Abstract
Circulating tumor cells (CTC) harvested from peripheral blood have received significant interest as sources for serial sampling to gauge treatment efficacy. Nanotechnology and microfluidic based approaches are emerging to facilitate such analyses. While of considerable clinical importance, there is little information on how similar or different CTC are from their shedding bulk tumors. In this clinical study, paired tumor fine needle aspirate and peripheral blood samples were obtained from cancer patients during image guided biopsy. Using targeted magnetic nanoparticles and a point-of-care micro-NMR system, we compared selected biomarkers (EpCAM, EGFR, HER-2 and vimentin) in both CTC and fine needle biopsies of solid epithelial cancers. We show a weak correlation between each paired sample, suggesting that use of CTC as ‘liquid biopsies’ and proxies to metastatic solid lesions could be misleading.
Keywords: Circulating tumor cells, magnetic nanoparticles, micro-NMR (µNMR), point of care diagnostics
Introduction
Circulating tumor cells (CTC) have been examined as predictors of clinical trajectory and as sources of easily accessible cancer material for subsequent testing1–6. Validation studies of various CTC detection technologies are ongoing to determine the true prognostic value of CTC counts at diagnosis and during therapy. Enumeration alone, however is insufficient to tap into the rich potential of CTC as sources for genomic and proteomic analyses7. With the advent of targeted drugs and individualized treatments, repeated molecular profiling becomes essential8. As the need for rapid tumor profiling increases, it is becoming clear that integrating easily accessible tissues, such as blood, with robust, operator-independent analyses would serve the purpose of current research platforms. Yet significant unknowns remain including i) how similar or divergent are CTC to their respective bulk tumors at the proteomic level, ii) are key cancer related proteins sufficiently elevated to harvest enough CTC in the majority of patients and iii) could CTC be used to unequivocally obviate the need for biopsies during serial testing?
The FDA-approved CTC assay, CellSearch, relies heavily on positive selection for the epithelial surface marker, EpCAM and differentiation from leukocytes via absence of CD459. EpCAM (CD326, tumor associated calcium signal transducer-1) is a pan-epithelial differentiation antigen and believed to be present in most primary epithelial cancers9. More recently, it has been shown that ~30% of epithelial cancers have low EpCAM levels10. In addition, EpCAM may be further down-regulated in cells undergoing epithelial-mesenchymal transition (EMT) during increased cell proliferation11,12. Hence, EpCAM based methods could underestimate true CTC numbers. Techniques that extend beyond a single marker for CTC enumeration are under active investigation and are generally believed to be more accurate13. Various platforms for CTC investigation have been developed, based on mass spectrometry14, fluorescence microscopy15, microfluidic sorting16, surface plasmon resonance17, electrical impedance18, and field-effect gating19. Drawbacks of these approaches include the need for extensive sample processing thus leading to cell loss and biomarker decay20.
The goal of this study was to compare key biomarker levels in CTC and cancer cells procured through image-guided fine needle biopsies. Both sample types were obtained at the same visit and within 1 hour of each other. We enrolled 34 patients with advanced cancers and identified 21 patients with sufficient numbers of CTC for further analyses. Specifically, matched CTC and biopsy samples were used to compare EGFR, EpCAM, HER-2 and vimentin. We used magnetic nanoparticles and a miniaturized micro-NMR (µNMR) system for analysis of these cancer targets. Overall, we found a weak correlation between the protein markers of both sample types. From a diagnostics standpoint, this suggests that CTC and needle biopsies are not interchangeable.
Methods
µNMR measurements
The µNMR technology and magnetic nanoparticle labeling have previously been extensively tested in cell lines13,21–23, mouse models of cancer23 and validated in clinical trials10,13,24,25. In this study, sample labeling and µNMR measurements (Figure 1) were performed as previously described13,21. Briefly, the transverse relaxation rates (R2) were determined in an effective sample volume of 1µL. µNMR measurements and analysis of cellular expression were performed as previously descried13. Briefly, the following Carr-Purcell-Meiboom-Gill pulse sequences were used: echo time, 4 ms; repetition time, 6 s; number of 180° pulses per scan, 500; number of scans, 8. R2 values from samples were subtracted from those of PBS buffer to calculate ΔR2. The ΔR2 of samples were normalized to ΔR2 obtained from negative control to account for background signal. Negative controls for each sample were prepared similarly to the test samples without the antibody incubation step. For each sample, measurement was done in triplicate and the average value was obtained for analysis.
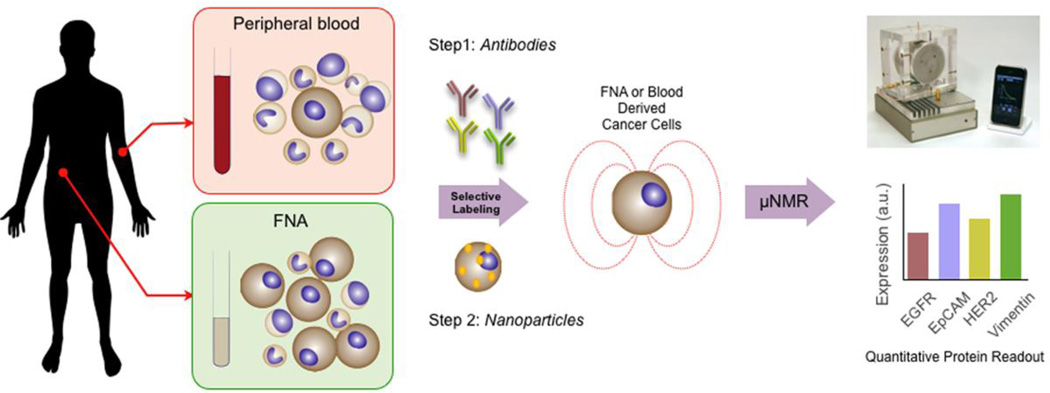
Figure 1.
