Abstract
The oxidative modification of apolipoprotein A-I ‘s methionine148(M148) is associated with defective HDL function in vitro. Multiple Reaction Monitoring (MRM) is a mass spectrometric technique that can be used to quantitate post-translational modifications. In this study, we developed an MRM assay to monitor the abundance ratio of the peptide containing oxidized M148 to the native peptide in Apo A-I. Measurement of the oxidized-to-unoxidized-M148 ratio was reproducible (CV<5%). The extent of methionine M148 oxidation in the HDL of healthy controls, and type 2 diabetic participants with and without prior cardiovascular events (CVD) were then examined. The results suggest a significant increase in the relative ratio of the peptide containing oxidized M148 to the unmodified peptide in the HDL of participants with diabetes and CVD (p<0.001), compared to participants without CVD. Monitoring the abundance ratio of the peptides containing oxidized and unoxidized M148 by MRM provides a means of examining the relationship between M148 oxidation and vascular complications in CVD.
Keywords: HDL, Multiple Reaction Monitoring, Proteomics, Diabetes, Cardiovascular Disease
Introduction
Cardiovascular disease (CVD) is the leading cause of mortality in the U.S, and diabetes is an important risk factor (1). Recent trials examining the impact of intensive glycemic control on the reduction of CVD endpoints (2–4) highlight the challenges for reducing the increased CVD risk, and the need for new biomarkers of diabetic complications. Several studies suggest that the function of HDL is defective in diabetes(5). Shao and colleagues demonstrated that the ability of apolipoprotein A-I (Apo A-I) to activate LCAT is impaired by methionine oxidation of residue 148, M148(O) (6). LCAT esterifies cholesterol on HDL and is an important component for reverse cholesterol transport(7). Thus, Apo A-I methionine oxidations are potential markers of diabetic complications.
Mass spectrometry (MS)-based applications are particularly well-suited to measure post-translational modifications of proteins (8). Conventional MS-based quantitation work flows using spectral counting or extracted ion chromatograms involve lengthy MS data acquisition and analysis times and are often limited to quantifying differences between small sample sets. Hence, there is a need to develop high-throughput assays with simple sample preparation and reduced MS analysis time. Multiple Reaction Monitoring (MRM) is a tandem MS (MS/MS) scan mode unique to triple quadrupole MS instrumentation that is capable of rapid, sensitive, and specific quantitation of peptides in highly complex sample matrices, such as plasma (9,10). MRM is a targeted approach that requires knowledge of the molecular weight the peptide of interest and its fragmentation pattern, leading to the generation of target “transitions” for monitoring protein levels.
In this study, we defined the transitions for monitoring the ratio of oxidized M148 to its unmodified peptide in Apo A-I using MRM. We applied this technology to HDL samples from the plasma of participants with and without diabetes and prior cardiovascular events to determine if this ratio was higher in diabetic participants with vascular complications.
Methods
The study was approved by the University of Arizona Institutional Review Board, and all participants provided written informed consent prior to testing.
Clinical Samples
The plasma samples were collected at University of Arizona diabetes Clinics and from the community. 34 participants (8 healthy controls, 11 with type 2 diabetes and 15 with both diabetes and a prior CVD event) reported to the Center for Clinical and Translational Sciences (CaTS) after an overnight fast. CVD events were defined by a prior history of coronary artery bypass surgery (CABG), percutaneous transluminal angioplasty (PTCA), prior MI, or thrombotic stroke as previously defined in major clinical trials (11). The study excluded subjects if they met any of the following criteria: had type 1 diabetes, were on an active weight loss program, history of cancer, HIV, or steroid use. All study participants had oral glucose tolerance tests (OGTTs). New diagnosis of diabetes was based on fasting blood sugar >125 mg/dL, 2 hour OGTT>200 mg/dl or HbA1c>6.5%. Established diabetes was defined by clinical history. All non-diabetic participants participating underwent oral glucose testing. The subjects were asked to fill a physical activity questionnaire (12) regarding if they participated in a structured exercise program, the type of exercise and its frequency per day or week. None of the patients recruited were participating in a structured exercise program. In their questionnaires, the majority of subjects did not report daily exercise activities. Participants did not engage in high intensity exercise for at least two days prior to testing. Plasma samples were collected in EDTA tubes between 2008 and 2009, and were immediately frozen at −80 °C. Sample Analysis by mass spectrometry was done in 2011 at University of Arizona and University of Victoria proteomics cores.
