Abstract
IMPORTANCE
Epigenetic studies present unique opportunities to advance schizophrenia research because they can potentially account for many of its clinical features and suggest novel strategies to improve disease management.
OBJECTIVE
To identify schizophrenia DNA methylation biomarkers in blood.
DESIGN, SETTING, AND PARTICIPANTS
The sample consisted of 759 schizophrenia cases and 738 controls (N = 1497) collected in Sweden. We used methyl-CpG–binding domain protein-enriched genome sequencing of the methylated genomic fraction, followed by next-generation DNA sequencing. We obtained a mean (SD) number of 68 (26.8) million reads per sample. This massive data set was processed using a specifically designed data analysis pipeline. Critical top findings from our methylome-wide association study (MWAS) were replicated in independent case-control participants using targeted pyrosequencing of bisulfite-converted DNA.
MAIN OUTCOMES AND MEASURES
Status of schizophrenia cases and controls.
RESULTS
Our MWAS suggested a considerable number of effects, with 25 sites passing the highly conservative Bonferroni correction and 139 sites significant at a false discovery rate of 0.01. Our top MWAS finding, which was located in FAM63B, replicated with P = 2.3 × 10−10. It was part of the networks regulated by microRNA that can be linked to neuronal differentiation and dopaminergic gene expression. Many other top MWAS results could be linked to hypoxia and, to a lesser extent, infection, suggesting that a record of pathogenic events may be preserved in the methylome. Our findings also implicated a site in RELN, one of the most frequently studied candidates in methylation studies of schizophrenia.
CONCLUSIONS AND RELEVANCE
To our knowledge, the present study is one of the first MWASs of disease with a large sample size using a technology that provides good coverage of methylation sites across the genome. Our results demonstrated one of the unique features of methylation studies that can capture signatures of environmental insults in peripheral tissues. Our MWAS suggested testable hypotheses about disease mechanisms and yielded biomarkers that can potentially be used to improve disease management.
The methylation of DNA cytosine residues at the carbon 5 position is a common epigenetic modification that is often found in the sequence context CpG. Investigations of these markings provide a promising complement to schizophrenia studies of DNA sequence variation. First, methylation can directly affect gene expression, so it may capture additional variation in disease susceptibility. Indeed, specific epimutations have already been associated with human diseases, including psychiatric disorders.1 Second, methylation studies may advance our understanding of schizophrenia. For example, they can potentially account for a variety of features, such as its episodic nature.2,3 Third, the translational potential is considerable. For example, epigenetic markings are modifiable by pharmaceutical interventions, making them possible new drug targets.4
The pathogenic processes for psychiatric disorders likely involve the brain. However, brain tissue is not readily accessible in living patients, so blood is typically used in biomarker studies. There are 2 models explaining how methylation studies in blood can advance schizophrenia research.5 Neither model assumes that methylation in blood directly affects disease susceptibility, although this is possible, in principle, because blood provides a biological environment for other tissues, including the brain. In the “signature” model, associations occur between schizophrenia and methylation markings because the factors that increase disease susceptibility leave a biomarker signature in blood. Thus, the methylation markings in blood implicate a cause of the disease, which may affect schizophrenia through processes that are unrelated to methylation in the brain. In contrast, the “functional mirror site” model assumes a causal role of methylation sites in the brain. When the methylation status of these sites in the brain is mirrored by the corresponding sites in the blood, we will observe associations between schizophrenia and methylation markings at the same loci in blood. Compared with tissue-specific differentially methylated regions,6 correlated methylation profiles across tissues are common. Mirror sites occur because peripheral tissues may reveal methylation markings predating or resulting from the epigenetic reprogramming events affecting the germ line and embryogenesis,7 and environmental factors and genetic polymorphisms can affect methylation levels in multiple tissues.8,9 To study the 2 models,5 we administered haloperidol decanoate to inbred mice and then performed whole-methylome profiling in the blood, cortex, and hippocampus. More than 65% of the sites showed correlated changes where the concordance rates were similar between blood and brain vs between the 2 brain tissues. This showed that factors affecting brain processes (eg, haloperidol) can leave biomarker signatures in blood and that the methylation status of many sites in the brain is mirrored in the blood.
Current knowledge about the role of DNA methylation in schizophrenia is mainly acquired from relatively small studies of peripheral blood10–17 and postmortem brain tissue.18–26 Most studies focused on specific genes, such as RELN,19,22 HTR2A,20 COMT,13,18 SOX10,23 and FOXP2.26 Two studies1,12 investigated a broader set of sites. One investigated approximately 12 000 regulatory regions in postmortem brain tissue samples from 35 patients with schizophrenia and 35 controls.1 It reported differences in the vicinity of loci that can be functionally linked to disease etiology. The second study12 investigated approximately 27 000 CpG sites in peripheral blood from 11 pairs of monozygotic twins discordant for schizophrenia. Dempster et al12 observed significant epigenetic disruptions in biological networks relevant to psychiatric disease and neurodevelopment.
The goal of the present study is to identify schizophrenia methylation biomarkers in blood through a methylome-wide association study (MWAS). The most comprehensive method involves the use of next-generation sequencing after bisulfite conversion of unmethylated cytosines. Currently, however, this is not economically feasible considering the sample sizes required for an MWAS.27 As a cost-effective alternative, we first captured the methylated DNA fragments and then sequenced this methylation-enriched portion of the genome28 (see Aberg et al29 for a discussion of the merits of methyl-CpG binding domain [MBD]protein-enriched genome sequencing [MBD-seq]). Our “discovery” MWAS sample consisted of almost 1500 schizophrenia cases and controls. Critical findings were replicated in an independent group of participants using targeted bisulfite pyrosequencing.
Methods
Detailed descriptions of the method can be found else-where.29–31 Our study was approved by the institutional review board at Karolinska Institutet, Stockholm, Sweden, and written informed consent was obtained from all participants.
Sample
Table 1 describes the “discovery” MWAS and replication samples. All participants were selected from national population registers in Sweden and are part of a larger study.32 Because 3 participants withdrew their consent during the study, we report results for 1497 participants. Key findings were replicated in an independent group of 1144 participants and at other sites in an independent group of 360 participants. For all participants, DNA was extracted from the buffy coat of whole blood.
Table 1.