Schematic of the detection and characterization study of CTC and metastatic tumor using magnetic nanoparticle labeling and quad-µNMR detection system. Peripheral blood (7 ml) and fine needle biopsy from the site of metastasis were obtained. Samples were labeled by incubating cells with primary antibodies followed by TCO-modified secondary antibodies (Step 1). Cells were then pelleted by centrifugation and resuspended directly with Tz-nanoparticles that selectively targeted TCO-antibodies (Step 2). The process of labeling and targeting required 40 minutes. Cancer marker measurements were taken using the µNMR device to produce quantitative protein readouts.
Marker Selection
CTC detection
We used a previously identified cocktail of four (quad) markers (EGFR, EpCAM, HER-2, MUC-1) for CTC detection. The combined application of these markers allows more accurate CTC counting than a single marker (EpCAM) based detection13. Profiling: Given the pilot nature of the study, we selected only four clinically and/or biologically relevant protein markers (EGFR, EpCAM, HER-2, vimentin) for the molecular profiling of paired CTC-biopsies. The rationale for this set was as follows. 1) EGFR, EpCAM, and HER-2 are abundant in tumor biopsies10 and collectively integral to a panel set with better diagnostic performance than conventional pathology. Yet, the individual heterogeneity of these markers across human specimen types is not well characterized. 2) Vimentin is a key EMT marker26 and reportedly expressed in CTC27,28.
Preparation of transcyclooctene (TCO)-modified antibodies
Monoclonal antibodies against EpCAM, MUC-1, HER-2, EGFR and vimentin were used for the primary labeling of antigens. To enhance the labeling efficiency, TCO modification of antibodies were performed on secondary IgG antibodies instead of primary antibodies, (i.e. primary antibodies were incubated with cells per manufacturer’s protocol). Specifically, secondary IgG antibodies were conjugated with (E)-cyclooct-4-enyl 2,5-dioxopyrrolidin-1-yl carbonate (TCO-NHS), as previously reported13,21,21,29. TCO-conjugation was performed with 0.5 mg of mouse or rabbit IgG antibody in the presence of 1000 equivalents of TCO-NHS in phosphate-buffered saline (PBS) and 10% dimethylformamide at room temperature for 3 hours. Unreacted TCO-NHS was subsequently washed using 2 ml of Zeba desalting columns (Thermo Fisher, Rockford, IL), and antibody concentrations were determined by absorbance measurement.
Preparation of tetrazine (Tz)-modified nanoparticles
Cross-linked iron oxide (CLIO) nanoparticles were prepared as previously described21. Briefly, Tz-modified nanoparticles were created by reacting NH2-MNPs with 500 equivalents of 2,5-dioxopyrrolidin-1-yl 5-(4-(1,2,4,5-tetrazin-3-yl)benzylamino)-5-oxopentanoate (Tz-NHS). The reaction was performed in PBS containing 5% dimethylformamide for 3 hours at room temperature, as previously described13,21,29. Excess Tz-NHS was removed by gel filtration using Sephadex G-50 (GE Healthcare, Pittsburgh, PA).
Sample processing and labeling with Tz-modified nanoparticles
Each peripheral blood sample (7 ml) was lysed and the cell pellet resuspended in buffer (100 µl of 1× PBS/1% FBS). For CTC detection, we used the quad-labeling method in which primary antibodies against EpCAM, MUC-1, HER-2, EGFR were added as a cocktail. FNA samples were fixed as described previously10 and about 100 cells were used for each labeling experiment. For molecular profiling experiments of both blood and FNA specimens, EpCAM, HER-2, EGFR and vimentin were separately added to aliquots of parent samples. Cell pellets were incubated with the above antibodies for 20 minutes. TCO-modified secondary IgG antibodies (10 µg/ml) were added to the pellet and incubated for 20 minutes. Cell pellets were then washed twice with 1 × phosphate buffered saline (PBS) and incubated with magnetic nanoparticles (100 nM Tz-CLIO) for 10 minutes. Excess Tz-CLIO was subsequently removed by washing the pellet twice with 1×PB, before being resuspended in 20 µl of 1×PBS for µNMR measurements. Labeling experiments were performed at room temperature.
Clinical subjects
Thirty-four subjects with confirmed epithelial malignancies and receiving care at the Massachusetts General Hospital (MGH) (Boston, MA) were enrolled in this institutional review board approved study. Subjects had been initially referred for clinical biopsy of a suspicious lesion under computed tomography or ultrasound guidance at the MGH Abdominal Imaging and Intervention suites. On the day of enrollment, both peripheral blood (7 mL) and fine needle biopsy from a metastatic site were collected from each subject. Of the 34 paired samples, 21 were selected for further analyses based on positive CTC results from µNMR and the presence of ample cell quantity (~150–200 cells) within the biopsy. The median age of the 21 subjects was 65 years and comprised various cancer subtypes (see Table 1). Three clinician investigators (C.M.C., S. M. and R.W.) blinded to µNMR results, reviewed each subject’s documented clinical, imaging, and pathology data. Clinical trajectory (worsening, stable, improved) was established by reviewing clinical status at enrollment and integrating any ensuing data (e.g. scans, tumor markers, treatment changes, death) within the study period. Pathology data from cases of clinical interest were examined and interpreted separately by an experienced pathologist (M.M.K). All treating providers were blinded to any data generated from this pilot study.