HDL Isolation technique
HDL isolation by centrifugation was based on a modification of a previously published protocol (13). In brief, KBr (~55 mg) was added to 310 µL of plasma samples to create a density of 1.21 g/mL. The sample was overlaid with 200 µL of 1.21 g/mL density solution for a total volume of 500 µL. Samples were then spun at 120,000 rpm, at 16°C for 2 hours (Beckman TLX ultracentrifuge with a type 120.1 fixed angle rotor using thick-walled 500 µL Polycarbonate tubes, item 343776. The upper 125 µL solution that had a density of less than 1.21 g/mL was removed and 150µL of NaCl/EDTA solution (0.9% (w/v) NaCl, 0.1% (w/v) EDTA, pH 7.4) was added to each tube for a final density of 1.063 g/mL. Subsequently, 225 µL of 1.06 KBr solution in NaCl/EDTA was added creating a final volume of 500 µL for a second 2 hour spin at the same parameters listed. The bottom 125 µL (HDL fraction) of solution was removed for further analysis. We confirmed that albumin and apolipoprotein B (apo B) were depleted in the HDL isolate using MRM and four HDL samples were sent to Myriad RBM to externally validate our measurements using an immunoassay in a CLIA certified laboratory. HDL proteins were measured using the method of Lowry. HDL samples were submitted in a 96 well plate in 100 µL aliquots with a final protein concentration of 0.3 mg/mL.
MRM Analysis
1. Developing Apo A-I M148 Peptide Transitions for MRM
This method was developed at the University of Arizona’s proteomics core facility. An HDL isolate from a control participant was screened for methionine oxidations (M148: oxidation +16 defined as M148(O)) and the transitions from theoretical fragmentation patterns using Prospector were obtained. Three transitions for M148(O) peptide had signal-to-noise (S/N) ratio>3. The modified peptides of M148(O) were then synthesized (New England Peptides, Gardner, MA). By infusing the peptides into the mass spectrometer, the transitions of the modified peptides were then optimized and the peaks were confirmed in MRM mode as previously described (8). Following confirmation of in vivo peaks, stable-isotope-labeled standard (SIS) peptides for M148(O) were synthesized. In addition to monitoring the methionine containing Apo A-I peptides, a second Apo A-I peptide (ATEHLSTLSEK), a peptide for Apo B100 (FPEVDLIK) and albumin peptide (LVNEVTEFAK) were monitored to assess the quality of the HDL isolate. These experiments were completed at University of Arizona proteomics core. An extensive list of plasma protein transitions for MRM use has been previously published (10).
2. MRM analyses
One control sample and thirty five HDL samples were sent to University of Victoria - Genome BC Proteomics Centre which has a dedicated MRM service. Eight replicate MRM runs of a control sample were done to determine the coefficient of variation (CV) of the target peptides measurements. All HDL samples were run once and analyzed in one batch in 2011.
Sample Preparation prior to LC/MRM-MS
Samples were first diluted by the addition of 140µL of 37.5 mM ammonium bicarbonate to each 100 µL of sample. Each diluted HDL sample was denatured by adding 30 µL of 10% w/v sodium deoxycholate (NaDOC) in 37.5mM ammonium bicarbonate. Disulphide bonds were reduced by the addition of 7.46 µL of 50 mM tris (2-carboxyethyl) phosphine (TCEP, in 37.5mM ammonium bicarbonate), and incubation at 60°C for 30 min in a dry-air incubator. Free sulfhydryl groups were alkylated by the addition of8.28 µL of 100 mM iodoacetamide (in 37.5mM ammonium bicarbonate), and incubation at 37°C for 30 min in a dry-air incubator. Any remaining iodoacetamide was quenched by the addition of 8.28 µL of 100 mM DTT (in 37.5mM ammonium bicarbonate) and incubation at 37°C for 30 min in a dry-air incubator. Six µL of sequencing-grade trypsin (0.4 µg/µL (Promega) in 37.5mM ammonium bicarbonate) was added to each sample. The final volume of each digest was 300 µL, and digestion was conducted at 37°C for 16 hours in a dry air incubator.
SIS peptide addition & solid phase extraction
Digestion was stopped by the addition of an acidified SIS peptide mixture in formic acid, to give a final formic acid concentration of 0.5 % v/v and to reduce the pH to <3, which inactivates trypsin and precipitates NaDOC). Two ng of methionine oxidized SIS peptides were spiked into the sample per MRM run. Samples were centrifuged for 10 min at 12,000 × g (23°C) to remove the NaDOC precipitate. The supernatant containing the peptides was desalted and concentrated by solid phase extraction using Waters Oasis HLB 1cc columns (10 mg). The eluted samples were frozen and lyophilized to dryness overnight. Prior to the LC/MRM-MS analysis, samples were rehydrated in a volume of Solvent A (0.1% v/v formic acid) to obtain a concentration of 0.5 µg/µL of original sample.
