1-IVNV), with predicted unique signaling capabilities, have been cloned in fetal brain. Because the developmental expression and genetic regulation of NRG1-IVNV in human brain and relationship to schizophrenia is unknown, the authors investigated the temporal dynamics of NRG1-IVNV transcription, compared to the major NRG1 isoforms (types I-IV), across human prenatal and postnatal prefrontal cortical development and examined the association of rs6994992 with NRG1-IVNV expression.
METHOD
NRG1, types I-IV and NRG1-IVNV isoform expression was evaluated using quantitative real-time PCR in prefrontal cortex during human fetal brain development (14-39 weeks gestation: N=41) and postnatally through aging (age range 0-83 years: N=195). The association of rs6994992 genotype with NRG1-IVNV expression was determined. In-vitro assays were performed to determine the subcellular distribution and proteolytic processing of NRG1-IVNV isoforms.
RESULTS
Expression of NRG1, types I, II, III was temporally regulated during human prenatal and postnatal neocortical development and the trajectory of NRG1-IVNV was unique, being expressed from 16 weeks gestation until 3 years of age, after which it was undetectable. NRG1-IVNVs expression was associated with rs6994992 genotype, whereby homozygosity for the schizophrenia-risk allele (T) conferred lower cortical NRG1-IVNV levels. Finally, in-vitro cellular assays demonstrate that NRG1-IVNV is a novel nuclear enriched, truncated NRG1 protein that is resistant to proteolytic processing.
CONCLUSION
This study provides the first quantitative map of NRG1 isoform expression during human neocortical development and aging and identifies a potential mechanism of early developmental risk for schizophrenia at the NRG1 locus, involving a novel class of NRG1 proteins.
Introduction
Neuregulin 1 (NRG1) is a key developmental growth factor that binds to and activates the ErbB class of receptor tyrosine kinases (1). Differential promoter usage and extensive alternative splicing generates several distinct isoforms of the NRG1 gene, namely types I-VI (1, 2). NRG1 is a key mediator of multiple neurodevelopmental processes including cell migration, synaptic formation and plasticity and myelination (3). Despite growing evidence demonstrating NRG1’s essential role in the developing murine brain (4-7) and its involvement in disorders of neurodevelopment and maturation, including schizophrenia (8-11) and bipolar disorder (12, 13), the developmental expression trajectories of individual NRG1 isoforms during human pre- and postnatal neocortical development are unknown.
Polymorphisms in the NRG1 gene have been associated with risk for schizophrenia in multiple populations. The original risk haplotype (HapICE) was first isolated in the Icelandic population and is comprised of several single nucleotide polymorphisms (SNPs), including SNP8NRG243177 (rs6994992) located in the 5’ end of the NRG1 gene; an association subsequently shown to be relevant to schizophrenia in Scottish, English, Irish and Northern Indian populations (8-11). Although, NRG1 polymorphisms have yet to be identified in large genome-wide association studies (GWAS) of schizophrenia, likely because of heterogeneity within the gene and across populations (14, 15), support for association of the NRG1 HapICE region has additionally come from meta-analyses of published data (16, 17) and three GWA schizophrenia datasets (18).
rs6994992 is located proximal to the 5’ exon (E187), central to a cis- regulatory element in the NRG1 promoter (19). Experimental examination of rs6994992, using luciferase promoter assays and site directed mutagenesis of human DNA constructs, suggests that the SNP is functional (19). rs6994992 predicts expression levels of NRG1-IV in brain, whereby the schizophrenia risk allele (T) is associated with increased levels in the hippocampus and dorsolateral prefrontal cortex (20, 21). In addition, convergent data from human neurocognitive and imaging studies show that rs6994992 is associated with impairments in neurocognitive function, sensory processing, abnormal brain morphology and reduced neural connectivity (22-26). Recently allelic variation at the locus has been associated with cortical and subcortical neuroanatomical structure in human neonates (27), indicative of a role for NRG1 in early human brain development. Although the exact neural mechanisms by which the polymorphism is associated with human brain development and function remain unclear, previous data implicates at the molecular level, the mechanism involves the NRG1- IV class of neuregulins (19, 20).
Emerging evidence suggests that variation in genes that are temporally regulated during neurodevelopment and expressed preferentially in fetal brain are related to the genetic architecture of schizophrenia (19, 27-30). Unlike other NRG1 isoforms, NRG1-IV is expressed exclusively in brain in humans, expression of which is three-fold higher in early fetal life (19). Recently, NRG1-IV has been confirmed as a bioactive transmembrane pro-protein which is susceptible to proteolytic processing (31) and regulated by neuronal activity (4). Novel transcripts of NRG1-IV have been cloned exclusively in human fetal brain, including fetal variants E and F (19); (referred to herein as NRG1-IVNV) that share the same E187 leader exon and conserved promoter as NRG1-IV, but lack a functional epidermal growth factor and matrix metalloprotease cleavage domain. These findings highlight NRG1-IVNV isoforms as a biologically novel class of early developmental NRG1 proteins, potentially relevant to risk for schizophrenia (19).
During normal brain development, genes are expressed in patterns specific to developmental stage (32). Although evidence suggests that total levels of NRG1 mRNA are correlated with postnatal age in the human prefrontal cortex (33), the expression profiles of individual major NRG1 isoforms (types I-IV) in human fetal and postnatal neocortical development are unknown. Furthermore, little is known about the transcriptional regulation of fetal-brain derived splice variants of the NRG1-IV class and whether NRG1-IVNV expression is expressed in the postnatal human brain and associated with rs6994992 genotype (as would be predicted given the conserved genetic promoter (19, 20)).
In this study we examined quantitative gene expression profiles of NRG1 Types I, II, III and IV in the prefrontal cortex during human fetal brain development (second trimester) and Types I, II, III across the postnatal lifespan through aging (age range 0-83 years) and sought to determine if expression of novel NRG1-IV isoforms, (NRG1-IVNVs), are temporally regulated during fetal development, detectable after birth, and associated with genetic variation at the schizophrenia risk polymorphism rs6994992. Examination of the subcellular localization and biochemical processing of NRG1-IVNV and NRG1-IV isoforms was explored using recombinant DNA technologies and heterologous expression systems.