Key Findings of Methylome-Wide Association Study of Schizophreniaa
| Variable | % of Participants (% of Participants With Missing Data) | ||
|---|---|---|---|
| Cases | Controls | Total | |
| Discovery sample | |||
| Participants, No. | 759 | 738 | 1497 |
| Male | 54.9 (0) | 54.1 (0) | 54.5 (0) |
| Non-Finnish Nordic parents | 89.4 (19.1) | 93 (13.1) | 91.2 (16.2) |
| Alcohol use | 58.1 (15) | 93.6 (13.8) | 75.7 (14.4) |
| Smoker | 50.4 (82.2) | 53.8 (85.6) | 51.9 (83.9) |
| Narcotic use | 25.5 (1.7) | 6.8 (0.1) | 16.2 (0.9) |
| Epilepsy | 5.1 (1.8) | 0.5 (0.8) | 2.8 (1.3) |
| Diabetes | 8.8 (1.4) | 3.4 (0.8) | 6.1 (1.1) |
| Hyperthyroid | 0.5 (3) | 1.7 (3.8) | 1.1 (3.4) |
| Hypothyroid | 4.5 (2.9) | 2.7 (3.5) | 3.6 (3.2) |
| Autoimmune disorder | 1 (9.6) | 1.8 (2.4) | 1.4 (6.1) |
| Age, mean (SD), y | 53.2 (11.6) | 55.1 (11.8) | 54 (11.7) |
| Replication sample 1 | |||
| Participants, No. | 178 | 182 | 360 |
| Male | 61.8 (0) | 65.9 (0) | 63.9 (0) |
| Alcohol use | 56.1 (16.9) | 95.4 (4.4) | 77.3 (10.6) |
| Smoker | 40.1 (17.4) | 13.2 (0) | 25.2 (8.6) |
| Narcotic use | 19.1 (0) | 7.1 (0) | 13.1 (0) |
| Autoimmune disorder | 1.2 (3.9) | 0.6 (4.9) | 0.9 (4.4) |
| Age, mean (SD), y | 56.4 (10.7) | 58.5 (10.3) | 57.5 (10.6) |
| Replication sample 2 | |||
| Participants, No. | 561 | 582 | 1144 |
| Male | 63.3 (0) | 60.5 (0) | 61.9 (0.1) |
| Alcohol use | 55.2 (10) | 94.9 (2.2) | 76.3 (6.1) |
| Smoker | 42.7 (38.7) | 12 (0) | 23.4 (19.1) |
| Narcotic use | 20.3 (0.9) | 6.5 (0.2) | 13.3 (0.6) |
| Autoimmune disorder | 0.6 (5.2) | 0.2 (2.7) | 0.4 (4) |
| Age, mean (SD), y | 54.5 (11.4) | 57.1 (10.9) | 55.8 (11.2) |
Because of screening, 100% of the participants in the replication samples have Non-Finnish Nordic parents, and 0% of the replication samples have participants who received a diagnosis of epilepsy, diabetes mellitus, hyperthyroid, or hypothyroid.
Whole-Methylome Profiling
We used MethylMiner (Invitrogen), which employs MBD protein-based enrichment of the methylated DNA fraction, followed by single-end sequencing (50 base-pair reads) on the SOLiD platform (Life Technologies). We eluted the captured methylated fraction with 0.5M sodium chloride to increase the relative number of fragments from CpG-poorregions,29 which otherwise would not be as well covered.33 To avoid batch effects, samples were processed in random order.
eTable 1 in the Supplement gives descriptive statistics for a variety of sequencing parameters. In summary, after deleting reads with more than 2 missing calls, we obtained a mean (SD) number of 68(26.8) million reads per sample. Reads were aligned (build hg19/GRCh37) using BioScope 1.2 (Life Technologies). We deleted all samples with less than 40% alignment. For the remaining samples, the mean (SD) percentage of mapped reads was 69.2% (6.2%). We eliminated 32.1% of the mapped reads because they were low-quality multireads (reads aligning to multiple locations) or duplicate reads (reads with identical start positions). We excluded 38 participants because less than 15 million reads remained after quality control. This left 1459 participants with a mean (SD) number of 32.4 (13.7) million quality-control reads. Using data from 73 technical replicates, we observed a mean/median correlation of 0.90/0.92 between the methylation profiles from the replicates.29 This supported the reproducibility of our assay.
The MBD protein only binds to methylated CpG sites, so we only consider the 26 752 702 autosomal CpG sites in the reference genome for our analysis. The 10.5 million CpG sites (36%) located in regions showing alignment problems were eliminated.29 Most (71.8%) of these were in regions flagged as repetitive elements by RepeatMasker (http://www.repeatmasker.org/). Methylation measurements were obtained by estimating how many fragments covered each CpG site.31 Highly intercorrelated coverage estimates at adjacent CpG sites were combined to obtain more reliable measurements.34 Rather than using a sliding window of an arbitrary fixed length, we combined sites adaptively based on their observed intercorrelations.30 Using the 99th percentile of the coverage estimates at non-CpG sites29 as the threshold for background noise, we excluded 730 522 blocks with low coverage (likely unmethylated) from further analysis (eFigure 1 in the Supplement). This left 4 344 016 blocks for association testing.
Association Testing, Confounders, and Tissue Heterogeneity
A variety of efforts were made to control for confounders. First, we regressed out possible assay-related technical artifacts such as the quantity of genomic DNA starting material, the quantity of methylation-enriched DNA captured, and the sample batch. In addition, we controlled for age and sex.
Second, after regressing out the measured confounders, we performed principal component analysis to capture the major remaining unmeasured confounders. Because existing soft-ware cannot handle the ultrahigh-dimensional MWAS data, we used our own software30 that allows for parallel processing, that uses C++ for CPU-intensive and input/output-intensive calculations, and that follows Gower35 by performing the eigen-decomposition of a much smaller transposed variant of the data matrix. Based on a scree test (eFigure 2 in the Supplement), the first 7 principal components were regressed out of the association analysis.
Third, we correlated principal component scores with a variety of variables to check whether additional covariates were required (see Table 2 in Aberg et al29). For example, these analyses showed that, in this fairly homogeneous sample, ancestry did not contribute substantially to variation in the methylome, and it was therefore not included as a covariate.
Table 2.