LC/MRM-MS method
The MS analyses were performed on an AB/MDS Sciex 4000 QTRAP equipped with an Eksigent NanoLC-1Dplus LC system. The trapping column used was a 5 × 0.3 mm C18 PepMap column, with 5 µm particles (Dionex/LC Packings). The analytical column was a 75 µm × 150 mm Reprospher 100 C18 Aqua column, packed with 3µm particles, 100 Å pore size, packed in-house under argon. The solvent system consisted of solvent A (100% H2O, 0.1% v/v formic acid), and solvent B (90% aqueous acetonitrile, 0.1% v/v formic acid). The on-line analyses were 43 min in length and the gradient was constructed as follows: samples were loaded onto the trapping column at 10 µL/min (2% aqueous acetonitrile, 0.1% v/v aqueous formic acid) for 3 min, followed by a 2 min linear gradient from 3% to13% solvent at 300 nL/min, a10 min linear gradient at 300 nL/min from 13% to20% solvent B, a 9 min linear gradient at 300 nL/min from 20% to 27% solvent B, and a final 6 min linear gradient at 300 nL/min from 27% to44% solvent B before high organic column flushing and re-equilibration. A blank solvent injection was run between all samples to prevent sample carryover on the HPLC column. An AB/MDS Sciex 4000 QTRAP with a Michrom Captive Spray source, controlled by Analyst 1.5 software (Applied Biosystems) was used for all of the LC/MRM-MS analyses. All acquisition methods used the following instrument parameters: 1300–1500 V ion spray voltage, a 110°C interface heater temperature, an MS operating pressure of 3.5 × 10−5Torr, and Q1 and Q3 set to unit resolution (0.6 – 0.8 Da FWHH). MRM acquisition methods were constructed using 1 or 2 ion pairs per peptide with empirically-tuned DP and CE voltages for each transition. A default collision cell exit potential of 23 V was used for all MRM ion pairs, with a target cycle time of 2 seconds.
MRM data acquisition
All MRM data was processed using MultiQuant 1.2 (Applied Biosystems) with the MQL algorithm for peak integration. Automatic peak detection,3-point Savitsky-Golay smoothing, a peak-splitting factor of 2, and default MultiQuant values for the noise percentage and baseline subtraction window were used. All integrated peaks were manually inspected to ensure correct peak detection and integration.
Statistical Analysis
The statistical program R was used. The M148 oxidation ratios, and the triglyceride were not normally distributed and the analysis was completed on log transformed data. The transitions for the modified and unmodified Apo A-I peptides were correlated using pearson coefficients. M148 y7 2+ transition was used for data analysis. Biochemical and MRM measures were compared using ANOVA with p-value < 0.05. Linear models were used to calculate the p values of the correlated variables. The M148 oxidation ratio and HDL Apo A-I were the primary endpoint and the statistical significance was assessed at the 0.05 level. For the other measurements or recorded data, a p value of 0.005 or less was considered significant to adjust for multiple comparisons.