Table 1.
Summary of patient data
| Characteristic | Number | Percentage | CTC/mL (mean/range) |
|---|---|---|---|
| Number of patients | 21 | 8 (3–27) | |
| Age | |||
| Median | 65 | ||
| Range | 30–87 | ||
| Gender | |||
| Male | 11 | 52 | 7 (3–21) |
| Female | 10 | 48 | 8 (3–27) |
| Stage | |||
| Locally Advanced | 3 | 14.5 | 5 (4–6) |
| Metastatic | 18 | 85.5 | 8 (3–27) |
| Tumor Subtypes | |||
| Breast | 3 | 14.5 | 18 (9–27) |
| Gastrointestinal | 7 | 33 | 5 (3–7) |
| Gynecologic | 1 | 5 | 4 |
| Lung | 3 | 14.5 | 5 (3–7) |
| Pancreatobiliary | 7 | 33 | 8 (3–21) |
| Clinical Status | |||
| Response | 3 | 14.5 | 4 (3–5) |
| Progression | 8 | 38 | 10 (3–27) |
| Treatment naive | 6 | 28.5 | 8 (3–21) |
| No active therapy | 4 | 19 | 7 (3–9) |
| Treatment Status | |||
| Active therapy | 3 | 14.5 | 12 (4–27) |
| Previously treated | 5 | 23.5 | 6 (3–9) |
| Treatment Naive | 13 | 62 | 7 (3–21) |
Statistical analysis
Paired non-parametric student t tests were performed to evaluate the statistical significance between each marker’s expression values between peripheral blood and biopsy.
Results
Detection of CTC in peripheral blood
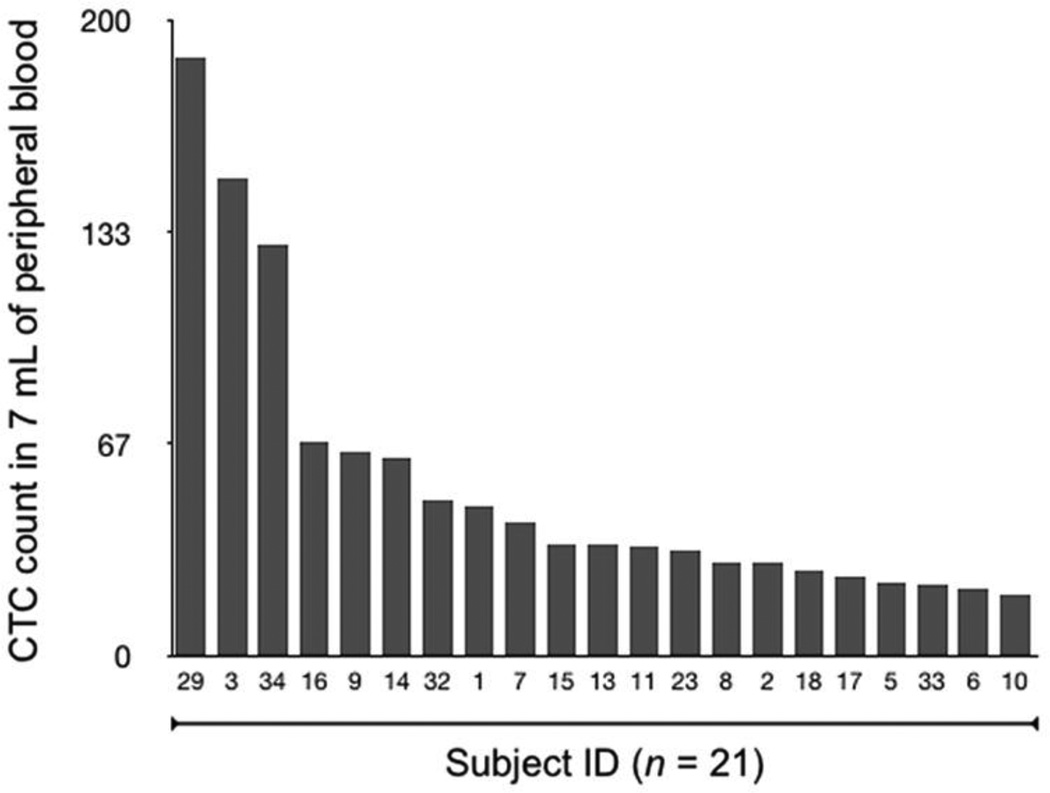
The distribution of CTC of peripheral blood obtained from the 21 subjects with matched samples ranged from 19 to 188 cells/7mL (~3–27 CTC/mL; mean 54 cells/mL and median 35 cells/mL) (Figure 2). CTC count was notably higher in subjects with established metastatic cancer (85.5%) than in subjects with locally advanced cancer (14.5%) (Table 1). The average CTC count was also higher in subjects with progressive disease (10 CTC/mL, range 3–27 CTC/mL) compared to subjects who were clinically responding to therapy (4 CTC/mL, range 3–5 CTC/mL). Finally, subjects who were previously but not currently exposed to chemotherapy for their cancers (i.e. >6 months prior) had a similar average CTC count but narrower range (6 CTC/mL, range 3–9 CTC/mL) compared to subjects who were newly diagnosed and not yet treated (i.e. treatment naive; 7 CTC/mL, range 3–21 CTC/mL).