Gene Network Analyses of 139 Findings From MWAS of Schizophrenia
| Variable | Significant Genes in MWAS | Totala | Genes,b No. | Enrichmentc | P Value |
|---|---|---|---|---|---|
| Protein-protein interaction | |||||
| IgA1 | FCAR, RUNX3, CREB1, SMAD3 | 19 | 4 | 23.8 | 4.6 × 10−6 |
| elf5 | FRZB, SS18L1, RPS15A | 10 | 3 | 34.0 | 2.5 × 10−5 |
| EPAS1 | ARNT, SMAD3 | 2 | 2 | 113.2 | 3.7 × 10−5 |
| RhoG |
CHN2, TIAM1, ARHGAP15, PIK3R3, ARHGAP26, RHOH GDI2 |
153 | 7 | 5.2 | 4.1 × 10−5 |
| ETS1 | RUNX2, ETS2, SMAD3, CREB1, ARNT | 76 | 5 | 7.4 | 9.8 × 10−5 |
| Chp | TIAM1, CHN2, ARHGAP15, ARHGAP26, RHOH, GDI2 | 127 | 6 | 5.3 | 1.2 × 10−4 |
| mtf2 | PRKD2, CREB1, SMAD3 | 17 | 3 | 20.0 | 1.4 × 10−4 |
| PEBPB2 | RUNX2, RUNX3, SMAD3 | 20 | 3 | 17.0 | 2.3 × 10−4 |
| PKC-delta |
PPM1D, INPP5D, TIAM1, PPARA, RUNX2, CHN2, CXCR4 PHLPP1 |
274 | 8 | 3.3 | 2.6 × 10−4 |
| EP300 |
RUNX2, TRERF1, ARNT, PPARA, CREB1, SMAD3, ETS2 RUNX3, SS18L1, SATB1, TCF12, BRD4 |
616 | 12 | 2.2 | 3.5 × 10−4 |
| Pathway | |||||
| Signaling by Rho GTPases | TIAM1, CHN2, ARHGAP15, ARHGAP26, RHOH, GDI2 | 126 | 6 | 3.3 | 1.1 × 10−4 |
| PIP3 activates AKT signaling | MAPKAP1, PHLPP1, CREB1 | 28 | 3 | 7.5 | 6.2 × 10−4 |
| HIF1A transcription factor network |
SMAD3, CXCR4, CREB1, ARNT | 66 | 4 | 4.3 | 6.6 × 10−4 |
| Regulated nuclear SMAD2/3 signaling |
RUNX2, RUNX3, CREB1, SMAD3 | 78 | 4 | 3.6 | 1.2 × 10−3 |
| Signaling by SCF-KIT | MAPKAP1, PHLPP1, PIK3R3, CREB1 | 78 | 4 | 3.6 | 1.2 × 10−3 |
| Pyrimidine salvage reactions | UPP2, UCK2 | 9 | 2 | 15.6 | 1.3 × 10−3 |
| Androgen receptor | RUNX2, SMAD3, KDM3A, CREB1, ARNT | 146 | 5 | 2.4 | 1.9 × 10−3 |
| Wnt core | FRZB, CREB1, SMAD3 | 44 | 3 | 4.8 | 2.3 × 10−3 |
| Fcγ-mediated phagocytosis | FCGR2B, PIK3R3, INPP5D, FCGR2C | 95 | 4 | 3.0 | 2.6 × 10−3 |
| Erythropoietin neuroprotective NF-κB |
CREB1, ARNT | 13 | 2 | 10.8 | 2.7 × 10−3 |
| microRNA | |||||
| miR-376c | ARNT, FAM63B, BRD4, MYT1L, TIAM1, SATB1, ACSL1 | 155 | 7 | 5.2 | 4.0 × 10−4 |
| miR-217 |
GDI2, TCF12, PPM1D, AFF3, SATB1, RUNX2, BRP44L CHN2 |
234 | 8 | 3.9 | 9.9 × 10−4 |
| miR-19 |
RUNX3, PIK3R3, MBD6, NDFIP2, DAAM1, MYT1L, TIAM1 PPARA, SATB1, RBMS3, FOXP1, FNDC3B, NAALADL2 ACSL1, NUS1, CSPP1 |
923 | 16 | 2.0 | 7.7 × 10−3 |
| miR-218 |
CSDE1, ARL3, MBD6, CHFR, DAAM1, FOXN3, RCOR1 TCF12, FAM63B, MYT1L, RASSF2, ACSL1, RUNX2 |
689 | 13 | 2.2 | 8.4 × 10−3 |
| miR-9 |
PIK3R3, BTBD10, TRAPPC6B, FOXN3, RCOR1, FAM63B PPARA, TBC1D22A, RBMS3, FOXP1, FNDC3B, IPO11 NUS1, MTHFD1L, AUH |
926 | 15 | 1.8 | 2.3 × 10−2 |
| miR-544 | PIK3R3, CSDE1, PELI2, RCOR1, SATB1, FOXP1, MLL3 | 339 | 7 | 2.4 | 2.9 × 10−2 |
| miR-504 | ARNT, FAM63B, MYT1L, FOXP1 | 138 | 4 | 3.3 | 3.3 × 10−2 |
| miR-181 |
PIK3R3, ARL3, RCOR1, TMEM131, CREB1, RASSF2 SS18L1, PPARA, FOXP1, NAALADL2, GNB4, ACSL1, NUS1 MLL3 |
883 | 14 | 1.8 | 3.4 × 10−2 |
| miR-135 |
PIK3R3, ARNT, MBD6, PELI2, FOXN3, RCOR1, MYT1L NAALADL2, MAN1A1 |
505 | 9 | 2.0 | 4.2 × 10−2 |
| miR-203 |
MBD6, TRAPPC6B, SMAD3, SS18L1, ETS2, FOXP1 NSUN7, ACSL1, RUNX2 |
548 | 9 | 1.9 | 5.7 × 10−2 |
Abbreviation: MWAS, methylome-wide association study.
Total number of genes in the network.
Number of genes in the network with q < 0.01 in the MWAS.
Calculated as the proportion of genes observed in the network vs the proportion of genes expected assuming no enrichment.
Blood consists of a variety of cell types. By using whole-blood samples, we are studying an “average” methylation pattern that will be dominated by the common types. This can produce false positives only if both (1) the relative abundance of common cell types differs across cases and controls, and (2) methylation patterns of common cell types differ. Ideally, we would have case-control MBD-seq data obtained from separated white blood cells36 to identify sites that are at risk for creating false positives. The principal component analysis provides an alternative in situations where cell-type heterogeneity affects many methylation sites.36–38 Participants with a similar cell-type composition will have more similar multilocus methylation patterns, and these patterns will be captured by the principal components. However, situations where few methylation sites are involved will remain uncorrected. We note, however, that most tissue samples will be heterogeneous, so similar risks are present when studying other tissues too.