Results
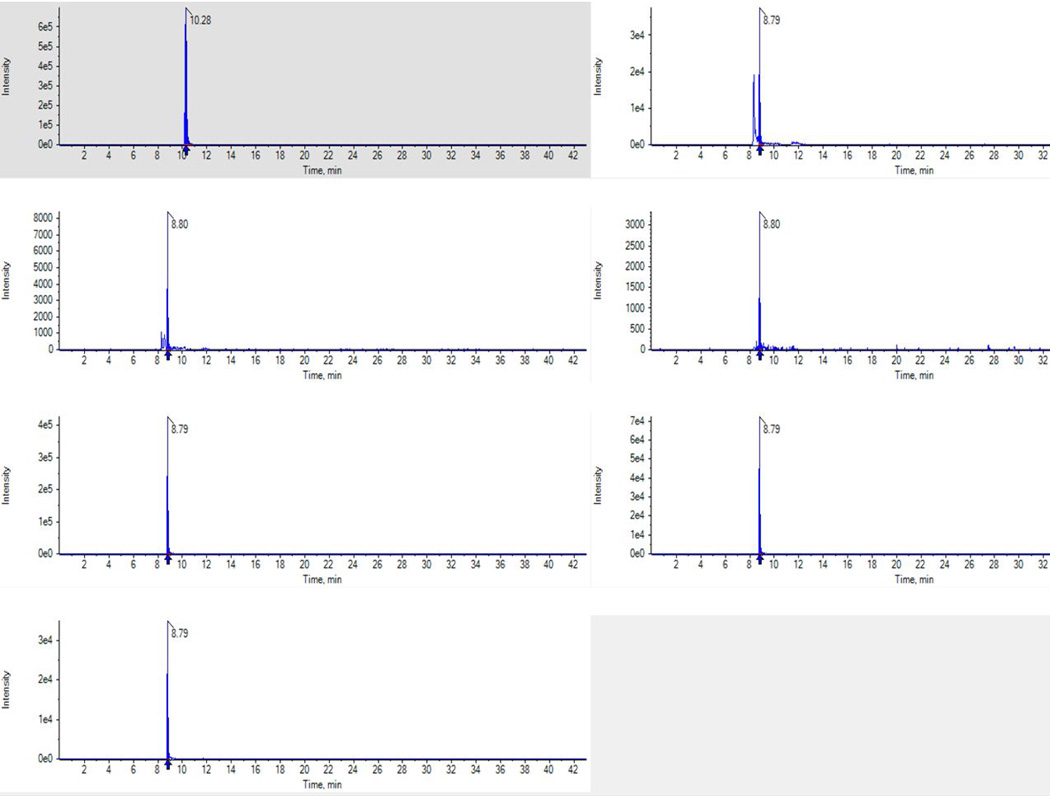
Using theoretical transitions derived from MS Prospector, an HDL sample was screened for M148 and M148(O) peptides and a signal-to-noise (S/N) ratio> 3 was observed. To validate these peaks, modified and SIS peptides for M148(O) were synthesized and the transitions were optimized. The MRM transitions are summarized in Table 1. An HDL sample was then monitored for M148 oxidations. As shown in Fig. 1, the in vivo oxidized peaks had near identical retention times to the heavy peptides (SIS) providing peak validation with three transitions for M148(O) peptide. Of the three M148(O) transitions, the y72+ ion showed the highest peak intensity, but had two peaks. Only one of these peaks eluted at the same retention time of the other M148(O) transitions and that of the added SIS peptides. This coeluting peak was selected for quantitation. This finding highlights the need for multiple transitions per peptide in addition to added SIS peptides for correctly identifying the peak of interest when using MRMs. Although we used a S/N ratio of > 3 to screen for the oxidized M148 peptide, the in vivo M148(O) peak observed after MRM transitions optimization was several fold (~ 10-fold) greater than the noise background. As expected, the peak areas among the three M148 transitions were highly correlated with each other (r≥0.95, p<0.001). To determine the reproducibility of the MRM assay, 8 replicates of a control sample were analyzed. The MRM assay for M148(O) ratio was highly reproducible (table 1, M148(O) CV<5%), and the transition with the best CV (y7 2+) was used to compare the relative ratio of the oxidized methionines among the study groups.
Table 1.
peptides monitored by MRM
| Target | Q1 | Q3 | DP | CE | Peptide | Ion | CV(%) |
|---|---|---|---|---|---|---|---|
| Albumin | 575.3 | 937.5 | 70 | 25 | LVNEVTEFAK | y8 | 5.2 |
| Apo B-100 | 524.3 | 450.8 | 55 | 28 | FPEVDVLTK | y82+ | 20 |
| Apo A-I | 405.9 | 522.3 | 52 | 18 | ATEHLSTLSEK | y9 | 3.2 |
| Apo A-I | 405.9 | 572.8 | 52 | 19 | ATEHLSTLSEK | y102+ | 1.2 |
| M148 | 516.3 | 416.2 | 60 | 30 | LSPLGEEMR | y72+ | 1.5 |
| M+148(O)=M148 + 16 | 524.3 | 424.4 | 60 | 27 | LSPLGEEM(O)R | y72+ | 3.8 |
| M+148(O)=M148 + 16 | 524.3 | 637.3 | 60 | 33 | LSPLGEEM(O)R | y5 | 4.2 |
| M+148(O)=M148 + 16 | 524.3 | 750.4 | 60 | 33 | LSPLGEEM(O)R | y6 | 4.7 |
The CVs were calculated from 8 replicate analyses of a control sample.
DP: Declustering Potential.
CE: Collision Energy.
CV: Coefficient of variation.
Fig. 1.
Representative chromatogram of M148(LSPLGEEMR) peptides in a control HDL sample showing the intensity of the peaks (y axes) and retention time (x axes). Unmodified M148 (y72+) together with in vivo oxidized peptides (y72+, y5 and y6) and the corresponding heavy peptides (y72+, y5 and y6) are shown. Note the M148(O) y72+ shows two peaks. The larger of these two peaks co-elutes with the other modified and heavy peptide peaks and this was selected for quantification.
