Abstract
Reducing the incidence and mortality rates for clear cell renal cell carcinoma (ccRCC) remains a significant clinical challenge with poor 5-year survival rates. A unique tissue cohort was assembled of matched ccRCC and distal non-tumor tissues (n=20) associated with moderate risk of disease progression, half of these from individuals who progressed to metastatic disease and the other half who remained disease free. These tissues were used for MALDI imaging mass spectrometry profiling of proteins in the 2–20 kDa range, resulting in a panel of 108 proteins that had potential disease specific expression patterns. Protein lysates from the same tissues were analyzed by tandem mass spectrometry, resulting in identification of 56 proteins of less than 20 kDa molecular weight. The same tissues were also used for global lipid profiling analysis by MALDI-FTICR mass spectrometry. From the cumulative protein and lipid expression profile data, a refined panel of 26 proteins and 39 lipid species were identified that could either distinguish tumor from non-tumor tissues, or tissues from recurrent disease progressors from non-recurrent disease individuals. This approach has the potential to not only improve prognostic assessment and enhance post-operative surveillance, but also to inform on the underlying biology of ccRCC progression.
Keywords: MALDI imaging, MALDI-FTICR, renal cancer, lipid profiling, tissue proteomics
Introduction
Incidence and mortality rates for renal carcinoma (RCC) continue to rise [1–3], and hallmark features of RCC include a predominance of the clear cell subtype (ccRCC), a cellular metabolism genotype and Warburg effect phenotype, poor 5-year survival (~60%) and a characteristically unpredictable clinical course [4–6]. A portion of the increase in incidence can be attributed to the increased use of advanced imaging modalities and as such, the largest increase in incidence has been among, smaller (< 4cm), asymptomatic, organ confined tumors [7–9]. To date, the only widely accepted treatment for patients with localized ccRCC remains surgical excision [10,11]. Related to this, while surgical excision is curative for the majority of patients with localized ccRCC, approximately 25–30% of these patients will experience disease recurrence (i.e. progression to metastasis) and most of these will occur within 1–3 years following surgery. Given the lack of effective therapeutics for metastatic ccRCC, less than 10% of patients survive for 5 years following the diagnosis of a metastatic lesion. [3,12,13]. Taken together, the aforementioned features of ccRCC underscore the pressing need to identify tumor-based biomarkers that can help to forecast which patients will experience progression after surgery as well as give insight in to innovative treatment strategies for this increasingly common cancer.
Unfortunately, tumor based prognostic biomarkers for ccRCC that have real clinical value are still lacking, despite the ready access to well-annotated tissue samples of ccRCC obtained by nephrectomy. This is not for lack of effort, as multiple approaches across the breadth of genomic and proteomic technologies have been applied to primary ccRCC tissues [6,14–19], many of these linked to the most common mutation in the von Hippel-Lindau tumor suppressor gene and its effect on signaling cascades [6,13–16]. Complicating matters further, authors of recent genomic studies have demonstrated that contradictory prognostic results were obtained from whole exome sequencing analysis of different regions of the same patient tumor [20]. Thus, the homogenized tissue analysis of proteomic targets that have dominated to date would be expected to represent an average or pooled effect of these genomic differences and would result in potential masking of signals from a particular biomarker.
Matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS) has emerged as a tissue-based approach that can address the inherent heterogeneity of a tumor by maintaining tissue integrity, histopathology features and analyte distribution across the tissue [21–25]. For proteins, this approach has found wide-spread application to different tumor types [24,25], including ccRCC [26,27]. However, the application of MALDI-IMS to analyze the tissue distribution of lipids, drugs/small molecule metabolites, or N-glycans [28–32] has not been previously applied to ccRCC tissues. For the current study, a unique tissue cohort was assembled of matched tumor and distal non-tumor (i.e normal kidney) tissues (n =10 pairs) obtained by nephrectomy from individuals with ccRCC and moderate risk of disease progression [33,34]. Half of these tissues represent individuals who progressed to metastatic disease within 3 years of nephrectomy, and the other half remained disease free for at least five years. At the histopathology level, these tissues are essentially identical to clinical pathology assessment, and there are no obvious cellular and histology features that would predict disease progression. These tissues were used for MALDI-IMS profiling of protein and lipid expression. Additional tandem mass spectrometry analyses of proteins from each tissue were also done. From these analyses, combined panels of proteins and lipids have been identified that distinguish tumor from non-tumor tissues, as well as provide lead candidates to distinguish recurrent disease progressors with ccRCC from non-progressors.
Materials and Methods
Materials
Sinapinic acid, 2,5-Dihydroxybenzioc Acid (DHB) and trifluoroacetic acid were obtained from Sigma-Aldrich (St. Louis, MO). HPLC-grade methanol, ethanol and water were obtained from Fisher Scientific. Immobilized titanium oxide (ITO) slides were purchased from Bruker Daltonics (Billerica, MA) for MALDI-IMS experiments. Mass spectrometry-grade trypsin gold was from Promega (Madison, WI, USA).
ccRCC Tumor and Non-tumor Tissues
Samples were selected from individuals treated with nephrectomy for newly diagnosed, histologically confirmed, localized ccRCC at Mayo Clinic Florida between January 1, 2004 and December 31, 2008 who are classified to be at intermediate risk of progression based on the Mayo SSIGN scores between 3–6 [33,34]. One group of individuals had disease recurrence and metastasis within 3 years of surgery. Conversely, the other group were from ccRCC patients who remain progression free more than 5 years following surgery. For both groups individuals with any of the following characteristics were excluded: bilateral synchronous tumors, non-clear cell histology, multiple ipsilateral tumors of different histological subtypes, von Hippel-Lindau syndrome or Wilms’ disease, less than 18 years old or refused access to medical records. Further clinical information for each subject including age, race, gender and pathology details is provided in Supplementary Table 1. The non-tumor tissues associated with each tumor sample were harvested from the cortex region of the kidneys that were most distal from the primary tumor. Fresh-frozen tumor tissue was obtained from the biorepository at Mayo Clinic Florida. Tissue sections (10 microns) were prepared using a Thermo Microm HM550 cryostat and stored at 80oC. For each section analyzed, a serial section was collected for histological analysis and staining with hematoxylin and eosin (H & E). At least 4 slices per tissue were analyzed for protein or lipid profiling.
MALDI-IMS analysis of proteins
Tissues were processed and analyzed as previously described [35]. Briefly, kidney tissues on ITO coated slides were washed and fixed with 70% ethanol and 95% ethanol for 30s each. A water wash was performed to remove residual embedding media followed by a repeat of the ethanol washes of 70% and 95%. Slides were air dried and stored in a dessicator for 1 hour before matrix deposition. A matrix solution of sinapinic acid (10 mg/ml) containing 75% acetonitrile and 0.13% TFA was sprayed uniformly over the tissue using an automated spraying device, ImagePrep workstation (Bruker Daltonics, Billerica, MA) which controls matrix deposition and thickness of the matrix layer. Digital images of the sprayed tissue sections were acquired with a flatbed scanner prior to MALDI analysis. Spectra were collected across the entire tissue area using an AutoFlex III or Ultraflex III MALDI-TOF/TOF instrument (Bruker Daltonics) with a SmartBeam laser operating at 200 Hz in linear mode over a mass range of 2,000 to 20,000 Daltons. A laser spot diameter of 50 µm and a raster width of 100 µm were employed. A total of 200 laser shots were accumulated and averaged from each laser spot rastered across the tissue section. Calibration was performed externally using a Protein Calibration Standard I (Bruker Daltonics) in the mass range of 5000–17500 Da, and images were generated and visualized using Flex Imaging 4.0.