Figure 2.
Enumeration of CTC in whole blood. Quad-µNMR was used to identify CTC directly in whole blood. Twenty one subjects demonstrated positive CTC values, ranging from 19 to 188 counts.
Molecular characterization and heterogeneity of CTC
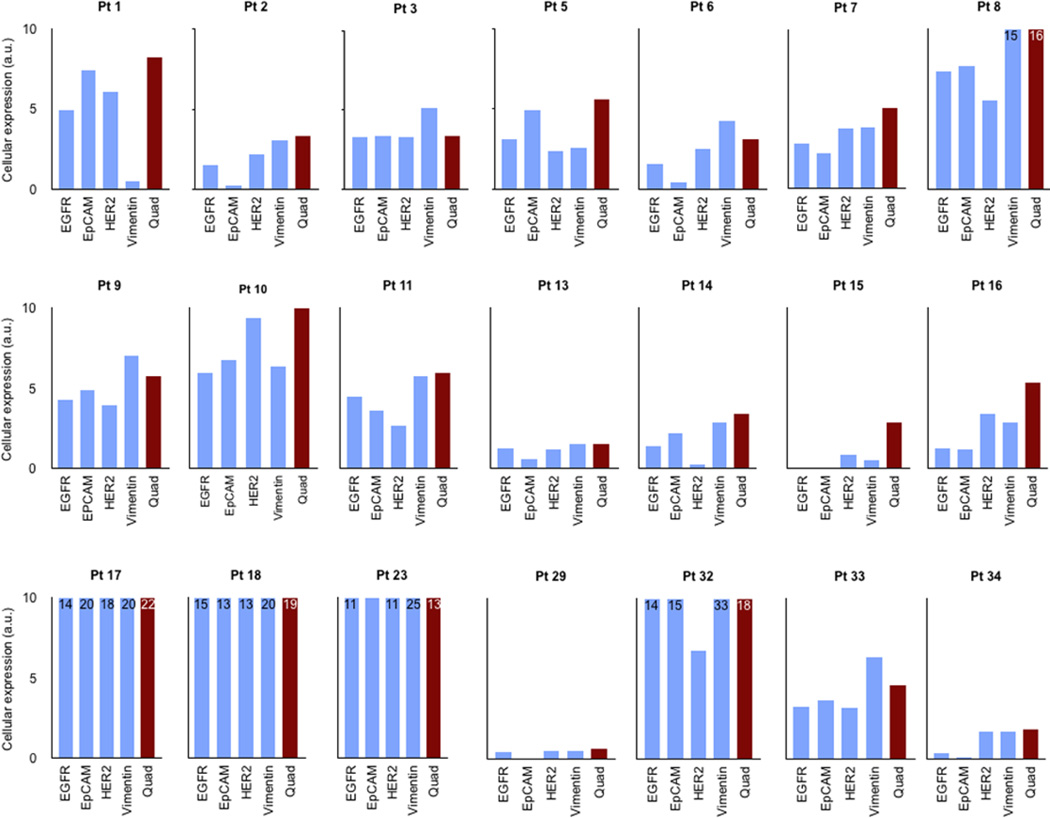
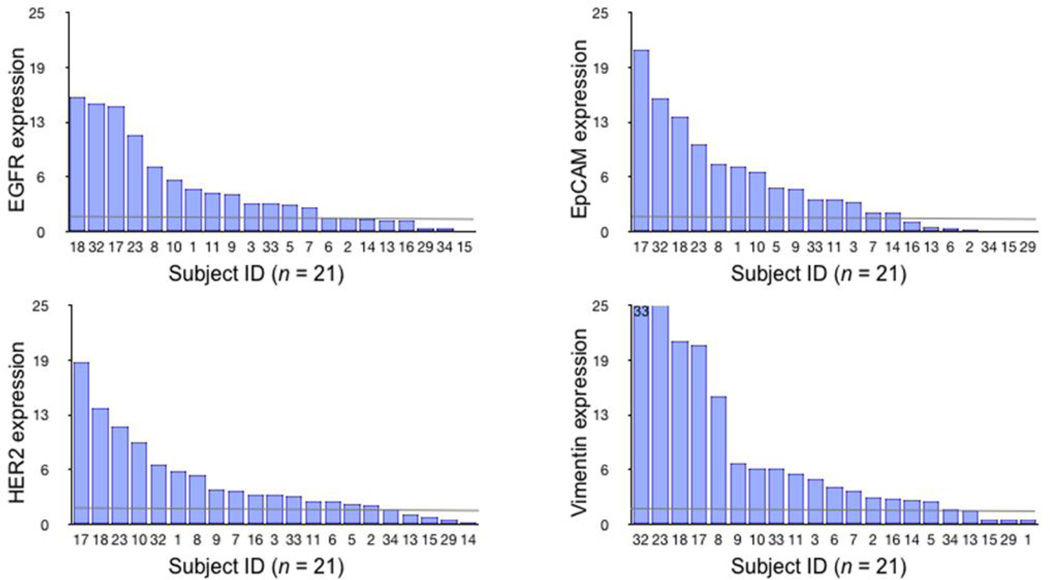
Quantitative comparison of the molecular profiles obtained from EpCAM, EGFR, HER-2 and vimentin across subjects’ CTC demonstrated considerable heterogeneity of marker expression (Figure 3). EpCAM alone was positive in 67% of the CTC samples and negative in the remainder (Figure 4). EGFR was positive in 62%, HER-2 in 76%, and vimentin in 76%. Among the individual markers, the average expression of vimentin in CTC was higher than EGFR (30.3%), EpCAM (19.2%) and HER-2 (30.1%) (Figure 4). The average CTC vimentin expression levels of subjects with worsening clinical trajectories was 50% lower than subjects with stable or improving trajectories (6.40 vs 14.50 a.u., respectively; Table S1). CTC to biopsy vimentin ratios were then calculated for each subject; they demonstrated similar patterns based on clinical trajectory. The signal from a quad-marker set was consistently higher than any single marker (EGFR, EpCAM, HER-2). This confirms more efficient loading of magnetic nanoparticles on CTC through multi-marker targeting.