Our results provide novel evidence that NRG1 isoform expression is temporally regulated in the human prefrontal cortex during the second trimester of development and postnatally throughout the lifespan. Specifically, NRG1-I expression was most abundant during the first two weeks of the second trimester and declined with fetal gestational age, becoming stable at birth, while NRG1-III increased significantly with fetal gestational age and decreased dramatically after birth to reach stable levels in early adolescence. Additionally, we demonstrate that NRG1- IVNV isoforms are uniquely regulated at the transcriptional level during prenatal and early postnatal development, being expressed after gestational age 15 weeks, through birth and until 3 years of age, after which they are transcriptionally undetectable. Moreover, expression levels of NRG1-IVNV are associated with rs6994992 genotype, identifying a potential molecular mechanism of early developmental risk for schizophrenia at the NRG1 locus. Finally, we show preliminary, qualitative data that suggest that NRG1-IVNVs, unlike full-length NRG1-IV (and other transmembrane pro-protein NRG1’s), are resistant to classic proteolytic processing of the extracellular domain and enriched within the nucleus of both neurons and human embryonic cells, suggesting that NRG1-IVNV splice isoforms are biologically distinct NRG1-IV proteins that may be key regulators of early cortical development relevant to the pathophysiology of schizophrenia.
Methods
Human postmortem tissue
Postmortem human brains from the Clinical Brain Disorders Branch were obtained at autopsy from the Washington, D.C. and Northern Virginia Medical Examiners’ Offices, all with informed consent from the legal next of kin (protocol #90-M-0142 approved by the NIMH/NIH Institutional Review Board). Additional postmortem fetal, infant, child, and adolescent brain tissue samples were provided by the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders [www.BTBank.org] under contracts NO1-HD-4-3368 and NO1-HD-4-3383. A total of 195 control samples ranging in age from 0-83 years were available for this study (mean age 30.73 y; 59 female, 136 male; 101 African American, 85 Caucasian, 5 Hispanic; 4 Asian; postmortem interval 30.46; (SD=16.54 h); pH 6.54 (SD=0.31); RNA Integrity Number 8.3 (SD=0.86). A total of 41 fetal brain samples were available (gestational age weeks 14-39, 21 male, 20 female; 39 African American, 2 Caucasian; postmortem interval, 2.47 (SD=2.1); RNA Integrity Number 8.85 (SD=1.1)). The Institutional Review Board of the University of Maryland (UMD) at Baltimore and the State of Maryland approved the protocol, and the tissue was donated to the NIMH under the terms of a Material Transfer Agreement. Processing of tissue from both tissue banks was handled in the NIMH laboratory by the same team of investigators, as described previously (32). Control subjects were defined as individuals with no history of psychiatric symptoms, psychiatric diagnosis, psychiatric admissions or substance abuse. All subjects were also without a neuropathological diagnosis and had negative toxicology (34). Further details of brain dissections and sample preparation are described in Supporting Text.
Real-time quantitative PCR
Quantitative gene expression levels were measured by real-time RT-PCR using an ABI Prism 7900 sequence detection system with 384-well format (Applied Biosystems, Foster City, CA, USA) and quantification via the standard curve method, as described previously (20). Specific primer and isoform- specific TaqMan® probe combinations were used to assess mRNA expression levels of NRG1 types I-IV in prefrontal cortex, as previously described (20). Additional primer and isoform specific probe sets were designed to amplify NRG1-IVNV isoforms based on the structure of novel fetal variants E and F cloned in Tan et al., 2007 (19) (accessions EF372277 and EF3372275, respectively). Details of NRG1-IVNV primers and probe design and genetic location are shown in Table S1 and Supplemental Figure 1. No other known NRG1 transcript lacks the β and stalk coding exons, therefore the real-time PCR probe designed to span these exons selectively detects NRG1-IVNVs. Expression levels of NRG1 isoforms were normalized to the geometric mean of the expression of three control genes (β-actin, GAPDH and PBGD), as described previously (35) and in Supporting Text.
Determination of rs6994992 genotype
rs6994992 was genotyped because of its previous association with schizophrenia and with brain expression of full length NRG1-IV (8-11, 20, 21). Genotyping was performed using the Taqman 5’-exonuclease allelic discrimination assay (details available on request). Genotype reproducibility was routinely assessed by re-genotyping all samples and was generally >99%. Overall genotyping failure rate was <1%. DNA genotype was determined from cerebella of fetal and postnatal samples up to 3 years of age, using previously described methods (20) and the following custom designed SNP assay: forward primer 5’AATTAGTAGGATTGGATGTTTGAACCA 3’, reverse primer 5’ GATGGAGCGCTTCAGGAGAA 3’, probe 1 5’ FAM-CCAGTATACgTTCACTTG-MGB 3’, probe 2 5’ VIC-CCAGTATACaTTCACTTGA-MGB 3’. Genotype analysis was restricted to NRG1-IVNV mRNA. Fetal samples aged 14 and 15 weeks were excluded from the analysis because of low detectable expression of NRG1-IVNV (see Figure 2A). Thus 34 individuals (gestational age 16 weeks and above) were used for genotypic analysis.
Figure 2. Temporal dynamics of NRG1-IVNV mRNA expression in the human prefrontal cortex during fetal and postnatal life.
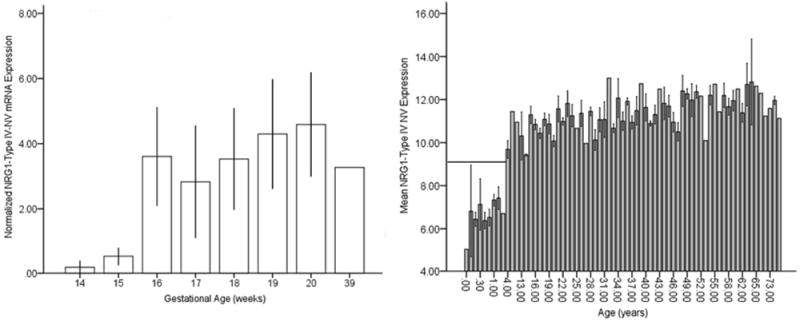
Quantitative PCR analysis of NRG1-IVNV mRNA expression in human fetal prefrontal cortex from gestational ages 14-39 weeks (Gestational age (weeks) 14, n=4; 15, n=3; 16, n=2; 17, n=5; 18, n=12; 19, n=12; 20, n=2; 39, n=1) (Panel A), and dorsolateral prefrontal cortex from postnatal samples ranging in age from 0-83 years (n=195) (Panel B). In Panel A data represent quantitative mean mRNA normalized to the geometric mean of three housekeeping genes (β-actin, GAPDH, PBGD), in Panel B data represent normalized ΔCT values. In Panel B, the bar graph is inverse to the relative amounts of NRG1-IVNV expression; higher bars indicate lower expression since the figure uses Ct values. Samples with Ct values greater than 31 were denoted as having undetectable levels and not included in the regression analysis. Absence of detectable NRG1-IVNV expression is observed after 3 years of age and denoted by the black bar.