MALDI-IMS of lipids
Kidney tissues on ITO coated slides were desiccated at room temperature for 20 minutes, followed by addition of 2,5-Dihydroxybenzoic acid (DHB) matrix at a concentration of 0.2M in 50% acetonitrile (ACN) .01% trifluoroacetic acid (TFA) sprayed on to the slide using the ImagePrep. Spectra in positive ion mode were acquired across the entire tissue section on a Solarix 70 dual source 7T FTICR mass spectrometer (Bruker Daltonics, Billerica, MA) to detect the lipid species of interest (m/z 200–2000) with a SmartBeam II laser operating at 1000 Hz, a laser spot size of 25 µm, and a raster width of 200 µm for general profiling, and 75 µm for high resolution images. For each laser spot, 800 spectra were averaged. Images of differentially expressed lipids were generated using FlexImaging 4.0 software (Bruker Daltonics). Following MS analysis, data was loaded into FlexImaging Software focusing on the m/z = 200–2000 range and reduced to 0.98 ICR Reduction Noise thresholds. All data was normalized using root means square. For structural analysis, specific ions of interest were collected using CASI (continuous accumulation of selected ions). CASI allows for trapping of a specific ion of interest within the quadrupole of the Solarix 70 mass spectrometer. Following accumulation, the ion was fragmented using CID (collision-induced fragmentation). Structural assignments were made following detection of specific fragmentation patterns, like the loss of a phosphocholine head group (m/z =184.1) for phosphatidylcholine species, and cross-validated with the Lipid Maps database.
Data processing
Automated analysis of the spectral data was performed to identify all differentially expressed peaks between samples. Spectra derived from defined Regions of Interest (ROI) in each tissue protein or lipid set were exported using the FlexImaging software for profile analysis. Raw spectra were processed with Progenesis MALDI (version 1.4, Nonlinear Dynamics, Durham, NC) using a noise filter of 4 and top hat filter of 60. Peaks were selected between m/z = 400–1200 with a 200,000 threshold. Peak heights were normalized to total ion current (for proteins) or root means square (for lipids) and used for further analysis.
Tissue homogenization and protein digestion
Tissues were sliced at 30 µm and placed in 1.5 ml tubes, and resuspended in 0.1 ml 50 mM ammonium bicarbonate (AmBic) and 0.1 ml trifluoroethanol (TFE) [36]. The tissues were then sonicated three times each for 20 s at 30% power and allowed to sit on ice for 30 s in between sonication. After sonication, the samples were heated at 60 °C for 1 hr with vortexing every 15 min. The samples were sonicated again, followed by addition of 0.05 ml 50 mM dithiothreitol and 20 mM TCEP in 100 mM AmBic. Following a 30 min incubation at 60 °C, 0.05 ml 100mM iodoacetamide was added for 20 min. Lastly, 0.7 ml 100mM AmBic and 2 µg trypsin was added to each tube followed by an overnight incubation at 37°C. Peptides were isolated by C18 column purification and concentrated as previously described [37].
LC-MS/MS Analysis
Concentrated digests were resuspended in 1 mL buffer A (97.8% HPLC-grade H2O, 2% acetonitrile, 0.2% formic acid), and 15 µL of digest was loaded onto a 25cm×0.0075mm C-18 reversed phase LC column (packed in house, Waters ODS C18). Liquid chromatography (LC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) on a linear ion trap mass spectrometer (LTQ, ThermoFischer) coupled to a LC Packings nano-LC system was performed utilizing a 120 min linear gradient from 5% acetonitrile, 0.2% formic acid to 50% acetonitrile, 0.2% formic acid. Data dependent analysis was selected to perform MS/MS on the 10 most intense ions between m/z = 400 to 2000 in each MS spectra with a minimum signal of 500 cps. All samples were run in duplicate. Dynamic exclusion was used with a repeat count of two and an exclusion duration of 120 seconds. Acquired data were searched using Mascot (version 2.4.1, Matrix Science, London, UK) followed by post-processing with ProteoIQ (version 2.3.08). The data were searched against a UniProt SwissProt Homo sapien database (2012_10 release) containing both canonical and isoform entries, which was concatenated with entries from the common Repository of Adventitious Proteins (37,031 entries total) as well as a separate search against a reverse decoy database of the same entries. Precursor mass tolerance was set to 2 Da and fragment mass tolerance to 1 Da. Enzyme specificity was trypsin, allowing for two missed cleavages. Carbamidomethyl (Cys) was specified as a fixed modification and protein N-term acetylation, deamidation (Asn, Gln), pyro-Glu (N-term Gln), and oxidation (Met) were set as variable modifications. Using ProteoIQ a protein false discovery rate (FDR) cutoff of 5% was applied and 2 peptides were required for protein identification. Spectral counts were normalized across the experiment and average normalized spectral counts between duplicate runs were used for downstream analysis.
Principal Component Analysis and Statistics
Wilcoxon signed-rank or rank-sum test was used to compare MALDI peak intensities between tumor versus normal and recurrent versus non-recurrent, respectively. Principal component analysis was performed on MALDI data that was first standardized by dividing each peak intensity by the standard deviation of all the peak intensities for that given m/z. Loadings were evaluated using the coefficients of the linear combinations of the original variables that generated each principal component. A heat map was generated from average protein peak intensities that were log10 transformed. All analysis was performed using MATLAB (R2013a; Mathworks).
Results
Protein profiling by MALDI-IMS of matched ccRCC tumor and non-tumor tissues
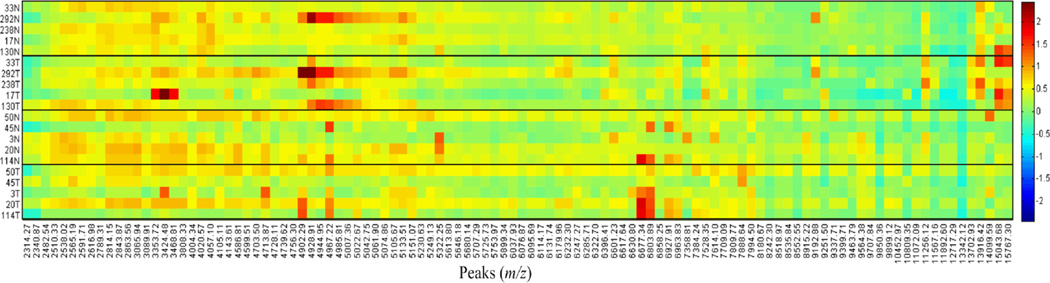
A tissue cohort was assembled representing patient matched ccRCC and distal non-tumor tissues (10 pairs, n = 20) with moderate risk of disease progression. Fresh frozen 10 µm slices from each of the 20 tissue samples were processed for MALDI-IMS analysis of protein and peptide expression as described in the Materials and Methods. Shown in Figure 1 are representative images of different tissue pairs, showing differences in expression between matched tumor and normal tissue pairs. However, initial visual comparisons of protein expression differences indicated significant heterogeneity across the tissue pairs, either in tumor versus normal or recurrent versus non-recurrent disease comparisons. Therefore, representative regions of interest for each tissue were selected that included 50 to 100 spectra, and peak intensities were normalized for expression across all 20 tissues. Average intensities of 108 identified peaks were used to compare conditions and are shown in the intensity heatmap in Figure 2. Peak intensities were compared between tumor and normal and recurrent and non-recurrent using a signed rank test and rank sum test, respectively, and 27 were significantly different (p < 0.05). Additionally, ROC curves were utilized to evaluate the ability of each peak to discriminate tumor versus normal or recurrent versus non-recurrent, and 44 had an AuROC > 0.70. From these comparisons, 26 protein peaks with p < 0.05 and AuROC > 0.70 were selected. The individual statistics for each of these m/z values are provided in the Supplementary Data Table 2. As illustrated in Figure 2, there were clear peak clusters that were tumor specific and highly abundant in individual sample pairs, but overall there was much heterogeneity in protein expression patterns across the groups.