Figure 3.
Detection and characterization of CTC in whole blood. Cellular expression obtained from single and quad-µNMR values are shown in each 21 subjects. Quad-µNMR method (red bars, cocktail of (EGFR, EpCAM, HER-2, MUC-1) were used to detect the presence and abundance of CTC in whole blood. Single µNMR (blue bars) for markers EGFR, EpCAM, HER-2 and vimentin were used for CTC profiling. In each patient, the level of cancer biomarkers are heterogenous in a given patient. Note that cellular expression level obtained from quad-µNMR is higher than that of individual markers.
Figure 4.
Distribution of cellular expression markers of EGFR, EpCAM, HER-2 and vimentin across 21 subjects. Waterfall plots showing the expression levels of each of the different biomarkers sorted from high (left) to low (right). Each column represents a different patient sample. Dashed lines represent threshold of detection.
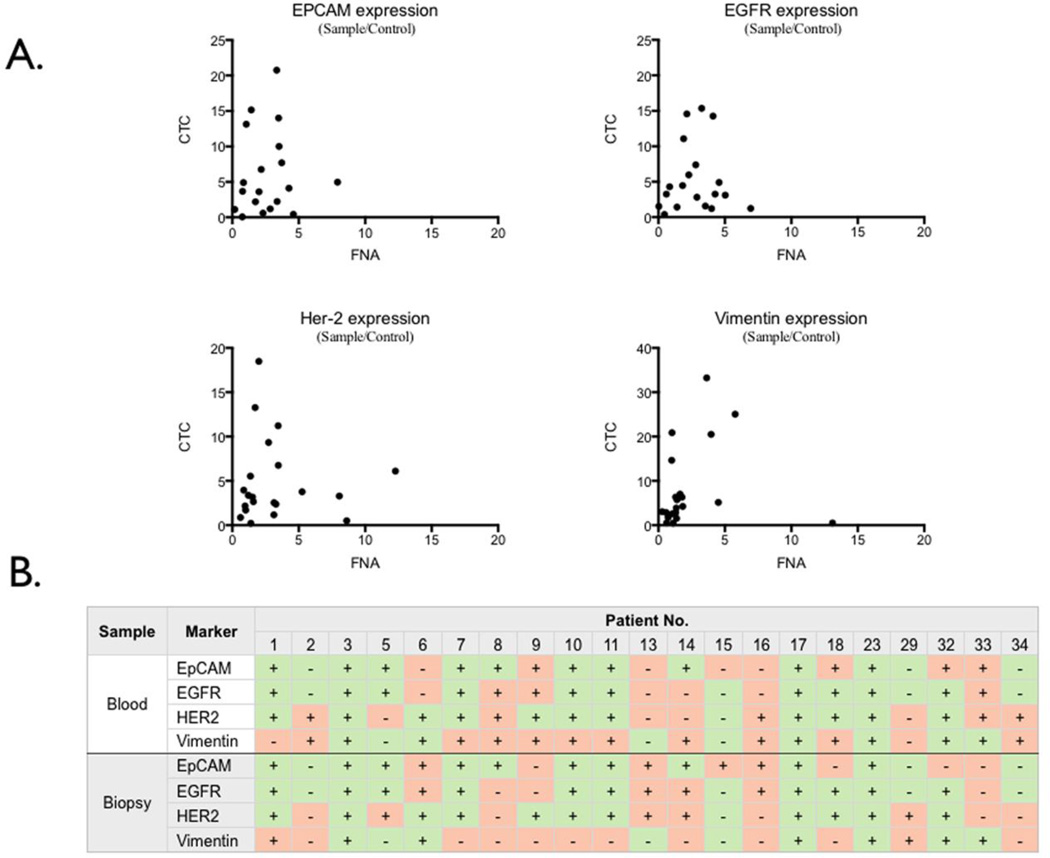
Molecular profile comparisons of paired CTC-biopsies
The molecular profiles between subjects’ paired CTC-biopsies were compared by both cellular expression levels and concordance patterns of EGFR, EpCAM, HER-2 and vimentin. The correlation for all markers was poor: EpCAM (p = 0.7604), EGFR (p = 0.1894), and HER-2 (p = 0.2242) (Figure 5A). However, paired non-parametric student t test indicated a statistical difference in vimentin expression levels (p = 0.0112) between CTC and their respective biopsies. For concordance analyses, all CTC and biopsy marker values were first scored as positive or negative (Figure 5B). Positive (+) scores were assigned to µNMR expression levels that exceeded a previously established experimental threshold in control samples13. Negative (−) scores were assigned to values below threshold. Positive concordance (+/+ or −/−) between CTC and biopsy results occurred in 48 of 84 (57%) tests compared to 36 of 84 (43%) discordant tests (+/− or −/+). Stratifying by clinical trajectory (i.e., worsening or stable/improving disease) did not appreciably increase or decrease concordance (Table S2).