A second Apo A-I peptide (ATEHLSTLSEK) was used to measure Apo A-I concentrations on HDL. The correlation between the peak area of ATEHLSTLSEK to unmodified M148 peptides was weak (r=0.42, p<0.001), possibly due to the susceptibility of methionine residues to oxidation. To validate the measurement of protein concentrations using MRM, four HDL samples were sent to Myriad RBM that has a CLIA certified laboratory with the ability of running multiplexed immunoassays. Concentrations of albumin, Apo B100, and Apo A-I (ATEHLSTLSEK) measured using the multiplexed immunoassays at Myriad were strongly correlated to measurements by MRM (r > 0.95, p <0.001 for all three proteins). The ratio of ATEHLSTLSEK peptide to the corresponding SIS peptide was used to calculate the concentrations of Apo A-I on HDL (table 2) in the clinical samples. SIS peptides for the unmodified M148 was not synthesized, and thus we were unable to determine Apo A-I concentrations based on the M148 peptide.
Table 2. Study Characteristics and Measurements.
The p value in the table refers to all three groups being compared with each other. Pairwise comparisons are shown by the superscripts, and are explained in the footnote below.
| Characteristic | Non-Diabetic Controls (n=8) Group 1 |
Participants with Diabetes (n=11) Group 2 |
Participants with Diabetes and CVD (n=15) Group 3 |
p (for comparison) |
|---|---|---|---|---|
| Demographic profile | ||||
| Sex | 3 males, 5 females | 4 males, 7 females | 8 males, 7 females* | 0.03 |
| Race | 6 Caucasians, 2 Hispanics | 8 Caucasians, 2 Hispanics, 1 Asian | 10 Caucasians, 5 Hispanics | 0.24 |
| Age (years) | 48±13 | 56 ± 13 | 53 ± 12 | 0.38 |
| Body mass index (kg/m2) | 25±3 | 34 ± 6 | 39 ± 12* | 0.005 |
| Vital signs | ||||
| Systolic blood pressure (mm Hg) | 110±14 | 123 ± 12 | 132 ± 13* | 0.002 |
| Diastolic blood pressure (mm Hg) | 76±8 | 77 ± 7 | 79 ± 11 | 0.71 |
| Medications (# of participants) | ||||
| Statins | 0 | 0# | 7* | <0.001 |
| Metformin | 0 | 2 | 5 | 0.1 |
| Insulin | 0 | 2 | 2 | 0.2 |
| Aspirin | 0 | 1 | 7* | 0.001* |
| Beta blockers | 0 | 2 | 6* | 0.002* |
| ACE inhibitors or ARBs | 0 | 4 | 7* | 0.003* |
| Biochemical profile | ||||
| Fasting Glucose (mg/dL) | 95± 12 | 142 ± 57# | 202± 88* | <0.001 |
| OGTT (mg/dl) $ | Fasting=93± 10 2 hour post=135 ± 31 |
|||
| Hemoglobin A1c (%) | 5.1±0.27 | 7.6 ± 2.9# | 8.4 ± 2.9* | 0.009 |
| Fasting Insulin (IU) | 7 ± 3 | 12 ± 7 | 16 ± 13* | 0.04* |
| HDL cholesterol (mg/dL) | 70 ± 14 | 53 ± 16#^ | 39 ± 8 * | <0.001 |
| LDL cholesterol (mg/dL) | 117±31 | 125 ± 21 | 111 ± 43 | 0.67 |
| Triglycerides (mg/dL) | 116±87 | 146 ± 64 | 212 ± 138 | 0.11 |
| C-reactive protein (mg/L) | 4 ± 4 | 7 ± 5 | 13 ± 12 | 0.23 |
| HDL Apolipoprotein A-I (mg/dL) | 127.5±25 | 90 ± 40 | 84 ± 39* | 0.029* |
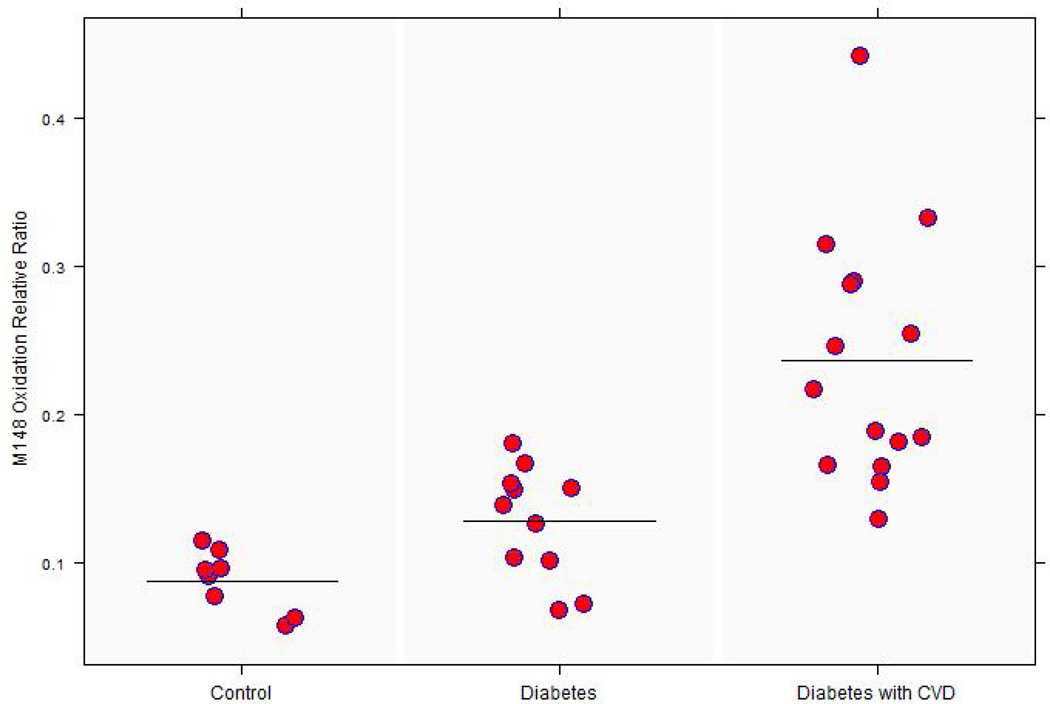
| M148 oxidation relative ratio $$ | 0.087±0.02 | 0.127 ± 0.037# | 0.236 ± 0.084* | <0.001 |
The results were not corrected for possible ex vivo oxidation of methionine
Values are means ± SD
ANOVA Group comparisons were followed by Tukey group-wise comparisons when the p for the three-comparison was <0.05:
, p<0.05 Type diabetics with CVD vs. non-diabetics;
, p<0.05 Type 2 diabetics without CVD vs. non-diabetics.
, p<0.05 Type diabetics with CVD vs. Type 2 diabetics without CVD
OGTTs were only performed in non-diabetic subjects
The results were not corrected for possible ex vivo oxidation of methionine
M148 oxidation and HDL Apo A-I were the primary endpoint and the statistical significance was assessed at the 0.05 level. For the other measures, a p value of 0.005 or less was considered significant to adjust for multiple comparisons.
ACE: Angiotensin Converting Enzyme
ARB: Angiotensin Receptor Blocker
Thirty four participants were recruited to examine the impact of disease on Apo A-I methionine oxidations. As shown in Table 2, controls were leaner, and had a lower systolic blood pressure (p<0.005). Participants with diabetes and heart disease were taking more statins, aspirin, and blood pressure medication compared to controls or diabetics without a prior history of a cardiac event. Participants with diabetes and CVD had significantly decreased HDL Apo A-I concentrations compared to participants with diabetes but without CVD (p=0.029 for the group comparison by ANOVA, and p=0.027 for the group with CVD vs diabetes without CVD). The relative ratio of oxidized to native M148 peptide in HDL was three times as high in the diabetes and CVD group, and 1.5 times as high in the diabetic group without prior CVD, compared to the control group (p <0.001 for the group comparison by ANOVA, with p<0.001 for both diabetes and CVD vs control, and for diabetes without CVD vs control, Fig. 2).
Fig. 2.
The distribution of oxidized M148 relative ratio among the three disease groups. The mean M148(O) ratio to native peptide in diabetes and CVD group was significantly greater than both the control and diabetes group without CVD (* p<0.001 for both comparisons).
Discussion
In this study, we defined MRM transitions to monitor the relative ratio of M148 oxidations compared to M148 peptide on Apo A-I. Our results demonstrated that monitoring the relative ratio of the M148(O)-to the M148-containing peptide was highly reproducible with a CV <5% using MRM. We did not measured the molar % oxidized M148 in this proof-of-concept study, because this would have required absolute quantitation of both forms of this peptide. Clinically, HDL isolated from participants with diabetes and CVD had a significantly increased ratio of oxidized M148 to unoxidized M148. These proof-of-concept findings suggest a role for M148(O) as a biomarker for CVD; however, larger clinical studies are needed to validate this role. M148 lies at the center of LCAT activation domain. Shao et el demonstrated that oxidation of M148(O) was associated with decreased capacity to activate LCAT (6). In addition, reversing M148 oxidation using methionine sulfoxide reductase restored the ability of Apo A-I to activate LCAT. We monitored the changes in Apo A-I M148 oxidations in HDL isolates rather than whole serum since HDL is the site where LCAT activity esterifies cholesterol, and changes of oxidation status in this local environment may have a functional relevance.