Generation of NRG1-IV splice variant specific recombinant DNA constructs
Human NRG1-I-β1, full-length NRG1-IV (NRG1-IV-β1a), Genbank accession (EF372273) and NRG1-IVNV (accession EF372275) N-terminal c-Myc tagged DNA constructs were generated as described in detail in Supporting Text. NRG1-I-β1 was generated as a positive control for comparison with full length NRG1-IV-β1a, given that both isoforms are single pass NRG1 transmembrane pro-proteins and have previously been shown to have similar subcellular distributions (31).
Analysis of membrane targeting and proteolytic processing of human NRG1-IV isoforms in heterologous cells
Proteolytic processing of transmembrane neuregulins (i.e. NRG1-I-β1) and subsequent release of the extracellular epidermal growth factor domain is regulated by activation of protein kinase c (PKC) with the phorbol ester, phorbol 12-myristate 13-acetate (PMA) (36, 37). The susceptibility of NRG1–IV variants to stimulated proteolytic processing was studied in HEK293 cells transiently transfected with NRG1-IV-β1a or NRG1-IVNV as described in Supporting Text.
Subcellular localization of NRG1- IV and NRG1-IVNV in rat primary neurons and HEK293 cells
Rat hippocampal neurons and human embryonic kidney (HEK293) cells were transiently transfected with c-Myc tagged DNA constructs encoding NRG1-I-β1, NRG1-IV-β1a or NRG1-IVNV and immunostained using an antibody to human c-Myc (Calbiochem), as described in Supporting Text. Qualitative comparison of the subcellular localization of NRG1 variants was determined in neurons and heterologous cells using confocal microscopy.
Statistical analyses
Statistical analyses were performed using IBM Statistic SPSS, version 21. The relationship between expression levels of NRG1 types and demographic variables including age, postmortem interval, pH and RNA Integrity Number were tested using Spearman’s test of correlation, separately in fetal and postnatal study cohorts. Effects of sex and race on NRG1 isoform expression were examined using ANCOVA controlling for age. Effects of genotype at rs6994992 on NRG1-IVNV expression levels were explored using non-parametric Kruskal Wallis with genotype as an independent factor. To explore potential confounds on rs6994992 association with NRG1-IVNV expression, post hoc ANOVAs were conducted in the 3 genotypic groups separately to assess effects of age, sex (and race where warranted).
Results
Developmental expression profiles of NRG1 Types I-IV and NRG1-IVNV isoforms in the human fetal prefrontal cortex
Developmental profiling of transcripts encoding NRG1 isoforms I-IV in the prefrontal cortex during human neocortical development (gestational age weeks 14-39) revealed that NRG1 Types I-IV are tightly regulated and somewhat distinct. NRG1-I mRNA expression was highest at the beginning of the second trimester and subsequently decreased with gestational age (r=-0.49, p=0.01, n=41). In contrast, NRG1-III exhibited an opposite trajectory, being lowest at the beginning of the second trimester and significantly increasing with gestational age (r=0.61, p=<0.0001, n=41) (Figure 1A, C). Expression of NRG1-II and NRG1-IV showed no correlation with gestational age (r=-0.25, p=0.12, n=41; r=0.063, p=0.70, n=41, respectively) (Figure 1B, D). We note that the observed lack of developmentally regulated changes in NRG1-IV expression is potentially confounded in fetal development because the primer and probe used spanning the E187 and Ig exons will additionally detect NRG1-IVNV. To wit, targeted amplification of NRG1-IVNV isoform expression during fetal prefrontal cortex development revealed that this novel class of NRG1-IV isoforms, increase significantly with age during the second trimester (r=0.63, p=<0.00001, n=41) (Figure 2A), with the highest levels observed after 15 weeks gestation. No correlations were observed between RNA Integrity Number and NRG1 isoform expression (range; r= 0.15 to-0.15; p=0.24-0.92); postmortem interval and NRG1 isoform expression (range; r= 0.05-0.14; p=0.69-0.97) or RNA Integrity Number and gestational age(r=-0.13; p=0.93). No effects of sex were observed (p>0.2). Ct (cycle threshold) ranges for each isoform are reported in supplementary results.
Figure 1. Temporal dynamics of NRG1 type I-IV mRNA expression in the human prefrontal cortex during fetal brain development.
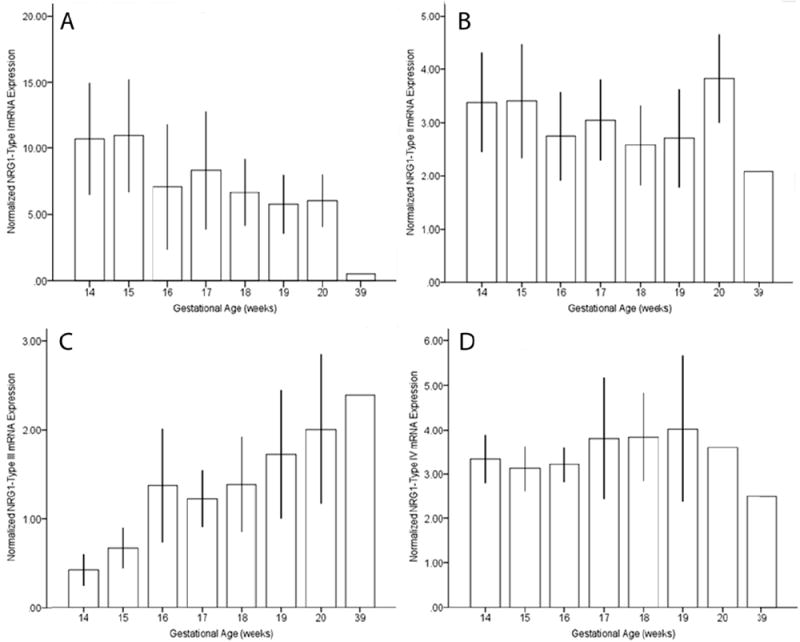
Quantitative PCR analysis of NRG1 type I (Panel A), type II (Panel B), type III (Panel C) and type IV (Panel D) mRNA expression in human fetal prefrontal cortex from gestational ages 14-39 weeks. (Gestational age (weeks) 14, n=4; 15, n=3; 16, n=2; 17, n=5; 18, n=12; 19, n=12; 20, n=2; 39, n=1). Data represent mean and standard deviation. Data are normalized to the geometric mean of three housekeeping genes (β-actin, GAPDH, PBGD).