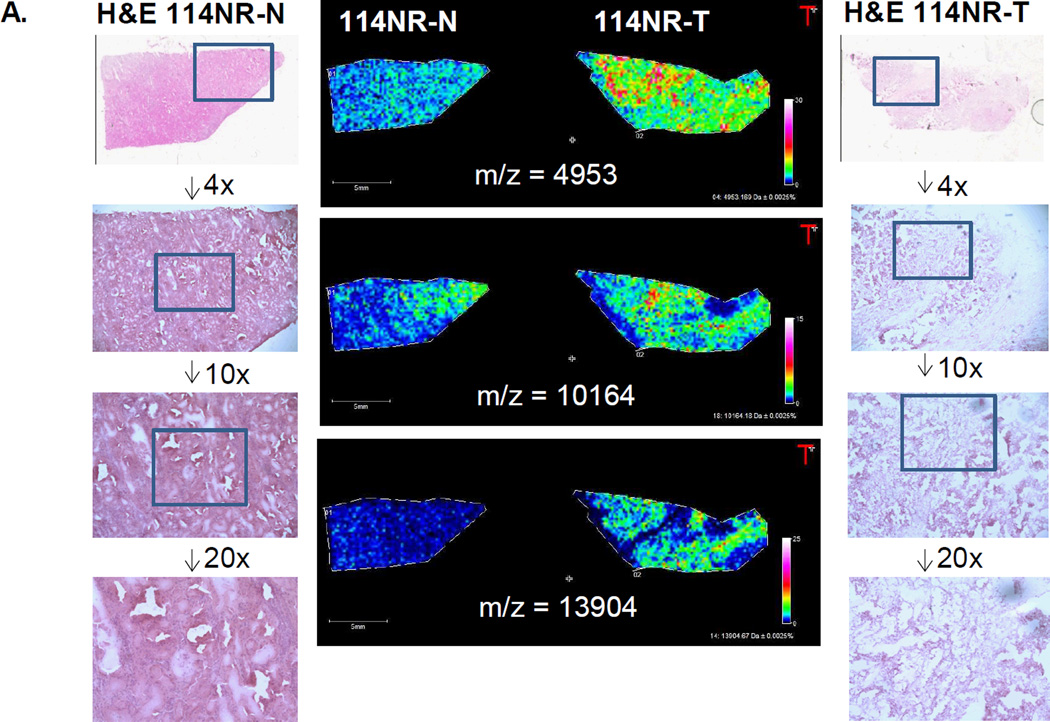
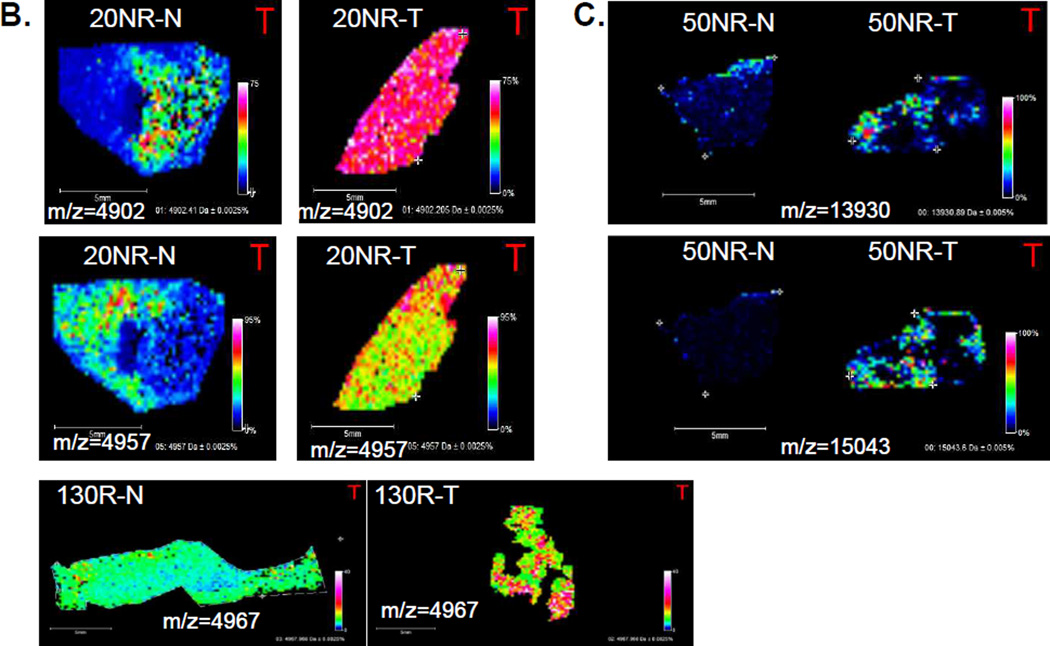
Figure 1. Representative MALDI-MS images of proteins in matched ccRCC tumor and non-tumor tissues.
A. Three images of ions (m/z = 4953, 10164, 13904) with increased expression in non-recurrent tumor tissues (114NR-T). H&E stained images of both tissues are also shown, including representative magnifications of regions at 4×, 10× and 20× magnification, indicated by the inset boxes. B. Two images of two ions at m/z = 4901 and m/z = 4967 enriched in tumor for a non-recurrent pair (20NR), and a m/z = 4967 with higher tumor expression. C. Representative images of increased detection of m/z= 15043 and 13930 in a non-tumor/tumor pair.
Figure 2. Heat map of 108 selected protein peaks.
Protein peak intensities for a representative region of interest for each sample (n=20) were normalized and average peak intensities were log10 transformed before plotting as a heatmap. The scale is log10 average normalized peak intensity. Also, a Wilcoxon signed-rank or rank-sum test was used to compare peak intensities between tumor versus normal and recurrent versus non-recurrent, respectively (Supplemental Table 2).
A limitation of MALDI-IMS of intact small proteins is the direct ability to determine amino acid sequence and protein identity of peaks of interest, a process which usually requires specific isolation and enrichment of individual protein targets, followed by LC-MS/MS. Recently, a database of 106 individual proteins identified by MALDI-IMS has been reported, and can be accessed as a searchable database resource, termed MaTisse DB [25]. The peak list of the 108 ccRCC mass values generated in the MALDI-IMS profiles was queried directly, and there were some protein classes that matched or were near the reported m/z values in the MaTisse DB. This included thymosin β4 and β10, S100 A8-A11, cytochrome c oxidase subunits, histones, defensins and hemoglobins, and may include the ions shown in Figure 1. To complement this, 30 micron slices of all 20 tissues were used to generate lysates for trypsin digestions, followed by tandem mass spectrometry protein identification. While over 750 total proteins were detected in this global proteomic tissue analysis, we refined the list to only those proteins that were of 20,000 predicted molecular mass and lower (n =122). This list was further refined for protein abundance based on averaged normalized spectral counts, and is provided in Table 1, for 56 proteins. The protein identities from this list were also cross-referenced to the MaTisse DB lists, and if present in the database, are indicated with an asterisk in the table, with 25 being shared in the reference database. We used the relative abundance of the proteins in Table 1 to match with the most abundant protein peaks detected by MALDI-IMS, cross-referenced to the MaTisse DB information. From the data in the MALDI-IMS database, we did not expect to get direct matches with expected molecular masses from the tandem MS data, relative to the observed m/z values in MALDI-IMS. There were few proteins that had perfect correlation across the groups, but it is likely that the 4900–4970 peak cluster in the MALDI data represents thymosin beta-4 and thymosin beta-10. The image shown in Figure 1A for the m/z = 10164 ion that is overexpressed in the tumor sample, exactly matches to a S100A8 peak in the MaTisse DB. It is also on average overexpressed in tumors as shown in Table 1 and Supplementary Table 3. In the tandem MS data, there was one protein in particular that was discriminatory for both tumor vs normal and recurrent vs non-recurrent, fatty acid binding protein, liver (FABP7). This native mass was not detected in the tissues, nor was there any indication of it being detected in other MALDI-IMS studies as listed in the MaTisse DB.