Figure 5.
Comparison of molecular profiles of CTC and biopsy from the site of metastasis. (A) Correlation studies between the expression levels in CTC and in FNA biopsy for each marker of EGFR, EpCAM, HER-2 and vimentin are shown. For each marker, the expression values in CTC and in FNA biopsy showed poor correlation.
(B) Comparison of similarity of pattern of cellular expression between CTC and that of biopsy. Each column represents a patient sample. CTC results (top) are expressed as positive or negative. Results from the biopsy from the site of metastasis (bottom) are similarly indicated as positive or negative. In each subject, concordant test result (i.e. both CTC and biopsy positive or both negative) is designated in green. Discordant results are shown in pink.
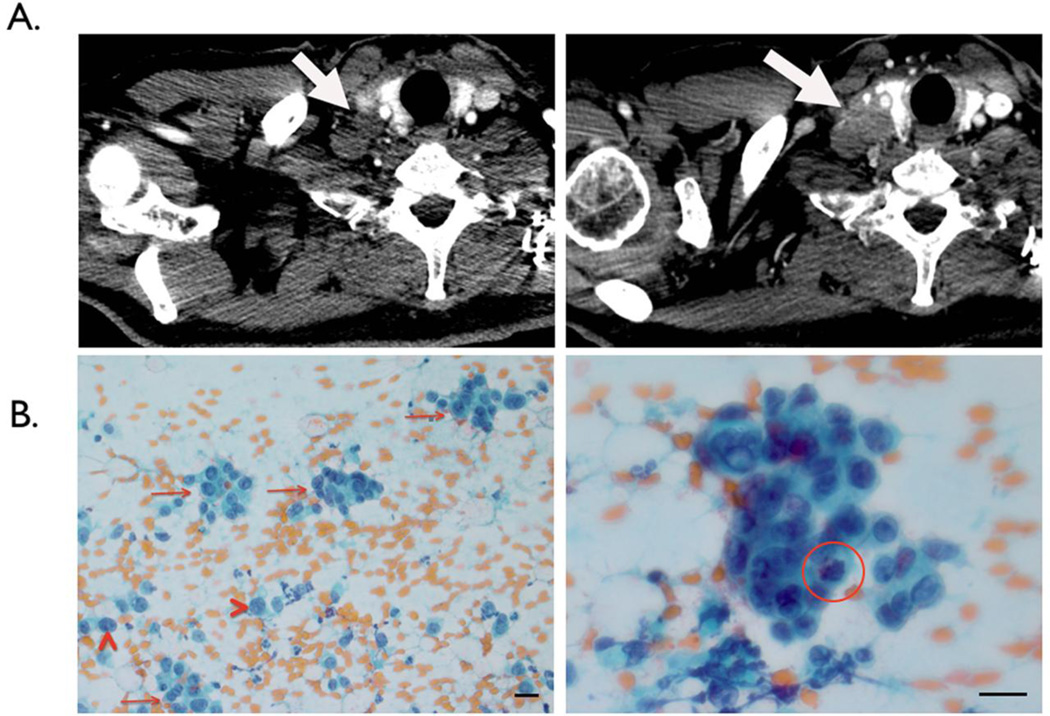
Subject 7 further illustrates µNMR’s potential. Initial clinical assessment rendered a right supraclavicular node suspicious but not certain for malignancy. At the same time, µNMR analysis unequivocally favored supraclavicular malignancy and tumor seeding into circulation (CTC count = 42 in 7 mL of whole blood). Follow-up imaging two months later showed an enlarging supraclavicular lesion and a subsequent biopsy revealed adenocarcinoma (Figure 6).
Figure 6.
A representative clinical case illustrating the potential use of magnetic nanoparticle labeling and µNMR-based detection in early cancer diagnostics. (A) CT imaging of the right supraclavicular lymph node (1.4 cm) in subject 7 contributed to the clinical impression of suspicious but not certain of underlying malignancy (left). µNMR analysis, however, identified 42 CTC per 7 mL sample of whole blood obtained from this subject and unequivocally classified malignancy. Molecular characterization of CTC also identified expression of EpCAM, EGFR, HER-2 in circulating cells, supporting the presence of cancer biomarkers in the subject’s whole blood (Figure 3). After 2 months, CT imaging revealed a significant enlargement of the lesion (2.5 cm) consistent with a metastases (right). (B) Repeat biopsy showed scattered clusters of tumor cells (arrows) and single tumor cells (arrow heads) in the background of hemorrhage (image is 200×). A high-power view (image is 600×) shows the 3-dimensional cluster with a mitosis (red circle). The scale bar is 20µm in size
Discussion
The overall goal of this study was to determine 1) whether the nanotechnology driven µNMR approach could be used for rapid molecular profiling of CTC and FNA and 2) whether CTC testing could bypass fine needle biopsies of solid tumor lesions? Specifically, we were interested in determining the congruency of important cancer markers between two key sources of cancer cells. Additionally, we were interested in CTC expression levels of EpCAM (and other markers) in patients with various underlying epithelial cancers. The study was conducted as a prospective trial into which we enrolled late stage cancer patients with high likelihood of disseminated disease. Fine needle biopsies of visceral lesions were obtained under image guidance. Matched peripheral blood samples were obtained at the same time to minimize temporal variation and all samples were processed in parallel. We show that the chosen biomarker levels were discordant between CTC and primary cancer cells in 43% of samples and in 86% of patients. EpCAM based identification of CTC alone would have missed 33% of CTC. Notably, average CTC/biopsy vimentin ratios were 50% lower in subjects with worsening clinical trajectories. This association is supported by studies suggesting that only circulating non-EMT cells attach to the vessel wall, extravasate, and reestablish distant secondary sites30,31. However, further research is needed to confirm if CTC with low vimentin ratios (non-EMT) confer worse clinical outcomes and potentially serve as prognostic markers.