Temporal dynamics of NRG1 isoform expression in human prefrontal cortex throughout the postnatal lifespan and aging
Analysis of quantitative expression trajectories of NRG1-I, II and III across postnatal development revealed significant inverse correlations of age and expression of NRG1-II and NRG1-III, whereby expression was highest at birth and significantly decreased with age (Type II, r=-0.26, p<0.001 and Type III, r=-0.47, p<0.000001; Figure 3B, C). Expression of NRG1-I, in contrast to early fetal development, was essentially stable across postnatal development and aging, showing no correlation with age (Figure 3A). Previous investigations indicate that NRG1-IVNV isoforms may be exclusively transcribed in the fetal brain, as informed from absence of expression in library screens of adult human brain (19). Remarkably, postnatal expression of NRG1-IVNV was detected at birth and in early infancy and decreased dramatically with age, becoming undetectable after 3 years (cut off CT>31; r=0.64, p=0.001; Figure 2B). There were no significant effects of race or sex on any NRG1 isoform mRNA expression (p>0.2) and no correlations of pH or postmortem interval with individual NRG1 isoform expression (p>0.1). RNA Integrity Number correlated weakly, but significantly with individual NRG1 isoform expression (range r=0.17-0.23; p<0.01). However, RNA Integrity Number did not correlate with age (r=-0.33, p-0.68) and controlling for RNA Integrity Number in a partial correlational analysis confirmed significant effects of age on Type II (r=-0.34, p<0.001) and Type III (r=-0.53, p<0.000001) NRG1 expression. A secondary post-hoc analysis using ANOVA examining the effects of age, controlling for sex, race, pH, PMI and RNA Integrity number on NRG1 types I-III expression across postnatal development, confirmed significant effects of age on NRG1 types II and III (see supplemental results). There were no significant correlations of age with individual housekeeping gene expression (GAPDH, β-Actin or PBGD), or with the geometric mean (r=-0.079-0.099, p=>0.18). Ct (cycle threshold) ranges for each isoform are reported in supplementary results.
Figure 3. Temporal dynamics of NRG1 isoform expression during postnatal human brain development and aging.
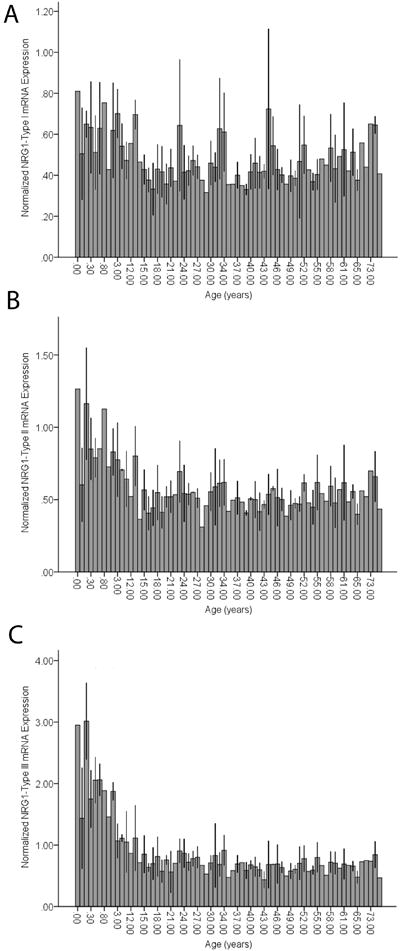
Quantification of human NRG1 Type I (Panel A), II (Panel B) and III (Panel C) isoform expression by real-time PCR in human dorsolateral prefrontal cortex across the postnatal lifespan, ranging in age from 0-83 years (n=189, Note. RNA from 6 samples used in Fig.2 panel B were not available for study). Data represent quantitative mean and standard deviation mRNA expression normalized to the geometric mean of three housekeeping genes (β-actin, GAPDH, PBGD).
Effect of rs6994992 genotype on expression of NRG1-IVNV mRNA in human prefrontal cortex
Given the previous association of rs6994992 with full length NRG1-IV mRNA expression in adult human brain (20, 21), we investigated for association with expression of the novel NRG1-IVNV splice isoform in the developing brain. The effect of rs6994992 genotype on NRG1-IVNV mRNA expression was examined in the prefrontal cortex of fetal samples from gestational age weeks 16-39 (n=34) and postnatal samples ranging from birth to 3 years of age (n=23), when NRG1-IVNV expression was apparent (Figure 2A, B). rs6994992 genotype was significantly associated with NRG1-IVNV expression in fetal prefrontal cortex, with trend significance in the same allelic directionality in early postnatal life (main effect of genotype, H(2)=7.17, p=0.028; H(2) = 4.78, p=0.08, respectively). Individuals homozygous for the risk allele (T) exhibited lower cortical levels of NRG1-IVNV mRNA compared to individuals carrying the non-risk allele C (Figure 4A, B). Post-hoc ANOVA exploring the impact of age and sex on NRG1-IVNV expression separately in the 3 genotypic groups revealed no significant effects of either factor in either the fetal (sex, p>0.12; age, p>0.29) or postnatal cohorts (sex, p>0.19; age, p>0.2). No significant effects of race (p>0.07) were observed on NRG1-IVNV expression across genotype.
Figure 4. Association between rs6994992 genotype and NRG1-IVNV variant expression.