Table 1.
Tandem mass spectrometry identified list of the 56 most abundant tissue proteins of <20 kDa.
| Avg NSc's (± S.D.) | Avg NSc's (± S.D.) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence ID | Description | Gene Name | Avg MW | normal | tumor | p-value | nonrecurrent | recurrent | p-value |
| P63313 | Thymosin beta-10* | TMSB10 | 4894.48 | 2.18 ± 2.1 | 4.90 ± 3.1 | 0.027 | 4.42 ± 3.3 | 2.66 ± 2.4 | 0.199 |
| P62328 | Thymosin beta-4* | TMSB4X | 4921.46 | 4.79 ± 2.2 | 7.43 ± 2.7 | 0.049 | 6.92 ± 2.8 | 5.30 ± 2.5 | 0.241 |
| P59665 | Neutrophil defensin 1* | DEFA1 | 6306.36 | 2.57 ± 1.2 | 4.01 ± 4.9 | 0.922 | 1.87 ± 1.3 | 4.71 ± 4.5 | 0.064 |
| P62987 | Ubiquitin-60S ribosomal protein L40 | UBA52 | 8564.84 | 4.13 ± 0.9 | 5.39 ± 2.4 | 0.160 | 5.09 ± 1.8 | 4.44 ± 2.1 | 0.450 |
| P02652 | Apolipoprotein A-II* | APOA2 | 8707.91 | 2.83 ± 1.7 | 2.61 ± 1.6 | 0.770 | 2.80 ± 1.3 | 2.65 ± 2.0 | 0.571 |
| P07602 | Proactivator polypeptide | PSAP | 9107.55 | 3.30 ± 1.3 | 2.16 ± 1.7 | 0.137 | 2.29 ± 1.3 | 3.17 ± 1.8 | 0.162 |
| P14854 | Cytochrome c oxidase subunit 6B1* | COX6B1 | 10061.2 | 5.02 ± 1.8 | 1.01 ± 1.7 | 0.006 | 2.34 ± 2.2 | 3.69 ± 3.1 | 0.363 |
| P10606 | Cytochrome c oxidase subunit 5B, mitochondrial * | COX5B | 10613.04 | 2.26 ± 0.7 | 0.69 ± 0.9 | 0.004 | 1.33 ± 1.2 | 1.62 ± 1.1 | 0.544 |
| P61604 | 10 kDa heat shock protein, mitochondrial* | HSPE1 | 10800.5 | 5.99 ± 2.3 | 3.76 ± 2.1 | 0.084 | 5.31 ± 2.3 | 4.43 ± 2.6 | 0.473 |
| P05109 | Protein S100-A8* | S100A8 | 10834.51 | 1.36 ± 1.7 | 3.13 ± 2.0 | 0.039 | 2.46 ± 2.4 | 2.03 ± 1.7 | 0.791 |
| P60903 | Protein S100-A10* | S100A10 | 11071.91 | 1.36 ± 1.0 | 1.75 ± 1.4 | 0.322 | 1.59 ± 1.3 | 1.52 ± 1.1 | 0.849 |
| P62805 | Histone H4* | HIST1H4A | 11236.15 | 16.43 ± 3.3 | 16.61 ± 5.6 | 0.770 | 18.08 ± 4.5 | 14.96 ± 4.2 | 0.212 |
| P0CG05 | Ig lambda-2 chain C region | IGLC2 | 11294 | 4.32 ± 4.2 | 4.41 ± 5.0 | 0.492 | 3.55 ± 2.9 | 5.09 ± 5.7 | 0.910 |
| O75964 | ATP synthase subunit g, mitochondrial | ATP5L | 11297.28 | 2.00 ± 0.9 | 1.17 ± 0.8 | 0.084 | 1.76 ± 1.1 | 1.41 ± 0.7 | 0.405 |
| P10599 | Thioredoxin* | TXN | 11606.3 | 1.62 ± 0.7 | 1.38 ± 1.3 | 0.557 | 1.60 ± 1.2 | 1.40 ± 0.9 | 0.791 |
| P01834 | Ig kappa chain C region | IGKC | 11609 | 32.36 ± 12.3 | 32.27 ± 12.2 | 0.770 | 29.40 ± 8.0 | 35.23 ± 14.7 | 0.345 |
| P31949 | Protein S100-A11 * | S100A11 | 11609.25 | 3.34 ± 1.6 | 4.29 ± 1.7 | 0.242 | 4.10 ± 1.0 | 3.53 ± 2.2 | 0.473 |
| P35754 | Glutaredoxin-1 | GLRX | 11644.55 | 2.67 ± 1.1 | 1.86 ± 1.8 | 0.275 | 1.98 ± 1.8 | 2.56 ± 1.2 | 0.596 |
| P01620 | Ig kappa chain V-III region SIE | - | 11775 | 1.60 ± 1.3 | 2.15 ± 1.7 | 0.098 | 1.68 ± 1.6 | 2.07 ± 1.5 | 0.704 |
| P14174 | Macrophage migration inhibitory factor* | MIF | 12345.11 | 7.42 ± 1.7 | 9.49 ± 4.7 | 0.193 | 9.39 ± 4.1 | 7.51 ± 2.9 | 0.473 |
| P20674 | Cytochrome c oxidase subunit 5A, mitochondrial * | COX5A | 12501.17 | 2.70 ± 1.6 | 0.32 ± 0.7 | 0.004 | 1.28 ± 1.6 | 1.74 ± 1.9 | 0.785 |
| P58546 | Myotrophin* | MTPN | 12763.6 | 2.04 ± 0.5 | 2.29 ± 1.2 | 0.420 | 2.40 ± 1.1 | 1.93 ± 0.7 | 0.226 |
| P06702 | Protein S100-A9* | S100A9 | 13110.8 | 1.21 ± 1.4 | 1.80 ± 0.8 | 0.375 | 1.38 ± 1.1 | 1.62 ± 1.3 | 0.733 |
| O75348 | V-type proton ATPase subunit G 1 | ATP6V1G1 | 13626.33 | 2.35 ± 1.7 | 0.42 ± 0.5 | 0.014 | 0.90 ± 1.1 | 1.87 ± 1.9 | 0.378 |
| P02766 | Transthyretin* | TTR | 13761.41 | 6.71 ± 3.4 | 5.41 ± 2.7 | 0.492 | 5.54 ± 2.6 | 6.58 ± 3.6 | 0.650 |
| P62807 | Histone H2B type 1-C/E/F/G/I | HIST1H2BC | 13774.95 | 3.11 ± 1.0 | 3.63 ± 1.5 | 0.361 | 3.51 ± 1.6 | 3.23 ± 0.9 | 0.880 |
| P33778 | Histone H2B type 1-B* | HIST1H2BB | 13819 | 16.73 ± 10.1 | 18.99 ± 9.5 | 0.695 | 16.74 ± 7.9 | 18.98 ± 11.3 | 0.850 |
| P20671 | Histone H2A type 1-D | HIST1H2AD | 13976.29 | 4.24 ± 1.4 | 5.19 ± 1.6 | 0.275 | 5.11 ± 1.7 | 4.32 ± 1.3 | 0.273 |
| P04908 | Histone H2A type 1-B/E | HIST1H2AB | 14004.3 | 15.72 ± 6.4 | 18.26 ± 6.5 | 0.625 | 16.42 ± 6.0 | 17.58 ± 7.1 | 0.521 |
| Q96A08 | Histone H2B type 1-A | HIST1H2BA | 14036.31 | 8.11 ± 2.1 | 8.26 ± 2.9 | 1.000 | 8.29 ± 2.0 | 8.08 ± 3.0 | 0.910 |