Measuring oxidized M148 using conventional mass spectrometry methods that utilize spectral counting or extracted ion chromatograms can be lengthy and challenging in large sample sizes. In contrast, MRM is a promising technique that allows multiplexing of several targets and has been successfully applied to quantitate plasma proteins (10,14). The addition of SIS peptides in MRM allows for absolute quantitation. Since our goal was to develop an assay to assess the relative ratio of oxidized M148 to the native peptide rather than the absolute concentrations, the SIS peptide for the unmodified M148 was not synthesized. M148(O) SIS peptide was used to correctly identify the peaks of the in vivo M148 peaks, and optimize the transitions. The rationale for not determining the absolute concentration of M148(O) in plasma was that this concentration can vary because of variations in the Apo A-I concentration. In contrast, monitoring the relative ratio of oxidized M148 to the non-oxidized peptide represents the “quality” of this peptide, is cost-effective and simple with less inherent variability. Thus, this approach is better suited for comparing M148 oxidation ratios among different patient groups. One advantage of MRM is that different peptide variants can be selected, depending on the goal of the particular project. The M148-containing Apo A-I peptide would not normally be selected for Apo A-I quantitation because of its susceptibility to methionine oxidation. The “ATEHLSTLSEK” Apo A-I peptide likely gives a better estimate of total Apo A-I concentration.
Several limitations of the study deserve mention. We have not measured the molar % oxidized of M148, as such measure would require calibration of both forms of the peptide. Methionines are also susceptible to ex vivo oxidation that can result from inadequate or prolonged freezing, repeated thawing, or centrifugations (15). Because an anti-oxidant solution was not immediately added before freezing the samples or after HDL isolations, this might have permitted additional oxidation. In addition, the HDL samples were stored at −80 °C after isolation by ultracentrifugation. The ratios of methionine oxidation observed in our study were higher than those reported in an earlier study on diabetes where the samples were preserved in an anti-oxidant solution prior freezing (16). In this earlier study, however, younger patients with type 1 diabetes were recruited. We have recently demonstrated that immediate freezing of samples at −80 °C without the use of an anti-oxidant solution results in low levels of Apo A-I oxidation that are stable for up to 2 years of storage (17). Since all samples were immediately frozen at −80 °C following collection and centrifugations, and processed at the same time and using the same approach, any interference from ex vivo oxidations would be limited and would apply equally to each clinical group of samples.
A second limitation of our study was that the limit of detection and the recovery rate of M148(O) concentrations on Apo A-I by MRM were not determined. We used a S/N ratio cut off of > 3 as the detection limit for all of the analyzed peptides. However, the M148(O) oxidation peak area was well above this ratio (as shown in Fig. 1). A third limitation is batch-to-batch variation or auto digestion that can result from using different lots of trypsin. We have used multiple transitions per peptide and fresh trypsin match to minimize this source of variation. Finally, our clinical findings are a proof-of-concept demonstration, and need to be validated in larger clinical studies.
Conclusion
We conclude that MRM can be applied to monitor the relative abundance of M148 Apo A-I oxidation. This approach would facilitate examining the relationship between M148 oxidation and vascular complications in CVD studies.
Highlights.
Methionine (M148) of Apolipoprotein A-I is critical for LCAT function on HDL
We developed a method to monitor modified M148 on HDL using Multiple Reaction Monitoring (MRM)
An increase in oxidized M148 relative to the native peptide was observed in HDL isolated from research participants with diabetes and evidence of past cardiovascular disease
Monitoring of M148 oxidations using MRM can serve as a potential biomarker for diabetes complications
Acknowledgments
We would also like to recognize Tyra J. Cross and Suping Zhang of the University of Victoria - Genome British Columbia Proteomics Centre for the synthesis of all of the SIS peptides, and Juncong Yang, also of the Proteomic Centre, for exemplary technical support. We also thank Dr. George Tsaprailis with his assistance in running MRMs at the Arizona Proteomics Consortium.
Sources of funding: Dr. Yassine was supported by K23HL107389, AHA12CRP11750017. Drs. Nelson, Reaven, Lau and Yassine were supported by R24DK090958. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. MRM method development was done by the Arizona Proteomics Consortium, which is supported by NIEHS grant P30ES06694 to the Southwest Environmental Health Sciences Center (SWEHSC to Dr. Lau), NIH/NCI grant P30CA023074 to the Arizona Cancer Center (AZCC), and by the BIO5 Institute of the University of Arizona. CHB and AMJ would also like to thank Genome Canada and Genome British Columbia for their support of the University of Victoria - Genome BC Proteomics Centre through Science and Technology Innovation Centre funding.