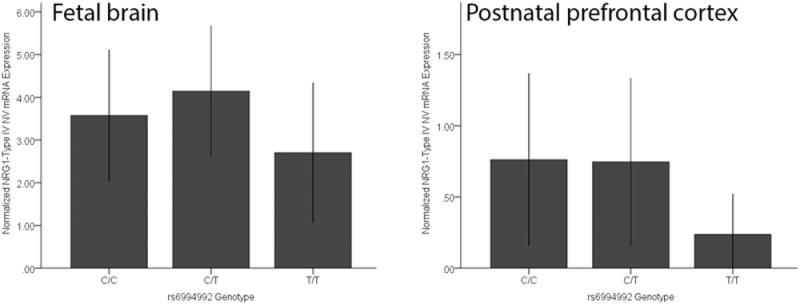
Effect of rs6994992 genotype on NRG1-IVNV expression in fetal brain (Genotype C/C, n=8; C/T, n=20; T/T, n=6) is shown in Panel A and in postnatal prefrontal cortex (ages 0-3 years), Panel B (Genotype C/C, n=7; C/T, n=14; T/T, n=2). Subjects homozygous for the risk allele at rs6994992 (T) in both age groups have the lowest level of NRG1-IVNV mRNA expression. Data represent the mean and standard deviation NRG1-IVNV mRNA expression. Panel B shows normalized expression relative to CC genotype (fold change=2-ΔΔCT).
Subcellular localization of NRG1- IV and NRG1-IVNV isoforms in rat hippocampal neurons in-vitro
NRG1-IVNV isoforms represent a novel class of NRG1 isoforms that are predicted to give rise to truncated NRG1 proteins that lack a functional transmembrane domain (19). However, direct experimental evidence for this is lacking. We used confocal microscopy to qualitatively compare the subcellular distribution of 5’ c-Myc tagged human DNA constructs encoding NRG1-I-β1, NRG1-IV-β1a or NRG1-IVNV in transiently transfected hippocampal neurons. Immunofluorescence revealed that NRG1-IV-β1a and NRG1-I-β1 proteins are targeted to the cell membrane and distributed in a punctate manner throughout the cell soma and proximal dendrites (Figure 5A, B). In contrast, NRG1-IVNV was more diffusely present throughout the cell body and dendrites and highly localized to the nucleus (Figure 5C). Immunofluorescent staining confirmed enrichment of NRG1-IVNV within the cell nucleus of transiently transfected HEK293 cells, as observed by a high degree of co-localization with the nuclear marker DAPI (Figure S2).
Figure 5. Subcellular localization of NRG1-IV-β1a and NRG1-IVNV in rat primary hippocampal neurons.
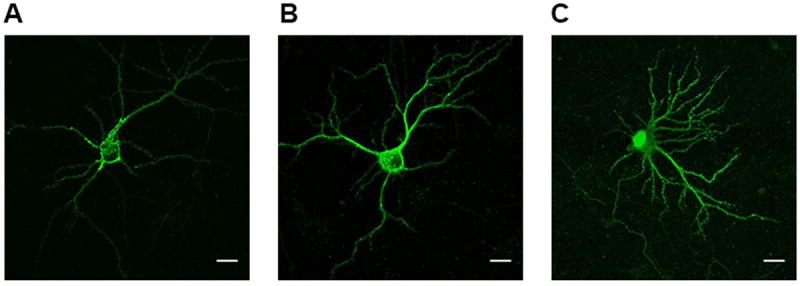
Rat hippocampal neurons expressing N-terminal c-Myc tagged human NRG1-I-β1 (Panel A), NRG1-IV-β1a (Panel B), or NRG1-IVNV (Panel C) were stained with a c-Myc antibody to determine subcellular protein distribution. NRG1-I-β1 and NRG1-IV-β1a proteins are expressed predominantly at the cell membrane and dendritic processes; whereas NRG1-IVNV is highly localized to the nucleus. Scale bar represents 20μM.
Effect of proteolytic processing mediated via phorbol ester (PMA) stimulation on cell surface protein expression of NRG1-IV and NRG1-IVNV
HEK293 cells transiently expressing NRG1-IV-β1a or NRG-IVNV were established. Western blot analysis of whole cell protein extracts revealed an immunoreactive band of approximately 66kDa for NRG1-IV-β1a, consistent with the predicted molecular weight of NRG1-IV-β1a (19) (Figure S3A, lane 3). Lysates from cells transfected with NRG1-IVNV confirmed translation of truncated protein products of the predicted size (approximately 29kDa- 31kDa, respectively; Figure S3A, lanes 4 and 5), in line with the fact that these NRG1-IV variants contain a premature 3’ stop codon (19). No immunoreactive bands were present from lysates of untransfected HEK cells or cells transfected with an empty vector (Figure S3A, lanes 1 and 2). NRG1-IVNV isoforms, unlike the NRG1-IV pro-protein, lack a matrix metalloproteinase domain which is necessary for proteolytic processing of the extracellular domain (19). To experimentally assess the proteolytic processing susceptibility of NRG1-IV transcripts, HEK293 cells were transiently transfected with either 5’c-Myc tagged NRG1-IV-β1a or NRG1-IVNV (type IV fetal variants E and F; (19)), stimulated with PMA and the conditioned media collected. PMA stimulation induced release of a 30kDa extracellular NRG1 fragment in cells transfected with NRG1-IV-β1a (Figure S3B, lane 7). In contrast, no immunoreactive bands were observed in media collected from PMA treated cells transfected with NRG1-IVNV (Figure S3B, lanes 7 and 8). Vehicle treated cells also showed no immunoreactive band, confirming PMA stimulation as the mechanism of extracellular fragment release (Figure S3B, lane 5).
Discussion
The trajectory of normal brain development occurs in multiple stages that span the embryonic, neonatal and early postnatal periods into adolescence and adulthood (38, 39), with each critical window of development being genetically determined, epigenetically regulated and environmentally influenced. Research has generated many putative psychiatric risk genes some of which fall into critical neurodevelopmental pathways (4-7, 27-30) but the transcriptional mapping of the normal developmental expression trajectories of these genes across human brain development, and the mechanisms by which risk genetic variants could alter these courses largely remains unclear.
This is the first study, to our knowledge, to describe the individual temporal dynamics of NRG1 isoform (i.e. types I-IV and NRG1-IVNV) expression during human fetal and postnatal neocortical development and reveals potential novel mechanisms of the role of NRG1 in early cortical development. Furthermore, our study characterizes a novel class of NRG1-IV variants, NRG1-IVNV, which in human brain are developmentally and genetically regulated, expressed exclusively during critical early developmental periods and represent a biologically distinct subclass of NRG1 proteins, with relevance to risk for schizophrenia.