Network Analyses
We used ConsensusPathDB39–41 to generate protein-protein interaction (PPI) networks and perform pathway analyses based on the Reactome,42 Kyoto Encyclopedia of Genes and Genomes,43 and BioCarta databases. To create microRNA (miRNA) networks, we used the University of California, Santa Cruz, genome track TS miRNA site for GRCh37/hg19, which is based on TargetScan 5.1 (Bioinformatics and Research Computing). All blocks with q < 0.01 in the MWAS were matched to the closest gene ±20 kilobases. For each of the 4601 reference pathways present in ConsensusPathDB, incorporating 9859 known genes, a hypergeometric test was performed to study whether the overlap between the top MWAS genes and those present in each reference pathway was higher than expected by chance.
Replication
For the replication, we used targeted bisulfite pyrose-quencing.44,45 We replicated the top 5 MWAS findings and 10 sites selected from the network analyses. Controlling the familywise error rate at the α level of .05 through a Bonferroni correction therefore gives a threshold of .05/15 = 3.3 × 10−3. We conservatively used the highest (least significant) P value if there were multiple (correlated) CpG sites in the same assay. Finally, we added a negative control by assaying a site with a high nonsignificant MWAS P value, and to assess the efficacy of the principal component analysis, we selected the 2 most significant findings obtained after performing the MWAS without principal components.
For network/pathways findings, a second “replication” opportunity existed by testing whether, after excluding the (top) findings used to identify the networks, the remaining genes from that network are also associated with case-control status in the MWAS (for miRNA networks, these tests are not suitable because miRNA likely regulates genes that may have different functions). For this purpose, we performed permutation tests.
Results
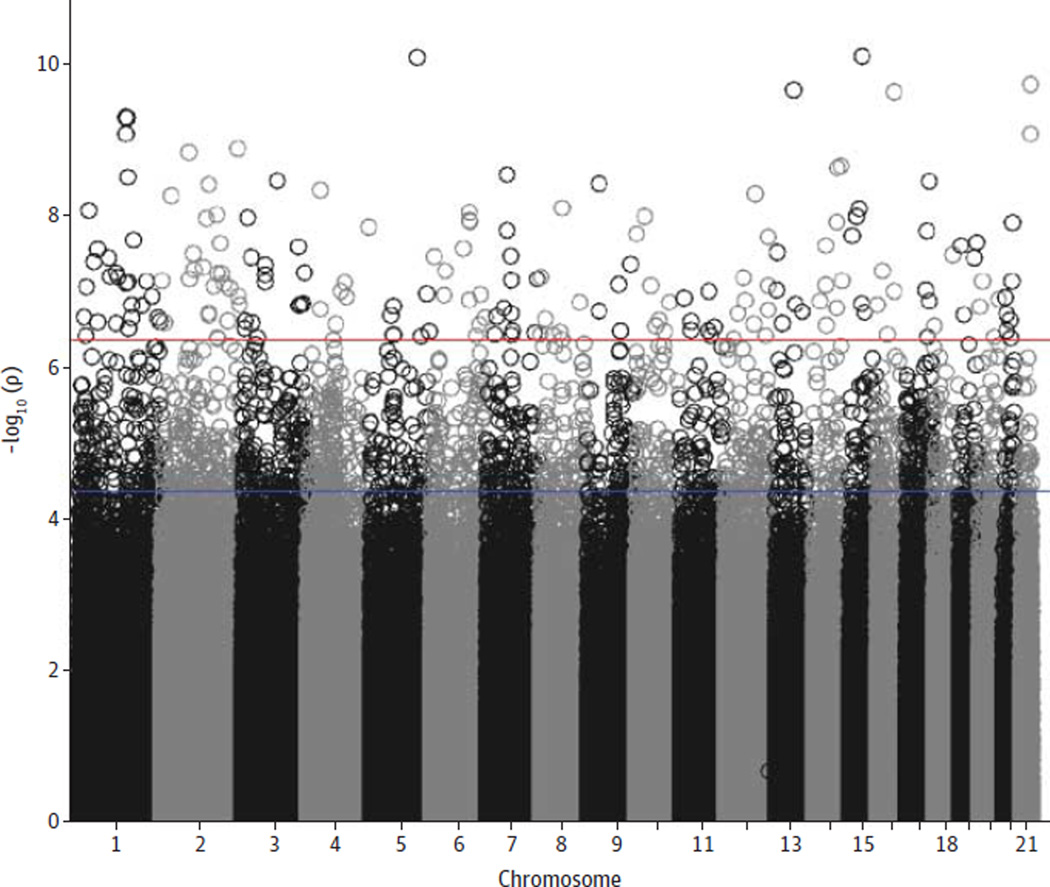
Our Figure shows the MWAS Manhattan plot with 139 tests with q < 0.01, meaning that less than 1% of the 139 findings are expected to be false discoveries (eFigure 3 in the Supplement).46,47 The P values for these sites ranged from 10−7 to 10−11, with 25 sites reaching significance after we used the highly conservative Bonferroni correction (threshold P = 1.15 × 10−8). Our test statistic inflation parameter λ of 1.12 was higher compared with what is commonly observed in genome-wide association studies. This λ value is unlikely an artifact. After we performed a square root transformation to normalize the data and mitigate the effects of possible outliers, λ did not change. Furthermore, increasing the stringency of the quality control resulted in higher rather than lower λ values (eFigure 4 in the Supplement). Instead, this λ value reflects that methylation studies are more akin to gene expression studies that typically show many correlated effects with relatively large effect sizes.
Figure. Methylome-Wide Association Study Manhattan Plot.
The 22 autosomes are displayed along the x-axis, with the negative logarithm of the association P value for each block displayed on the y-axis. All P values above the upper (red) line have q values of less than 0.01, and those above the lower (blue) line have q values of less than 0.1.
Of the 139 MWAS findings, 112 overlapped with genes. Table 2 shows that regardless of whether we used PPI networks, pathway databases, or miRNA target networks, hypoxia was the dominant theme. For example, the PPI network centered on EPAS1 (previously known as hypoxia-inducible factor 2) includes 2 genes, both of which were detected in our MWAS. EPAS1 encodes a transcription factor induced as oxygen levels fall and is known to specifically interact with ETS1, another center for a PPI network among our findings, which is involved in the regulation of vascular development in the neonatal mouse brain.48 Furthermore, transcription coactivator EP300 is necessary for hypoxia-induced transcriptional activation and is upregulated in low-oxygen conditions.49 Using reference biological pathways, we detected the hypoxia-inducible factor 1 alpha (HIF1A) transcription factor network. HIF1A, together with ARNT, forms hypoxia-inducible factor (HIF), which regulates hypoxia-inducible genes.50 In addition, AKT signaling is an important modulator of HIF activity,51 and signaling by Rho GTPases has been linked to hypoxia response, particularly in the vascular system.52 Finally, miRNA miR-217 regulates heme oxygenase 1, an enzyme responsive to hypoxic conditions.53
Other findings shown in Table 2 converge on immune sys-tem themes. A prominent example is IgA1 (encoded by IGHA1), which is highlighted by our PPI network analyses. Although several genes associated with this network (RUNX3, CREB1, and SMAD3) are involved in multiple pathways, FCAR is highly specific to IgA because it encodes the receptor for the Fc fragment of IgA. In blood, FCAR interacts with IgA to initiate inflammatory reactions and phagocytosis. Fcγ-mediated phagocytosis, related to the action of IgG, was also among the top findings in our pathway analysis.