Abbreviations
- HDL
High Density Lipoprotein
- CVD
Cardiovascular Disease
- Apo A-I
Apolipoprotein A1
- MRM
Multiple Reaction Monitoring
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A, Lloyd-Jones DM, Nelson SA, Nichol G, Orenstein D, Wilson PW, Woo YJ American Heart Association Advocacy Coordinating, C., Stroke, C., Council on Cardiovascular, R., Intervention, Council on Clinical, C., Council on, E., Prevention, Council on, A., Thrombosis, Vascular, B., Council on, C., Critical, C., Perioperative, Resuscitation, Council on Cardiovascular, N., Council on the Kidney in Cardiovascular, D., Council on Cardiovascular, S., Anesthesia, Interdisciplinary Council on Quality of, C., and Outcomes, R. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 2.Duckworth W, Abraira C, Moritz T, Reda D, Emanuele N, Reaven PD, Zieve FJ, Marks J, Davis SN, Hayward R, Warren SR, Goldman S, McCarren M, Vitek ME, Henderson WG, Huang GD. Glucose control and vascular complications in veterans with type 2 diabetes. The New England journal of medicine. 2009;360:129–139. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 3.Gerstein HC, Miller ME, Byington RP, Goff DC, Jr, Bigger JT, Buse JB, Cushman WC, Genuth S, Ismail-Beigi F, Grimm RH, Jr, Probstfield JL, Simons-Morton DG, Friedewald WT. Effects of intensive glucose lowering in type 2 diabetes. The New England journal of medicine. 2008;358:2545–2559. doi: 10.1056/NEJMoa0802743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel A, MacMahon S, Chalmers J, Neal B, Billot L, Woodward M, Marre M, Cooper M, Glasziou P, Grobbee D, Hamet P, Harrap S, Heller S, Liu L, Mancia G, Mogensen CE, Pan C, Poulter N, Rodgers A, Williams B, Bompoint S, de Galan BE, Joshi R, Travert F. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. The New England journal of medicine. 2008;358:2560–2572. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 5.Kontush A, Chapman MJ. Why is HDL functionally deficient in type 2 diabetes? Curr Diab Rep. 2008;8:51–59. doi: 10.1007/s11892-008-0010-5. [DOI] [PubMed] [Google Scholar]
- 6.Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc Natl Acad Sci U S A. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabresi L, Franceschini G. Lecithin:cholesterol acyltransferase, high-density lipoproteins, and atheroprotection in humans. Trends Cardiovasc Med. 2010;20:50–53. doi: 10.1016/j.tcm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Yassine H, Borges CR, Schaab MR, Billheimer D, Stump C, Reaven P, Lau SS, Nelson R. Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: Potential applications to cardiovascular disease and diabetes. Proteomics. Clinical applications. 2013;7:528–540. doi: 10.1002/prca.201200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittes J, Lakatos E, Probstfield J. Surrogate endpoints in clinical trials: Cardiovascular diseases. Statistics in Medicine. 1989;8:415–425. doi: 10.1002/sim.4780080405. [DOI] [PubMed] [Google Scholar]
- 12.Carels RA, Darby LA, Cacciapaglia HM, Douglass OM. Reducing cardiovascular risk factors in postmenopausal women through a lifestyle change intervention. Journal of Women's Health. 2004;13:412–426. doi: 10.1089/154099904323087105. [DOI] [PubMed] [Google Scholar]
- 13.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. The Journal of clinical investigation. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis GA. High density lipoprotein oxidation: in vitro susceptibility and potential in vivo consequences. Biochimica et biophysica acta. 2000;1483:217–235. doi: 10.1016/s1388-1981(99)00181-x. [DOI] [PubMed] [Google Scholar]
- 16.Brock JW, Jenkins AJ, Lyons TJ, Klein RL, Yim E, Lopes-Virella M, Carter RE, Research G, Thorpe SR, Baynes JW. Increased methionine sulfoxide content of apoA-I in type 1 diabetes. Journal of lipid research. 2008;49:847–855. doi: 10.1194/jlr.M800015-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Borges CR, Rehder DS, Jensen S, Schaab MR, Sherma ND, Yassine H, Nikolova B, Breburda C. Elevated Plasma Albumin and Apolipoprotein AI Oxidation under Suboptimal Specimen Storage Conditions. Molecular & Cellular Proteomics. 2014 doi: 10.1074/mcp.M114.038455. mcp. M114. 038455. [DOI] [PMC free article] [PubMed] [Google Scholar]