Abstract
STING has emerged in recent years as an important signalling adaptor in the activation of type I interferon responses during infection with DNA viruses and bacteria. An increasing body of evidence suggests that STING also modulates responses to RNA viruses, though the mechanisms remain less clear. In this review, we give a brief overview of the ways in which STING facilitates sensing of RNA viruses. These include modulation of RIG-I-dependent responses through STING's interaction with MAVS, and more speculative mechanisms involving the DNA sensor cGAS and sensing of membrane remodelling events. We then provide an in-depth literature review to summarise the known mechanisms by which RNA viruses of the families Flaviviridae and Coronaviridae evade sensing through STING. Our own work has shown that the NS2B/3 protease complex of the flavivirus dengue virus binds and cleaves STING, and that an inability to degrade murine STING may contribute to host restriction in this virus. We contrast this to the mechanism employed by the distantly related hepacivirus hepatitis C virus, in which STING is bound and inactivated by the NS4B protein. Finally, we discuss STING antagonism in the coronaviruses SARS coronavirus and human coronavirus NL63, which disrupt K63-linked polyubiquitination and dimerisation of STING (both of which are required for STING-mediated activation of IRF-3) via their papain-like proteases. We draw parallels with less-well characterised mechanisms of STING antagonism in related viruses, and place our current knowledge in the context of species tropism restrictions that potentially affect the emergence of new human pathogens.
Abbreviations: BiFC, bimolecular fluorescence complementation; CARD, caspase activation and recruitment domain; cGAMP, cyclic GMP-AMP; cGAS, cyclic GMP-AMP synthase; DC, dendritic cell; DUB, deubiquitinase; hSTING, human STING; IFN, interferon; IRF-3, interferon regulatory factor 3; ISG, interferon-stimulated gene; ISRE, interferon-stimulated response element; MAM, mitochondria-associated membrane; MAVS, mitochondrial antiviral signalling protein; MDA-5, melanoma differentiation-associated gene 5; MDDC, monocyte-derived dendritic cell; mKG, monomeric Kusabira Green; mSTING, murine STING; PBMC, peripheral blood mononuclear cells; PI(3)K, phosphatidylinositol-3-OH kinase; PLP, papain-like protease; PLpro, papain-like protease; RIG-I, retinoic acid-inducible gene I; STAT2, signal transducer and activator of transcription 2; STING, stimulator of interferon genes; TBK1, tank-binding kinase 1; TRIM32, tripartite motif-containing 32; TRIM56, tripartite motif-containing 56
Keywords: STING, Immune evasion, Dengue, Hepatitis C virus, SARS coronavirus
The production of the type I interferons (IFNs) IFN-α and IFN-β is one of the most critical early events in the induction of an antiviral innate immune response. Type I IFN is induced during viral infection and signals in an autocrine and paracrine manner to activate the transcription of a number of IFN-stimulated genes (ISGs) that establish an antiviral state to limit the spread of the virus. Crucial to the induction of type I IFN is the recognition of viral pathogen-associated molecular patterns (PAMPs) by cellular pattern recognition receptors (PRRs). The most well-characterised PRRs relevant to RNA virus infection are retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5). RIG-I and MDA-5 sense cytoplasmic viral double-stranded RNA (dsRNA) replication intermediates and associate with mitochondrial antiviral signalling protein (MAVS) (also known as IFN-β promoter stimulator 1, IPS-1, virus-induced signalling adapter, VISA, and CARD-adapter inducing IFN-β, CARDIF) at the mitochondrial membrane to activate the transcription factor IFN regulatory factor 3 (IRF-3) [1], [2].
More recently, stimulator of interferon genes (STING) was identified as a novel and important modulator of type I IFN induction via IRF-3 [3], [4], [5]. STING is also sometimes referred to as transmembrane protein 173 (TMEM173), mediator of IRF-3 activation (MITA), endoplasmic reticulum IFN stimulator (ERIS) or MPYS (named after its four N-terminal amino acids). STING is probably best known for its role in sensing bacteria and DNA viruses [6], [7]. However, it has become increasingly apparent that STING also plays an important role in restricting RNA virus replication. The replication of diverse positive- and negative-sense RNA viruses is enhanced in the absence of STING in in vivo, ex vivo and in vitro models [3], [4], [5], [8], [9], [10]. Furthermore, STING becomes activated, and its expression is increased, during RNA virus infection [4], [5], [9], [11], [12]. Finally, the fact that several RNA viruses have been shown to antagonise STING implies that STING is an important restriction factor during RNA virus infection [8], [9], [10], [11], [12], [13], [14], [15].
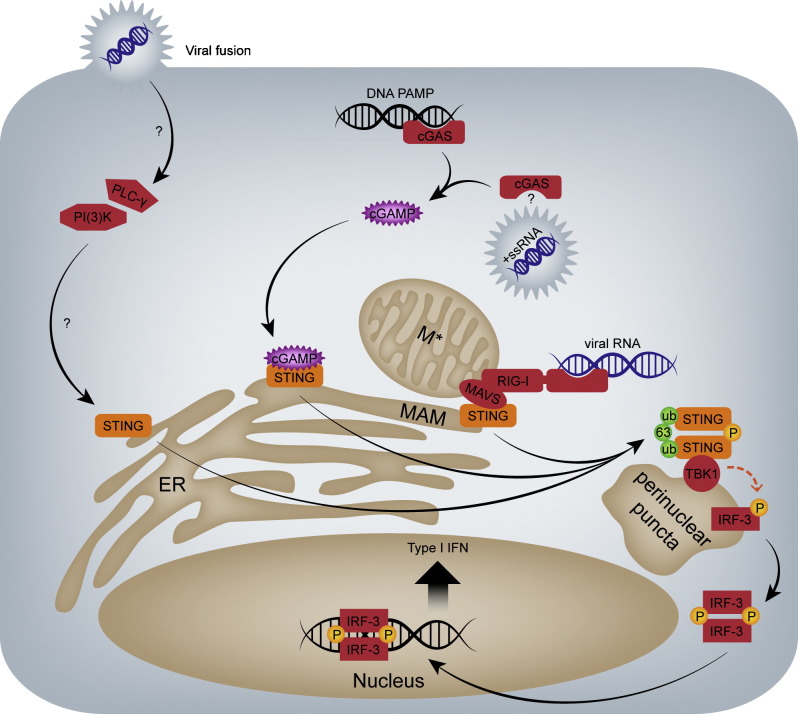
STING is membrane-associated via its four N-terminal transmembrane domains, and within the cell STING is localised to the endoplasmic reticulum (ER), with a partial localisation to mitochondria and mitochondria-associated membranes (MAMs) [3], [4], [5], [8]. Following stimulation, STING dimerises and translocates to punctate perinuclear vesicles, where it interacts with tank-binding kinase 1 (TBK1) and IRF-3 (Fig. 1 ) [reviewed in 7]. The subsequent phosphorylation of STING and IRF-3 by TBK1 leads to the nuclear translocation of IRF-3 and the induction of type I IFN and other cytokines [7]. The dimerisation and relocalisation of STING is essential for the recruitment of TBK1 to these perinuclear sites, as well as for the downstream activation of IRF-3 and type I IFN production [5], [7], [16]. The interaction between STING and TBK1, and possibly STING dimerisation, is promoted by K63-linked polyubiquitination of STING at various residues by tripartite motif-containing 32 (TRIM32) and TRIM56 [17], [18]. STING can also induce NF-κB signalling, though the mechanism remains to be fully elucidated [7].
Fig. 1.
STING signalling during RNA virus infection. Inactive STING resides in membranes of the ER, MAM and mitochondria (M*). Following its activation, STING dimerises and relocalises to perinuclear punctate domains, where it interacts with TBK1 to phosphorylate IRF-3. Activated STING is also modified by phosphorylation and K63-linked polyubiquitination. Following its activation, IRF-3 dimerises and translocates to the nucleus, where it induces the transcription of the type I interferons IFN-α and IFN-β. RNA viruses have been proposed to be sensed by three distinct mechanisms in relation to STING activation. (1) Viral dsRNA replication intermediates and stem-loop structures, and the 5′-triphosphates of uncapped viral RNAs are recognised by RIG-I, which associates with MAVS and STING at MAMs to activate STING. (2) The DNA sensor cGAS has been linked to the sensing of positive-sense RNA viruses by an as-yet undefined mechanism. In this scenario, cGAS produces the messenger molecule cGAMP after detecting RNA virus infection, and cGAMP binds and activates STING. (3) Viral fusion events at the plasma membrane have been shown to activate STING by a poorly-defined mechanism involving PI(3)K and phospholipase C-γ (PLC-γ). For simplicity, only signalling molecules referred to in the main text are shown.