| Q96QV6 | Histone H2A type 1-A | HIST1H2AA | 14102.32 | 10.80 ± 1.8 | 16.16 ± 6.5 | 0.084 | 14.66 ± 5.9 | 12.31 ± 4.8 | 0.678 |
| A6NHG4 | D-dopachrome decarboxylase-like protein | DDTL | 14195.29 | 3.58 ± 1.1 | 2.45 ± 1.8 | 0.064 | 3.00 ± 1.4 | 3.04 ± 1.7 | 1.000 |
| P07148 | Fatty acid-binding protein liver | FABP1 | 14208.38 | 3.82 ± 3.0 | 0.53 ± 1.3 | 0.016 | 1.34 ± 2.3 | 3.01 ± 3.1 | 0.219 |
| P52758 | Ribonuclease UK114 | HRSP12 | 14362.49 | 5.37 ± 2.7 | 2.88 ± 2.2 | 0.045 | 4.02 ± 2.9 | 4.23 ± 2.6 | 1.000 |
| P09382 | Galectin-1* | LGALS1 | 14584.51 | 6.13 ± 3.8 | 10.45 ± 5.0 | 0.105 | 9.08 ± 4.8 | 7.49 ± 5.0 | 0.427 |
| P61626 | Lysozyme C* | LYZ | 14700.67 | 3.67 ± 2.3 | 1.11 ± 1.3 | 0.006 | 1.33 ± 1.5 | 3.44 ± 2.4 | 0.037 |
| P05413 | Fatty acid-binding protein, heart | FABP3 | 14726.84 | 3.50 ± 2.3 | 2.42 ± 3.7 | 0.193 | 3.66 ± 3.9 | 2.25 ± 1.8 | 0.570 |
| O15540 | Fatty acid-binding protein, brain | FABP7 | 14757.72 | 2.01 ± 6.2 | 11.67 ± 17.6 | 0.031 | 11.73 ± 17.6 | 1.95 ± 6.0 | 0.055 |
| P07737 | Profilin-1* | PFN1 | 14923.04 | 6.31 ± 2.3 | 8.05 ± 4.0 | 0.232 | 8.24 ± 3.6 | 6.12 ± 2.7 | 0.241 |
| P30049 | ATP synthase subunit delta, mitochondrial | ATP5D | 15019.93 | 2.48 ± 1.4 | 1.10 ± 1.3 | 0.049 | 1.27 ± 1.5 | 2.30 ± 1.4 | 0.081 |
| P69905 | Hemoglobin subunit alpha* | HBA1 | 15126.36 | 34.77 ± 11.3 | 101.93 ± 56.8 | 0.010 | 47.92 ± 28.2 | 88.77 ± 64.4 | 0.307 |
| P00167 | Cytochrome b5 | CYB5A | 15198.91 | 3.30 ± 1.4 | 2.46 ± 1.8 | 0.322 | 2.73 ± 1.8 | 3.03 ± 1.4 | 0.940 |
| Q16695 | Histone H3.1t | HIST3H3 | 15377.06 | 4.89 ± 1.1 | 6.71 ± 5.3 | 0.492 | 6.95 ± 5.1 | 4.65 ± 1.6 | 0.364 |
| O60361 | Putative nucleoside diphosphate kinase | NME2P1 | 15529.06 | 1.41 ± 0.6 | 1.80 ± 1.7 | 0.504 | 1.48 ± 0.7 | 1.72 ± 1.6 | 0.880 |
| P00441 | Superoxide dismutase [Cu-Zn] | SOD1 | 15804.55 | 5.97 ± 2.6 | 4.59 ± 2.8 | 0.922 | 4.97 ± 2.7 | 5.58 ± 2.8 | 0.880 |
| P68871 | Hemoglobin subunit beta* | HBB | 15867.22 | 148.25 ± 58.3 | 372.74 ± 198.5 | 0.014 | 191.86 ± 116.9 | 329.14 ± 216.1 | 0.186 |
| P02042 | Hemoglobin subunit delta | HBD | 15924.29 | 9.81 ± 2.2 | 16.43 ± 6.1 | 0.020 | 11.86 ± 4.9 | 14.38 ± 6.3 | 0.521 |
| P69891 | Hemoglobin subunit gamma-1 | HBG1 | 16009.28 | 3.55 ± 4.9 | 12.93 ± 27.5 | 0.027 | 2.52 ± 1.9 | 13.96 ± 27.4 | 0.121 |
| P60660 | Myosin light polypeptide 6 | MYL6 | 16798.86 | 8.22 ± 2.8 | 7.91 ± 3.5 | 0.750 | 8.59 ± 3.5 | 7.53 ± 2.7 | 0.521 |
| P30044 | Peroxiredoxin-5, mitochondrial | PRDX5 | 17030.75 | 4.09 ± 1.4 | 2.83 ± 1.8 | 0.275 | 3.53 ± 1.8 | 3.39 ± 1.7 | 0.910 |
| P62937 | Peptidyl-prolyl cis-trans isomerase A* | PPIA | 18012.49 | 20.10 ± 3.2 | 19.98 ± 6.9 | 1.000 | 21.07 ± 4.1 | 19.01 ± 6.2 | 0.571 |
| O75947 | ATP synthase subunit d, mitochondrial | ATP5H | 18360.02 | 4.36 ± 1.9 | 1.12 ± 1.1 | 0.010 | 2.83 ± 2.2 | 2.66 ± 2.3 | 0.970 |
| P23528 | Cofilin-1* | CFL1 | 18371.3 | 6.15 ± 2.0 | 6.06 ± 3.6 | 0.770 | 6.76 ± 3.2 | 5.45 ± 2.4 | 0.571 |
| B9A064 | Immunoglobulin lambda-like polypeptide 5 | IGLL5 | 19278.73 | 6.77 ± 4.0 | 6.54 ± 2.7 | 0.846 | 6.15 ± 1.4 | 7.16 ± 4.5 | 1.000 |
| P02792 | Ferritin light chain | FTL | 19888.48 | 9.94 ± 5.5 | 11.07 ± 5.9 | 0.232 | 9.97 ± 5.0 | 11.03 ± 6.2 | 0.791 |
| Q99497 | Protein DJ-1 | PARK7 | 19891.05 | 4.76 ± 1.7 | 2.97 ± 1.9 | 0.105 | 3.79 ± 1.5 | 3.94 ± 2.5 | 0.850 |
Indicates overlap with proteins listed in the MaTisse database of identified proteins and peptides by MALDI-IMS [25]; (http://129.187.44.58:7171/MALDI/protein)
MALDI Imaging of Lipids
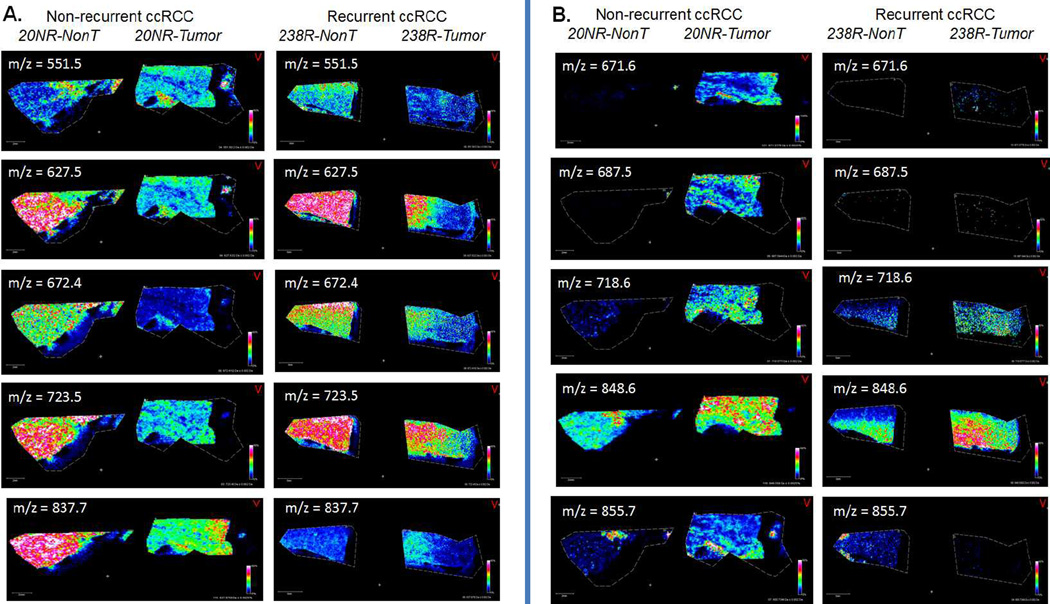