Table 3 shows the replication results (for design features of pyrosequencing assays and for full replication results, see eTables 2 and 3, respectively, in the Supplement). Except for the control sites, the direction of effects was the same in the replication as in the MWAS. Although all 5 top findings had replication P values of less than .05, only FAM63B remained significant after applying our very conservative correction for multiple testing. FAM63B was our top MWAS finding with a P = 6.3 × 10−11 (q = 2.1 × 10−4). The replication assay contained 3 CpG sites. The highest P value (2.3 × 10−10) of these 3 CpG sites was below our multiple testing threshold of P = 3.3 × 10−3. Table 2 shows that FAM63B is part of 4 networks regulated by miRNA. Three types of these miRNA (miR-218, miR-9, and miR-504) can be linked to neuronal differentiation and dopaminergic gene expression.54–56
Table 3.
Replication Results for MWAS Top Findings, Network Findings, Candidate Gene, and Control Sitesa
| Chr | Geneb | Featurec | MWAS | Replication | |||||
|---|---|---|---|---|---|---|---|---|---|
| P Value | q Value | Samples,d No |
Position, bp | P Value | Cohende | Mean (SD) Methylation Level, % |
|||
| Top 5 MWAS findings | |||||||||
| 15 | FAM63Bf | Exon, 3′ UTR | 6.28 × 10−11 | 2.08 × 10−4 | 1075 | 59 146 738 | 3.76 × 10−12 | −0.447 | 35.55 (8.73) |
| 1065 | 59 146 744 | 7.72 × 10−12 | −0.447 | 37.11 (9.01) | |||||
| 1033 | 59 146 756 | 2.31 ×10−10 | −0.424 | 34.72 (8.49) | |||||
| 5 | ARHGAP26 | 8.19 × 10−11 | 2.08 × 10−4 | 333 | 142 138 110 | 2.96 × 10−3 | −0.274 | 28.99 (8.17) | |
| 331 | 142 138 121 | 1.03 × 10−2 | −0.216 | 25.83 (7.40) | |||||
| 315 | 142 138 140 | 5.54 × 10−3 | −0.284 | 26.76 (7.24) | |||||
| 13 | CTAGE11P | 2.05 ×10−10 | 2.27 × 10−4 | 344 | 75 795 508 | 2.71 × 10−3 | −0.285 | 40.07 (8.28) | |
| 330 | 75 795 510 | 8.04 × 10−3 | −0.355 | 44.58 (8.52) | |||||
| 22 | TBC1D22A | Intron | 2.08 ×10−10 | 2.27 × 10−4 | 335 | 47 170 345 | 6.30 × 10−3 | −0.328 | 15.25 (6.80) |
| 328 | 47 170 354 | 2.03 × 10−2 | −0.301 | 12.41 (6.46) | |||||
| 323 | 47 170 371 | 3.24 × 10−2 | −0.255 | 10.78 (5.48) | |||||
| 16 | Intergenic | 2.23 ×10−10 | 2.27 × 10−4 | 351 | 54 209 779 | 8.07 × 10−2 | −0.187 | 51.49 (8.79) | |
| 353 | 54 209 801 | 4.85 × 10−2 | −0.197 | 37.08 (8.86) | |||||
| Network findings | |||||||||
| 22 | PKDREJ/PPARAf | Exon | 1.46 × 10−9 | 7.42 × 10−4 | 1094 | 46 655 187 | 5.94 × 10−10 | −0.390 | 31.11 (8.65) |
| 14 | RCOR1 | Intron | 3.83 × 10−9 | 1.13 × 10−3 | 1082 | 103 130 134 | 1.26 × 10−10 | −0.383 | 44.55 (9.39) |
| 1069 | 103 130 138 | 2.35 × 10−8 | −0.340 | 52.26 (8.51) | |||||
| 1027 | 10 313 0147 | 5.34 × 10−1 | −0.042 | 86.87 (4.25) | |||||
| 3 | SATB1 | Intron | 8.61 × 10−9 | 1.98 × 10−3 | 330 | 18 443 092 | 3.08 × 10−2 | −0.270 | 25.64 (7.40) |
| 329 | 18 443 106 | 5.64 × 10−2 | −0.210 | 26.16 (7.11) | |||||
| 327 | 18 443 135 | 3.71 × 10−2 | −0.269 | 25.00 (6.71) | |||||
| 21 | ETS2 | Intron | 2.28 × 10−8 | 2.92 × 10−3 | 349 | 40 183 132 | 3.98 × 10−3 | −0.304 | 28.38 (7.10) |
| 19 | FCAR | Promoter | 2.36 × 10−8 | 2.92 × 10−3 | 1093 | 55 384 107 | 6.61 × 10−3 | −0.181 | 77.05 (4.77) |
| 1058 | 55 384 104 | 2.75 × 10−2 | −0.147 | 84.71 (4.30) | |||||
| 3 | FNDC3B | Intron | 4.60 × 10−8 | 3.89 × 10−3 | 189 | 171 788 002 | 9.58 × 10−4 | −0.478 | 35.51 (9.75) |
| 188 | 171 788 032 | 3.65 × 10−3 | −0.398 | 32.98 (8.27) | |||||
| 2 | CREB1/METTL21Af | Exon/intron | 1.28 × 10−7 | 6.87 × 10−3 | 1100 | 208 461 648 | 2.33 × 10−10 | −0.395 | 38.88 (9.24) |
| 1086 | 208 461 657 | 1.03 × 10−10 | −0.412 | 39.48 (9.44) | |||||
| 15 | SMAD3f | Intron | 2.09 × 10−7 | 8.69 × 10−3 | 1109 | 67 396 559 | 1.81 ×10−14 | −0.503 | 17.47 (6.45) |
| 1099 | 673 965 64 | 6.82 × 10−14 | −0.508 | 21.53 (7.34) | |||||
| 1 | ARNTf | Intron | 2.30 × 10−7 | 9.17 × 10−3 | 1087 | 150 825 039 | 1.43 × 10−9 | −0.364 | 30.38 (8.69) |
| 1011 | 150 825 079 | 7.87 × 10−4 | −0.166 | 46.70 (5.87) | |||||
| Candidate gene | |||||||||
| 7 | RELNf | Intron | 5.18 × 10−6 | 4.36 × 10−2 | 1017 | 103 576 641 | 3.14 × 10−3 | 0.107 | 92.99 (3.29) |
| 944 | 103 576 649 | 6.28 × 10−4 | 0.187 | 71.12 (6.25) | |||||
| Control sites | |||||||||
| 6 | PARK2 | Intron | 1.28 × 10−1 | 8.47 × 10−1 | 333 | 162 054 935 | 5.24 × 10−1 | 0.022 | 90.57 (2.10) |
| 330 | 162 054 989 | 3.18 × 10−1 | −0.065 | 69.26 (8.55) | |||||
| 4 | HTRA3 | Intron | 1.90 × 10−12 | 4.70 × 10−6 | 344 | 8281 202 | 9.07 × 10−1 | 0.010 | 33.93 (13.38) |
| 339 | 8281 204 | 8.40 × 10−1 | 0.042 | 33.53 (11.93) | |||||
| 6 | GMPR/ATXN1 | 4.40 × 10−12 | 4.70 × 10−6 | 1120 | 16 295 927 | 9.61 × 10−1 | −0.017 | 94.45 (4.40) | |
| 1089 | 16 295 949 | 3.81 × 10−1 | 0.005 | 95.65 (4.55) | |||||
Abbreviations: Chr, chromosome; MWAS, methylome-wide association study.