In order to establish a productive infection, successful viruses must evade recognition by the innate immune system, and antagonism of STING signalling has been identified in several divergent positive-sense RNA viruses [8], [9], [10], [11], [12], [13], [14], [15]. Curiously, to our knowledge, STING antagonism has not yet been described for any negative-sense RNA virus. In the remainder of this article, we will briefly discuss the putative ways by which STING facilitates sensing of RNA viruses, and provide an in-depth review of the known mechanisms RNA viruses use to evade STING signalling.
1. Every breath you take: how STING may sense multiple stages of the RNA virus life cycle
STING is activated by several distinct mechanisms. For detailed information on the mechanisms of STING activation in response to DNA PAMPs, the reader is directed to two recent reviews by Unterholzner, and Ran et al. [6], [7]. Fig. 1 summarises the mechanisms by which RNA virus infection is thought to activate STING. While STING has been shown to directly bind DNA, it does not interact with the dsRNA mimic poly(I:C) [19], suggesting that STING itself does not function as a PRR for the recognition of RNA virus PAMPs. However, STING has been shown to function as an adapter in the sensing of RNA viruses via RIG-I [reviewed in 7]. STING interacts with RIG-I, but not MDA-5, and consequently STING is activated by ligands that bind RIG-I, but not those detected by MDA-5 [3], [4], [7]. MDA-5 senses long dsRNA molecules, while RIG-I senses shorter dsRNA elements and the uncapped 5′-triphosphate of exogenous RNAs [reviewed in 20]. STING can also be activated through the exogenous expression of a C-terminally truncated RIG-I construct (ΔRIG-I) that is constitutively active because it encodes only the N-terminal CARD signalling domains of RIG-I [3], [21]. Ultimately this results in STING participating in immune responses mounted against RNA viruses sensed by RIG-I, including Sendai virus (SeV), vesicular stomatitis virus (VSV), Newcastle disease virus (NDV) and Japanese encephalitis virus (JEV), but not those sensed exclusively by MDA-5 [7]. Following activation via RIG-I, STING interacts with the adapter protein MAVS at the MAM or mitochondria, and after its translocation to perinuclear vesicles, STING then acts as a scaffold for the recruitment of TBK1 and other signalling components required for IRF-3 activation and type I IFN induction [3], [4], [7], [8]. This role in modulating RIG-I-dependent antiviral responses is likely one of the major ways in which STING contributes to immune sensing of RNA viruses. However, several recent reports suggest that there may be additional RIG-I-independent ways by which STING helps sense RNA virus infection [22], [23], [24].
Cyclic GMP-AMP synthase (cGAS) is a recently-discovered cytosolic PRR that detects double-stranded DNA (dsDNA) and produces the signalling molecule cyclic GMP-AMP (cGAMP), which binds and activates STING during DNA virus infection [6], [7]. Interestingly, in a recent screen for the antiviral activity of known ISGs, Schoggins et al. identified cGAS as one of the most potent restriction factors for all positive-sense RNA viruses tested [22], [23]. The antiviral effects of cGAS were still evident in the absence of RIG-I [23]. Importantly, mortality and virus replication were increased in cGAS knock-out mice infected with the positive-sense RNA virus West Nile virus (WNV) [22]. Whether cGAS acts as a bona fide RNA sensor remains controversial. While, cGAS does not bind poly(I:C) in vitro [6], [7], [25], a recent report showed that cGAS was able to bind a synthetic 50mer dsRNA, although this did not result on the generation of cGAMP [26]. Since cGAMP is required for cGAS-mediated STING activation [6], [7], [25], it remains somewhat mysterious how cGAS contributes to RNA virus sensing. Schoggins et al. proposed that, because basal expression levels were reduced for a subset of ISGs in the absence of cGAS, cGAS might modulate the overall refractoriness of cells to RNA viruses [22]. Nevertheless, negative-sense RNA viruses are not restricted by cGAS [22], and this exquisite specificity of cGAS for positive-sense RNA viruses suggests that cGAS might be directly involved in sensing positive-sense RNA virus infection.
One of the characteristics shared by positive-sense RNA viruses is that they modify cellular membranes to form cytoplasmic membranous replication compartments required for viral RNA synthesis [27]. Direct evidence that such membrane modifications activate STING and/or cGAS is lacking. However, STING signalling has been shown to be induced by fusion events at the plasma membrane [24]. Both cationic liposomes and herpesvirus-derived virus-like particles that lack nucleic acids were shown to cause the relocalisation of STING to TBK1-containing perinuclear vesicles, as well as the induction of cytokines in a manner that was dependent upon STING, TBK1 and IRF-3 [24]. This activation of STING did not require the RIG-I signalling pathway [24]. Instead, the fusion-mediated activation of STING involved an as-yet poorly-characterised mechanism involving phospholipase C-γ and phosphatidylinositol-3-OH kinase (PI(3)K) [24]. Therefore, membrane-remodelling events during viral entry or later on in the viral life cycle may also be sensed by STING to induce STING activation. It is interesting to speculate that STING might also sense other disruptions to the normal cellular equilibrium, such as the release of self-DNA into the cytoplasm or mitochondrial dysregulation during infection with certain viruses.
Further work will be needed to clarify whether positive-sense RNA viruses are detected by a multitude of mechanisms converging on STING, including RIG-I-dependent sensing of cytoplasmic viral RNA, fusion events during viral entry, poorly-defined sensing via cGAS, or a combination of these and other potential mechanisms.
2. Don’t stand so close to me: how RNA viruses take the STING out of innate immune recognition
Despite the fact that the role of STING in sensing of DNA viruses is more clearly elucidated than for RNA viruses, the first STING antagonist was discovered in an RNA virus, the flavivirus yellow fever virus (YFV) [8]. Just one year after first identifying STING, the Barber group went on to show that the YFV non-structural protein NS4B inhibits IFN-β induction by STING [8]. However, it was not until several years later that the first fully-characterised mechanism of STING antagonism by a viral protein was described for the coronavirus papain-like proteases (PLPs) [11], followed shortly thereafter by the NS2B/3 protease complex from the flavivirus dengue virus (DENV) [9], [12]. The remainder of this review will focus on the known STING antagonists encoded by positive-sense RNA viruses, from the families Flaviviridae and Coronaviridae, and the mechanisms by which they counteract the STING signalling pathway (summarised in Table 1 ).
Table 1.
Mechanisms of STING antagonism by positive-sense RNA viruses.
| Family | Genus | Species | Protein | Mechanism of STING Antagonism | References |
|---|---|---|---|---|---|
| Flaviviridae | Flavivirus | DENV | NS2B/3 | Binds and cleaves hSTING, but not mSTING, inactivating STING function. Transgenic mice expressing hSTING might be improved animal models for DENV pathogenesis. Protease activity is essential and sufficient. | [9], [12], [39] |
| YFV | NS4B | Unknown | [8] | ||
| Hepacivirus | HCV | NS4B | Binds STING at MAMs via N-terminal STING-homology domain. May disrupt STING signalling complexes. Acts independently of NS3/4A-dependent MAVS antagonism. | [10], [13], [57], [58] | |
| Coronaviridae | Alphacoronavirus | HCoV-NL63 | PLP2 | Binds STING to disrupt K63-linked polyubiquitination and dimerisation of STING, and formation of multicomponent signalling complexes. Protease activity not required. | [11], [71] |
| PEDV | PLP2 | Binds STING and disrupts K63-linked polyubiquitination of STING. DUB activity essential?a | [15] | ||
| Betacoronavirus | SARS-CoV | PLpro | Binds STING to disrupt K63-linked polyubiquitination and dimerisation of STING, and formation of multicomponent signalling complexes. Protease activity not required. | [11], [14], [70], [71] | |
The role of DUB activity in STING antagonism by coronavirus PLPs remains controversial and may be affected by the presence of the transmembrane domain.