Although we have refrained from inferring direct comparative abundance of the individual NRG1 isoforms at different stages of development, primarily because even small differences in PCR efficiencies across targets can distort relative expression measurements (40), we note that the Ct values for NRG1-IVNV in the fetal brain (post 15 weeks) are in the range of 25-27. Cts <29 represent strong positive reactions indicative of abundant target nucleic acid, suggesting that NRG1-IVNV represent relatively abundant isoforms of NRG1, with key roles during early neurodevelopment. Further studies are needed to determine the regional and cell-type specific expression of NRG1-IVNV during human brain development.
Regarding NRG1 isoforms, type I, II, III expression during human brain development we found that NRG1-I expression peaks during the early second trimester of brain development and decreases with fetal gestational age. NRG1-1 was stable from birth throughout the postnatal lifespan. In contrast, NRG1-II is stable throughout fetal development and decreases after birth to reach stable levels in early adolescence across aging, whereas NRG1-III increases dramatically during the second trimester, is expressed prominently at birth and declines dramatically in the periadolescent period to become stable across the lifespan. Overall our observations are broadly consistent with expression patterns observed in developing rat cerebral cortex, whereby NRG1-II and III were shown to be highest in the early neonatal period (P5) declining by P15, and becoming stable levels across development (4), and those seen in mouse (41).
NRG1 has a diverse range of functions during neurodevelopment (1, 3) and our findings provide further support that NRG1 isoforms, in particular NRG1-I and NRG1-III, may play distinct roles in human cortical development and maturation, including neuronal migration, synaptogenesis, gliogenesis, myelination and the development of glutamatergic, GABAergic and cholinergic neurotransmission (42, 43). For example, the steep trajectory of increase in NRG1-III expression in the second/third trimester and the decreasing levels after adolescence is potentially consistent with the onset of myelination, which in humans occurs around week 28 and continues into adolescence (39). Interestingly, the temporal dynamics of NRG1-IVNV expression was similar to that of NRG1-III, with the exception that NRG1-IVNV was undetectable after three years of age, a postnatal period critical to the elaboration of neuronal processes and synaptogenesis (39).
rs6994992 risk genotype predicts elevated expression of full length NRG1- IV in the adult brain (20, 21). Here we explored the genetic association of developmentally regulated NRG1-IV splice isoforms, i.e. NRG1-IVNV by rs6994992. Unexpectedly, we observed that the schizophrenia-risk genotype is associated with lower expression of NRG1-IVNV during the fetal and early neonatal critical period. The mechanisms behind the differential regulation of NRG1-IV isoform expression as it relates to the rs6994992 locus, remain unknown, but may involve interaction of the cis effects of rs6994992 with epigenetic regulation of splicing of the NRG1-IV pre-mRNA, a mechanism which plays key roles in regulation of splicing during early development (44). The dichotomous expression association suggests that a balance between NRG1-IVNV expression and full length NRG1- IV expression is required throughout neocortical development. This suggestion is consistent with a potential molecular mechanism underlying the association of rs6994992 with cortical and subcortical structure in the human neonate (27). While initial data suggests that rs6994992 is a functional polymorphism (19), we also note that rs6994992 may be in linkage disequilibrium with an unknown causal variant in the NRG1 promoter, which modulates expression of NRG1-IV and NRG1-IVNV and risk for schizophrenia. Finally, in demonstrating a transcriptional association of a schizophrenia susceptibility variant during early fetal brain development, we identify a potential molecular mechanism of early developmental risk at the rs6994992 NRG1 locus and provide support for schizophrenia as a disorder of neurodevelopment (45).
Proteolytic cleavage of the extracellular domain of neuregulins allows paracrine activation of ErbB receptors (1) and we provide evidence consistent with this for NRG1-IV. Remarkably, NRG1-IVNV isoforms are insensitive to PMA-induced cleavage, as predicted from their structure (19). The differential localization and resistance to proteolytic cleavage of NRG1-IVNV, suggests these variants have a unique function within the cell compared to NRG1-β1 isoforms. Like NRG1-IVNV, other truncated C-terminal NRG1 isoforms also display resistance to proteolytic processing and are confined to the nucleus and intracellular organelles (46-49). Intriguingly, some isoforms of NRG1 have been shown to translocate to the nucleus where they act as transcriptional regulators, altering the activity and expression of target genes (50, 51). Given its biochemical structure and nuclear enrichment, we propose that NRG1-IVNV may act as a transcription factor, rather than a bioactive NRG1. In keeping with the hypothesis, alternative splicing of β and stalk exons in NRG1-IVNV transcripts alters the amino acid sequence so that only 4 of the 6 cysteine epidermal growth factor residues are present and thus epidermal growth factor bioactivity is predicted to be lost (19).
Taken together, our observations provide novel insights into the transcriptional and genetic regulation of NRG1’s during normal human brain development, and identify NRG1-IVNV as a novel, diverse class of developmentally regulated growth factors associated with schizophrenia susceptibility. Future studies are required to determine the neurobiological role of NRG1-IV and NRG1-IVNV and mechanistically how they relate to the development of schizophrenia.
Supplementary Material
Exonic organization of NRG1-IV and NRG1-IVNV transcripts (fetal variants E and F) is shown. The exon nomenclature is stated in the top row, with the corresponding functional domain specified underneath. The location of transcript specific real-time PCR forward and reverse primers are depicted by right or left pointing arrows, respectively. The location of the exon spanning, transcript specific, internal TaqMan® probe is denoted by solid red lines (the adjoining dashed red line depicts the intronic sequence not detected). Note that the assay design is identical for Fetal E and F transcripts; therefore, this assay will detect expression levels of both variants herein collectively named NRG1-IVNV. Exons and primer or probe lengths are not drawn to scale. Abbreviations: Ig, Immunoglobulin; s, spacer; EGFc, epidermal growth factor-like domain; TMc, transmembrane domain.
HEK293 cells transiently expressing N-terminal c-Myc tagged human NRG1-IV-β1a (Panels A-C) or NRG1-IVNV constructs (Panels D-F) were stained with a c-Myc antibody to determine subcellular protein distribution (Panels A, D) and mounted onto slides with medium containing a stain to detect the nuclear marker DAPI (Panels B, E). Merged images of both cellular markers show NRG1-IV-β1a proteins are expressed predominantly in a cell membrane bound pattern with little overlap with DAPI (Panel C), whereas NRG1-IVNV expression is more diffuse with a high level of co-localization with DAPI expression, confirming nuclear enrichment of expression (Panel F). Scale bar represents 20μM.