The 20 renal tissues were also evaluated for changes in levels of lipid species in positive ion mode using a Bruker Solarix 70 MALDI-FTICR. Because of the large number of samples and large file sizes that result from high resolution FTICR tissue analyses, each tissue was initially run at lower spatial resolution to acquire a broader profile of the major lipid specie differences. The resulting spectra were analyzed using Progenesis software to align and identify 141 peaks. Normalized (TIC) intensities of the 141 identified peaks were used to compare conditions. Peak intensities were compared between tumor and normal and recurrent and non-recurrent using a signed rank test and rank sum test, respectively, and 39 were significantly different (p < 0.05). Additionally, ROC curves were utilized to evaluate the ability of each peak to discriminate tumor versus normal or recurrent versus non-recurrent, and 68 had an AuROC > 0.70 (Supplementary Table 4). From these comparisons, 39 lipid peaks with p < 0.05 and AuROC > 0.70 were selected. Representative tissue pairs from recurrent and non-recurrent tumors were further analyzed at higher spatial resolution, and different images of ions matched to the 141 lipid species list are shown in Figure 3. Representative histology images of each tissue is provided in Supplementary Figure 1. Five of these are more abundant in non-tumor tissues (Figure 3A), and the other five species are more abundant in tumor tissues (Figure 3B). Each of these lipids is part of the 39 lipids highlighted in the data analysis, and these tissues were also used for specific continuous accumulation of selected ions (CASI) isolation and collision-induced dissociation (CID) to attempt to determine their structures. Three peaks were identified as phosphatidylcholines, PC 26:0 +Na at m/z = 672.4; PC 30:3 +Na at m/z = 718.6; PC 30:2 at m/z = 723.5. These structures were also correlated with structures in the Lipid MAPS database (www.lipidmaps.org). Confirmation of these assignments using standards, and structural determination of the other lipid species, is ongoing using LC-MS/MS approaches.
Figure 3. Representative MALDI-MS images of lipid species in matched ccRCC tumor and non-tumor tissues.
Image profiles of ten lipids are shown for two matched tissues, a non-recurrent (20NR) and recurrent (238R) tumor/non-tumor pair. The representative lipids were selected as part of the 39 lipids that were most discriminatory across conditions (Supplemental Table 4). A. Five lipid ions that are predominantly over-expressed in non-tumor tissues. B. Five lipid ions that are predominantly expressed in tumor tissues.
Distinguishing tumor from non-tumor and non-recurrent from recurrent disease
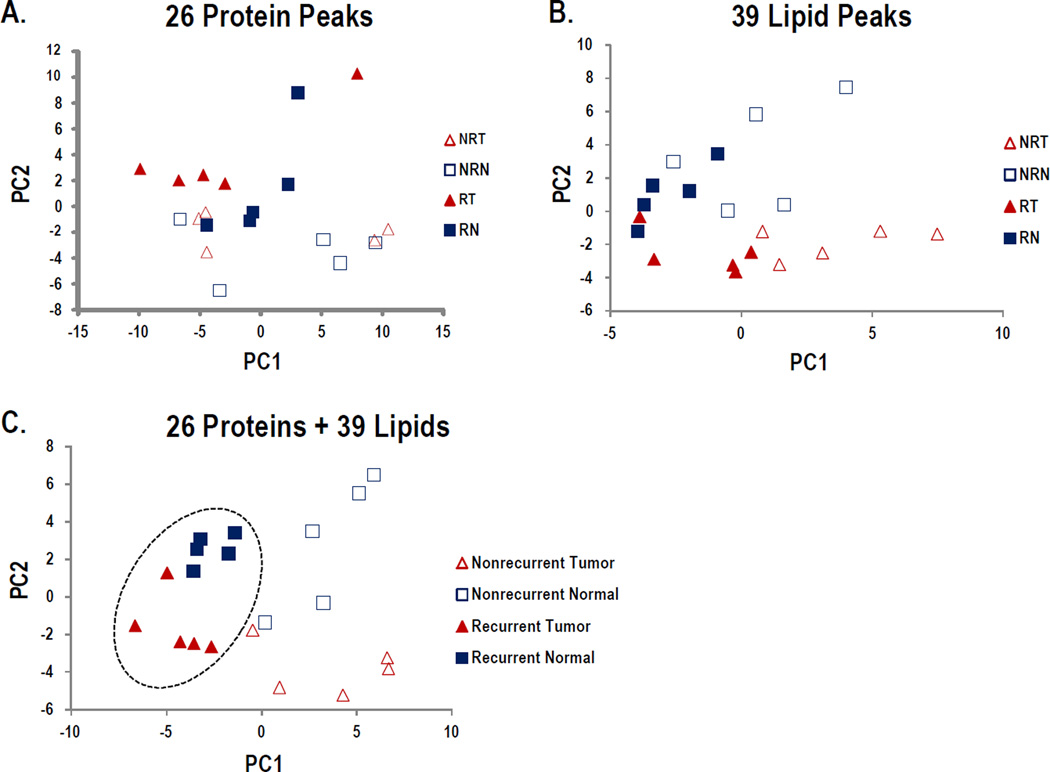
Protein and lipid peaks of interest, n = 26 and 39, respectively, were selected based on an individual peaks ability to distinguish groups (AuROC > 0.70) and statistical significance (p < 0.05; Supplemental Tables 2 and 4). Principal component analysis was used to evaluate the data in multidimensional space to detect whether the peaks separate the data in an unsupervised analysis. Using only the protein peaks of interest, or the lipid peaks of interest alone, all four groups are not separated (Figure4A and 4B). The protein peaks have some discriminatory ability to separate recurrent versus non-recurrent ccRCC (driven by PC2; Fig 4A), while the lipid peaks seem to better separate tumor versus normal (PC2; Fig 4B), but neither is clearly separated. By combining both sets of peaks of interest (Figure 4C), there is much clearer group separation, such that PC2 is driving tumor versus normal, and PC1 is driving recurrent versus non-recurrent. The negative loadings for PC1 (so driving recurrence), include 9 lipid and 6 protein peaks, with the main loadings coming from lipid 837.68 and protein peaks 13916.42 and 15043.68. The other loadings are more evenly distributed across protein and lipid peaks. Overall this highlights that using both lipid and protein data in tandem may identify a panel of peaks that could correctly classify tumor versus nontumor or recurrent versus nonrecurrent ccRCC.
Figure 4. Principal Component Analysis of Protein and Lipid Species.
Protein and lipid peaks of interest were selected based on an individual peak's ability to distinguish groups (AuROC > 0.70) and statistical significance (p < 0.05; Supplemental Tables 2 and 4), then evaluated separately and combined using PCA. A. Plot of PCA results using 26 proteins. B. Plot of PCA results using 39 lipids. C. Plot of PCA results using combined 26 proteins and 39 lipids.