2.1. Flaviviridae
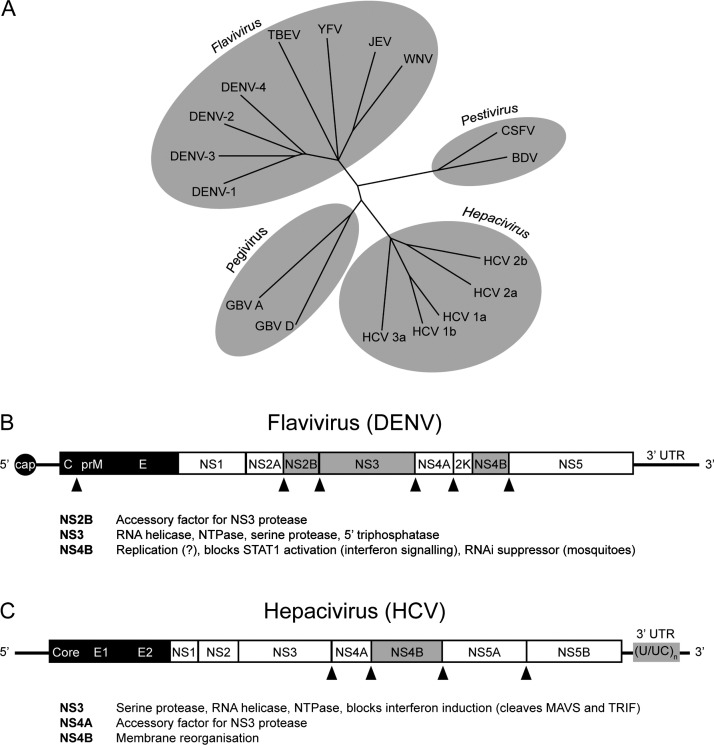
The family Flaviviridae encompasses four genera, Flavivirus and Hepacivirus (discussed here), and Pegivirus and Pestivirus (Fig. 2A) [28]. Members of the family Flaviviridae are enveloped viruses that encode a non-segmented positive-sense single-stranded RNA (ssRNA) genome that lacks a poly(A) tail (Fig. 2B and C) [28]. Immediately upon entry into the cytoplasm, translation of the viral genome produces a polyprotein that is proteolytically processed by viral and cellular proteases to yield the structural and non-structural (NS) proteins [28]. Viral replication occurs via a negative-sense RNA intermediate in cytoplasmic membranous compartments derived from the ER, with maturation and release of infectious viral progeny proceeding through the secretory pathway [28].
Fig. 2.
Flaviviridae phylogenetic tree and genome organisation. (A) Flaviviridae family tree. Viruses of relevance to this review, and other representative viruses, shown. (B) Mosquito-borne flavivirus (∼11 kb) and (C) hepacivirus (∼9.6 kb) genome organisation (not to scale). Structural proteins are in black. STING antagonists are highlighted in grey. A STING antagonist function has been described for the YFV NS4B protein, but not for the DENV NS4B. Additional known functions for proteins discussed in this review are also given. Triangles indicate cleavage sites for the flavivirus NS2B/3 (B) or hepacivirus NS3/4A (C) proteases. The HCV polyuridine tract ((U/UC)n) is a well-known PAMP that induces type I IFN production. Note that the hepacivirus genome is uncapped. Members of the Flaviviridae lack a poly(A) tail. Viruses not defined elsewhere; CSFV, classical swine fever virus; BDV, bovine diarrhoea virus; GBV, GV virus.
We will begin by describing the STING antagonists encoded by the genus Flavivirus, the human pathogens of which are all arthropod-borne viruses (arboviruses) transmitted by mosquitoes or ticks [29]. Flaviviruses pathogenic to humans include DENV, JEV, WNV, YFV and the tick-borne encephalitis virus (TBEV) complex. All of these flaviviruses cause potentially severe disease in humans, and many are considered emerging or re-emerging pathogens [30]. YFV, JEV and TBEV are the only flaviviruses for which vaccines are currently approved, and there are no licensed specific antiviral therapies available [30], [31].
2.1.1. Every little thing she does is magic: STING antagonism by the dengue virus NS2B/3 protease complex
Of the flaviviruses, the mechanism of STING antagonism has been studied in most detail for DENV, which is also the focus of research in our lab. DENV is arguable the most significant arboviral pathogen of humans with up to 2.5 billion people at risk of infection in the tropics and subtropics, and an estimated 50 million cases of dengue fever and the more severe dengue haemorrhagic fever annually, resulting in over 20,000 deaths [29], [30], [32], [33]. The ways in which DENV evades STING signalling is of particular interest because the dysregulated production of cytokines and chemokines during DENV infection is known to contribute to symptoms associated with severe dengue disease [29], [34]. Furthermore, developing a DENV vaccine has so far proven challenging [35], [36], and understanding how DENV modulates the immune response may lead to improved vaccine strategies.
In vivo, DENV replicates predominantly in myeloid cells, including monocytes, dendritic cells (DCs) and macrophages [29], [37]. We were the first to show that DENV fails to induce a robust IFN response in primary human monocyte-derived DCs (MDDCs), despite the fact that the production of other pro-inflammatory cytokines and chemokines is unimpaired [38]. Type I IFN is also poorly induced in DENV-infected plasmacytoid and blood-circulating DCs, B cells, monocytes and in whole peripheral blood mononuclear cells (PBMCs) [9]. Furthermore, DENV-infected MDDCs failed to induce Th1 responses in allogeneic T-cells [38], suggesting that inappropriate activation of DCs may have down-stream consequences for adaptive immune responses mounted during DENV infection.
We later went on to identify the viral protease, consisting of the non-structural proteins NS3 and its cofactor NS2B, which together are also responsible for cleaving the viral polyprotein [29], as an antagonist of type-I IFN production [39]. Specifically, we demonstrated that the NS2B/3 protease complex inhibited activation of a luciferase reporter controlled by the IFN-β promoter in 293T cells stimulated with poly(I:C) or after infection with SeV [39]. We verified these findings by measuring endogenous IFN-α production in MDDCs infected with an NDV mutant expressing the DENV NS2B/3 protein. This NDV virus exhibited impaired IFN-α induction relative to its parental wild-type (wt) [39]. Yu et al. later showed that NS2B/3 inhibits type I IFN induction above the level of IRF-3 phosphorylation [12], which we had also observed during DENV infection in MDDCs [39]. In addition to its protease activity, NS3 by itself also functions as a helicase, NTPase and 5′ triphosphatase during viral RNA replication [29]. We were able to distinguish between these diverse functions of NS3 by showing that a construct encoding a truncated NS3 containing only its proteolytic core, as well as the minimal NS2B accessory region, was just as efficient at inhibiting IFN-β promoter activation as wt NS2B/3 [39]. Importantly, we also confirmed that a proteolytically-inactive NS2B/3 mutant harbouring a serine-to-alanine mutation at position 135 (S135A) did not inhibit type I IFN induction [39].
In 2012, IFN antagonism by the DENV NS2B/3 protease was linked to its ability to bind and cleave STING in two concurrent papers originating from our lab and that of Yi-Ling Lin [9], [12]. The stories presented by both groups are broadly in agreement of each other. While our work characterised the mechanism of STING antagonism by NS2B/3 in primary human MDDCs [9], highly relevant to DENV infection in vivo, work by the Lin group provided additional mechanistic insights into the fate of STING after its cleavage by NS2B/3 [12].
STING was shown to interact with both wt NS2B/3 and the catalytically inactive S135A mutant in over-expression experiments [9], [12]. Furthermore, wt NS2B/3 expressed exogenously or in the context of DENV infection rapidly degraded endogenous and over-expressed STING, by 24 h post-infection (hpi) in tissue culture and in MDDCs infected at high multiplicity with DENV [9], [12]. Catalytically inactive NS2B/3 did not degrade STING [9], [12]. The Lin group also reported that STING and NS2B/3 colocalise when over-expressed in A549 cells, and that STING-NS2B/3 binding, colocalisation and cleavage is enhanced when STING signalling is stimulated with a dsDNA PAMP such as poly(dA:dT) [12]. It is known that exogenous over-expression of STING activates type I IFN production [3], and this induction was inhibited in the presence of the wt, but not catalytically inactive, DENV protease [9], [12]. In combination, these findings demonstrate that NS2B/3 binds and degrades STING to inhibit IFN induction, and that this function requires NS2B/3's protease activity. Importantly, although there are four antigenically distinct serotypes of DENV (DENV-1 to -4), these observations were not limited to any particular serotype as STING degradation was observed during infection with DENV-1 and several different DENV-2 strains [9], [12].