The technologies underpinning this study include a point-of-care, handheld µ-NMR system10,32 and bioorthogonal magnetic nanoparticle based tagging29 to identify cells of interest. The approach has a number of advantages including fast turn around, high sensitivity and the fact that measurements can be performed in whole blood without major purification. The technology has previously been shown to be more sensitive than the current clinical standard in identifying patients with CTC13. In its latest iteration, µ-NMR harnesses a cocktail approach to maximize cell detection by targeting multiple biomarkers (Figure 1). The technology thus distinguishes itself from others through its more comprehensive approach for CTC enumeration and profiling and because it does not require cell isolation to render accurate results.
The current study contains a few caveats. Perhaps most important, the read-outs rely on bulk instead of single cell measurements and thus the number of biomarkers chosen can be limited by sample size. On average, about 100 cells were required for each biomarker comparison. Second, we chose not to study a single cancer type but rather sample a typical cohort of patients referred for image guided biopsies to examine general relevance. Third, we decided to focus on protein biomarkers rather than genetic analyses since the latter is relatively more established33,34 and shown to correlate better between CTC and primary cancers35. Despite these caveats we feel that our findings are relevant for designing future basic and clinical studies and developing next generation technologies.
Magnetic cell tagging and analytical techniques are rapidly advancing with the development of superior nanomaterials24,36–38, conjugation technologies29,39,40 and sensing strategies41–43. Regarding more sensitive cellular analysis, recent magnetic technologies enable single cell profiling44,45 at fairly high count rates (106 cells/sec). By combining these new sensors with other alternative yet complementary labeling methods46, it is conceivable to screen for broader biomarker diversity within and between cell type populations.
The described findings have important implications for comparing molecular profiles between CTC and metastatic lesions and tracking their respective changes. Notably, implementing a quantitative nanosensing approach into preclinical studies and early phase drug trials could facilitate unmasking novel biomarker sets of early response and informing ‘go-no go’ decisions. Achieving this in peripheral blood and/or limited biopsies - specimens amenable to repeat interrogation - has significant advantages over the use of a single baseline biopsy. The latter does not capture the dynamic nature of tumors and response to emerging targeted therapies. Point-of-care and operator-independent µNMR protein analyses of patient-derived specimens could thus function as timely surrogates of an individual’s underlying biology and help meet the goals of precision medicine.
Supplementary Material
Acknowledgements
The authors thank N. Sergeyev for synthesizing cross-linked dextran iron oxide nanoparticles, T. Reiner for preparing TCO, S. Hilderbrand and S. Agasti for synthesizing the tetrazine precursor, D. Issadore for µNMR developments.
This work was funded in part by 2R01EB004626, U54-CA151884, DOD W81XWH-11-1-0706, and K12CA087723
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
Authors declare no conflict of interest.
References
- 1.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Botteri E, Sandri MT, Bagnardi V, Munzone E, Zorzino L, Rotmensz N, et al. Modeling the relationship between circulating tumour cells number and prognosis of metastatic breast cancer. Breast Cancer Res Treat. 2010;122(1):211–217. doi: 10.1007/s10549-009-0668-7. [DOI] [PubMed] [Google Scholar]
- 3.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192(3):373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(23):7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 5.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 6.Okegawa T, Nutahara K, Higashihara E. Prognostic significance of circulating tumor cells in patients with hormone refractory prostate cancer. J Urol. 2009;181(3):1091–1097. doi: 10.1016/j.juro.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Lang JM, Casavant BP. Circulating Tumor Cells: Getting More from Less. … Translational Medicine. 2012 doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 8.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. New England Journal of Medicine. 2012;366(6):489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 9.Coumans FAW, Doggen CJM, Attard G, De Bono JS, Terstappen LWMM. All circulating EpCAM+ CK+ CD45− objects predict overall survival in castration-resistant prostate cancer. Annals of oncology. 2010;21(9):1851–1857. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 10.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, et al. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3(71):71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connelly M, Wang Y, Doyle GV, Terstappen L, McCormack R. Re: Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101(12):895. doi: 10.1093/jnci/djp117. author reply 896–895; author reply 897. [DOI] [PubMed] [Google Scholar]
- 13.Ghazani AA, Castro CM, Gorbatov R, Lee H, Weissleder R. Sensitive and direct detection of circulating tumor cells by multimarker micro-nuclear magnetic resonance. Neoplasia. 2012;14(5):388–395. doi: 10.1596/neo.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendall SC, Simonds EF, Qiu P, Amir E-D, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science Signalling. 2011;332(6030):687. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan DJ, O’Connor DP, Rexhepaj E, Ponten F, Gallagher WM. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nature Reviews Cancer. 2010;10(9):605–617. doi: 10.1038/nrc2902. [DOI] [PubMed] [Google Scholar]