Discussion
This project initially began as a MALDI-IMS proteomic profiling analysis of the unique cohort of ccRCC tissues (Supplementary Table 1). While there are distinct proteins that are detected at elevated levels within tumor/normal pairs, there was no clear correlation of expression differences for tumor versus normal or recurrent versus non-recurrent conditions at the tissue levels. The proteomic analysis clearly highlights the inherent heterogeneity associated with ccRCC tumors [20] at the individual peak levels (Figure 2). Comparative protein identification strategies on the same tissues for proteins under 20 kDa indicated several proteins that were overexpressed in tumors, including thymosin β4, thymosin β10, fatty acid binding proteins and hemoglobin alpha and beta. Using a new database resource, MaTisse (http://129.187.44.58:7171/MALDI/protein) [25], the tandem MS proteomic data and MALDI-IMS profiling data could be cross-referenced to identify tissue ions that correlated with protein identities. Thymosin β4 and thymosin β10 illustrate the complexities of the approach, as they are most likely represented by the peaks in the m/z = 4900–4970 clusters. There are multiple values for these proteins in the reference database in this range, i.e. m/z = 4963–4965 for thymosin β4 and m/z = 4933–4937 for thymosin β10, but these do not align with the major peaks detected at m/z = 4902, 4957, 4967 shown in Figure 1B in individual tissues, or for m/z = 4902, 4928, 4944, 4967 in the averaged and normalized comparisons (Figure 2; Supplementary Table 2). This will need to be resolved by direct tryptic peptide analysis of these proteins following specific isolation of them directly from tissues. In Table 1, both of these proteins were more abundant in tumor tissues relative to non-tumor tissues, but did not discriminate recurrent from non-recurrent tumors.
Of the four S100 proteins detected (Table 1), A8, A9, A10 and A11, only S100A8 had any differential expression on average in tumors versus non-tumors. Hemoglobin alpha and hemoglobin beta were the most differential for distinguishing both groups, and were elevated in recurrent tumors relative to non-recurrent tumors and all non-tumor tissues. There were again discrepancies between the masses of the averaged spectra ions (Figure 2), database values [25] and expected molecular weight values (Table 1). There was one protein at m/z = 13916 in the MALDI-IMS data that was potentially discriminatory for recurrent tumors (AuROC = 0.95), and along with the putative hemoglobin alpha peak at m/z = 15043 (AuROC = 0.75), were responsible for 20% of the negative loadings on PC1, which appears to separate recurrent and non-recurrent (Figure 4C). There are no obvious correlates for identifying this m/z = 13916 protein in the tandem MS data or reference database, and certainly this ion may be an as yet unidentified post-translationally modified variant of one of the database proteins. Conversely, there is an example protein from the tandem MS data, fatty acid binding protein 7 (FABP7), that is discriminatory for expression in non-recurrent tumors versus the other three groups, being elevated in 4 of 5 non-recurrent tumors (Table 2, Supplemental Table 2). This protein has been reported to be differentially expressed in ccRCC tissues in previous proteomic studies [38–40]. Based on the expected molecular weight of FABP7 of 14,757, there is no correlate to a peak ion in the MALDI-IMS data or value for this protein previously identified in the MaTisse database. Based on expression patterns alone in the MALDI-IMS data for non-recurrent tumors, candidates could be truncated variants at the m/z = 6777 or 6803 ions (Figure 2). Specific isolation and protein identification strategies for these different tissue target proteins are ongoing.
Compared to the protein analyses, the lipid profiling by MALDI-IMS of the same ccRCC tissues identified multiple lipid species that were more consistently discriminatory across each tissue group (Figure 3, Supplemental Table 4). Because of the high resolution capability of the FTICR platform, the number of potential species detected and large data file sizes, make this data more challenging from a bioinformatics perspective [41]. We chose to select regions of interest from each tissue and normalize peak data using Progenesis software. This strategy stratified the data to identify the most abundant ions (n=141; Supplemental Table 4), and when combined with Wilcoxon exact rank sum and AuROC analyses, facilitated the group comparisons across this complex data set. From this, 39 lipid species were identified that were most discriminatory for tumor versus non-tumor or recurrent versus non-recurrent tumor conditions. Initial analysis of these lipid species by targeted isolation in situ using the CASI (continuous accumulation of selected ions) feature of the FTICR instrument have primarily identified phosphatidylcholine species based on loss of choline phosphate and accurate mass comparisons to Lipid MAPS. When used alone, these 39 lipid species were more discriminatory than the corresponding proteins (Figure 4B) in the principal component analyses. These 39 lipid ions represent an initial target population, and will be further assessed in larger tissue cohorts. Continued data analysis is ongoing with this large, promising data set, and it is likely that other biomarker candidates may be identified as these analyses progress.
A wealth of new genomic insight related to ccRCC development, progression and therapeutics is emerging [4,15–17], and shows great promise in defining the molecular pathways that represent new therapeutic targets. Because a genetic hallmark of ccRCC is altered cellular metabolism [4,17], the approach described herein of linking protein and lipid profiling data across 20 tissues could aid in delineating the phenotypes of these molecular pathways. The genetic microheterogeneity that has been described within the same ccRCC tumors [20] highlights the significant challenges that remain for improving diagnostics and therapeutics for ccRCC. An emerging therapeutic approach is to target the fundamental metabolic processes of ccRCC, especially to lipid pathway targets like inhibitors of fatty acid synthase, or cholesterol metabolism via statins [42,43]. Another target, stearoyl-CoA desaturase (SCD1) has also been recently described [44]. SCD1 catalyzes the biosynthesis of C16 and C18 Δ9 monounsaturated fatty acids from the saturated fatty acids stearic and palmitic acid. The ability to effectively profile the many lipid species that could be affected by metabolic targeted therapies for ccRCC, as well as larger studies linking their expression to tumor stage and outcomes, are particularly well suited to MALDI-IMS imaging using the FTICR platform.