The consensus cleavage site for the DENV protease is located towards the N-terminus of the 379-residue STING molecule (94RR↓GA97) [9], [12], close to a cysteine-rich redox motif (88CXXC91) that is required for STING dimerisation and the induction of type I IFN [9], [40]. Using a STING construct containing an N-terminal HA tag and a C-terminal V5 tag, the Lin group demonstrated that the STING cleavage products co-migrate on western blots with STING truncation mutants designed around the predicted cleavage site [12], suggesting that this sequence is in fact the target site for NS2B/3. In contrast to full-length STING, neither of the truncation mutants were functional in a reporter assay that measures the ability of over-expressed STING to induce type I IFN [12], confirming that cleavage of STING by the DENV protease disrupts STING functionality as an adaptor for type I IFN induction.
Interestingly, unlike the 94RR↓GA97 NS2B/3 cleavage site found in human STING (hSTING), the equivalent sequence in murine STING (mSTING) is quite different (94HCMA97) [9], [12]. Consequently, while NS2B/3 is able to interact with both human and murine STING in over-expression experiments, only hSTING is cleaved by the DENV protease [9], [12]. When the NS2B/3 cleavage site in hSTING is replaced with its murine counterpart, the DENV protease loses its ability to degrade hSTING and inhibit STING function [9], [12]. In the reciprocal experiment, introducing the cleavage site of hSTING into mSTING does not make mSTING susceptible to the action of the DENV protease [9], [12], suggesting that additional distal sequences within STING are required for STING cleavage by NS2B/3. These observations were initially made in over-expression experiments in 293T and A549 cells (both human cell lines) in which luciferase reporter activity or endogenous IRF-3 phosphorylation was induced by STING over-expression [9], [12]. We further verified these findings in a more biologically-relevant context by using Semliki Forest virus (SFV)-based protein expression vectors to infect human MDDCs and murine bone marrow-derived DCs [9]. In this experiment, SFV expressing the catalytically inactive S135A protease mutant induced type I IFN to a higher degree than SFV expressing wt NS2B/3 in human MDDCs, while similar levels of IFN induction were observed with both SFV constructs in murine DCs [9]. Furthermore, when we introduced wt hSTING and the uncleavable hSTING mutant containing the murine sequence into human MDDCs by lentivirus transduction we observed more pronounced IFN-β induction during DENV infection, and a concomitant reduction in viral RNA levels, with the uncleavable hSTING mutant [9].
The development of a vaccine and antiviral therapies against DENV has been hampered in part by the lack of an immunocompetent mouse model, largely because immunocompetent mice are refractory to DENV infection unless the virus is first adapted for growth in mice [41], [42]. This restriction is thought to arise due to species-specific differences in immunological restriction factors [43], and potentially also in virus–host interactions required for viral entry into cells [44]. We and others have proposed that an immunocompetent mouse model for DENV could be developed by humanising mice through the replacement of key immunological restriction factors with their human counterparts [9], [12], [43]. Regarding the potential utility of STING in such transgenic mice, we observed enhanced levels of DENV replication in Sting −/− mouse embryonic fibroblasts (MEFs) compared to Sting +/+ (wt) MEFs using human strains from DENV serotypes 2, 3 and 4, as well as the mouse-adapted strain New Guinea C [9]. Relative to human cells, we consistently observe a more robust induction of type I IFN during DENV infection in wt MEFs and murine bone marrow-derived DCs due to the inability of the DENV protease to antagonise mSTING, and importantly IFN-β levels during DENV infection were reduced in Sting −/− MEFs in comparison to Sting +/+ MEFs [9]. Taken together, these data confirm the importance of STING signalling for the induction of type I IFN during DENV infection, as well as identifying STING as one of the species-specific DENV restriction factors in mice.
We next went on to show that hSTING is able to functionally replace mSTING in Sting −/− MEFs [9]. Specifically, only the uncleavable hSTING mutant, and not wt hSTING, rescued type I IFN induction during DENV infection in Sting −/− MEFs, with a concomitant reduction in DENV replication [9]. This is because wt hSTING, but not the uncleavable mutant, is rendered non-functional by the DENV NS2B/3 protease. These promising preliminary observations suggest that transgenic mice expressing hSTING in place of mSTING might be productively infected with non-mouse-adapted DENV strains (more relevant to human clinical infection), while retaining their ability to mount a more authentic immune response than current mouse models for DENV, which are more immunocompromised. In reality it would likely be beneficial to engineer such transgenic (knock-in) mice to also express human version of other known immunological restriction factors, such as STAT2 [43]. If successful, these immunocompetent humanised mouse models would be hugely beneficial to the development of vaccines and targeted therapies against DENV.
2.1.2. Synchronicity I: conservation of STING antagonism among flaviviruses
Although somewhat similar, the cleavage specificities of the flavivirus proteases are nevertheless distinct [45], [46]. This becomes of interest when considering the potential conservation of NS2B/3's alternative function as a STING antagonist. In fact, the Lin group did not observe STING cleavage during JEV infection or when JEV NS2B/3 was over-expressed in isolation [12]. Furthermore, we were unable to detect STING cleavage with the YFV NS2B/3 (unpublished observations), although it should be noted that we have only tested the YFV vaccine strain 17D and it is possible that part of the attenuation in this virus is due to an altered ability of the protease to antagonise STING. Nevertheless, YFV is known to antagonise STING's ability to induce type I IFN with its NS4B protein, which binds and colocalises with STING in transient transfection experiments [8]. Further mechanistic details remain to be elucidated. Interestingly, we did not observe an inhibition of type I IFN induction with the DENV-2 NS4B protein using various stimuli in human MDDCs and in 293T cells [39]. However, we did not test STING-specific stimuli in our assays. It is nevertheless interesting to postulate that DENV and YFV have evolved distinct mechanisms to antagonise STING because the different cleavage specificities of their proteases prohibit the YFV protease from degrading STING. It is surprising then that JEV does not cleave STING, especially considering that STING has been shown to be important for orchestrating the innate immune response to JEV in neuronal cells [47]. However, we cannot rule out that JEV has evolved different mechanisms for blocking STING signalling, either upstream or downstream of STING itself, or that the observations made by Yu et al. are virus strain-specific.
Finally, STING is known to be conserved across divergent vertebrate species [3], [48], [49], as well as in Drosophila [50]. It would be highly interesting to perform a systematic comparison of STING antagonism (and the diversity of mechanisms underlying it) across the Flavivirus genus in the diverse vertebrate hosts and invertebrate vectors infected by these viruses. Understanding the functions of invertebrate STING is of particular interest because how arboviruses are sensed, and how they might counteract inducible immune responses, remains poorly understood in the vector. These virus-vector immune interactions likely have a large impact on arbovirus transmission, and understanding them better may lead to new strategies for blocking arbovirus transmission.
2.1.3. End of the game: STING antagonism by the hepatitis C virus NS4B protein
Hepatitis C virus (HCV) belongs to the Hepacivirus genus within the Flaviviridae family, and is only distantly related to the flaviviruses discussed so far [28]. World-wide, an estimated 170 million people are chronically infected with HCV, with 3–4 million new infections and more than 350,000 deaths annually [51], [52]. HCV is a blood-borne virus that primarily infects hepatocytes [53]. The virus becomes persistent in 60–80% of patients, eventually causing progressive liver disease, including fibrosis, cirrhosis and in some cases hepatocellular carcinoma [51], [53], [54]. There is currently no licensed vaccine for HCV [55]. Up until recently, HCV infection was treated primarily with a combination of pegylated IFN-α and ribavirin, with a 50% success rate, however the development of specific antivirals targeting the HCV polymerase and protease has dramatically improved patient outcomes [52], [54]. Understanding the mechanisms by which HCV antagonises STING is of interest because it may help explain why the immune system spontaneously clears acute HCV infection in some patients, but not others, as well as why viral genotype-specific differences in the success of antiviral therapies are observed in the clinic [54].