All sites selected based on MWAS top findings and network findings, as well as the RELN candidate gene, showed the same direction of effect in the MWAS and the replication. All control sites showed a positive direction of effect in the MWAS.
Gene name(s) for loci located in or within 20 kilobase (kb) from the gene.
Indicates if the loci overlap with introns, exons, untranslated region (UTR), or promoters (8 kb upstream of transcription start). None of the included sites overlapped with CpG islands (CGI) or were located within 2 kb (CGI shore) of a CGI.
Number of investigated samples in replication analysis after stringent quality control.
The mean difference between the cases and controls divided by a pooled estimate of the standard deviation, which indicates the effect size and the direction of effect. A negative d value indicates a decreased methylation level in the schizophrenia cases.
The highest (least significant) P value in the replication was significant after correction for multiple testing.
All genes selected from hypoxia pathways had a nominal P < .05. Whereas the most hypoxia-specific gene (ARNT) replicated after correcting for multiple testing, the most specific immune response–related gene, FCAR, was only nominally significant. To perform our second “replication” effort, we first removed the top MWAS findings that were used to detect the networks/pathways in the initial analyses and then performed 10 000 permutations to test whether the other genes in the implicated networks showed enrichment for small P values in the MWAS. For hypoxia networks created using PPIs, none of the test statistics obtained after permutation had a value more extreme than the observed test statistic (eFigure 5 in the Supplement). This implies a P < 1.0 × 10−4 (= 1/10 000), indicating that the MWAS results for the remaining group of network genes were more significant than expected under the null hypothesis. For the pathway analyses, the permutation test was also highly significant (P <.001; eFigure 5 in the Supplement). For the immune system, we combined PPI network and pathway results to avoid small sets of genes. None of the permutation test statistics had a value more extreme than the observed test statistic (P < 1.0 × 10−4; eFigure 6 in the Supplement).
Interestingly, a site in RELN had an MWAS q value of less than 0.1. RELN has previously been associated with schizophrenia via messenger RNA expression studies57,58 and, although some inconclusive results exist,25 is one of the most prominent schizophrenia candidate genes in methylation studies.19,22 Furthermore, support for a strong inverse correlation between RELN expression and promoter methylation has been observed in mice59 and humans.60 Table 3 shows that the RELN site also replicated. Similar to previous findings,19,22 we observed increased levels of methylation in schizophrenia cases. Traditionally, methylation studies of RELN have focused on the promoter region. Our best finding was located in the first intron and did not directly overlap the previous findings.
Table 3 shows that our negative control did not replicate, nor did the 2 most significant sites obtained after we performed an MWAS without regressing out the principal components. This suggests that the principal components were useful to prevent false positives. Regressing out the covariates from Table 1 did not alter results (eTables 4, 5, and 6 in the Supplement). For example, cigarette smoking can result in impaired oxygen release to tissues,61 and nicotine can upregulate HIF1A.62 However, we did not observe correlations between the methylation of genes in hypoxia networks and smoking, nor did the inclusion of smoking status as a covariate change the replication results (eTable 4 in the Supplement).
Discussion
Our top MWAS finding (FAM63B) replicated with a P = 2.3 × 10−10. It was part of the networks regulated by miRNA that can be linked to neuronal differentiation and dopaminergic gene expression,54–56 functions of potential relevance for schizophrenia. Many of our other top MWAS results could be linked to hypoxia and sometimes infection. Replicated findings also implicated RELN, one of the most frequently studied candidates in methylation studies of schizophrenia.22 Interestingly, RELN is regulated by HIF1/2a and can therefore also be linked to hypoxia.63,64
The hypoxia findings were very robust. Regardless of whether we used PPI networks, pathway databases, or an miRNA target gene database, hypoxia was a dominant theme. The hypoxia genes replicated in independent samples using a different technology. Furthermore, genes that were not among the top findings in the MWAS but were in the hypoxia pathways were also significantly enriched for small P values. Although the scope and quality of our phenotype data were limited, smoking or other covariates did not account for the hypoxia findings. Although we can only speculate about the cause, we note that a substantial amount of literature exists showing that hypoxia during fetal development increases the risk of schizophrenia.3 It is known that environmentally induced methylation changes can be preserved over a prolonged period of time.65,66 One intriguing hypothesis is that early hypoxia events alter methylation profiles in blood DNA, traces of which are preserved in the adult patient.