HEK293 cells transiently expressing N-terminus c-Myc tagged human NRG1-IV-β1a or NRG1-IVNV (fetal variants E and F) constructs were assessed for protein translation from whole cell lysate samples (Panel A) and proteolytic processing of the extracellular domain from conditioned media samples (Panel B). Expression of NRG1-IV-β1a (IV) resulted in a protein of 66kDa (lanes 3 and 7, Panel A), whereas NRG1-IVNV E (FV E) and F (FV F) resulted in protein products of 29-31kDa (lanes 4-5 and 8-9, Panel A). Treatment of HEK293 cells expressing NRG1-IV-β1a with PMA (+) induced the release of a 30kDa fragment in the conditioned media (lane 6, Panel B); whereas no protein products were identified in the conditioned media of PMA treated HEK293 cells expressing NRG1-IVNV (lanes 7 and 8, Panel B). Untransfected HEK cells (HEK) and empty vectors (CON) were used as negative controls.
Acknowledgments
This work was funded by a NARSAD Young Investigator Award held by Dr. Amanda Law, and funds from the NIMH Intramural Research Program.
The authors are grateful to Dr. Wei Tan, for construction of the human cDNA constructs, Dr. Tian Zhang Ye for computational support and Dr. Daniel Weinberger for research support.
Footnotes
Drs. Paterson, Wang, Kleinman and Law report no competing interests.
References
- 1.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 2.Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Bates R, Yin DM, Shen C, Wang F, Su N, Kirov SA, Luo Y, Wang JZ, Xiong WC, Mei L. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci. 2011;31:8491–8501. doi: 10.1523/JNEUROSCI.5317-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deakin IH, Law AJ, Oliver PL, Schwab MH, Nave KA, Harrison PJ, Bannerman DM. Behavioural characterization of neuregulin 1 type I overexpressing transgenic mice. Neuroreport. 2009;20:1523–1528. doi: 10.1097/WNR.0b013e328330f6e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen L, Lu YS, Zhu XH, Li XM, Woo RS, Chen YJ, Yin DM, Lai C, Terry AV, Jr, Vazdarjanova A, Xiong WC, Mei L. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1211–1216. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fazzari P, Paternain AV, Valiente M, Pla R, Lujan R, Lloyd K, Lerma J, Marin O, Rico B. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- 8.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, Ingason A, Gulcher JR, Stefansson K, St Clair D. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corvin AP, Morris DW, McGhee K, Schwaiger S, Scully P, Quinn J, Meagher D, Clair DS, Waddington JL, Gill M. Confirmation and refinement of an ‘at-risk’ haplotype for schizophrenia suggests the EST cluster, Hs.97362, as a potential susceptibility gene at the Neuregulin-1 locus. Mol Psychiatry. 2004;9:208–213. doi: 10.1038/sj.mp.4001412. [DOI] [PubMed] [Google Scholar]
- 11.Kukshal P, Bhatia T, Bhagwat AM, Gur RE, Gur RC, Deshpande SN, Nimgaonkar VL, Thelma BK. Association study of neuregulin-1 gene polymorphisms in a North Indian schizophrenia sample. Schizophr Res. 2013;144:24–30. doi: 10.1016/j.schres.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson PA, Christoforou A, Morris SW, Adie E, Pickard BS, Porteous DJ, Muir WJ, Blackwood DH, Evans KL. Association of Neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population. Mol Psychiatry. 2007;12:94–104. doi: 10.1038/sj.mp.4001889. [DOI] [PubMed] [Google Scholar]
- 13.Walker RM, Christoforou A, Thomson PA, McGhee KA, Maclean A, Muhleisen TW, Strohmaier J, Nieratschker V, Nothen MM, Rietschel M, Cichon S, Morris SW, Jilani O, Stclair D, Blackwood DH, Muir WJ, Porteous DJ, Evans KL. Association analysis of Neuregulin 1 candidate regions in schizophrenia and bipolar disorder. Neurosci Lett. 2010;478:9–13. doi: 10.1016/j.neulet.2010.04.056. [DOI] [PubMed] [Google Scholar]
- 14.Stefansson H, Thorgeirsson TE, Gulcher JR, Stefansson K. Neuregulin 1 in schizophrenia: out of Iceland. Mol Psych. 2003;8:639–640. doi: 10.1038/sj.mp.4001384. [DOI] [PubMed] [Google Scholar]
- 15.Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol Psych. 2006;60:132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psych. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 18.Agim ZS, Esendal M, Briollais L, Uyan O, Meschian M, Martinez LA, Ding Y, Basak AN, Ozcelik H. Discovery, validation and characterization of Erbb4 and Nrg1 haplotypes using data from three genome-wide association studies of schizophrenia. PloS One. 2013;8:e53042. doi: 10.1371/journal.pone.0053042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan W, Wang Y, Gold B, Chen J, Dean M, Harrison PJ, Weinberger DR, Law AJ. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007;282:24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- 20.Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5’ SNPs associated with the disease. Proc Nat Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon E, Rollins B, Mesen A, Sequeira A, Myers RM, Akil H, Watson SJ, Barchas J, Jones EG, Schatzberg A, Bunney WE, DeLisi LE, Byerley W, Vawter MP. Lack of association to a NRG1 missense polymorphism in schizophrenia or bipolar disorder in a Costa Rican population. Schizophr Res. 2011;131:52–57. doi: 10.1016/j.schres.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, Thomson PA, Porteous DJ, Cunningham-Owens DG, Johnstone EC, Lawrie SM. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 23.Stefanis NC, Trikalinos TA, Avramopoulos D, Smyrnis N, Evdokimidis I, Ntzani EE, Ioannidis JP, Stefanis CN. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 24.McIntosh AM, Moorhead TW, Job D, Lymer GK, Munoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D, Evans KL, Johnstone EC, Lawrie SM, Hall J. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- 25.Mata I, Perez-Iglesias R, Roiz-Santianez R, Tordesillas-Gutierrez D, Gonzalez-Mandly A, Berja A, Vazquez-Barquero JL, Crespo-Facorro B. Additive effect of NRG1 and DISC1 genes on lateral ventricle enlargement in first episode schizophrenia. Neuroimage. 2010;53:1016–1022. doi: 10.1016/j.neuroimage.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Roussos P, Giakoumaki SG, Adamaki E, Bitsios P. The influence of schizophrenia-related neuregulin-1 polymorphisms on sensorimotor gating in healthy males. Biol Psychiatry. 2011;69:479–486. doi: 10.1016/j.biopsych.