The HCV protease complex, consisting of the non-structural protein NS3 and its cofactor NS4A, is well-known to disrupt the RIG-I-dependent induction of type I IFN by cleaving MAVS at residue 508 [reviewed in 52]. MAVS and NS3/4A colocalise at MAMs, and following its cleavage, MAVS loses its ability to dimerise, associate with intracellular membranes and signal the induction of type I IFN [52]. Cheng et al. were the first to report an NS3/4A-independent mechanism by which HCV counteracts IFN production [56]. In this article, inhibitors of the NS3/4A protease were shown to block the ability of HCV to antagonise IFN-β induced by MAVS over-expression, but not when IFN-β production was stimulated by poly(I:C) transfection [56]. Later, two reports published in quick succession independently demonstrated that transiently over-expressed NS4B inhibited activation of an IFN-β reporter stimulated with poly(I:C) [57] or by over-expression of the constitutively active ΔRIG-I construct [21], [58]. A concomitant reduction in downstream IFN signalling was also observed [58]. Importantly, these observations were not viral strain-specific [58].
While this early work identified an important novel role for NS4B in blocking type I IFN responses during HCV infection, it was not until 2013 that the mechanism of this antagonism was published. Following up on their initial findings [58], research led by Mamoru Watanabe demonstrated that NS4B inhibits IFN-β production and IRF-3 phosphorylation stimulated by poly(dA:dT) treatment, or STING, MAVS or ΔRIG-I over-expression [10]. Ding et al. independently made similar observations, and also showed that NS4B blocks type III IFN induction during SeV infection [13]. NS4B did not inhibit IFN-β production stimulated by over-expression of IRF-3 or NF-κB, suggesting that NS4B acts upstream of IRF-3 [13]. The NS3/4A protease complex did not block STING-mediated IFN-β induction or IRF-3 phosphorylation [10], confirming that NS3/4A and NS4B antagonise IFN production by distinct mechanisms. In addition, NS4B, but not NS3/4A, inhibited IFN production induced by over-expression of a MAVS construct that mimics the product of NS3/4A-mediated MAVS cleavage [10], [13]. This MAVS cleavage product loses its ability to dimerise and localise to the mitochondrial membrane [59], [60], [61], but residual levels of IFN-β induction are still detectable [10], [13], [59]. Therefore, HCV has evolved distinct and additive mechanisms involving NS4B and the NS3/4A protease for blocking IFN induction via RIG-I and STING.
Importantly, STING was confirmed to be important for type I and type III IFN induction by the HCV 3′ untranslated region (UTR) [13], which contains a well-characterised PAMP recognised during HCV infection [62]. Silencing STING by RNA interference (RNAi) or antagonising STING by over-expressing NS4B enhanced replication of an HCV replicon or HCV virus respectively [10]. Taken together, these data highlight the important role STING plays in signalling the induction of antiviral effectors that limit HCV replication.
Work by Ding et al. and the Watanabe group demonstrated that NS4B co-immunoprecipitates with STING when both proteins are over-expressed [10], [13]. Ding et al. further verified these findings with endogenous STING in the PH5CH8 hepatocyte cell line [13]. In addition, colocalisation of NS4B and STING was observed when both proteins were over-expressed [10], [13], with over-expressed NS4B and endogenous STING in PH5CH8 cells [13], and when STING was over-expressed during HCV infection in the hepatocyte cell line Huh7 [13]. These experiments are complicated by the fact that HCV only readily infects Huh7 cells, in which endogenous STING is undetectable by western blot [13]. Nevertheless, it is encouraging that findings from over-expression experiments have been validated with endogenous STING and during viral infection. Colocalisation and interaction of STING with NS4B was further verified using bimolecular fluorescence complementation (BiFC), in which the activity of a split fluorescent protein (in this case monomeric Kusabira Green; mKG) is reconstituted when two proteins tagged with complementary mKG fragments interact in close proximity [10]. The interaction between NS4B and STING was shown to occur adjacent to and partially colocalised with MAMs [10], which is also the site at which STING associates with MAVS [8], [10]. However, NS4B interacted and colocalised only with STING, and not with MAVS [10], [13].
The precise mechanism by which NS4B inhibits STING signalling remains to be fully elucidated. Transient over-expression of NS4B does not appear to affect levels of endogenous STING or STING dimerisation [13]. There is conflicting evidence as to whether NS4B might inhibit the interaction between STING and MAVS [10], [13], and whether NS4B targets TBK1 to inhibit IFN production [13], [58]. These contradictory findings might stem from the limitations of over-expression experiments, and until better tissue culture models for HCV are developed it will remain challenging to validate these studies in the context of viral infection. Although the exact mechanism of STING antagonism remains unclear, the N-terminus of NS4B, which shares homology with STING's dimerisation domain, appears to be important [8], [10]. While this domain is not important for the ability of NS4B to colocalise with STING, it is essential for NS4B's ability to inhibit IFN-β induction [10]. Interestingly, this STING homology domain is conserved in the NS4B proteins of other members of the Flaviviridae family [8], suggesting that the YFV NS4B might also inhibit STING signalling through a mechanism that is conserved between flaviviruses and hepaciviruses. The DENV NS4B also shares this homology domain [8]. In the future, it will be important to further clarify the mechanism and conservation of STING antagonism by Flaviviridae NS4B proteins. This is of particular interest because several antiviral compounds targeting the HCV NS4B are under development or in the early stages of clinical testing [reviewed in 63], and it has been suggested that they function at least in part by blocking the ability of HCV to inhibit innate immune responses important for viral clearance [10].
2.2. Coronaviridae
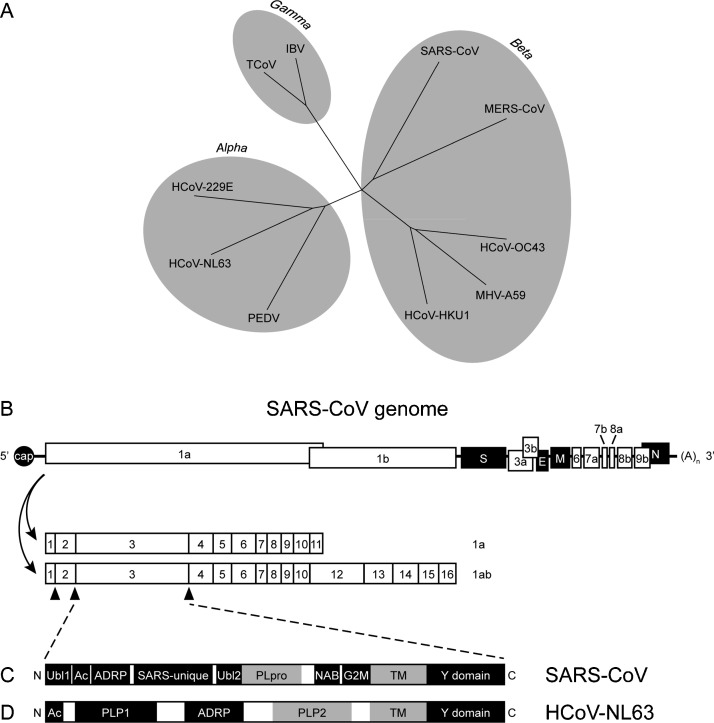
Coronaviruses are the largest RNA viruses, with non-segmented positive-sense ssRNA genomes of up to 26–32 kb in length [64]. The family Coronaviridae is further subdivided into the subfamilies Coronavirinae, which we will focus on here and which encompasses the alpha-, beta- and gammacoronaviruses (Fig. 3A), and Torovirinae [64]. Following viral entry into the cell, the replicase-transcriptase polyprotein is translated directly from the viral genome by a frame-shifting mechanism (Fig. 3B) [64]. The polyprotein is autocatalytically processed to produce 16 non-structural proteins (nsp1-nsp16), which help form cytoplasmic vesicles in which viral genome replication and the production of multiple nested subgenomic mRNAs encoding the structural and additional non-structural proteins occurs via negative-sense RNA intermediates [64]. Viral particles are assembled at the ER-Golgi intermediate compartment (ERGIC) and released via exocytic vesicles [64].