- 16.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chemical reviews. 2008;108(2):462. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 18.Sun T, Holmes D, Gawad S, Green NG, Morgan H. High speed multi-frequency impedance analysis of single particles in a microfluidic cytometer using maximum length sequences. Lab Chip. 2007;7(8):1034–1040. doi: 10.1039/b703546b. [DOI] [PubMed] [Google Scholar]
- 19.Gu B, Park TJ, Ahn J, Huang X, Lee SY, Choi Y. Nanogap Field-Effect Transistor Biosensors for Electrical Detection of Avian Influenza. small. 2009;5(21):2407–2412. doi: 10.1002/smll.200900450. [DOI] [PubMed] [Google Scholar]
- 20.Sorger PK. Microfluidics closes in on point-of-care assays. Nat Biotechnol. 2008;26(12):1345–1346. doi: 10.1038/nbt1208-1345. [DOI] [PubMed] [Google Scholar]
- 21.Haun JB, Yoon TJ, Lee H, Weissleder R. Molecular detection of biomarkers and cells using magnetic nanoparticles and diagnostic magnetic resonance. Methods Mol Biol. 2011:72633–72649. doi: 10.1007/978-1-61779-052-2_3. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14(8):869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, Sun E, Ham D, Weissleder R. Chip–NMR biosensor for detection and molecular analysis of cells. Nature medicine. 2008;14(8):869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc Natl Acad Sci U S A. 2009;106(30):12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao H, Min C, Issadore D, Liong M, Yoon T-J, Weissleder R, et al. Magnetic nanoparticles and microNMR for diagnostic applications. Theranostics. 2012;2(1):55. doi: 10.7150/thno.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McInroy L, Maatta A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun. 2007;360(1):109–114. doi: 10.1016/j.bbrc.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 27.Barriere G, Riouallon A, Renaudie J, Tartary M, Rigaud M. Mesenchymal characterization: alternative to simple CTC detection in two clinical trials. Anticancer Res. 2012;32(8):3363–3369. [PubMed] [Google Scholar]
- 28.Connelly L, Barham W, Onishko HM, Chen L, Sherrill TP, Zabuawala T, et al. NF-kappaB activation within macrophages leads to an anti-tumor phenotype in a mammary tumor lung metastasis model. Breast Cancer Res. 2011;13(4):R83. doi: 10.1186/bcr2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5(9):660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, et al. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res. 2008;68(24):10377–10386. doi: 10.1158/0008-5472.CAN-08-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69(18):7135–7139. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issadore D, Min C, Liong M, Chung J, Weissleder R, Lee H. Miniature magnetic resonance system for point-of-care diagnostics. Lab Chip. 2011;11(13):2282–2287. doi: 10.1039/c1lc20177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolke E, Orth K, Gerber PA, Lammering G, Mota R, Peiper M, et al. Gene expression of circulating tumour cells and its correlation with tumour stage in breast cancer patients. Eur J Med Res. 2009;14(8):359–363. doi: 10.1186/2047-783X-14-8-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strati A, Markou A, Parisi C, Politaki E, Mavroudis D, Georgoulias V, et al. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 2011;11422 doi: 10.1186/1471-2407-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smirnov DA, Zweitzig DR, Foulk BW, Miller MC, Doyle GV, Pienta KJ, et al. Global gene expression profiling of circulating tumor cells. Cancer research. 2005;65(12):4993–4997. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, Song HT, et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2007;13(1):95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 37.Yoon TJ, Lee H, Shao H, Weissleder R. Highly magnetic core-shell nanoparticles with a unique magnetization mechanism. Angew Chem Int Ed Engl. 2011;50(20):4663–4666. doi: 10.1002/anie.201100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon TJ, Lee H, Shao H, Hilderbrand SA, Weissleder R. Multicore assemblies potentiate magnetic properties of biomagnetic nanoparticles. Adv Mater. 2011;23(41):4793–4797. doi: 10.1002/adma.201102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agasti SS, Liong M, Tassa C, Chung HJ, Shaw SY, Lee H, et al. Supramolecular host-guest interaction for labeling and detection of cellular biomarkers. Angew Chem Int Ed Engl. 2012;51(2):450–454. doi: 10.1002/anie.201105670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liong M, Tassa C, Shaw SY, Lee H, Weissleder R. Multiplexed magnetic labeling amplification using oligonucleotide hybridization. Adv Mater. 2011;23(36):H254–H257. doi: 10.1002/adma.201101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinberg I, Ben-David M, Gannot I. A new method for tumor detection using induced acoustic waves from tagged magnetic nanoparticles. Nanomedicine: Nanotechnology. 2011 doi: 10.1016/j.nano.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Boubeta C, Balcells L, Cristòfol R, Sanfeliu C, Rodríguez E, Weissleder R, et al. Self-assembled multifunctional Fe/MgO nanospheres for magnetic resonance imaging and hyperthermia. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6(2):362–370. doi: 10.1016/j.nano.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Brullot W, Valev VK, Verbiest T. Magnetic-plasmonic nanoparticles for the life sciences: calculated optical properties of hybrid structures. Nanomedicine: Nanotechnology, Biology and Medicine. 2012;8(5):559–568. doi: 10.1016/j.nano.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4(141):141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loureiro J, Andrade PZ, Cardoso S, da Silva CL, Cabral JM, Freitas PP. Magnetoresistive chip cytometer. Lab Chip. 2011;11(13):2255–2261. doi: 10.1039/c0lc00324g. [DOI] [PubMed] [Google Scholar]
- 46.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301(5641):1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.