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Knickmeyer RC, Wang J, Zhu H, Geng X, Woolson S, Hamer RM, Konneker T, Lin W, Styner M, Gilmore JH. Common Variants in Psychiatric Risk Genes Predict Brain Structure at Birth. Cereb Cortex. 2013 doi: 10.1093/cercor/bhs401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinman JE, Law AJ, Lipska BK, Hyde TM, Ellis JK, Harrison PJ, Weinberger DR. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry. 2011;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Group PS, Nimgaonkar VL, Go RC, Savage RM, Swerdlow NR, Gur RE, Braff DL, King MC, McClellan JM Consortium on the Genetics of S, Group PS. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamir A, Buonanno A. Molecular and cellular characterization of Neuregulin-1 type IV isoforms. J Neurochem. 2010;113:1163–1176. doi: 10.1111/j.1471-4159.2010.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, Law AJ. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Nat Acad Sci USA. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozaki M, Itoh K, Miyakawa Y, Kishida H, Hashikawa T. Protein processing and releases of neuregulin-1 are regulated in an activity-dependent manner. Journal of neurochemistry. 2004;91:176–188. doi: 10.1111/j.1471-4159.2004.02719.x. [DOI] [PubMed] [Google Scholar]
- 37.Esper RM, Loeb JA. Neurotrophins induce neuregulin release through protein kinase Cdelta activation. J Biol Chem. 2009;284:26251–26260. doi: 10.1074/jbc.M109.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neurosci Biobehav Rev. 2003;27:3–18. doi: 10.1016/s0149-7634(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 39.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brookman-Amissah N, Packer H, Prediger E, Sabel J. qPCR application guide. Experimental overview, protocol, troubleshooting. 3. Integrated DNA technologies and Biogazalle NV; [Google Scholar]
- 41.Meyer D, Yamaai T, Garratt A, Riethmacher-Sonnenberg E, Kane D, Theill LE, Birchmeier C. Isoform-specific expression and function of neuregulin. Development. 1997;124:3575–3586. doi: 10.1242/dev.124.18.3575. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Bendito G, Cautinat A, Sanchez JA, Bielle F, Flames N, Garratt AN, Talmage DA, Role LW, Charnay P, Marin O, Garel S. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, Emmetsberger J, Talmage DA, Role LW. Type III neuregulin 1 is required for multiple forms of excitatory synaptic plasticity of mouse cortico-amygdala circuits. J Neurosci. 2013;33:9655–9666. doi: 10.1523/JNEUROSCI.2888-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 46.Holmes WE, Sliwkowski MX, Akita RW, Henzel WJ, Lee J, Park JW, Yansura D, Abadi N, Raab H, Lewis GD, et al. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256:1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- 47.Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, Janssen AM, Ben-Baruch N, Trollinger DB, Jacobsen VL, et al. Structural and functional aspects of the multiplicity of Neu differentiation factors. Mol Cell Biol. 1994;14:1909–1919. doi: 10.1128/mcb.14.3.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Prentiss L, Heitzman D, Stahl RC, DiPino F, Jr, Carey DJ. Neuregulin isoforms in dorsal root ganglion neurons: effects of the cytoplasmic domain on localization and membrane shedding of Nrg-1 type I. J Neurosci Res. 2006;84:1–12. doi: 10.1002/jnr.20861. [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Hwang H, Cao L, Wen D, Liu N, Graham RM, Zhou M. Release of the neuregulin functional polypeptide requires its cytoplasmic tail. J Biol Chem. 1998;273:34335–34340. doi: 10.1074/jbc.273.51.34335. [DOI] [PubMed] [Google Scholar]
- 50.Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golding M, Ruhrberg C, Sandle J, Gullick WJ. Mapping nucleolar and spliceosome localization sequences of neuregulin1-beta3. Exp Cell Res. 2004;299:110–118. doi: 10.1016/j.yexcr.2004.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exonic organization of NRG1-IV and NRG1-IVNV transcripts (fetal variants E and F) is shown. The exon nomenclature is stated in the top row, with the corresponding functional domain specified underneath. The location of transcript specific real-time PCR forward and reverse primers are depicted by right or left pointing arrows, respectively. The location of the exon spanning, transcript specific, internal TaqMan® probe is denoted by solid red lines (the adjoining dashed red line depicts the intronic sequence not detected). Note that the assay design is identical for Fetal E and F transcripts; therefore, this assay will detect expression levels of both variants herein collectively named NRG1-IVNV. Exons and primer or probe lengths are not drawn to scale. Abbreviations: Ig, Immunoglobulin; s, spacer; EGFc, epidermal growth factor-like domain; TMc, transmembrane domain.
HEK293 cells transiently expressing N-terminal c-Myc tagged human NRG1-IV-β1a (Panels A-C) or NRG1-IVNV constructs (Panels D-F) were stained with a c-Myc antibody to determine subcellular protein distribution (Panels A, D) and mounted onto slides with medium containing a stain to detect the nuclear marker DAPI (Panels B, E). Merged images of both cellular markers show NRG1-IV-β1a proteins are expressed predominantly in a cell membrane bound pattern with little overlap with DAPI (Panel C), whereas NRG1-IVNV expression is more diffuse with a high level of co-localization with DAPI expression, confirming nuclear enrichment of expression (Panel F). Scale bar represents 20μM.
HEK293 cells transiently expressing N-terminus c-Myc tagged human NRG1-IV-β1a or NRG1-IVNV (fetal variants E and F) constructs were assessed for protein translation from whole cell lysate samples (Panel A) and proteolytic processing of the extracellular domain from conditioned media samples (Panel B). Expression of NRG1-IV-β1a (IV) resulted in a protein of 66kDa (lanes 3 and 7, Panel A), whereas NRG1-IVNV E (FV E) and F (FV F) resulted in protein products of 29-31kDa (lanes 4-5 and 8-9, Panel A). Treatment of HEK293 cells expressing NRG1-IV-β1a with PMA (+) induced the release of a 30kDa fragment in the conditioned media (lane 6, Panel B); whereas no protein products were identified in the conditioned media of PMA treated HEK293 cells expressing NRG1-IVNV (lanes 7 and 8, Panel B). Untransfected HEK cells (HEK) and empty vectors (CON) were used as negative controls.