Fig. 3.
Coronavirinae phylogenetic tree, genome organisation, and nsp3 functional domains. (A) Coronavirinae family tree. Viruses of relevance to this review, and other representative viruses, shown. (B) SARS-CoV genome organisation (not to scale). Structural proteins are in black. The two polyproteins (1a and 1ab) translated by ribosomal frameshifting are shown. Triangles indicate PLpro cleavage sites; the remaining polyprotein cleavage sites are processed by the main 3C-like protease. The 3′ structural and additional accessory proteins differ among coronaviruses. Functional domains encoded by the SARS-CoV (C) and the HCoV-NL63 (D) nsp3 are also shown (not to scale). Domains linked to STING antagonism are highlighted in grey; the precise function of the transmembrane (TM) domain remains unknown. Terms not defined elsewhere; TCoV, turkey coronavirus; IBV, infectious bronchitis virus; Ubl, ubiquitin-like domain; Ac, acidic domain; ADRP, poly(ADP-ribose)-binding/ADP-ribose-1′-phosphatase macrodomain; NAB, nucleic acid-binding domain; G2M, betacoronavirus marker; Y domain, highly conserved coronavirus domain.
Coronaviruses cause several respiratory and enteric diseases of economic importance in domestic animals [64]. In addition, the alphacoronaviruses human coronavirus NL63 (HCoV-NL63) and HCoV-229E, and betacoronaviruses HCoV-OC43 and HCoV-HKU1 cause the ‘common cold’ in humans, as well as more severe disease in young, elderly and immunocompromised patients [64]. HCoV-NL63 in particular is associated with croup in children [64]. More recently, coronaviruses have attracted wide-spread media attention with the zoonotic emergence of two epidemic betacoronaviruses causing high mortality in humans; severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 [reviewed in 65]. There are no specific antivirals available to treat coronavirus infection, and no vaccines against the highly pathogenic SARS-CoV and MERS-CoV [65], [66].
Coronaviruses possess multiple mechanisms by which they evade the host innate immune response [67], [68], and it has been suggested that this immune evasion may contribute to severe coronavirus disease [69]. In addition, a better understanding of coronavirus immune evasion tactics may facilitate the development of antiviral therapies and vaccines. We will focus here on the recently characterised mechanism of STING antagonism by coronaviruses.
2.2.1. Wrapped around your finger: STING antagonism by the SARS-CoV papain-like protease
Coronavirus PLPs are contained within the nsp3 protein (Fig. 3C and D) and, along with the nsp5-encoded 3C-like protease, they are responsible for cleaving the replicase-transcriptase polyprotein [64], [68]. Antagonism of STING by coronavirus PLPs has been most extensively studied for the SARS-CoV PLP, called PLpro. SARS-CoV was first identified in China in 2003 and rapidly spread around Southeast Asia, with isolated cases reported around the globe [65], [68]. The virus is thought to have originated from bats, with palm civet cats as a potential intermediate host that transmitted the virus to humans in the crowded live-animal markets of China [65]. SARS-CoV was an air-borne pathogen that caused a severe lower respiratory tract infection with an estimated case-fatality rate of around 10% [64], [65], [68]. The epidemic spread from Southeast Asia via international air travel and trade, and was contained largely by strict quarantine and travel restrictions [65]. More than 8000 SARS cases were reported in 2003, and since then no further human infections have been reported [65]. Nevertheless, the pertinence of studying the pathogenic mechanisms of this severe respiratory pathogen was highlighted when MERS-CoV emerged in the Arabian peninsula in 2012 [65].
The ability of SARS-CoV PLpro to block type I IFN production was first reported by Devaraj et al. in 2007, when PLpro was shown to inhibit IRF-3 phosphorylation, dimerisation and nuclear translocation, and IFN-β induction in HeLa cells stimulated with poly(I:C) or SeV [70]. Interestingly, this IFN antagonism was independent of PLpro's catalytic activity, as several inactive PLpro mutants were still able to block IFN-β promoter activation with varying efficiencies [70]. Unless specified, these and subsequent studies described herein were performed with constructs encompassing just the PLP and its transmembrane domain, distinguishing between PLP function and nsp3's other activities.
Subsequent reports linking the ability of PLPs to inhibit type I IFN induction with their ability to antagonise STING all originate from the lab of Zhongbin Chen [11], [14], [15]. Initially, PLpro was shown to inhibit the activation of an interferon-stimulated response element (ISRE)-linked luciferase reporter stimulated by the exogenous over-expression of STING [11]. In line with previous reports [70], [71], PLpro still retained some ability to antagonise STING-mediated IFN induction when its catalytic activity was inactivated by point mutation [11].
PLpro was then shown to co-immunoprecipitate with STING when both proteins where over-expressed, and this interaction was also observed with catalytically inactive PLpro mutants [11]. In a follow-up paper, the STING-PLpro association was found to be dependent on the four N-terminal transmembrane domains of STING [14]. However, whether this region actively facilitates STING-PLpro binding or whether it is only necessary for localisation of STING to intracellular membranes, presumably required for interaction with membrane-bound PLpro, remains unclear, as deletion of either transmembrane domains 1 and 2, or 3 and 4 did not affect STING-PLpro binding [14].
Importantly, the interaction between PLpro and STING was found to inhibit STING function. Dimerisation of over-expressed STING, which was enhanced by treatment with SeV, was reduced in the presence of PLpro [11]. Additionally, dimerisation of over-expressed STING was reduced during SARS-CoV infection [11]. Later, PLpro was also found to disrupt the formation of signalling complexes involved in the induction of type I IFN [14]. Specifically, PLpro co-immunoprecipitated with over-expressed TBK1 and (to a lesser extent) IRF-3, and also inhibited the binding of STING to MAVS and IRF-3 [14]. In addition, PLpro dramatically inhibited the activation of luciferase reporter constructs driven by the IFN-β promoter or IRF-3-responsive elements when these were induced through the over-expression of STING or TBK1 [14]. IRF-3 phosphorylation and dimerisation induced by STING or TBK1 over-expression was also reduced in the presence of PLpro [14]. In combination, these findings demonstrate that PLpro associates with the signalling complexes assembled around STING, disrupting their formation to block downstream signalling via IRF-3. It remains to be determined whether PLpro interacts with STING directly, or whether other components of the signalling complex are required. However, other viral proteins are not necessary for mediating the interaction between PLpro and STING [11], [14].
It has been proposed that PLpro inhibits IRF-3 activation by virtue of its deubiquitinase (DUB) activity, interfering with K63-linked ubiquitination events known to be important for downstream type I IFN induction [11], [14]. Initially, X-ray crystallography studies highlighted structural similarities between PLpro and cellular DUBs [72]. Subsequently, purified PLpro was shown to hydrolyse K48- and K63-linked polyubiquitin chains, as well as the ubiquitin-like molecule ISG15, in vitro [73], [74], [75]. Deubiquitinating activity was also observed in cultured cells [71], [73], [75]. Of note, the modification of over-expressed STING, TBK1, RIG-I and IRF-3 with an HA-tagged ubiquitin construct that is only capable of forming K63-linked bonds (HA-Ub-K63) was also reduced in the presence of PLpro [14]. Surprisingly then, chemical inhibitors of PLpro's DUB activity do not affect PLpro's ability to antagonise type I IFN induction [71]. Furthermore, inactivation of PLpro's DUB domain by point mutation failed to abrogate its ability to block IFN-β induction by SeV [70]. A confounding factor in these experiments is that the importance of functional DUB activity was not always measured in the presence of STING-specific stimuli. Therefore DUB-inactive PLpro may inhibit type I IFN induction by a STING-independent mechanism, for example by targeting IRF-3 directly [70]. In addition, the Chen group has proposed that, in the absence of a functional DUB domain, PLpro is still able to block K63-linked polyubiquitination of STING signalling complex components by preventing access to the cellular ubiquitination machinery [11], [71], [76]. Alternatively, since catalytically inactive PLpro mutants still interact with STING [11], they may function by sequestering STING away from other molecules required for the induction of type I IFN. While the precise mechanism of action remains to be fully elucidated, an increasing body of work has clearly demonstrated that the SARS-CoV PLpro protein interacts with STING to prevent its dimerisation and association with other signalling molecules required for the activation and nuclear translocation of IRF-3.