Many MWAS results reflected environmental insults. Because environmental effects cannot alter sequence variation, these phenomena cannot be detected with genome-wide association studies or exome-sequencing studies. Although there was some thematic overlap (eg, genome-wide association studies have implicated genes involved in immune response67), this likely explains why we found genes that were different from those found in studies of sequence variants. To find overlapping loci, different analytical strategies may be required. For example, the DNA sequence can regulate methylation patterns,9,68–70 and we are currently conducting analyses to find loci where these regulatory mechanisms may be disrupted in schizophrenia. Because these analyses combine sequence variants with methylation patterns, they are more likely to yield results that overlap with genome-wide association study findings. Methylation signatures of environmental insults may not impact gene expression in blood. Thus, whole transcriptome studies may not be able to capture the phenomena detected in this study; therefore, methylation studies provide unique possibilities compared with other technologies.
Our results demonstrate how methylation studies in whole blood can advance schizophrenia research. First, they suggest that a record of pathogenic events may be preserved in the methylome. Etiologically distinct disease subtypes may be distinguishable from each other with respect to prognosis, course, or response to treatment.71 The possibility of identifying these subtypes using methylation markers that tend to have large effect sizes and can be measured with cost-effective assays, using DNA from blood that is stable and easy to collect, would be of great clinical importance. Second, methylation studies can generate testable hypotheses about disease mechanisms. For example, the hypoxia findings show how methylation studies can point to disease-causing factors. As postulated by the “signature model,”5 the causal factors may affect schizophrenia through processes that have nothing to do with methylation (eg, possible causal mechanisms include disruption of the laminar organization of the cerebral cortex72). Our RELN finding possibly demonstrates the second possible model, in which the methylation status of disease-relevant sites in the brain is mirrored by the corresponding sites in the blood. Thus, previous studies19,22 have implicated methylation sites in RELN in schizophrenia using postmortem brain samples. The fact that we find this gene in whole blood provides a possible illustration of the “functional mirror-site model.”5
A variety of efforts were taken to control for potential confounders. Our results suggested genes related to hypoxia, the immune system, and brain function rather than genes that, for example, are potentially relevant to medication and life style differences. This suggests that our efforts worked satisfactorily. For biomarkers other than genetic variants, there is always the inherent risk of confounding effects. Experiments studying model systems in controlled environments (eg, cell culture) would be the next step to rule out confounders completely.
Studies have suggested that 30 to 60 million reads per sample may be sufficient to reveal valuable information for whole-genome methylation analysis.33,73 We obtained, on average, 68.0 million reads, of which 32.4 million high-quality reads (47.6%) remained after stringent quality control. The MWAS was performed on “blocks” that summed reads across correlated CpG sites to improve the reliability of the measurements. This appeared to be sufficient to detect methylation markers that replicated in independent samples. It is possible, however, that increasing the number of reads would allow the detection of sites (eg, in CpG-poor regions) that could currently not be measured reliably.
Conclusions
In summary, to our knowledge, the present study is one of the first MWASs of disease with a large sample size using a technology that provided good coverage of methylation sites across the genome. Our results demonstrate how methylation studies can suggest new avenues to increase our understanding of disease and yield biomarkers that can be used to potentially improve disease management.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by the National Institute of Mental Health (grant RC2MH089996) and is part of a larger project entitled “A Large-Scale Schizophrenia Association Study in Sweden” that is supported by grants from the National Institute of Mental Health (grant MH077139) and the Stanley Medical Research Institute. The institutions involved in this project are the Karolinska Institutet, the Icahn School of Medicine at Mount Sinai in New York, the University of North Carolina at Chapel Hill, Virginia Commonwealth University, the Broad Institute in Cambridge, Massachusetts, and the US National Institute of Mental Health in Bethesda, Maryland.
Role of the Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplemental content at jamapsychiatry.com
Author Contributions: Dr van den Oord had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Aberg, McClay, Chen, Xie, Gao, Sullivan, van den Oord.
Acquisition of data: Aberg, Hudson, Harada, Hultman, Magnusson, van den Oord.
Analysis and interpretation of data: Aberg, McClay, Nerella, Clark, Kumar, Chen, Khachane, Xie, Hudson, Gao, Hultman, Magnusson, van den Oord.
Drafting of the manuscript: Aberg, McClay, Nerella, Clark, Kumar, Hudson, Gao, Hultman, van den Oord.
Critical revision of the manuscript for important intellectual content: Aberg, Clark, Chen, Khachane, Xie, Harada, Sullivan, Magnusson, van den Oord.
Statistical analysis: Aberg, McClay, Clark, Chen, Gao, van den Oord.
Obtained funding: Aberg, Hultman, Sullivan, Magnusson, van den Oord.
Administrative, technical, or material support: Aberg, McClay, Nerella, Khachane, Hudson, Harada, Hultman, Magnusson, van den Oord.
Study supervision: Aberg, Khachane, Hultman, van den Oord.
Conflict of Interest Disclosures: None reported.
Additional Contributions: Library construction and next-generation sequencing were performed by EdgeBio. We thank the Swedish Schizophrenia Consortium and Life Technologies for their advice.