This study represents a feasibility study for combining different MALDI-IMS target molecules for identifying biomarker candidates. The sample size is too small to draw any statistically meaningful clinical conclusions, but certainly different candidate molecules have been identified that can be assessed in larger sample cohorts. It is clear that a single molecular marker, either protein or lipid, is not going to be an effective discriminator. Principal component analysis indicated that the combined data from lipids and proteins could be used to separate tumor from non-tumor, and begin to differentiate recurrent disease progressors from non-recurrent subjects. Given the small sample set, cross-validation for performance measures was not utilized, but in future studies candidate markers will be qualified against independent test sets. Other biomolecule classes can also be incorporated into the MALDI-IMS strategy and are ongoing, in particular small molecule metabolites and membrane lipids detected in negative ion mode, as ccRCC exemplifies a metabolic Warburg effect tumor type. Analysis of on-tissue N-glycans is also ongoing, using a recently described new MALDI-IMS approach [32]. Combining differentially detected metabolites and glycans from the ccRCC groups with the continued lipid and protein analyses could further define biomolecular patterns reflective of the emerging underlying genetic pathways being reported for ccRCCs. Not only can all of these metabolites be profiled in ccRCC tissues, but these biomolecules could also be used as biomarker candidates for detection in urine or blood samples associated with ccRCC [45], or a newly reported tissue biopsy approach termed mPREF [46]. A novel intraoperative characterization of human tissues using rapid evaporative ionization mass spectrometry (REIMS) is an emerging technique that could also be applied to ccRCC tissues [47]. Overall, the underlying metabolic genotype and phenotype of ccRCC makes it an ideal tumor system for application with MALDI-IMS approaches.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health/National Cancer Institute grants R01CA135087, the Department of Defense grant W81XWH-10-1-0136, and the state of South Carolina SmartState Endowed Research program to R.R.D
ABBREVIATIONS
- AuROC
Area under Receiver Operating Characteristic
- ccRCC
clear cell Renal Cell Carcinoma
- CID
collision-induced dissociation
- CASI
continuous accumulation of selected ions
- FTICR
Fourier Transform Ion Cyclotron Resonance
- MALDI-IMS
Matrix-Assisted Laser Desorption Ionization Imaging Mass Spectrometry
- PC
phosphatidylcholine
- PCA
Principal Component Analysis
Footnotes
Conflict of Interest Statement
The authors have no conflict of interests to declare.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Moore LE, Wilson RT, Campleman SL. Lifestyle factors, exposures, genetic susceptibility, and renal cell cancer risk: a review. Cancer Invest. 2005;23:240–255. doi: 10.1081/cnv-200055962. [DOI] [PubMed] [Google Scholar]
- 3.Mourad WF, Dutcher J, Ennis RD. State-of-the-Art Management of Renal Cell Carcinoma. Am J Clin Oncol. 2012 doi: 10.1097/COC.0b013e31825d5522. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin JK, Lipworth L. Epidemiologic aspects of renal cell cancer. Sem Oncol. 2000;27:115–123. [PubMed] [Google Scholar]
- 6.Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparison of outcome and prognostic features among the histologic subtypes of renal cell carcinoma. Amer Journal of Surg Path. 2003;27:612–624. doi: 10.1097/00000478-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Dutcher JP, Mourad WF, Ennis RD. Integrating innovative therapeutic strategies into the management of renal cell carcinoma. Oncology. 2012;26:526–530. [PubMed] [Google Scholar]
- 8.Mohammed A, Shergill I, Little B. Management of metastatic renal cell carcinoma: current trends. Expert Rev Mol Diagn. 2009;9:75–83. doi: 10.1586/14737159.9.1.75. [DOI] [PubMed] [Google Scholar]
- 9.Jorns JJ, Thiel DD, Lohse C, Williams A. Kidney size and cancer-specific survival for patients undergoing nephrectomy for pT1 clear cell renal cell carcinoma. Urology. 2012;80:147–150. doi: 10.1016/j.urology.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Nelson EC, Evans CP, Lara PN., Jr Renal cell carcinoma: current status and emerging therapies. Cancer Treatment Reviews. 2007;33:299–313. doi: 10.1016/j.ctrv.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Wood CG. Multimodal approaches in the management of locally advanced and metastatic renal cell carcinoma: combining surgery and systemic therapies to improve patient outcome. Clin Cancer Res. 2007;13:697s–702s. doi: 10.1158/1078-0432.CCR-06-2109. [DOI] [PubMed] [Google Scholar]
- 12.Cella D, Cappelleri JC, Bushmakin A, Charbonneau C, et al. Quality of life predicts progression-free survival in patients with metastatic renal cell carcinoma treated with sunitinib versus interferon alpha. J Oncology Prac. 2009;5:66–70. doi: 10.1200/JOP.0922004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banumathy G, Cairns P. Signaling pathways in renal cell carcinoma. Cancer Biol Ther. 2010;10:658–664. doi: 10.4161/cbt.10.7.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper SJ, Tun HW, Roper SM, Kim Y, et al. Biomarkers for the identification and treatment of clear cell renal cell carcinoma. J Mol Biomarkers Diagnosis. 2012 [Google Scholar]
- 15.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 17.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seliger B, Dressler SP, Lichtenfels R, Kellner R. Candidate biomarkers in renal cell carcinoma. Proteomics. 2007;7:4601–4612. doi: 10.1002/pmic.200700415. [DOI] [PubMed] [Google Scholar]
- 19.Kim HL, Seligson D, Liu X, Janzen N, et al. Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. J Urol. 2005;173:1496–1501. doi: 10.1097/01.ju.0000154351.37249.f0. [DOI] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, Larkin J, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldwell RL, Caprioli RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Mol Cell Proteomics. 2005;4:394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Lemaire R, Desmons A, Tabet JC, Day R. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 23.Balluff B, Schöne C, Höfler H, Walch A. MALDI imaging mass spectrometry for direct tissue analysis: technological advancements and recent applications. Histochem Cell Biol. 2011;136:227–244. doi: 10.1007/s00418-011-0843-x. [DOI] [PubMed] [Google Scholar]
- 24.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry--looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 25.Maier SK, Hahne H, Moghaddas Gholami A, Balluff B, et al. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M113.027599. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oppenheimer SR, Mi D, Sanders ME, Caprioli RM. Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res. 2010;9:2182–2190. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan TM, Seeley EH, Fadare O, Caprioli RM, Clark PE. Imaging the clear cell renal cell carcinoma proteome. J Urol. 2013;189:1097–1103. doi: 10.1016/j.juro.2012.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaurand P, Cornett DS, Angel PM, Caprioli RM. From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O110.004259. O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willems SM, van Remoortere A, van Zeijl R, Deelder AM, et al. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J Pathol. 2010;222:400–929. doi: 10.1002/path.2771. [DOI] [PubMed] [Google Scholar]
- 30.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3:2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One. 2010;5:e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powers TW, Jones EE, Betesh LR, Romano P, et al. A MALDI Imaging Mass Spectrometry Workflow for Spatial Profiling Analysis of N-linked Glycan Expression in Tissues. Anal Chem. 2013;85:9799–9806. doi: 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank I, Blute ML, Cheville JC, Lohse CM, et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 34.Zigeuner R, Hutterer G, Chromecki T, Imamovic A, et al. External validation of the Mayo Clinic stage, size, grade, and necrosis (SSIGN) score for clear-cell renal cell carcinoma in a single European centre applying routine pathology. Eur Urol. 2010;57:102–109. doi: 10.1016/j.eururo.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 35.Cazares LH, Troyer D, Mendrinos S, Lance RS, et al. Imaging mass spectrometry of a specific fragment of mitogen activated protein kinase/extracellular signal-regulated kinase kinase kinase discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15:5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Qian WJ, Mottaz HM, Clauss TR, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roper SM, Zemskova M, Neely BA, Martin A, et al. Targeted glycoprotein enrichment and identification in stromal cell secretomes using azido sugar metabolic labeling. Proteomics Clin Appl. 2013;7:367–371. doi: 10.1002/prca.201300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raimondo F, Salemi C, Chinello C, Fumagalli D, et al. Proteomic analysis in clear cell renal cell carcinoma: identification of differentially expressed protein by 2-D DIGE. Mol Biosyst. 2012;8:1040–1051. doi: 10.1039/c2mb05390j. [DOI] [PubMed] [Google Scholar]
- 39.Takaoka N, Takayama T, Teratani T, Sugiyama T. Analysis of the regulation of fatty acid binding protein 7 expression in human renal carcinoma cell lines. BMC Mol Biol. 2011;12:31. doi: 10.1186/1471-2199-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domoto T, Miyama Y, Suzuki H, Teratani T. Evaluation of S100A10, annexin II and BFABP expression as markers for renal cell carcinoma. Cancer Sci. 2007;98:77–82. doi: 10.1111/j.1349-7006.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones EA, van Zeijl RJ, Andrén PE, Deelder AM, et al. High speed data processing for imaging MS-based molecular histology using graphical processing units. J Am Soc Mass Spectrom. 2012;23:745–752. doi: 10.1007/s13361-011-0327-1. [DOI] [PubMed] [Google Scholar]
- 42.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 43.Pinthus JH, Whelan KF, Gallino D, Lu JP, Rothschild N. Metabolic features of clearcell renal cell carcinoma: mechanisms and clinical implications. Can Urol Assoc J. 2011;5:274–282. doi: 10.5489/cuaj.10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–2380. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wettersten HI, Weiss RH. Potential biofluid markers and treatment targets for renal cell carcinoma. Nat Rev Urol. 2013;10:336–344. doi: 10.1038/nrurol.2013.52. [DOI] [PubMed] [Google Scholar]
- 46.Brown MV, McDunn JE, Gunst PR, Smith EM, et al. Cancer detection and biopsy classification using concurrent histopathological and metabolomic analysis of core biopsies. Genome Med. 2012;4:33. doi: 10.1186/gm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balog J, Sasi-Szabó L, Kinross J, Lewis MR, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med. 2013;5:194ra93. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.