2.2.2. Synchronicity II: conservation of STING antagonism among coronavirus papain-like proteases
Unlike SARS-CoV, most alpha- and betacoronaviruses encode two PLPs within their nsp3 protein (Fig. 3D) [64]. Interestingly, Clementz et al. demonstrated that one of the PLPs of the alphacoronavirus HCoV-NL63, PLP2, which, like SARS-CoV PLpro, is encoded at the C-terminal end of nsp3, also possesses DUB activity [71]. PLP2 was also shown to inhibit type I IFN production [71]. The Chen group later confirmed that PLP2 antagonises STING-induced IRF-3 nuclear translocation and ISRE activation [11].
Despite the low sequence conservation between SARS-CoV PLpro and HCoV-NL63 PLP2 [75], the mechanism of action of these two proteins appears to be very similar. PLP2 was shown to co-immunoprecipitate with STING in over-expression experiments, and this interaction was independent of PLP2's catalytic activities [11]. Furthermore, exogenously expressed PLP2 colocalised with STING in a pattern reminiscent of the ER [11]. A partial colocalisation between nsp3, which contains the PLP2 domain, and over-expressed STING was also observed in cells infected with HCoV-NL63 [11]. Importantly, PLP2 was shown to disrupt STING dimerisation, as well as the interaction between STING and TBK1 in over-expression experiments [11]. Interestingly, the DUB activity of HCoV-NL63 was found to be important for the inhibition of STING dimerisation [11]. Whether DUB activity is also important for the ability of SARS-CoV PLpro to inhibit STING dimer formation has not been published. Similarly to SARS-CoV PLpro though, the K63-linked polyubiquitination of STING was still inhibited in HCoV-NL63 PLP2 mutants that lacked DUB activity, albeit to a lesser degree [11]. As before, the authors speculated that STING-PLP2 binding may prevent the cellular ubiquitination machinery from modifying STING, regardless of whether PLP2 retains its ability to actively deubiquitinate STING [11]. Since certain PLP2 mutants that lack DUB activity are still capable of blocking STING-dependent IFN production, even though they do not prevent STING activation (dimerisation), these constructs may function by sequestering STING away from other proteins required for the induction of type I IFN [11].
The PLP2 protein of the alphacoronavirus porcine epidemic diarrhoea virus (PEDV) has also been shown to inhibit the STING-dependent activation of the IFN-β promoter and IRF-3-dependent reporters [15]. PEDV PLP2 also co-immunoprecipitates with STING in over-expression experiments, and reduces the modification of STING with K63-linked polyubiquitin chains [15]. However, unlike the coronavirus PLPs discussed so far, the ability of PEDV PLP2 to inhibit STING ubiquitination and type I IFN induction appears to be dependent on its DUB activity [15]. The authors speculated that, since the PLP constructs in the PEDV study differ from the others in that they are not linked to the C-terminal transmembrane domain of nsp3, the transmembrane domain may somehow modify PLP function [15]. In fact, the transmembrane domain was previously shown to augment IFN antagonism by both wt and catalytically-inactive HCoV-NL63 PLP2 [71]. It was proposed that the transmembrane domain may be required for the correct folding of PLP2 or for mediating interactions with components of the IFN pathway [71]. Therefore, while it is becoming increasingly clear that several coronavirus PLPs antagonise STING, certain mechanistic details, including the roles of the DUB and transmembrane domains, remain to be elucidated.
Whether the STING antagonist function of PLPs is conserved in all coronaviruses is unknown. Work led by Zhongbin Chen recently identified DUB activity in MERS-CoV PLP2, which was also shown to inhibit IRF-3 phosphorylation and nuclear translocation, and activation of the IFN-β promoter [76]. In addition, the PLP2 of the betacoronavirus murine hepatitis virus A59 (MHV-A59) has also been shown to have DUB and IFN antagonist activity [77], [78]. Whether MERS-CoV PLP2 and MHV-A59 PLP2 block type I IFN induction by targeting STING remains to be determined. It would be highly interesting to perform a systematic comparison of STING antagonism in different coronavirus PLPs using human STING as well as STING homologues from the animal reservoirs for these viruses. Such experiments would provide valuable insights into the evolutionary pressure posed by STING, the adaptations required for coronaviruses to emerge as zoonotic pathogens of humans, and the potential contribution of STING antagonism to disease severity in highly pathogenic coronaviruses. Such studies have gained in pertinence with the emergence of MERS-CoV as the most-recent highly pathogenic coronavirus with epidemic potential in humans.
3. Fields of gold: final thoughts and key questions for future research
STING has emerged as an important player in the cell's arsenal against RNA viruses. While we do not yet fully understand all of the mechanisms by which STING participates in the recognition of RNA virus infection, the multiple evasion tactics employed by divergent positive-sense RNA viruses imply that STING is a significant restriction factor for these pathogens. Exciting avenues for future research include the as-yet poorly defined mechanisms by which STING signalling is triggered during RNA virus infection. It will be highly interesting to investigate whether STING responds to RNA and DNA viruses via similar or distinct mechanisms. An intriguing possibility is that STING senses RNA viruses independently of the well-characterised nucleic acid PAMPs studied to-date, for example by responding to the aberrant modification of cellular membranes. Another key question regards the importance of STING in sensing and restricting negative-sense RNA virus infection. While improving our understanding of the mechanisms of STING activation during RNA virus infection will go some way towards answering this question, the clinching evidence will be to identify a STING antagonist in a negative-sense RNA virus, as viruses do not develop immune evasion mechanisms without a strong evolutionary imperative to do so.
In terms of flavivirus evasion of STING signalling, it will be important to address the evolutionary conservation of the various mechanisms of STING antagonism identified to-date. It will also be highly interesting to further explore the role STING plays in the host restriction of flaviviruses, and the putative roles STING homologues might play in the insect vectors of arboviral flaviviruses. Such studies will provide valuable insights into the potential for novel flaviviruses to emerge as human pathogens of the future, as well as potentially identifying new approaches for developing much-needed vaccines and antiviral therapeutics. For DENV, assessing the feasibility of using mice humanised for STING as immunocompetent animal models is of utmost importance to improve our understanding of DENV pathogenesis in vivo, and for testing vaccines and antiviral compounds.
For coronaviruses, the precise mechanism of STING antagonism remains to be clarified. It will also be important to define the contribution STING antagonism makes to the pathogenic potential of coronaviruses. The conservation of STING evasion among coronaviruses, and the role STING might play in restricting coronavirus host-switching are also exciting research avenues, especially given the emergence of two highly pathogenic coronaviruses in just over a decade.
The field of STING sensing of, and antagonism by, RNA viruses is still in its infancy, and the many important unanswered questions of relevance to diverse aspects of viral pathogenesis make this an exciting time to be working on this new player in the innate immune sensing of RNA viruses.
Acknowledgements
We acknowledge the contribution of all authors cited, and apologise if we inadvertently omitted publications related to this review. KM thanks Andrew Davidson (University of Bristol, UK) and Gillian Elliott (University of Surrey, UK) for their continued guidance and support. We also thank the entire Fernandez-Sesma group for their hard work and insightful discussions. Work in the laboratory of AF-S is funded by NIH/NIAID grants 1R01AI073450 and 1P01AI090935, by the NIH/NIAID Center for Research on Influenza Pathogenesis (CRIP) contract HHSN266200700010C as part of the CEIRS Network, and DARPA contract HR0011-11-C-0094. KM is supported by Wellcome Trust fellowship 096062. The funding bodies had no role in the preparation of this manuscript or the decision to publish.
Biographies

Kevin Maringer, PhD completed his PhD under the supervision of Prof. Gillian Elliott at Imperial College London (UK), where he studied protein interactions involved in herpesvirus assembly. Kevin currently holds a Sir Henry Wellcome postdoctoral fellowship from the Wellcome Trust. His work is focussed on improving our understanding of how dengue virus and other arboviruses interact with the immune system of the mosquito vector. Kevin's research interests include whether arboviruses evade immune recognition in their vertebrate hosts and invertebrate vectors using similar mechanisms, and how mosquito-virus interactions might affect disease processes in humans. His fellowship is based with Dr. Andrew Davidson at the University of Bristol (UK); however, Kevin is currently an Englishman in New York, performing his research with Dr. Ana Fernandez-Sesma at the Icahn School of Medicine at Mount Sinai (USA).