REFERENCES
- 1.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12(9):799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- 3.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374(9690):635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 4.Boks MP, de Jong NM, Kas MJ, et al. Current status and future prospects for epigenetic psychopharmacology. Epigenetics. 2012;7(1):20–28. doi: 10.4161/epi.7.1.18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aberg KA, Xie LY, McClay JL, et al. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics. 2013;5(4):367–377. doi: 10.2217/epi.13.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4(2):265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland JE, Costa M. Epigenetics and the environment. Ann N Y Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 9.Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40(7):904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 10.Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J Affect Disord. 2011;132(3):450–453. doi: 10.1016/j.jad.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Zhang J, Zhang L, Shen Y, Xu Q. Effects of MAOA promoter methylation on susceptibility to paranoid schizophrenia. Hum Genet. 2012;131(7):1081–1087. doi: 10.1007/s00439-011-1131-5. [DOI] [PubMed] [Google Scholar]
- 12.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24):4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lott SA, Burghardt PR, Burghardt KJ, Bly MJ, Grove TB, Ellingrod VL. The influence of metabolic syndrome, physical activity and genotype on catechol-O-methyl transferase promoter-region methylation in schizophrenia. Pharmacogenomics J. 2013;13(3):264–271. doi: 10.1038/tpj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012;26(6):2712–2718. doi: 10.1096/fj.11-202069. [DOI] [PubMed] [Google Scholar]
- 15.Petronis A, Gottesman II, Kan P, et al. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29(1):169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- 16.Zhang AP, Yu J, Liu JX, et al. The DNA methylation profile within the 5′-regulatory region of DRD2 in discordant sib pairs with schizophrenia. Schizophr Res. 2007;90(1–3):97–103. doi: 10.1016/j.schres.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Scarr E, Craig JM, Cairns MJ, et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl Psychiatry. 2013;3:e230. doi: 10.1038/tp.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdolmaleky HM, Cheng KH, Faraone SV, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 20.Abdolmaleky HM, Yaqubi S, Papageorgis P, et al. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophr Res. 2011;129(2–3):183–190. doi: 10.1016/j.schres.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet. 2006;7:10. doi: 10.1186/1471-2350-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grayson DR, Jia X, Chen Y, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(26):9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto K, Bundo M, Yamada K, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25(22):5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nohesara S, Ghadirivasfi M, Mostafavi S, et al. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 2011;45(11):1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Tochigi M, Iwamoto K, Bundo M, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63(5):530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Tolosa A, Sanjuán J, Dagnall AM, Moltó MD, Herrero N, de Frutos R. FOXP2 gene and language impairment in schizophrenia: association and epigenetic studies. BMC Med Genet. 2010;11:114. doi: 10.1186/1471-2350-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aberg KA, McClay JL, Nerella S, et al. MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case-control samples. Epigenomics. 2012;4(6):605–621. doi: 10.2217/epi.12.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Gao G, Nerella S, et al. MethylPCA: a toolkit to control for confounders in methylome-wide association studies. BMC Bioinformatics. 2013;14:74. doi: 10.1186/1471-2105-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Oord EJ, Bukszar J, Rudolf G, et al. Estimation of CpG coverage in whole methylome next-generation sequencing studies. BMC Bioinformatics. 2013;14(1):50. doi: 10.1186/1471-2105-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergen SE, O’Dushlaine CT, Ripke S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17(9):880–886. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28(10):1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollen KA. Structural Equations With Latent Variables. New York, NY: Wiley; 1989. [Google Scholar]
- 35.Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- 36.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Aryee MJ, Padyukov L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun YV, Turner ST, Smith JA, et al. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum Genet. 2010;127(6):651–658. doi: 10.1007/s00439-010-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39(Database issue):D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pentchev K, Ono K, Herwig R, Ideker T, Kamburov A. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics. 2010;26(21):2796–2797. doi: 10.1093/bioinformatics/btq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(Database issue):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft D, O’Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35(1):152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 45.Aparicio A, North B, Barske L, et al. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4(3):176–184. doi: 10.4161/epi.4.3.8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. [Google Scholar]
- 47.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elvert G, Kappel A, Heidenreich R, et al. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2003;278(9):7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- 49.Tan XL, Zhai Y, Gao WX, et al. p300 expression is induced by oxygen deficiency and protects neuron cells from damage. Brain Res. 2009;1254:1–9. doi: 10.1016/j.brainres.2008.11.092. [DOI] [PubMed] [Google Scholar]
- 50.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23(24):9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16(10):1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 52.Xue Y, Li NL, Yang JY, Chen Y, Yang LL, Liu WC. Phosphatidylinositol 3′-kinase signaling pathway is essential for Rac1-induced hypoxia-inducible factor-1(alpha) and vascular endothelial growth factor expression. Am J Physiol Heart Circ Physiol. 2011;300(6):H2169–H2176. doi: 10.1152/ajpheart.00970.2010. [DOI] [PubMed] [Google Scholar]
- 53.Beckman JD, Chen C, Nguyen J, et al. Regulation of heme oxygenase-1 protein expression by miR-377 in combination with miR-217. J Biol Chem. 2011;286(5):3194–3202. doi: 10.1074/jbc.M110.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2(3):152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 56.Huang W, Li MD. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol Psychiatry. 2009;65(8):702–705. doi: 10.1016/j.biopsych.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guidotti A, Auta J, Davis JM, et al. Decrease in reelin glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study [published correction appears in Arch Gen Psychiatry. Arch Gen Psychiatry. 2000;57(11):1061–1069. doi: 10.1001/archpsyc.57.11.1061. 2002;59(1):12] [DOI] [PubMed] [Google Scholar]
- 58.Impagnatiello F, Guidotti AR, Pesold C, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci U S A. 1998;95(26):15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci U S A. 2005;102(35):12578–12583. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatry. 2007;12(6):593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- 61.Sørensen LT, Jørgensen S, Petersen LJ, et al. Acute effects of nicotine and smoking on blood flow, tissue oxygen, and aerobe metabolism of the skin and subcutis. J Surg Res. 2009;152(2):224–230. doi: 10.1016/j.jss.2008.02.066. [DOI] [PubMed] [Google Scholar]
- 62.Guo L, Li L, Wang W, Pan Z, Zhou Q, Wu Z. Mitochondrial reactive oxygen species mediates nicotine-induced hypoxia-inducible factor-1α expression in human non-small cell lung cancer cells. Biochim Biophys Acta. 2012;1822(6):852–861. doi: 10.1016/j.bbadis.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt-Kastner R, van Os J, Steinbusch HWM, Schmitz C. Gene regulation by hypoxia and the neurodevelopmental origin of schizophrenia. Schizophr Res. 2006;84(2–3):253–271. doi: 10.1016/j.schres.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 64.Ralph GS, Parham S, Lee SR, et al. Identification of potential stroke targets by lentiviral vector mediated overexpression of HIF-1 alpha and HIF-2 alpha in a primary neuronal model of hypoxia. J Cereb Blood Flow Metab. 2004;24(2):245–258. doi: 10.1097/01.WCB.0000110532.48786.46. [DOI] [PubMed] [Google Scholar]
- 65.Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 66.Nestler EJ. Epigenetics: stress makes its molecular mark. Nature. 2012;490(7419):171–172. doi: 10.1038/490171a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schilling E, El Chartouni C, Rehli M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009;19(11):2028–2035. doi: 10.1101/gr.095562.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Rohde C, Reinhardt R, Voelcker-Rehage C, Jeltsch A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 2009;10(12):R138. doi: 10.1186/gb-2009-10-12-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schalkwyk LC, Meaburn EL, Smith R, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86(2):196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kennedy JL, Farrer LA, Andreasen NC, Mayeux R, St George-Hyslop P. The genetics of adult-onset neuropsychiatric disease: complexities and conundra? Science. 2003;302(5646):822–826. doi: 10.1126/science.1092132. [DOI] [PubMed] [Google Scholar]
- 72.Herr KJ, Herr DR, Lee CW, Noguchi K, Chun J. Stereotyped fetal brain disorganization is induced by hypoxia and requires lysophosphatidic acid receptor 1 (LPA1) signaling. Proc Natl Acad Sci U S A. 2011;108(37):15444–15449. doi: 10.1073/pnas.1106129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chavez L, Jozefczuk J, Grimm C, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20(10):1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.