Ana Fernandez-Sesma, PhD is an Associate Professor in the Department of Microbiology at Icahn School of Medicine at Mount Sinai. She has extensive experience on antiviral immunity using animal and primary human systems and her research focus is the modulation of innate immunity by viruses, with a special focus in type I interferons (IFN). The three areas of study in her laboratory are the modulation and control of innate immunity by dengue virus (DENV), influenza virus and HIV. Her group has optimised and developed several assays and techniques to study the initiation/modulation of immune responses in human primary immune cells, such as dendritic cells (DCs) and macrophages as well as primary lung epithelial cells and more recently human tonsil histocultures. She has over 45 publications in Virology and Immunology related Journals and is on the editorial board of several journals, including Journal of Virology and PLoS Pathogens. She is also a permanent member of the NIH study section Virology B and an active teacher for Medical and graduate students at Icahn School of Medicine at Mount Sinai (ISMMS).
References
- 1.Yoneyama M., Fujita T. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Sastre A., Biron C.A. Type 1 interferons and the virus–host relationship: a lesson in detente. Sci Rep. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong B., Yang Y., Li S., Wang Y.-Y., Li Y., Diao F. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Sun W., Li Y., Chen L., Chen H., You F., Zhou X. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unterholzner L. The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218:1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Ran Y., Shu H.-B., Wang Y.-Y. MITA/STING: a central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev. 2014:1–9. doi: 10.1016/j.cytogfr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitta S., Sakamoto N., Nakagawa M., Kakinuma S., Mishima K., Kusano-Kitazume A. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 11.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu C.-Y., Chang T.-H., Liang J.-J., Chiang R.-L., Lee Y.-L., Liao C.-L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Q., Cao X., Lu J., Huang B., Liu Y.-J., Kato N. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang S., Song X., Wang Y., Ru H., Shaw N., Jiang Y. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Hu M.-M., Wang Y.-Y., Shu H.-B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe T., Harashima A., Xia T., Konno H., Konno K., Morales A. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowzard J.B., Ranjan P., Sambhara S., Fujita T. Antiviral defense: RIG-Ing the immune system to STING. Cytokine Growth Factor Rev. 2009;20:1–5. doi: 10.1016/j.cytogfr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 22.Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Sci Rep. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol. 2013;2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach B.D., Thiel H.-J., Rice C.M. Flaviviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. 5th ed. vol. I. Lippincott Williams and Wilkins; Philadelphia, PA USA: 2007. pp. 1102–1152. (Fields virology). [Google Scholar]
- 29.Clyde K., Kyle J.L., Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol. 2006;80:11418–11431. doi: 10.1128/JVI.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 31.Heinz F.X., Stiasny K. Flaviviruses and flavivirus vaccines. Vaccine. 2012;30:4301–4306. doi: 10.1016/j.vaccine.2011.09.114. [DOI] [PubMed] [Google Scholar]
- 32.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver S.C., Barrett A.D.T. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubler D.J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halstead S.B. Dengue vaccine development: a 75% solution? Lancet. 2012;380:1535–1536. doi: 10.1016/S0140-6736(12)61510-4. [DOI] [PubMed] [Google Scholar]
- 36.Thisyakorn U., Thisyakorn C. Latest developments and future directions in dengue vaccines. Ther Adv Vaccines. 2014;2:3–9. doi: 10.1177/2051013613507862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride W.J.H., Bielefeldt-Ohmann H. Dengue viral infections pathogenesis and epidemiology. Microbes Infect. 2000;2:1041–1050. doi: 10.1016/s1286-4579(00)01258-2. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Madoz J.R., Bernal-Rubio D., Kaminski D., Boyd K., Fernandez-Sesma A. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol. 2010;84:4845–4850. doi: 10.1128/JVI.02514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Madoz J.R., Belicha-Villanueva A., Bernal-Rubio D., Ashour J., Ayllon J., Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J Virol. 2010;84:9760–9774. doi: 10.1128/JVI.01051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin L., Xu L.-G., Yang I.V., Davidson E.J., Schwartz D.A., Wurfel M.M. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meiklejohn G., England B., Lennette E. Propagation of dengue virus strains in unweaned mice. Am J Trop Med Hyg. 1952;1:51–58. doi: 10.4269/ajtmh.1952.1.51. [DOI] [PubMed] [Google Scholar]
- 42.Zompi S., Harris E. Animal models of dengue virus infection. Viruses. 2012;4:62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashour J., Morrison J., Laurent-Rolle M., Belicha-Villanueva A., Plumlee C.R., Bernal-Rubio D. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shresta S., Sharar K.L., Prigozhin D.M., Beatty P.R., Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol. 2006;80:10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 46.Shiryaev S.A., Kozlov I.A., Ratnikov B.I., Smith J.W., Lebl M., Strongin A.Y. Cleavage preference distinguishes the two-component NS2B–NS3 serine proteinases of Dengue and West Nile viruses. Biochem J. 2007;401:743. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazmi A., Mukhopadhyay R., Dutta K., Basu A. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep. 2012;2:1–10. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng X., Yang C., Zhang Y., Peng L., Chen X., Rao Y. Identification, characterization and immunological response analysis of stimulator of interferon gene (STING) from grass carp Ctenopharyngodon idella. Dev Comp Immunol. 2014;45:163–176. doi: 10.1016/j.dci.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Xie L., Liu M., Fang L., Su X., Cai K., Wang D. Molecular cloning and functional characterization of porcine stimulator of interferon genes (STING) Dev Comp Immunol. 2010;34:847–854. doi: 10.1016/j.dci.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Kemp C., Imler J.-L. Antiviral immunity in drosophila. Curr Opin Immunol. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 52.Horner S.M. Activation and evasion of antiviral innate immunity by hepatitis C virus. J Mol Biol. 2014;426:1198–1209. doi: 10.1016/j.jmb.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemon S.M., Walker C., Alter M.J., Yi M. Hepatitis C virus. In: Knipe D.M., Howley P.M., editors. 5th ed. vol. I. Lippincott Williams and Wilkins; Philadelphia, PA USA: 2007. pp. 1254–1305. (Fields virology). [Google Scholar]
- 54.Park S.-H., Rehermann B. Immune responses to HCV and other hepatitis viruses. Immunity. 2014;40:13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang T.J. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng G., Zhong J., Chisari F.V. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2006;103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moriyama M., Kato N., Otsuka M., Shao R.-X., Taniguchi H., Kawabe T. Interferon-beta is activated by hepatitis C virus NS5B and inhibited by NS4A, NS4B, and NS5A. Hepatol Int. 2007;1:302–310. doi: 10.1007/s12072-007-9003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tasaka M., Sakamoto N., Itakura Y., Nakagawa M., Itsui Y., Sekine-Osajima Y. Hepatitis C virus non-structural proteins responsible for suppression of the RIG-I/Cardif-induced interferon response. J Gen Virol. 2007;88:3323–3333. doi: 10.1099/vir.0.83056-0. [DOI] [PubMed] [Google Scholar]
- 59.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 60.Baril M., Racine M.-E., Penin F., Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X.-D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rai R., Deval J. New opportunities in anti-hepatitis C virus drug discovery: targeting NS4B. Antivir Res. 2011;90:93–101. doi: 10.1016/j.antiviral.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 64.Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. 6th ed. vol. I. Lippincott Williams and Wilkins; Philadelphia, PA USA: 2013. pp. 826–859. (Fields virology). [Google Scholar]
- 65.Coleman C.M., Frieman M.B. Coronaviruses: important emerging human pathogens. J Virol. 2014;88:5209–5212. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davidson A.D., Siddell S. Potential for antiviral treatment of severe acute respiratory syndrome. Curr Opin Infect Dis. 2003;16:565–571. doi: 10.1097/00001432-200312000-00009. [DOI] [PubMed] [Google Scholar]
- 67.Kindler E., Thiel V. To sense or not to sense viral RNA – essentials of coronavirus innate immune evasion. Curr Opin Microbiol. 2014;20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol. 2014;95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 77.Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS ONE. 2011;6:e17192. doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]